Significance
Psoriasis is an autoinflammatory disease characterized by cytokine-driven keratinocyte proliferation and infiltration of immune cells. While IL-17A and TNFα are established targets in psoriasis therapy, IL-36 is emerging as an important cytokine in this disease. The mechanisms of IL-36–driven proinflammatory responses are largely unknown. Here we identified IκBζ, a transcriptional regulator of selective NF-κB target genes, as a crucial mediator of IL-36 action. In keratinocytes, IκBζ was required for the expression of several psoriasis-related cytokines and chemokines. Moreover, genetic deletion of IκBζ prevented IL-36–mediated dermatitis induction in mice. Since IκBζ is essential not only for IL-36 but also for IL-17 signaling, our results suggest that inhibition of IκBζ function could be a future strategy in psoriasis therapy.
Keywords: NFKBIZ, IκBζ, IL-36, keratinocytes, psoriasis
Abstract
Proinflammatory cytokine signaling in keratinocytes plays a crucial role in the pathogenesis of psoriasis, a skin disease characterized by hyperproliferation and abnormal differentiation of keratinocytes and infiltration of inflammatory cells. Although IL-17A and TNFα are effective therapeutic targets in psoriasis, IL-36 has recently emerged as a proinflammatory cytokine. However, little is known about IL-36 signaling and its downstream transcriptional responses. Here, we found that exposure of keratinocytes to IL-36 induced the expression of IκBζ, an atypical IκB member and a specific transcriptional regulator of selective NF-κB target genes. Induction of IκBζ by IL-36 was mediated by NF-κB and STAT3. In agreement, IL-36–mediated induction of IκBζ was found to be required for the expression of various psoriasis-related genes involved in inflammatory signaling, neutrophil chemotaxis, and leukocyte activation. Importantly, IκBζ-knockout mice were protected against IL-36–mediated dermatitis, accompanied by reduced proinflammatory gene expression, decreased immune cell infiltration, and a lack of keratinocyte hyperproliferation. Moreover, expression of IκBζ mRNA was highly up-regulated in biopsies of psoriasis patients where it coincided with IL36G levels. Thus our results uncover an important role for IκBζ in IL-36 signaling and validate IκBζ as an attractive target for psoriasis therapy.
Transcription factor NF-κB has been implicated in several inflammatory diseases, including psoriasis, by activating various proinflammatory target genes (1). The classical activation of NF-κB is controlled by cytoplasmic inhibitory proteins, such as IκBα, which sequester NF-κB in the cytoplasm (2). Inflammatory stimulation of cells results in the rapid activation of IκB kinase (IKK), which triggers the phosphorylation-induced degradation of IκBα, leading to NF-κB’s nuclear translocation and transcriptional activation. Recent evidence, however, suggests that the activation of NF-κB target genes is more complex and is dependent on the particular gene context or stimulus, which is thought to facilitate selective gene regulation in distinct physiological settings (3). Whereas the rapid activation of primary response genes is directly induced by the classical NF-κB pathway, expression of so-called “secondary-response genes” requires prior protein synthesis of additional NF-κB regulators (4). In this context, we and others have identified IκBζ, an atypical nuclear IκB protein, which functions not only as a repressor but, more importantly, also as an activator of a selective subset of NF-κB target genes (5–8). The mechanisms of this differential gene regulation by IκBζ remain largely unknown, but increasing evidence suggests that the transcriptional activity of IκBζ is mainly mediated at the level of chromatin remodeling (6, 9, 10).
In keratinocytes (KCs), IL-17A and, more potently, its combination with TNFα induce IκBζ expression (11). Subsequently, IκBζ mediates the induction of important psoriasis-related gene products, including chemokines (e.g., CXCL8 and CCL20), cytokines (e.g., IL22 and IL17C), and antimicrobial proteins, such as S100 calcium-binding proteins (e.g., S100A9), β-defensin-2 (DEFB4A), or lipocalin-2 (LCN2). Antagonists of TNFα and IL-17A have therefore been approved for the treatment of psoriasis (12). Moreover, NFKBIZ, the gene encoding IκBζ, has been identified as a psoriasis-susceptibility locus (13). Global Nfkbiz-KO mice are resistant to imiquimod (IMQ)- or IL-23–induced psoriasis-like skin inflammation (11). In contrast, Tnfa- or Il17a-KO mice, which are only partially protected against IMQ-induced psoriasis, still show elevated IκBζ mRNA levels in inflamed skin areas (11). These observations imply an additional IL-17A/TNFα–independent pathway which drives IκBζ expression and thereby contributes to inflammatory gene expression in psoriasis.
Recently, IL-36 cytokines have received attention as therapeutic targets for psoriasis (14). This subfamily of IL-1–related cytokines consists of three proinflammatory members, IL-36α (encoded by IL1F6/IL36A), IL-36β (encoded by IL1F8/IL36B), and IL-36γ (encoded by IL1F9/IL36G) (15–17). All family members bind to a common heterodimeric receptor, composed of IL-36R (also termed “IL-1RL2”) and IL-1RAcP, leading to the recruitment of the adapter MyD88 and subsequent activation of NF-κB and MAPK (18). A fourth IL-36 member, IL-36RN, acts as a natural antagonist of IL-36 signaling, as it binds to IL-36R but does not recruit the coreceptor IL-1RAcP (19, 20). Importantly, while full-length IL-36 proteins seem to be biologically inactive, activation of IL-36 signaling requires their N-terminal proteolytic processing (19, 21).
IL-36 contributes to skin inflammation by acting on KCs and immune cells. Interestingly, IL-36 can induce a subset of proinflammatory target genes similar to those induced by IL-17A in KCs, including CXCL8, IL23A, DEFB4, or LCN2 (22–24). Vice versa, IL-17, which is typically expressed by immune cells, induces IL-36γ expression in KCs (25, 26). Therefore, IL-36 appears to have a central position in the interplay between immune cells and KCs. In patients with psoriasis vulgaris, IL-36α and IL-36γ are overexpressed, whereas inactivating mutations of IL36RN are enriched in a psoriasis subtype, called “generalized pustular psoriasis” (22, 23, 27, 28). In agreement, mice overexpressing IL-36α in basal KCs exert skin inflammation at 3 wk of age, which is augmented in an IL36RN-deficient background (20, 29). In contrast, mice deficient for the IL-36R are fully protected against IMQ-induced psoriasis (30).
Despite its involvement in psoriasis, little is known about IL-36 signaling and its transcriptional responses. In the present study, we found that IL-36α and IL-36γ are potent inducers of IκBζ expression. Moreover, we identified MyD88, NF-κB and STAT3 as crucial components for IL-36–induced IκBζ expression. Silencing of IκBζ in primary human KCs prevented IL-36–mediated up-regulation of multiple psoriasis-associated genes, while a global knockout of IκBζ protected against IL-36–mediated psoriasis-like dermatitis in mice. These results and our finding of a strong correlation of NFKBIZ and IL36G expression in psoriatic lesions uncover an important role for IκBζ in IL-36 signaling and thus validate IκBζ as an attractive target for psoriasis therapy.
Results
IL-36 Induces IκBζ Expression in KCs.
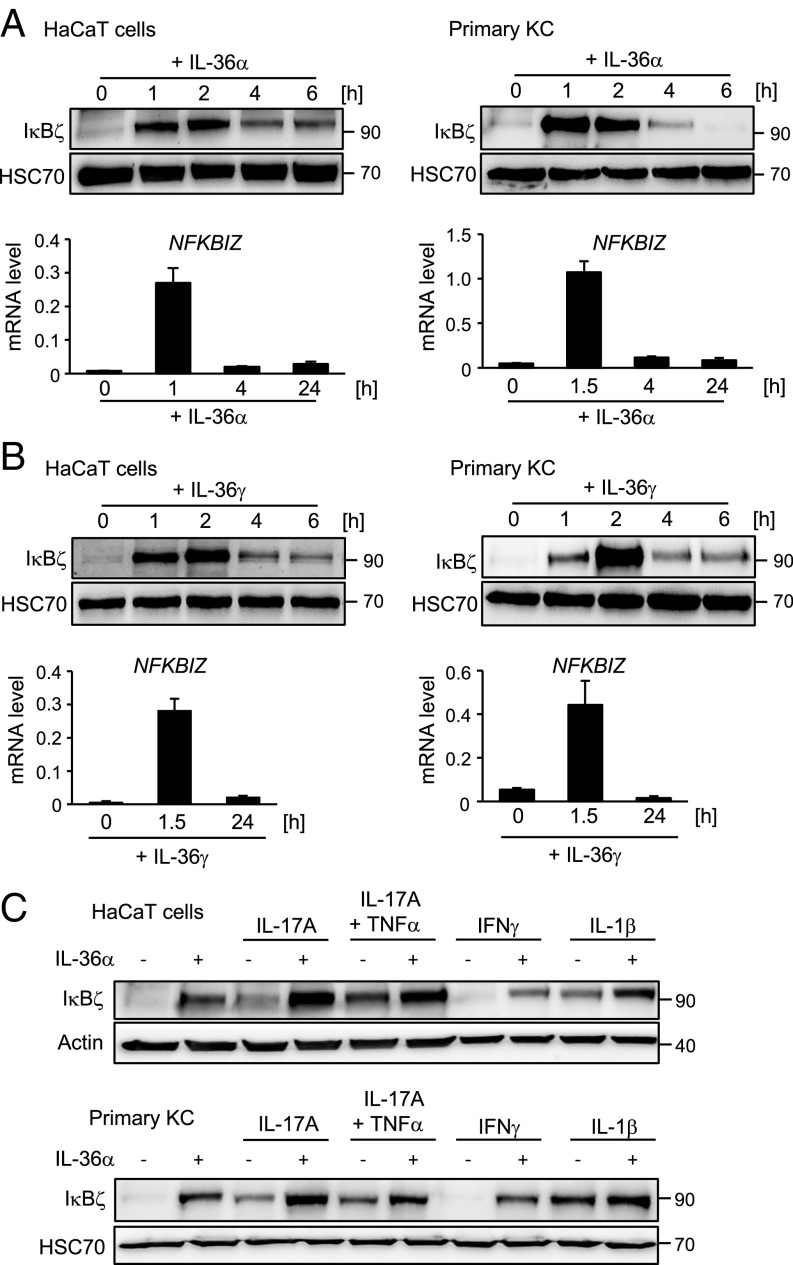
To investigate the relationship between IL-36 and IκBζ, we treated the keratinocyte cell line HaCaT and primary human KCs with recombinant IL-36α for 1–24 h. Whereas untreated KCs lacked IκBζ expression, 1 h of stimulation with IL-36α was sufficient to induce sustained IκBζ expression on the mRNA and protein level (Fig. 1A). As revealed by the addition of actinomycin D to IL-36α–treated cells, the increased NFKBIZ mRNA levels resulted from transcriptional up-regulation of NFKBIZ rather than from mRNA stabilization (SI Appendix, Fig. S1A). Importantly, full-length IL-36α, which supposedly lacks biological activity, failed to induce IκBζ expression, whereas IL-17A, either alone or combined with TNFα, induced IκBζ expression with kinetics similar to those of truncated IL-36α (Fig. 1A and SI Appendix, Fig. S1 B and C). As some reports implied distinct target gene regulation by the different IL-36 members (14, 24, 25), we also stimulated HaCaT cells and primary KCs with IL-36γ. IL-36γ induced NFKBIZ mRNA and protein expression with kinetics and potency similar to that of IL-36α (Fig. 1B).
Fig. 1.
IL-36 induces IκBζ expression in KCs. (A and B) HaCaT cells (Left) or human primary KCs (Right) were treated with 100 ng/mL IL-36α (amino acids 6–158) (A) or 100 ng/mL IL-36γ (amino acids 18–169) (B) for the indicated times. IκBζ protein was analyzed by Western blotting. Relative mRNA levels of NFKBIZ were measured in parallel and normalized to the reference RPL37A. (C) HaCaT cells (Upper) and primary KCs (Lower) were treated for 2 h with 100 ng/mL IL-36α alone or in combination with 100 ng/mL IL-17A, 10 ng/mL TNFα, 100 ng/mL IFNγ, or 100 ng/mL IL-1β. IκBζ was detected by Western blotting. HSC70 or β-actin served as loading controls.
We next investigated whether other psoriasis-associated cytokines, such as IL-1β, IL-17A, TNFα, or IFNγ, could potentiate the effect of IL-36α on IκBζ protein expression (Fig. 1C). Although certain differences were noted between HaCaT cells and primary KCs, most of the tested cytokines enhanced IL-36α–mediated IκBζ expression. Importantly, the combination of IL-17A and IL-36α was clearly more effective in triggering IκBζ expression than were the single cytokines alone.
Induction of IκBζ by IL-36 Is Mediated by MyD88, NF-κB, and STAT3.
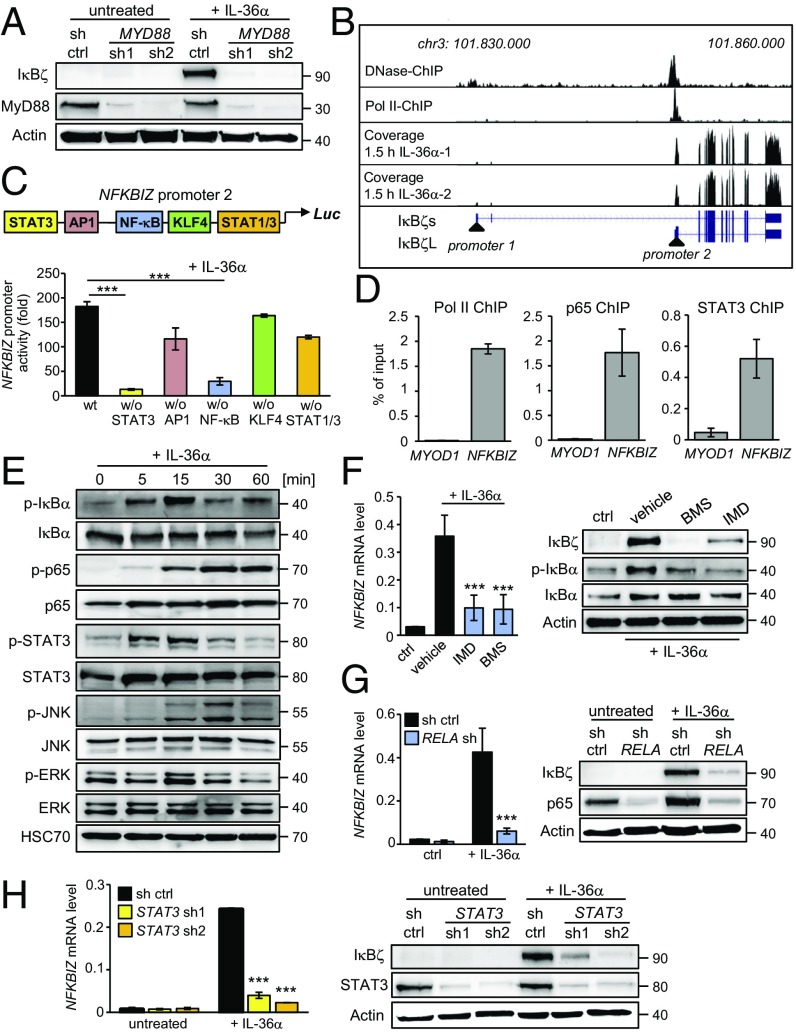
As IκBζ is also induced by IL-17A, we further dissected the mechanism of IκBζ expression induced by IL-36 compared to IL-17A. IL-17A binds and activates the IL-17RA/IL-17RC receptor, followed by the recruitment of the adapter protein Act1 and the activation of MAPK and NF-κB (31). In contrast, IL-36 utilizes a divergent proximal signaling cascade by binding to the IL-36 receptor complex, composed of IL1RL2 and its coreceptor IL1RAP, leading to the recruitment of MyD88 and activation of MAPK and NF-κB (17). Indeed, knockdown of MyD88 revealed that it was indispensable for IκBζ expression upon IL-36α stimulation, while it had no effect in IL-17A–treated cells (Fig. 2A and SI Appendix, Fig. S2A).
Fig. 2.
Molecular dissection of IκBζ induction by IL-36. HaCaT cells were stimulated for the indicated times with 100 ng/mL IL-36α. (A) Cells stably expressing a control (sh ctrl) or two different shRNAs targeting MyD88 were treated for 2 h with IL-36α and were analyzed by Western blotting. (B) Analysis of NFKBIZ promoter accessibility and structure. The genomic region around NFKBIZ was analyzed from a published DNase I dataset and a polymerase II ChIP-seq track (33). Exon reads of NFKBIZ were derived from our own RNA-seq data of HaCaT cells stimulated for 1.5 h with IL-36α. (C) Analysis of the NFKBIZ promoter 2 region in IL-36α–stimulated HaCaT cells using luciferase reporter constructs harboring deletions of transcription factor-binding sites. (D) P65, STAT3, and RNA polymerase II (Pol II) bind to NFKBIZ promoter region 2 in IL-36α–treated cells. ChIP was performed from HaCaT cells treated for 30 min with IL-36α. The promoter region of the muscle-specific gene MYOD1 represents a negative control. (E) Immunoblot analysis of IL-36α–induced signaling pathways. Active NF-κB and STAT3 were detected by the phosphorylated forms of IκBα (p-IκBα at Ser32), p65 (p-p65 at Ser536), and STAT3 (p-STAT3 at Tyr705). MAPK activation was detected by phosphorylated JNK (p-JNK at Thr183/Tyr185) and ERK (p-p44/42 at Thr202/Thr204). (F) Cells were treated for 1 h with IL-36α in the presence or absence of the vehicle DMSO or 10 µM of the IKK inhibitors BMS-345541 or IMD0354. NFKBIZ mRNA and IκBζ protein levels were measured after 2 h of IL-36α stimulation. Detection of p-IκBα served as a control for NF-κB inhibition. (G and H) Gene expression and Western blot analysis of IκBζ in control and RELA (p65)-knockdown (G) or STAT3-knockdown (H) cells after 1 h of IL-36α treatment. Knockdown was controlled by detection of p65 or STAT3. ***P < 0.001.
As IκBζ is transcriptionally induced by IL-36, we explored the NFKBIZ promoter region to identify relevant transcription factors. Two major IκBζ isoforms have been described, including a long isoform (IκBζL) of 718 aa and a N-terminally truncated isoform (IκBζS) of 618 aa that is thought to be generated by alternative splicing (8, 32). By analyzing published DNase I and Pol II ChIP-sequencing (ChIP-seq) data (33), we identified that the two isoforms arise not only from alternative splicing but also from two different promoter regions with distinct transcriptional start sites (Fig. 2B). Moreover, our own RNA-sequencing (RNA-seq) data revealed that KCs use only the proximal promoter 2 that is translated into the IκBζL isoform. Previous promoter analyses, however, had examined only promoter 1, which is located ∼20 kb upstream of promoter 2 (32, 34, 35). This distal promoter is used in several cell types for transcription of NFKBIZ variant 2, which lacks exon 3 and thus is translated to the IκBζS variant.
Bioinformatic analysis of the NFKBIZ promoter 2 revealed putative binding sites for STAT3, NF-κB, AP1, KLF4, and STAT1. To uncover the contribution of these sites to NFKBIZ induction, we cloned the promoter region (∼1.5 kb upstream of the transcription start site of IκBζL) into a luciferase construct and generated deletions lacking one of the predicted binding sites. Expression of the constructs was analyzed after transfection of HaCaT cells followed by stimulation with IL-36α. Indeed, expression of the NFKBIZ promoter 2 was significantly increased by IL-36α, whereas deletion of the STAT3- or the NF-κB–binding site inhibited NFKBIZ promoter expression (Fig. 2C). In accordance, ChIP identified a direct physical binding of NF-κB p65 and STAT3 to NFKBIZ promoter 2, along with the binding of phosphorylated RNA polymerase II as a marker for active transcription (Fig. 2D). IL-36α also triggered the early activation of STAT3, NF-κB, and MAPK in HaCaT cells or primary KCs (Fig. 2E and SI Appendix, Fig. S2B). Interestingly, a similar activation of STAT3 and NF-κB was detected in IL-17A–treated cells (SI Appendix, Fig. S2C). Whereas inhibition of MAPK did not affect IκBζ expression in IL-36α–treated HaCaT cells (SI Appendix, Fig. S2D), the blocking of NF-κB activation by IKK inhibition or knockdown of p65 efficiently prevented IκBζ expression upon IL-36α stimulation (Fig. 2 F and G). Moreover, depletion of STAT3 by two different shRNAs strongly inhibited IκBζ mRNA and protein expression (Fig. 2H). Similarly, depletion of p65 or STAT3 impaired IκBζ induction after stimulation with IL-17A (SI Appendix, Fig. S2 E and F). Thus, IL-36α and IL-17A both employ NF-κB and STAT3 for IκBζ induction.
IκBζ Is a Key Mediator of IL-36–Induced Gene Expression in KCs.
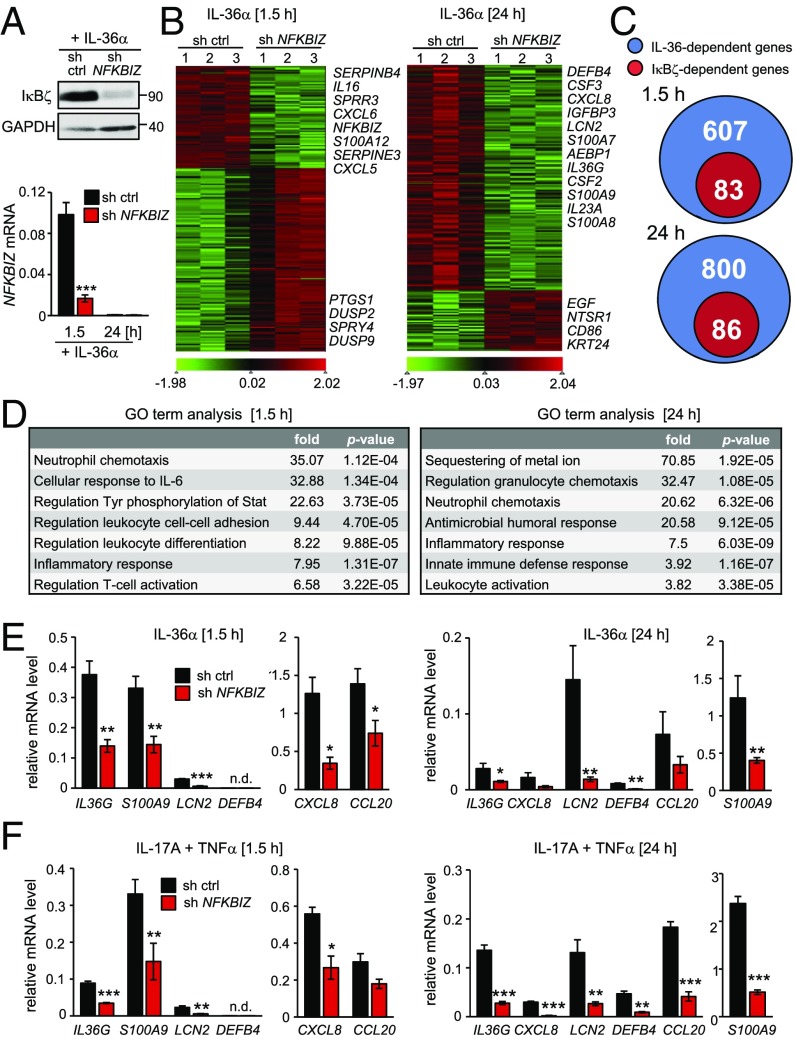
Next, we investigated the function of IκBζ in IL-36 signaling and therefore first explored the time course of IκBζ-modulated gene expression. We stimulated control and NFKBIZ-knockdown HaCaT cells for 0–24 h with IL-36α and analyzed selected IL-36 target genes. IL-36α stimulation led to the induction of IL36G, IL17C, CXCL5, or S100A9 with different kinetics (SI Appendix, Fig. S3A). Surprisingly, NFKBIZ silencing not only prevented the induction of late-responsive genes such as S100A9 but also affected early gene induction, e.g., of IL36G or IL17C.
To reveal a global picture of IL-36–driven gene expression by IκBζ, we generated control and NFKBIZ-depleted primary KCs and performed transcriptome analyses after 1.5 and 24 h of IL-36α stimulation (Fig. 3 A and B). Silencing of IκBζ resulted in the deregulation of several hundred target genes in IL-36α–stimulated primary human KCs (SI Appendix, Tables S1 and S2). Interestingly, early after IL-36α stimulation most genes were down-regulated by IκBζ, including genes for antiinflammatory phosphatases (DUSP2 and DUSP9). In contrast, after 24 h most IκBζ-modulated genes were positively regulated and hence were down-regulated by the NFKBIZ knockdown. Many of these IκBζ-inducible genes are typically overexpressed in psoriasis, including genes for antimicrobial proteins (DEFB4 and LCN2), S100 proteins (S100A7, S100A8, and S100A9), and chemo- and cytokines (CSF2, CSF3, CXCL8, IL23A, and IL36A).
Fig. 3.
IκBζ regulates a subset of psoriasis-related IL-36 target genes. Primary KCs or HaCaT cells were transduced with a control or NFKBIZ-specific shRNA. Triplicates of each time point and shRNA were analyzed by RNA-seq or qPCR and were normalized to the reference gene RPL37A. (A) Control of NFKBIZ-knockdown efficiency. (Upper) IκBζ protein was detected in primary KCs treated for 1 h with 100 ng/mL IL-36α. (Lower) NFKBIZ mRNA levels were measured after 1.5 h and 24 h of IL-36α stimulation. (B) After library preparation from total RNA, primary KC samples were sequenced, and reads were aligned to the human genome hg19. Depicted are two separate heatmaps with normalized z-scores of IκBζ target genes after 1.5 h and 24 h of IL-36α treatment. As a cutoff, genes with a minimum fold change of 1 and a P value < 0.05 were considered. (C) Venn diagrams showing the fraction of IκBζ target genes among IL-36α–regulated genes 1.5 and 24 h after stimulation of primary KCs. (D) GO term analysis of significantly enriched IκBζ-dependent gene sets after 1.5 and 24 h of IL-36α treatment. (E) Validation of selected IκBζ target genes by qPCR in primary KCs after 1.5 and 24 h of incubation with 100 ng/mL IL-36α. (F) Gene-expression analysis of IκBζ target genes in primary KCs stimulated with 100 ng/mL IL-17A and 10 ng/mL TNFα for 1.5 and 24 h. *P < 0.05; **P < 0.01; ***P < 0.001.
Principal component analysis (PCA) revealed that the gene-expression profile not only differed between untreated and IL-36α–stimulated cells but was also divergent after 1.5 and 24 h of IL-36α stimulation (SI Appendix, Fig. S3B). Moreover, as shown in the Venn diagrams in Fig. 3 C and D, only a subset of the IL-36α–regulated genes was IκBζ-dependent (83 of 607 genes after 1.5 h and 86 of 800 genes after 24 h of IL-36α stimulation). Gene ontology (GO) term analysis of the affected genes uncovered that IκBζ mostly regulated inflammatory responses, neutrophil chemotaxis, and leukocyte function downstream of IL-36 (Fig. 3D). We also compared our RNA-seq analyses with a previously defined IL-36 core signature comprising 182 genes that were regulated by IL-36 after 24 h in human KCs (14). The comparison not only revealed a high overlap with our RNA-seq analyses but also identified 39 of the 182 IL-36 core target genes as IκBζ-dependent (SI Appendix, Fig. S3 C and D).
The IκBζ-dependent gene regulation by IL-36α in primary KCs at early and late time points was confirmed by qPCR of selected genes, such as IL36G, S100A9, LCN2, DEFB4, CXCL8, and CCL20 (Fig. 3E). Importantly, regulation of these IκBζ target genes was conserved in IL-17A– and TNFα–treated primary KCs as well as in IL-36α–, IL-36γ–, and IL-1β–treated HaCaT cells (Fig. 3F and SI Appendix, Fig. S4 A–C). These findings thus implicate IκBζ as a master regulator of proinflammatory gene expression not only in IL-36–stimulated but also in IL-17A–,TNFα–, or IL-1β–treated KCs.
IκBζ Promotes IL-36–Driven Psoriasis-Like Disease in Vivo.
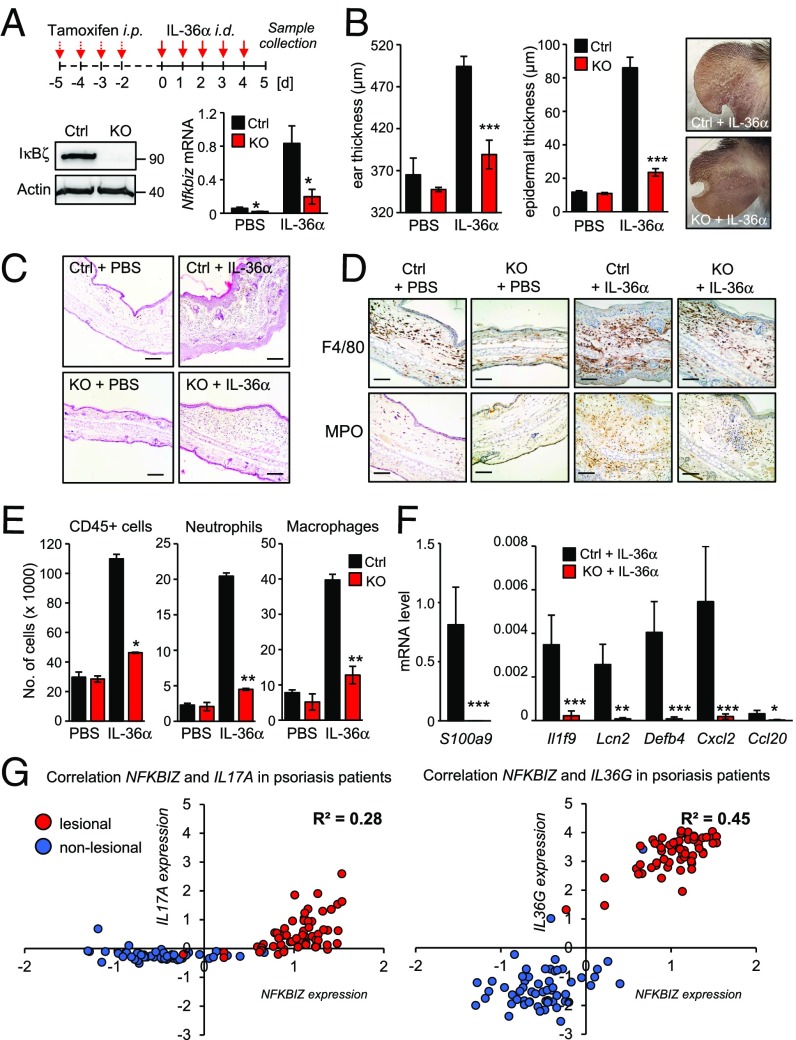
Global Nfkbiz-KO mice are protected against IMQ-induced psoriasis-like skin inflammation (11). Since the TLR7 agonist IMQ directly activates the innate immune response, it is difficult to discriminate between the contribution of IL-17 and IL-36 to the disease onset. Moreover, global Nfkbiz-KO mice develop an autoinflammatory phenotype in adulthood (36, 37), which could influence the skin inflammation of IMQ-treated mice. We therefore generated a mouse model using tamoxifen-inducible Nfkbiz-KO mice that received intradermal injections of active IL-36α into the ears. Tamoxifen-induced Cre recombinase activation just before IL-36α application led to an effective KO of IκBζ, thereby preventing potential congenital off-target effects (Fig. 4A). Intradermal injection of IL-36α into the ears of control animals induced Nfkbiz transcription (Fig. 4A) and, moreover, triggered ear swelling, scaling, epidermal thickening, KC hyperproliferation and increased infiltration of immune cells (Fig. 4 B and C and SI Appendix, Fig. S5A). These alterations were nearly absent in the IL-36α–treated KO mice. Histological and flow cytometric analyses revealed a marked increase in infiltrating CD45+ immune cells, macrophages, and neutrophils in the IL-36α–treated control animals, which was significantly blocked in the KO mice (Fig. 4 D and E). T cell infiltration was reduced in the KO animals as well, although the degree of T cell infiltration was generally low in the ears of IL-36α–treated mice (SI Appendix, Fig. S5B). Importantly, expression of several psoriasis-associated target genes, similar to those identified by transcriptome analysis of IL-36α–treated KCs (Fig. 3), was up-regulated in the ears of IL-36a-treated control but not in IL-36α–treated KO mice (Fig. 4E). Likewise, the expression of IκBζ-dependent proteins involved in granulocyte and leukocyte chemotaxis was also decreased in the KO mice (SI Appendix, Fig. S5C). Thus, IκBζ KO strongly protected against IL-36–driven psoriasis-like disease in vivo, which could be mediated by effects of Nfkbiz deficiency in KCs as well as in immune cells.
Fig. 4.
Characterization of the IL-36/IκBζ axis in vivo. (A, Upper) Scheme of tamoxifen and IL-36α treatment of control and inducible Nfkbiz-KO mice. (Lower) Verification of Nfkbiz deletion at the protein and mRNA level. For induction of IκBζ KO, Nfkbiz flox/flox (Ctrl) and Rosa-creERT2 Nfkbiz flox/flox (KO) mice received i.p. injections of tamoxifen (75 mg/kg) for four consecutive days to induce activation of Cre recombinase. Afterward, 1 µg murine IL-36α or PBS control was intradermally injected into one ear of the mice for five consecutive days. (B) Ear and epidermal thickness (± SEM) of PBS- and IL-36α–treated mice at day 5 from two (for PBS) or six (for IL-36α) animals per group. Pictures were taken at day 5 to show scaling at the treatment area. (C) H&E staining of ears from PBS- and IL-36α–treated control and KO mice. (Scale bars: 140 µM.) (D) Immunohistochemistry for the macrophage marker F4/80 and the neutrophil marker MPO. (Scale bars: 80 µM) (E) Characterization of CD45+ immune cell infiltrates by flow cytometry. Neutrophils were characterized as CD45+ Ly6G+ and macrophages as CD45+, CD11bhi and F4/80+. Error bars indicate results from two independent experiments. (F) Psoriasis-related gene expression in ears from IL-36α–treated mice. Results are shown as means ± SEM; n = 6 animals per group. (G) Expression data from skin biopsies of 64 healthy individuals and 58 psoriasis patients were analyzed from the Gene Expression Omnibus profile dataset GDS4602. Shown are normalized expression values of NFKBIZ and IL17A or NFKBIZ and IL36G, which were plotted against each other in every single nonlesional and lesional biopsy. Depicted is the regression coefficient (R2) from the expression values of the psoriatic skin biopsies. *P < 0.05; **P < 0.01; ***P < 0.001.
As previously reported (11, 13), we validated increased NFKBIZ expression in lesions from psoriasis patients, as compared with nonlesional skin areas or unaffected individuals (SI Appendix, Fig. S5D). Expression of IL17A and especially IL36G was elevated in psoriatic lesions. We then correlated the expression of NFKBIZ, IL36G, and IL17A in nonlesional and lesional samples in the individual patients to obtain an idea of the relevance of the two cytokines in driving NFKBIZ expression in psoriatic tissue. The correlation of IL36G and NFKBIZ was stronger than the link between IL17A and NFKBIZ, implicating IL-36 as an important driver of NFKBIZ expression in psoriasis (Fig. 4G). Moreover, as IL-36–mediated NFKBIZ induction could account for increased expression of psoriasis-related cytokines, we correlated the expression of LCN2, a bona fide IκBζ target gene (38), to IL36G, NFKBIZ, and IL17A expression. Indeed, the expression level of LCN2 matched strongly IL36G and NFKBIZ expression, whereas it was only weakly correlated to IL17A expression patterns in psoriatic lesions (SI Appendix, Fig. S5E). These findings support a major role of IκBζ in IL-36 signaling in KCs and psoriasis and suggest IκBζ as an attractive therapeutic target which mediates proinflammatory signaling downstream of IL-17A and IL-36.
Discussion
Previous studies by us and others found that IκBζ is overexpressed in psoriatic lesions, whereas Nfkbiz KO mice are protected against IMQ-induced psoriatic skin inflammation (11, 13). In these and follow-up studies, IκBζ was identified as a major mediator of IL-17A signaling, leading to the induction of proinflammatory signaling in KCs (11, 39). Interestingly, in Il17a- or Il17ra- KO mice neither induction of Nfkbiz nor skin inflammation were fully blocked after IMQ treatment (11, 40), implying additional pathways of NFKBIZ induction and promotion of psoriasis.
Recently, IL-36α and IL-36γ have been identified as being overexpressed in psoriatic lesions (22, 23). In agreement, IL-36 treatment of KCs induced proinflammatory signaling (14), whereas KO of the IL-36 receptor inhibited IMQ-induced skin inflammation in mice. Our results show that IκBζ provides an important link between IL-36 signaling and psoriasis-associated inflammatory gene expression. We revealed that IL-36 mediates IκBζ expression in HaCaT cells and primary KCs, which followed kinetics similar to those seen with IL-17A/TNFα treatment, implying similar signaling pathways in IκBζ induction.
By ChIP-seq data and our own RNA-seq analyses we identified that KCs induce transcription of NFKBIZ from the yet uncharacterized proximal promoter 2, which contains several conserved binding sites for proinflammatory transcription factors. Indeed, IL-36 and IL-17A stimulation led to the activation of NF-κB, whereas knockdown of the NF-κB subunit p65 prevented IκBζ induction.
Besides NF-κB, we identified STAT3 as a regulator of IκBζ expression, as its depletion was sufficient to block IL-36– and IL-17A–mediated induction of IκBζ. These findings are intriguing, as STAT3 itself can drive proinflammatory gene expression in psoriasis (41). Constitutively active STAT3 in the epidermis of psoriatic lesions is often detectable, whereas pharmacological inhibition of STAT3 ameliorated psoriasis-like skin lesions in mice (42, 43). Moreover, STAT3 was proposed to control IκBζ expression in T cells (44). As STAT3 is especially involved in IL-36–driven induction of IκBζ expression, STAT3 inhibitors could be promising agents for the effective treatment of general pustular psoriasis, which is caused by mutations of IL36RN and hyperactivation of the IL-36 pathway (27, 28).
Our gene-expression profiling revealed that IL-36 affected the expression of hundreds of genes at early and late stimulation time points. As early effects of IL-36 stimulation on gene expression have not been investigated before in KCs, we could not only validate defined IL-36 target genes (14) but also identify previously unknown IL-36 dependent genes (e.g., IL17C, CSF2, CSF3) that encode important psoriasis-promoting cytokines (45,46). Of note, NFKBIZ knockdown led to the deregulation of a specific subset of IL-36 target genes at early and late stimulation time points. Most of these IκBζ-dependent IL-36 target genes regulate antimicrobial and proinflammatory responses, neutrophil chemotaxis, and leukocyte activation and hence have been implicated in the pathogenesis of psoriasis. Moreover, IκBζ-dependent gene expression seems to be highly conserved, as we found similar changes in the expression of IκBζ-dependent genes (e.g., DEFB4, CCL20, S100A7, S100A9, and LCN2) in HaCaT cells and primary KCs as well as upon IL-36α, IL-36γ, or IL-17A/TNFα stimulation.
Employing an inducible Nfkbiz-KO model, we further demonstrate that the absence of IκBζ also impaired psoriasis-related gene expression under in vivo conditions of IL-36α stimulation. Nfkbiz-KO mice exhibited significantly reduced skin pathology, including less ear swelling and KC proliferation, and a strongly reduced infiltration of immune cells, in particular neutrophils. The results are consistent with findings in Il36r-deficient mice that are also protected in the IMQ psoriasis model (30). Notably, our previous study demonstrated that Nfkbiz-KO mice were even more protected than Il17a-deficient mice (11), supporting the idea that IκBζ might also be involved in IL-17–independent effects of psoriasis development. Of note, Il36r-deficient mice also showed stronger protection in the IMQ model than Il17a-KO mice (30).
In agreement with our findings in cultured KCs and Nfkbiz-KO mice, expression data from psoriasis patients validated elevated NFKBIZ and IL36G levels in psoriatic lesions as compared with nonaffected skin areas or skin from unaffected healthy individuals. Moreover, the expression of IL36G and NFKBIZ was strongly correlated with the IκBζ target gene LCN2 compared with a much weaker correlation of IL17A with NFKBIZ and LCN2 expression levels. These data strengthen the hypothesis that IL-36 is an initial driver for NFKBIZ expression in psoriasis.
While our results clearly position IκBζ downstream of IL-36, IL-17A and IL-36, in turn, are also transcriptional targets downstream of IκBζ (SI Appendix, Fig. S4D). Likewise, IL-17A, especially in combination with TNF, is a strong inducer of IκBζ expression but can be also induced downstream of IκBζ. Thus, the strong expression of NFKBIZ in psoriasis patients might be caused not only by elevated IL-36 expression but also by increased IL-17–type responses. The exact contribution of each cytokine is complicated by the existence of multiple members of the IL-17 and IL-36 families. Because IL-17 and IL-36 can mutually reinforce each other (25, 26), the two cytokines drive complex autoamplification loops in which IκBζ seems to have an integral role in promoting skin inflammation (for a scheme see SI Appendix, Fig. S6). In fact, our present and previous results (11) suggest a dual requirement for IκBζ in IL-36 signaling of innate epithelial cells, such as KCs, as well as in IL-17A signaling of T cells, both of which might be necessary to drive full-blown psoriasis.
In conclusion, our findings reveal that two major cytokines, IL-36 and IL-17A, promote psoriasis by inducing IκBζ expression. While IL-17A antibodies have proven therapeutic efficacy, blocking of IL-36 might represent an alternative for patients resistant to anti–IL-17A therapy. Moreover, targeting their common mediator IκBζ might lead to future approaches for efficient long-term treatment of psoriasis patients.
Materials and Methods
Detailed information on cell culture experiments, generation of knockdown cells, luciferase reporter assays, ChIP, analyses of RNA and protein expression, RNA-seq, cytokine antibody arrays, generation of Nfkbiz-KO mice, flow cytometry, histology, and analysis of patient data is provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank C. Schönfeld, C. Resch, and J. Loeffler for assistance and A. Witten and M. Stoll (University of Münster) for transcriptome profiling. Murine IL-36α was generously provided by Amgen. This work was supported by the grants Sonderforschungsbereich/Transregio SFB/TR 156 (to S.H. and D.K.), SFB/TR 209 (to K.S.-O. and S.H.), the Emmy-Noether program of the Deutsche Forschungsgemeinschaft (to S.H.), and the German Ministry for Education and Research Grant of the Network for Autoinflammatory Disorders in Children and Adolescents 01FP09104B (to K.S.-O.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. S.G. is a guest editor invited by the Editorial Board.
Data deposition: RNA-sequencing data have been deposited in the National Center for Biotechnology Information BioProject database (ID PRJNA465504; Sequence Read Archive accession no. SRP144926).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1801377115/-/DCSupplemental.
References
- 1.Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- 2.Renner F, Schmitz ML. Autoregulatory feedback loops terminating the NF-kappaB response. Trends Biochem Sci. 2009;34:128–135. doi: 10.1016/j.tibs.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 3.Smale ST. Hierarchies of NF-κB target-gene regulation. Nat Immunol. 2011;12:689–694. doi: 10.1038/ni.2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Annemann M, et al. Atypical IκB proteins in immune cell differentiation and function. Immunol Lett. 2016;171:26–35. doi: 10.1016/j.imlet.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 5.Totzke G, et al. A novel member of the IkappaB family, human IkappaB-zeta, inhibits transactivation of p65 and its DNA binding. J Biol Chem. 2006;281:12645–12654. doi: 10.1074/jbc.M511956200. [DOI] [PubMed] [Google Scholar]
- 6.Hildebrand DG, et al. IκBζ is a transcriptional key regulator of CCL2/MCP-1. J Immunol. 2013;190:4812–4820. doi: 10.4049/jimmunol.1300089. [DOI] [PubMed] [Google Scholar]
- 7.Yamamoto M, et al. Regulation of Toll/IL-1-receptor-mediated gene expression by the inducible nuclear protein IkappaBzeta. Nature. 2004;430:218–222. doi: 10.1038/nature02738. [DOI] [PubMed] [Google Scholar]
- 8.Kitamura H, Kanehira K, Okita K, Morimatsu M, Saito M. MAIL, a novel nuclear I kappa B protein that potentiates LPS-induced IL-6 production. FEBS Lett. 2000;485:53–56. doi: 10.1016/s0014-5793(00)02185-2. [DOI] [PubMed] [Google Scholar]
- 9.Kayama H, et al. Class-specific regulation of pro-inflammatory genes by MyD88 pathways and IkappaBzeta. J Biol Chem. 2008;283:12468–12477. doi: 10.1074/jbc.M709965200. [DOI] [PubMed] [Google Scholar]
- 10.Tartey S, et al. Akirin2 is critical for inducing inflammatory genes by bridging IκB-ζ and the SWI/SNF complex. EMBO J. 2014;33:2332–2348. doi: 10.15252/embj.201488447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johansen C, et al. IκBζ is a key driver in the development of psoriasis. Proc Natl Acad Sci USA. 2015;112:E5825–E5833. doi: 10.1073/pnas.1509971112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lowes MA, Suárez-Fariñas M, Krueger JG. Immunology of psoriasis. Annu Rev Immunol. 2014;32:227–255. doi: 10.1146/annurev-immunol-032713-120225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsoi LC, et al. Enhanced meta-analysis and replication studies identify five new psoriasis susceptibility loci. Nat Commun. 2015;6:7001. doi: 10.1038/ncomms8001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahil SK, et al. An analysis of IL-36 signature genes and individuals with IL1RL2 knockout mutations validates IL-36 as a psoriasis therapeutic target. Sci Transl Med. 2017;9:eaan2514, and erratum (2017) 9:eaar6600. doi: 10.1126/scitranslmed.aan2514. [DOI] [PubMed] [Google Scholar]
- 15.Dunn E, Sims JE, Nicklin MJ, O’Neill LA. Annotating genes with potential roles in the immune system: Six new members of the IL-1 family. Trends Immunol. 2001;22:533–536. doi: 10.1016/s1471-4906(01)02034-8. [DOI] [PubMed] [Google Scholar]
- 16.Dinarello CA. Overview of the IL-1 family in innate inflammation and acquired immunity. Immunol Rev. 2018;281:8–27. doi: 10.1111/imr.12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bassoy EY, Towne JE, Gabay C. Regulation and function of interleukin-36 cytokines. Immunol Rev. 2018;281:169–178. doi: 10.1111/imr.12610. [DOI] [PubMed] [Google Scholar]
- 18.Towne JE, Garka KE, Renshaw BR, Virca GD, Sims JE. Interleukin (IL)-1F6, IL-1F8, and IL-1F9 signal through IL-1Rrp2 and IL-1RAcP to activate the pathway leading to NF-kappaB and MAPKs. J Biol Chem. 2004;279:13677–13688. doi: 10.1074/jbc.M400117200. [DOI] [PubMed] [Google Scholar]
- 19.Towne JE, et al. Interleukin-36 (IL-36) ligands require processing for full agonist (IL-36α, IL-36β, and IL-36γ) or antagonist (IL-36Ra) activity. J Biol Chem. 2011;286:42594–42602. doi: 10.1074/jbc.M111.267922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blumberg H, et al. Opposing activities of two novel members of the IL-1 ligand family regulate skin inflammation. J Exp Med. 2007;204:2603–2614. doi: 10.1084/jem.20070157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Afonina IS, Müller C, Martin SJ, Beyaert R. Proteolytic processing of interleukin-1 family cytokines: Variations on a common theme. Immunity. 2015;42:991–1004. doi: 10.1016/j.immuni.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 22.D’Erme AM, et al. IL-36γ (IL-1F9) is a biomarker for psoriasis skin lesions. J Invest Dermatol. 2015;135:1025–1032. doi: 10.1038/jid.2014.532. [DOI] [PubMed] [Google Scholar]
- 23.Boutet MA, et al. Distinct expression of interleukin (IL)-36α, β and γ, their antagonist IL-36Ra and IL-38 in psoriasis, rheumatoid arthritis and Crohn’s disease. Clin Exp Immunol. 2016;184:159–173. doi: 10.1111/cei.12761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnston A, et al. IL-1F5, -F6, -F8, and -F9: A novel IL-1 family signaling system that is active in psoriasis and promotes keratinocyte antimicrobial peptide expression. J Immunol. 2011;186:2613–2622. doi: 10.4049/jimmunol.1003162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carrier Y, et al. Inter-regulation of Th17 cytokines and the IL-36 cytokines in vitro and in vivo: Implications in psoriasis pathogenesis. J Invest Dermatol. 2011;131:2428–2437. doi: 10.1038/jid.2011.234. [DOI] [PubMed] [Google Scholar]
- 26.Pfaff CM, Marquardt Y, Fietkau K, Baron JM, Lüscher B. The psoriasis-associated IL-17A induces and cooperates with IL-36 cytokines to control keratinocyte differentiation and function. Sci Rep. 2017;7:15631. doi: 10.1038/s41598-017-15892-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marrakchi S, et al. Interleukin-36-receptor antagonist deficiency and generalized pustular psoriasis. N Engl J Med. 2011;365:620–628. doi: 10.1056/NEJMoa1013068. [DOI] [PubMed] [Google Scholar]
- 28.Onoufriadis A, et al. Mutations in IL36RN/IL1F5 are associated with the severe episodic inflammatory skin disease known as generalized pustular psoriasis. Am J Hum Genet. 2011;89:432–437. doi: 10.1016/j.ajhg.2011.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blumberg H, et al. IL-1RL2 and its ligands contribute to the cytokine network in psoriasis. J Immunol. 2010;185:4354–4362. doi: 10.4049/jimmunol.1000313. [DOI] [PubMed] [Google Scholar]
- 30.Tortola L, et al. Psoriasiform dermatitis is driven by IL-36-mediated DC-keratinocyte crosstalk. J Clin Invest. 2012;122:3965–3976. doi: 10.1172/JCI63451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gaffen SL. Structure and signalling in the IL-17 receptor family. Nat Rev Immunol. 2009;9:556–567. doi: 10.1038/nri2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamazaki S, Muta T, Matsuo S, Takeshige K. Stimulus-specific induction of a novel nuclear factor-kappaB regulator, IkappaB-zeta, via toll/interleukin-1 receptor is mediated by mRNA stabilization. J Biol Chem. 2005;280:1678–1687. doi: 10.1074/jbc.M409983200. [DOI] [PubMed] [Google Scholar]
- 33.Bernstein BE, et al. The NIH roadmap epigenomics mapping consortium. Nat Biotechnol. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shiina T, et al. Genomic organization, chromosomal localization, and promoter analysis of the mouse Mail gene. Immunogenetics. 2001;53:649–655. doi: 10.1007/s00251-001-0376-x. [DOI] [PubMed] [Google Scholar]
- 35.Ishiguro-Oonuma T, Ochiai K, Hashizume K, Morimatsu M. The role of IFN-γ in regulating Nfkbiz expression in epidermal keratinocytes. Biomed Res. 2015;36:103–107. doi: 10.2220/biomedres.36.103. [DOI] [PubMed] [Google Scholar]
- 36.Okuma A, et al. Enhanced apoptosis by disruption of the STAT3-IκB-ζ signaling pathway in epithelial cells induces Sjögren’s syndrome-like autoimmune disease. Immunity. 2013;38:450–460. doi: 10.1016/j.immuni.2012.11.016. [DOI] [PubMed] [Google Scholar]
- 37.Ueta M, et al. Stat6-independent tissue inflammation occurs selectively on the ocular surface and perioral skin of IkappaBzeta-/- mice. Invest Ophthalmol Vis Sci. 2008;49:3387–3394. doi: 10.1167/iovs.08-1691. [DOI] [PubMed] [Google Scholar]
- 38.Karlsen JR, Borregaard N, Cowland JB. Induction of neutrophil gelatinase-associated lipocalin expression by co-stimulation with interleukin-17 and tumor necrosis factor-alpha is controlled by IkappaB-zeta but neither by C/EBP-beta nor C/EBP-delta. J Biol Chem. 2010;285:14088–14100. doi: 10.1074/jbc.M109.017129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muromoto R, et al. IL-17A plays a central role in the expression of psoriasis signature genes through the induction of IκB-ζ in keratinocytes. Int Immunol. 2016;28:443–452. doi: 10.1093/intimm/dxw011. [DOI] [PubMed] [Google Scholar]
- 40.El Malki K, et al. An alternative pathway of imiquimod-induced psoriasis-like skin inflammation in the absence of interleukin-17 receptor a signaling. J Invest Dermatol. 2013;133:441–451. doi: 10.1038/jid.2012.318. [DOI] [PubMed] [Google Scholar]
- 41.Sano S, et al. Stat3 links activated keratinocytes and immunocytes required for development of psoriasis in a novel transgenic mouse model. Nat Med. 2005;11:43–49. doi: 10.1038/nm1162. [DOI] [PubMed] [Google Scholar]
- 42.Andrés RM, Montesinos MC, Navalón P, Payá M, Terencio MC. NF-κB and STAT3 inhibition as a therapeutic strategy in psoriasis: In vitro and in vivo effects of BTH. J Invest Dermatol. 2013;133:2362–2371. doi: 10.1038/jid.2013.182. [DOI] [PubMed] [Google Scholar]
- 43.Miyoshi K, et al. Stat3 as a therapeutic target for the treatment of psoriasis: A clinical feasibility study with STA-21, a Stat3 inhibitor. J Invest Dermatol. 2011;131:108–117. doi: 10.1038/jid.2010.255. [DOI] [PubMed] [Google Scholar]
- 44.Okamoto K, et al. IkappaBzeta regulates T(H)17 development by cooperating with ROR nuclear receptors. Nature. 2010;464:1381–1385. doi: 10.1038/nature08922. [DOI] [PubMed] [Google Scholar]
- 45.Johnston A, et al. Keratinocyte overexpression of IL-17C promotes psoriasiform skin inflammation. J Immunol. 2013;190:2252–2262. doi: 10.4049/jimmunol.1201505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scholz T, et al. GM-CSF in murine psoriasiform dermatitis: Redundant and pathogenic roles uncovered by antibody-induced neutralization and genetic deficiency. PLoS One. 2017;12:e0182646. doi: 10.1371/journal.pone.0182646. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.