Significance
The human T cell leukemia virus I basic leucine zipper protein (HBZ) plays a central role in leukemogenesis and proliferation of transformed adult T cell leukemia cells. Through its interactions with the KIX domain of the transcriptional coactivators CBP and p300, HBZ deregulates transcriptional programs involved in hematopoietic differentiation. Here, we describe the crystal structure of the ternary complex formed between KIX and the activation domains of HBZ and the cellular transcription factor c-Myb. By binding to KIX to form the ternary complex, HBZ allosterically stabilizes the interaction between KIX and c-Myb. These studies provide molecular insights into the mechanism by which HBZ interferes with hematopoietic signaling pathways and promotes T cell proliferation.
Keywords: leukemogenesis, intrinsically disordered proteins, CREB-binding protein, T cell leukemia virus
Abstract
The human T cell leukemia virus I basic leucine zipper protein (HTLV-1 HBZ) maintains chronic viral infection and promotes leukemogenesis through poorly understood mechanisms involving interactions with the KIX domain of the transcriptional coactivator CBP and its paralog p300. The KIX domain binds regulatory proteins at the distinct MLL and c-Myb/pKID sites to form binary or ternary complexes. The intrinsically disordered N-terminal activation domain of HBZ (HBZ AD) deregulates cellular signaling pathways by competing directly with cellular and viral transcription factors for binding to the MLL site and by allosterically perturbing binding of the transactivation domain of the hematopoietic transcription factor c-Myb. Crystal structures of the ternary KIX:c-Myb:HBZ complex show that the HBZ AD recruits two KIX:c-Myb entities through tandem amphipathic motifs (L/V)(V/L)DGLL and folds into a long α-helix upon binding. Isothermal titration calorimetry reveals strong cooperativity in binding of the c-Myb activation domain to the KIX:HBZ complex and in binding of HBZ to the KIX:c-Myb complex. In addition, binding of KIX to the two HBZ (V/L)DGLL motifs is cooperative; the structures suggest that this cooperativity is achieved through propagation of the HBZ α-helix beyond the first binding motif. Our study suggests that the unique structural flexibility and the multiple interaction motifs of the intrinsically disordered HBZ AD are responsible for its potency in hijacking KIX-mediated transcription pathways. The KIX:c-Myb:HBZ complex provides an example of cooperative stabilization in a transcription factor:coactivator network and gives insights into potential mechanisms through which HBZ dysregulates hematopoietic transcriptional programs and promotes T cell proliferation.
Cooperative binding is a ubiquitous mechanism by which protein function is regulated in vivo (1). Eukaryotic transcriptional activation is exquisitely regulated by cooperative interactions between transcription factors and coactivators (2–4). The CREB-binding protein (CBP) and its paralog p300 are transcriptional coactivators that interact with numerous transcription factors through their modular protein-binding domains (5). Among these, the KIX domain integrates cellular signaling pathways by cooperatively interacting with the intrinsically disordered activation domains of transcription factors (6). Through its interactions with the transactivation domains of c-Myb, CREB, the mixed lineage leukemia protein (MLL), E2A, FOXO3, and other hematopoietic factors, KIX forms a critical node in the pathways that regulate hematopoietic differentiation and proliferation (7, 8).
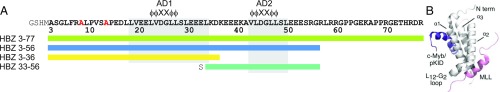
The KIX domain is also the target for viral transcription factors that compete with cellular transcription factors and result in subversion of cellular processes following infection. One such viral transcription factor is the human T cell leukemia virus type I (HTLV-1) basic leucine zipper protein (HBZ), which plays a crucial role in enhancing infectivity and maintaining chronic viral infection and leukemogenesis (9, 10). HTLV-1 is an oncogenic retrovirus associated with multiple lymphocyte-mediated diseases including adult T cell leukemia (ATL), a fatal neoplastic disease of CD4+ T lymphocytes (11). It is estimated that 10–20 million people worldwide are asymptomatic carriers of HTLV-1, with 1–5% of those expected to develop ATL (12). Although the role of HBZ in HTLV-1 pathogenesis is at present poorly understood, it appears to inhibit viral gene transcription by disrupting Tax-dependent viral transactivation. Upon HTLV-1 infection, the viral-encoded transcription factor Tax activates viral gene transcription in the early stages of leukemogenesis through recruitment of CBP by binding to the KIX domain, inducing a strong cytotoxic T cell response (13, 14). HBZ down-regulates Tax-induced transcription, allowing HTLV-1 to evade immunodetection (14, 15). Specifically, the isolated HBZ activation domain (HBZ AD, residues 1–77; Fig. 1A) directly competes with Tax by binding to KIX (16, 17).
Fig. 1.
(A) HBZ AD constructs used in the experiments. The KIX-binding regions of the AD1 and AD2 domains are highlighted with gray boxes and the conserved amphipathic ΦΦXXΦΦ (Φ is a bulky hydrophobic residue; X represent any hydrophilic amino acid) binding motifs within each domain are indicated. Two N-terminal alanine residues in red were mutated from cysteines. Additional N-terminal residues in gray are from recombinant protein production. (B) Structure of the KIX:cMyb:MLL ternary complex (6), showing the binding sites on KIX (gray) occupied by the activation domains of c-Myb (purple) and MLL (pink).
KIX consists of three α-helices (α1, α2, and α3) and two 310 helices (G1 and G2) that form two distinct binding surfaces: the binding site for c-Myb and phosphorylated CREB, composed of the α1 and α3 helices (18, 19), and the MLL binding site at the interface among α2, α3, and the L12-G2 loop that connects helices α1 and α2 (Fig. 1B) (3). The HBZ AD contains two similar amphipathic motifs (L/V)(V/L)DGLL (residues 23–28 and residues 43–48, Fig. 1A) that mediate binding to the MLL site on KIX, with the N-terminal motif contributing most to the binding affinity (16, 17). While the HBZ AD competes with the MLL activation domain for binding to KIX, it cooperatively enhances binding of c-Myb and the kinase inducible activation domain of CREB (pKID) to the opposite surface of KIX (16, 20). Little is known about the molecular mechanism through which HBZ AD interacts with KIX and allosterically modulates binding of other cellular factors.
Here, we report crystal structures of the ternary complex (denoted KIX:c-Myb:HBZ) formed by HBZ AD, the KIX domain of CBP, and the activation domain of c-Myb, a transcription factor that plays a crucial role in hematopoietic differentiation and oncogenesis (21). Surprisingly, the intrinsically disordered HBZ AD folds upon binding to form a single continuous α-helix, where the N-terminal (AD1) and C-terminal (AD2) subdomains each bind to one molecule of KIX. A number of biophysical techniques confirmed the 2:2:1 stoichiometry of the ternary complex and the reported cooperativity of c-Myb and HBZ AD binding to KIX (16, 20). By competing with MLL and other transcription factors for binding to KIX and by greatly enhancing binding of the c-Myb activation domain, HTLV-1 HBZ can efficiently hijack the cellular transcriptional machinery and deregulate CBP/p300-mediated hematopoietic pathways.
Results
Conformation of the Free HBZ AD.
The full-length HBZ AD [residues 3–77, henceforth abbreviated as HBZ (3–77)] and a truncated construct HBZ (3–56) were used for structure determination, affinity measurements, and NMR studies. To investigate the function of the individual (L/V)(V/L)DGLL motifs, shorter constructs spanning the AD1 and AD2 domains, designated HBZ (3–36) and HBZ (33–56), respectively, were expressed (Fig. 1A). Far-UV circular dichroism (CD) and NMR 1H-15N heteronuclear single quantum coherence (HSQC) spectra show that the full-length and truncated HBZ AD constructs are almost entirely disordered in their free forms (SI Appendix, Fig. S1).
Structure of HBZ AD in Complex with KIX and c-Myb.
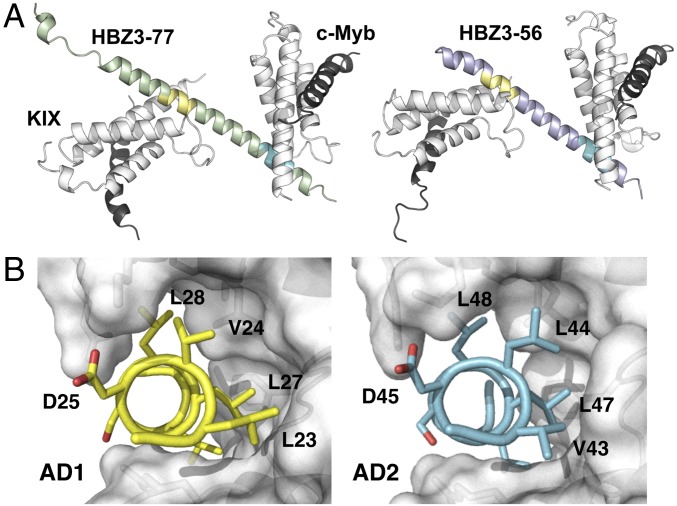
The KIX domain of CBP recognizes transcriptional activation domains of varying sequence through two distinct, conformationally dynamic binding sites (Fig. 1B). To elucidate the structural basis for the interactions among the HBZ AD, KIX, and the transactivation domain of c-Myb (residues 284–315), we determined the crystal structures of the ternary complexes KIX:c-Myb:HBZ (3–77) and KIX:c-Myb:HBZ (3–56) at 2.35 Å and 2.80 Å, respectively (SI Appendix, Table S1). The two structures have different crystal forms, allowing intramolecular and intermolecular contacts due to crystal packing to be distinguished from physiologically relevant interactions that are conserved between the two structures. The KIX:c-Myb:HBZ (3–77) complex crystallized in space group C2, with one molecule of HBZ (3–77) and two copies of the KIX:c-Myb complex in the asymmetric unit. The asymmetric unit of the KIX:c-Myb:HBZ (3–56) crystals, in space group P21, contains two molecules of HBZ (3–56) and four copies of the KIX:c-Myb complex. Nevertheless, the structures of the individual molecules and their binding modes are very similar (Fig. 2A), and the higher-resolution KIX:c-Myb:HBZ (3–77) structure will be used for further discussion. Inspection of 2Fo-Fc maps contoured at 1.0σ shows well-defined electron density for both AD1 and AD2 and residues A3 through G56 of HBZ are visible at 0.8σ (SI Appendix, Fig. S2).
Fig. 2.
Crystal structures of KIX:c-Myb:HBZ (3–77) and KIX:c-Myb:HBZ (3–56) complexes. (A) Overall structures of HBZ (3–77) (green) and HBZ (3–56) (purple) in ternary complexes with KIX:c-Myb, illustrating their similarity. AD1 is shown in yellow and AD2 in cyan. (B) The hydrophobic residues of the AD1 LVDGLL (yellow) and AD2 VLDGLL (cyan) motifs pack in the MLL-binding groove of KIX (gray surface).
In both crystal forms, residues 16–54 of HBZ fold to form a long α-helix that binds two KIX molecules through the (L/V)(V/L)DGLL motifs and flanking residues (Fig. 2A). The AD1 and AD2 domains bind in the hydrophobic groove that constitutes the MLL site, while c-Myb occupies the c-Myb/pKID binding site located on the opposite surface of KIX. The conserved hydrophobic residues of the AD1 (L23, V24, L27, L28, and L30) and AD2 domains (V43, L44, L47, L48, and L50) make similar interactions with hydrophobic side chains on the α2 and α3 helices and in the L12-G2 loop (residues 612–623) of KIX (Fig. 2B). These hydrophobic contacts are supplemented by a network of complementary electrostatic interactions, not all of which are conserved between the AD1 and AD2 binding sites. The differing lengths of the side chains at the N-terminal of the (L/V)(V/L)DGLL motifs result in a subtle reorientation of the HBZ helix axis, relative to KIX, in the AD1 and AD2 binding sites.
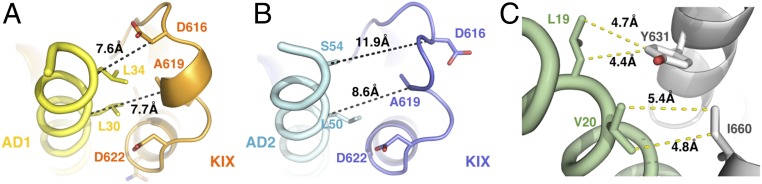
The structure of the L12-G2 loop differs between the KIX molecules bound at the AD1 and AD2 sites of HBZ (Fig. 3 A and B and SI Appendix, Fig. S3). Although larger-than-average B values indicate flexibility in both sites, the L12-G2 loop region in the AD1-bound KIX forms a well-defined 310-helix, whereas the loop is more flexible in the AD2 site and adopts a different conformation (Fig. 3 A and B). The L12-G2 loop is more closely packed against HBZ in the AD1 site, burying a larger surface area (210 Å2 on AD1 compared with 160 Å2 on AD2), with residues 614−619 of KIX contributing the most to the differences.
Fig. 3.
Differences in binding of AD1 and AD2. (A) AD1 (yellow) bound to the L12-G2 loop region of AD1-bound KIX (orange). (B) AD2 (cyan) bound to AD2-bound KIX (blue). (C) Additional intermolecular contacts between residues in the N-terminal region of the AD1 domain (green) and KIX (gray).
NMR Characterization of HBZ Interactions with KIX.
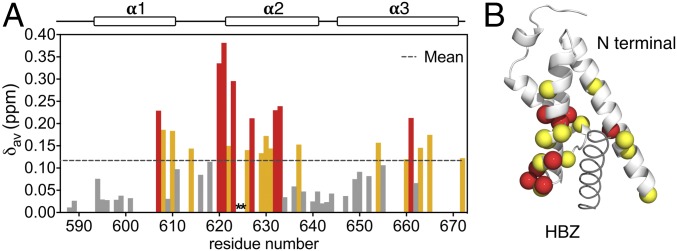
The interactions of the individual AD1 and AD2 domains of HBZ with KIX were characterized using NMR titrations. 1H-15N HSQC spectra show that binding of HBZ (33–56), which contains AD2, is in fast exchange on the chemical shift timescale (SI Appendix, Fig. S4A). In contrast, a construct containing the AD1 motif, HBZ (3–36) binds to KIX in intermediate exchange and extensive resonance broadening (SI Appendix, Fig. S5) precludes quantitative analysis of the binding affinity. The observation of intermediate-exchange interactions for HBZ (3–36) and fast-exchange interactions for HBZ (33–56) is consistent with higher affinity binding by AD1 (17). The 1H-15N HSQC spectrum of fully bound 15N-labeled HBZ (33–56) shows increased dispersion in the 1H dimension compared with the free peptide, indicating folding of AD2 upon binding to KIX (SI Appendix, Fig. S4A). Singular value decomposition (SVD) analysis (SI Appendix, Fig. S4C) of the 15N KIX titration with unlabeled HBZ (33–56) (SI Appendix, Fig. S4D) confirms the 1:1 stoichiometry of the AD2:KIX complex, while chemical shift changes and cross-peak broadening show that AD2 binds preferentially in the MLL site (Fig. 4). Fitting of the chemical shift titration curves (SI Appendix, Fig. S4 B and E) to a one-site binding model gives Kd values of 30 ± 3 and 21 ± 2 μM for titrations of unlabeled HBZ (33–56) into 15N-KIX and unlabeled KIX into15N-HBZ (33–56), respectively.
Fig. 4.
The primary binding site of the HBZ AD2 motif is the MLL site of KIX. (A) Histogram of the weighted average chemical shift differences Δδav for the backbone 1HN and 15N resonances of KIX between the free and AD2-bound state [ratio of KIX:HBZ (33–56) is 1:6]. Residues with Δδav greater than the mean (dotted line) plus one SD are colored red; residues with Δδav between mean and mean plus one SD are colored yellow; residues with Δδav between 0.05 and the mean are colored dark gray. The Δδav values of residues 624 and 625 cannot be calculated due to line broadening upon interaction with HBZ (33–56), and are therefore left blank and marked with asterisks in the histogram. (B) When mapped onto the structure of KIX bound to AD2, residues shown in red and yellow in A are primarily located in the MLL binding site.
Binding of KIX to 15N-HBZ (3–56), which contains both the AD1 and AD2 domains, is in intermediate exchange and results in extensive broadening of HBZ cross-peaks (SI Appendix, Fig. S6A). However, qualitative characterization of the interactions could be accomplished by inspection of peak intensity changes. At a low substoichiometric molar ratio of KIX to 15N-labeled HBZ (0.1:1), the intensities of the AD1 cross-peaks in the HSQC spectrum decrease drastically, whereas resonances of the AD2 motif are much less severely affected (SI Appendix, Fig. S6 B and C).
Whereas the binary interaction between 15N-labeled HBZ (3–56) and KIX leads to exchange broadening, interaction of HBZ with the KIX:c-Myb complex occurs in slow exchange on the chemical shift time scale. At 1:1:1 stoichiometry, the HSQC spectrum contains cross-peaks arising from both free HBZ and an KIX:c-Myb:HBZ complex (SI Appendix, Fig. S7); the cross-peak of G26 (AD1) is close to its position in the 1:2:2 complex (SI Appendix, Fig. S7), whereas the cross-peak of G46 (AD2) only undergoes a substantial shift after addition of the second aliquot of KIX:c-Myb, showing that binding preferentially occurs through the AD1 motif. At 1:2:2 HBZ:KIX:c-Myb stoichiometry, no cross-peaks associated with free HBZ are observed and new cross-peaks that indicate binding at the AD2 motif appear in the spectrum. Thus, the NMR data confirm that the 1:2:2 stoichiometry observed in the X-ray structures also occurs in solution.
Thermodynamic Basis for Cooperative Binding.
Previous GST pull-down and electrophoretic mobility shift assays indicated that HBZ and c-Myb bind cooperatively to KIX and that AD1 contributes more to KIX binding than does AD2 (16, 17). To quantitate the contributions of AD1 and AD2 to KIX binding and elucidate the thermodynamic basis of cooperativity, we measured binding affinities for the binary and ternary complexes using isothermal titration calorimetry (ITC) (Table 1 and SI Appendix, Fig. S8). Binding of peptides containing the isolated AD1 and AD2 motifs [HBZ (3–36) and HBZ (33–56), respectively] is relatively weak (Kd 8.2 μM for AD1; binding of AD2 is too weak to measure by ITC but is estimated to be 21–30 μM from the NMR titrations). Longer HBZ constructs containing both AD1 and AD2 bind to KIX with approximately three- to eightfold higher affinity than the isolated AD1 peptide. There is an increased entropic penalty for binding of the longer peptides [e.g., TΔΔS = −8.1 kcal/mol for binary complex formation by HBZ (3–56) relative to binding of HBZ (3–36)], which is presumably associated with folding and propagation of the helix spanning the AD1 and AD2 binding sites; this entropic penalty is offset by a more favorable binding enthalpy [ΔΔH = −8.7 kcal/mol for HBZ (3–56); Table 1].
Table 1.
Thermodynamics measurements (ITC) for binary and ternary complex formation between HBZ AD, KIX, and c-Myb
| Ligand | Binding partner | Kd, µM | ΔG, kcal·mol−1 | ΔH, kcal·mol−1 | −TΔS, kcal·mol−1 T = 301K | ΔΔHternary-binary, kcal·mol−1 | -TΔΔSternary-binary, kcal·mol−1 |
| HBZ (3–77) | KIX* | 1.0 ± 0.1 | −8.2 ± 0.1 | −14.9 ± 0.4 | −6.3 ± 0.6 | −4.2 | −2.7 |
| HBZ (3–77) | KIX:c-Myb* | 0.042 ± 0.005 | −10.2 ± 0.1 | −19.1 ± 0.3 | −9.0 ± 0.3 | −4.2 | −2.7 |
| HBZ (3–56) | KIX* | 2.4 ± 0.2 | −7.7 ± 0.05 | −11.1 ± 0.1 | −3.5 ± 0.2 | −4.6 | −2.4 |
| HBZ (3–56) | KIX:c-Myb* | 0.11 ± 0.01 | −9.6 ± 0.1 | −15.7 ± 0.2 | −5.9 ± 0.2 | −4.6 | −2.4 |
| HBZ (3–36) | KIX* | 8.2 ± 0.6 | −7.0 ± 0.04 | −2.4 ± 0.2 | 4.6 ± 0.3 | −4.2 | −2.2 |
| HBZ (3–36) | KIX:c-Myb* | 0.34 ± 0.05 | −8.9 ± 0.1 | −6.5 ± 0.9 | 2.4 ± 0.9 | −4.2 | −2.2 |
| c-Myb | KIX† | 0.19 ± 0.003 | −9.3 ± 0.01 | −3.2 ± 0.02 | −6.1 ± 0.02 | −6.6 | −5.4 |
| c-Myb | KIX:AD1† | 0.029 ± 0.004 | −10.4 ± 0.01 | −9.8 ± 0.1 | 0.63 ± 0.08 | −6.6 | −5.4 |
The Kd, ΔH, and −TΔS values are the average and SD of triplicate experiments. ΔG values were calculated with ΔG = −RTlnKd. All measurements were taken at T = 301 K.
Thermodynamic values are the average and SD of duplicate experiments.
Formation of ternary KIX:c-Myb:HBZ complexes from either of the binary complexes is strongly cooperative. The affinities of HBZ (3–56) and HBZ (3–77) binding to the preformed KIX:c-Myb complex are more than 20-fold higher than for binding to free KIX (Table 1). A similar enhancement in affinity is observed for the isolated AD1 peptide, HBZ (3–36). Conversely, binding of the c-Myb peptide to the preformed KIX:HBZ (3–36) complex is enhanced only eightfold relative to its affinity for free KIX. In all cases, ternary complex formation is driven by a decrease in enthalpy, while the entropy change is unfavorable (Table 1). The entropic penalties for binding of each of the three HBZ constructs to the KIX:c-Myb complex are similar (TΔΔS = −2.2 to −2.7 kcal/mol), whereas binding of c-Myb to the KIX:HBZ (3–36) complex incurs a larger entropic penalty (TΔΔS = −5.4 kcal/mol). Since the 32-residue c-Myb peptide spontaneously forms a high population of helix in the unbound state (22), the increased entropic cost of ternary complex formation may reflect local ordering of the KIX:HBZ (3–36) complex upon c-Myb binding.
Discussion
AD1 Dominates the Interactions Between KIX and the HBZ Activation Domain.
Using NMR titrations and ITC, we show that the isolated AD1 peptide has a higher affinity for KIX (8.2 ± 0.6 μM) than does AD2 (21–30 μM). This is consistent with previous findings that AD1 contributes most to the interactions between the HBZ AD and KIX as well as to the HBZ transcriptional activity (16, 17). Differential changes in cross-peak intensity in the 1H-15N HSQC spectrum of HBZ (3–56) upon binding to KIX (SI Appendix, Fig. S6) confirm that AD1 is indeed the primary interaction motif, even within the full-length HBZ activation domain. The differences in binding affinity of the AD1 and AD2 domains arise from sequence differences in the regions flanking the (L/V)(V/L)DGLL motifs. In AD1, the hydrophobic residues L19 and V20, replaced by glutamates E39 and E40 in AD2, are located one turn of helix before the LVDGLL motif, where they make additional hydrophobic interactions with Y631 and I660 of KIX (Fig. 3C). In addition, L34, located after the AD1 LVDGLL motif, makes more intimate interactions with the L12-G2 loop than does the corresponding residue (S54) in AD2 (Fig. 3). As a consequence, AD1 buries a larger total surface area (1,780 Å2 versus 1,600 Å2) than does AD2.
Cooperativity Between AD1 and AD2 in Binary Complex Formation.
Intrinsically disordered proteins frequently contain multiple conserved amphipathic or linear motifs that function synergistically to promote high-affinity binding, bridge signaling pathways, and facilitate assembly of higher-order signaling complexes (5, 23, 24). In the case of HBZ, the disordered activation domain utilizes the AD1 and AD2 domains synergistically to interact with KIX; HBZ (3–56), which contains both AD1 and AD2, binds to KIX with approximately threefold higher affinity than the isolated AD1 domain and 10-fold higher than AD2 alone (Table 1).
Thermodynamic Driving Force for Ternary Complex Formation.
The two binding sites on KIX exhibit positive cooperativity, such that binding of a ligand in the MLL site enhances binding in the c-Myb/pKID site and vice versa (3, 25). Allostery is mediated primarily through a decrease in the dissociation rate for a given ligand when a second ligand is bound at the other site (20, 26). With the pKID or c-Myb activation domains bound to KIX, the affinity for binding the MLL peptide is enhanced approximately twofold (3). Analogous complexes involving the HBZ activation domain instead of MLL show a much larger allosteric effect. The affinities of all HBZ constructs for KIX are increased by more than 20-fold in the presence of c-Myb. In parallel with the KIX, MLL, c-Myb system (3), cooperative formation of the KIX:c-Myb:HBZ ternary complexes is achieved through a favorable enthalpy change (ΔΔHternary-binary < 0) and an unfavorable entropy change (TΔΔSternary-binary < 0) for all constructs tested (Table 1). Similarly, cooperative binding of the c-Myb activation domain to KIX:HBZ (3–36) to form a ternary complex is also driven by a large decrease in enthalpy, which counteracts an unfavorable decrease in entropy. The thermodynamic parameters measured experimentally do not support proposed mechanistic models in which binding of either Myb or MLL at its cognate site stiffens the KIX domain to prepay the entropic cost of binding the second ligand (20, 27). Despite binding to the same site on KIX, c-Myb and pKID differ in the thermodynamic driving force for formation of ternary complexes with MLL. Whereas an entropic penalty is incurred in forming ternary complexes of c-Myb, cooperative binding of pKID and MLL is driven by a favorable increase in entropy and an unfavorable change in enthalpy (3).
Structural Basis for Allostery in KIX.
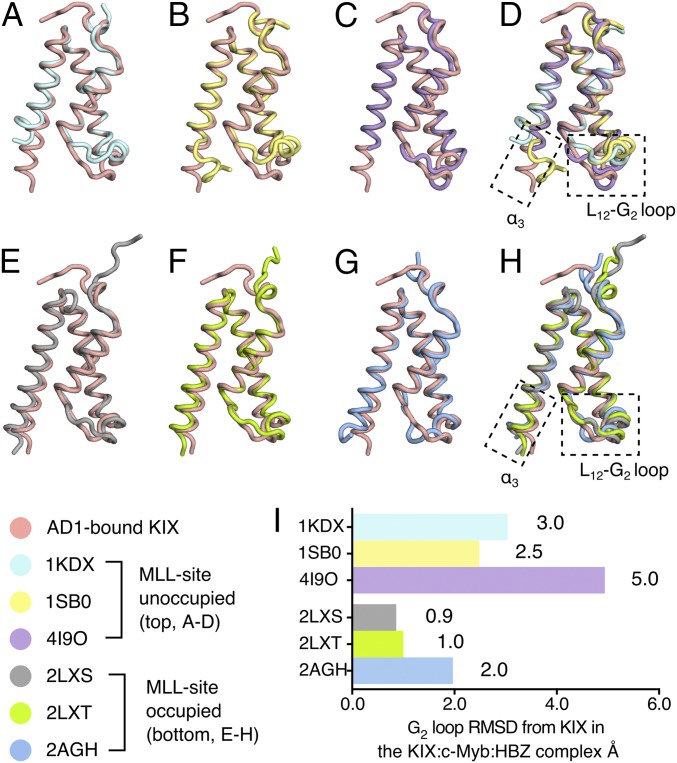
The structure of KIX bound to the AD1 domain in the KIX:c-Myb:HBZ (3–77) ternary complex is compared with published structures of KIX in binary complexes and ternary complexes with bound MLL in Fig. 5. Although the overall Cα rmsd values between the AD1-bound KIX and other KIX structures are relatively small (<1.5 Å in all cases shown in Fig. 5), the structures of the α3 helix and L12-G2 loop regions differ substantially in complexes that lack bound MLL. The C terminus of the α3 helix is disordered beyond E665 in the KIX:pKID and KIX:c-Myb binary complexes and in the covalently stabilized KIX:1–10 complex (19, 26, 28), whereas it is fully structured in HBZ-bound KIX (Fig. 5 B–D). Substantial differences are also observed in the L12-G2 loop; the local rmsd values computed using all backbone heavy atoms for residues 611–620 relative to AD1-bound KIX are 3.0 Å for KIX:pKID, 2.5 Å for KIX:c-Myb, and 4.9 Å for KIX:1–10 (Fig. 5I). In contrast, the α3 helices of all KIX complexes with HBZ or MLL bound in the MLL site are well defined and extend to R671 and the L12-G2 loop regions in each structure adopt similar conformations, with local rmsd values relative to AD1-bound KIX of 0.9 Å for the KIX:MLL binary complex, 1.0 Å for the KIX:pKID:MLL ternary complex, and 2.0 Å for the KIX:c-Myb:MLL ternary complex (Fig. 5 E–H).
Fig. 5.
The structure of KIX in ternary complexes resembles the MLL-bound form. (A–D) Structure of AD1-bound KIX in the KIX:c-Myb:HBZ (3–77) ternary complex (pink) superimposed with KIX:pKID binary complex (PDB ID code 1KDX) (A), KIX:c-Myb binary complex (PDB ID code 1SB0) (B), and KIX (C) covalently tethered to small molecule ligand 1–10 (PDB ID code 4I9O). (D) Superposition of the AD1-bound KIX structure with KIX structures in complexes in which the MLL site is unoccupied (1KDX, 1SB0) or occupied by a small molecule ligand (4I9O) reveals large structural differences in the C-terminal region of the α3 helix and in the L12-G2 loop. All ligands are omitted for clarity. (E–H) Superposition of the structure of AD1-bound KIX in the KIX:c-Myb:HBZ (3–77) complex (pink) with the structures of KIX in the KIX:MLL binary complex (PDB ID code 2LXS) (E), the KIX:pKID:MLL ternary complex (PDB ID code 2LXT) (F), and the KIX:c-Myb:MLL ternary complex (PDB ID code 2AGH) (G). (H) Superposition of the structure of KIX bound to the HBZ AD1 domain and the structures of KIX in ternary complexes with bound MLL (2LXS, 2LXT, and 2AGH). The α3 helix of KIX is well formed and extended and the L12-G2 loop adopts a similar conformation in all four structures with the MLL site occupied. (I) Local rmsd (Å) of the L12-G2 loop regions (residues 611–620, computed using all backbone heavy atoms) between AD1-bound KIX and the structure of KIX in other complexes. In making comparisons with solution structures, the local rmsd values were calculated using the average of all structures in the ensemble.
The α3 helix and L12-G2 loop are thought to play an important role in cooperative ligand binding to KIX (6, 27–31). Molecular dynamics simulations suggest that cooperativity is associated with a redistribution of the conformational ensemble of the L12-G2 loop and α3 helix upon binding of the first ligand to populate a state that favors binding of the second ligand (27, 30, 31). Binding of HBZ or MLL stabilizes the helical structure in the C-terminal region of the α3 helix of KIX (Fig. 5) and provides a more favorable binding surface for c-Myb (6, 29). The α3 helix plays an important role in maintaining the integrity of the MLL site; truncation of α3 at E665 removes part of the MLL contact surface and severely impairs binding of the HTLV-1 Tax oncoprotein in the MLL site while binding of pKID in the c-Myb site is unaffected (13). Changes in the conformation and dynamics of the L12-G2 loop and the C-terminal region of the α3 helix, implicated in allosteric communication between the Myb and MLL binding sites, have been observed directly by NMR relaxation measurements (28, 29).
Although the L12-G2 loop adopts a similar conformation in the KIX:c-Myb:MLL and KIX:c-Myb:HBZ ternary complexes (Fig. 5G), the HBZ and MLL helices bind in different orientations (SI Appendix, Fig. S9) such that HBZ side chains bury a greater surface area of the L12-G2 loop than do those of MLL (180 Å2 in the KIX:c-Myb:HBZ complex versus an average of 140 Å2 in the family of NMR structures of KIX:c-Myb:MLL). HBZ displays more than 10-fold greater cooperativity in binding to KIX:c-Myb than does MLL (Table 1 and ref. 3), lending support to mechanistic models that attribute allosteric communication between the Myb and MLL sites to changes in the conformational ensemble and dynamics of the L12-G2 loop and α3 helix (28, 30, 31). Importantly, the large differences in cooperativity exhibited by HBZ and MLL show that the ligand itself plays a functional role and that the magnitude of the allosteric effects cannot be explained by consideration of the properties of KIX alone.
Potential Selectivity in KIX-Mediated Cooperative Interactions.
The HBZ activation domain and c-Myb display exceptionally strong cooperativity in ternary complex formation with KIX. In the presence of c-Myb, the affinity with which the HBZ AD binds to KIX is increased by more than 20-fold, while c-Myb binds sevenfold more tightly when the HBZ AD1 is prebound to KIX. Although KIX is well-established as a promiscuous interaction site for numerous transcription factors and cooperativity is observed for several ligand pairs (3, 20, 26), the synergy between the HBZ AD and c-Myb appears to be much stronger than for other ligands. In a recent kinetic study by Shammas et al. (20), cooperativity in binding of c-Myb and HBZ to KIX was shown to be mediated by changes in dissociation rate, with an apparent cooperativity constant α of 18.7 for binding of the HBZ AD to the KIX:c-Myb complex and with α = 5.2 for binding of c-Myb to KIX:HBZ. The remarkably strong enhancement of c-Myb binding to KIX in the presence of the HBZ activation domain has important implications for leukemogenesis.
Implications for HTLV-1 Pathogenesis.
HBZ is the only HTLV-1 gene that is uniformly and constantly expressed in leukemic cells of ATL patients and asymptomatic carriers (32). HBZ plays a key role in leukemogenesis by repressing viral gene transcription and promoting T cell proliferation (33). The structure of the KIX:c-Myb:HBZ ternary complex provides insights into the molecular mechanism by which HBZ could hijack the basal transcriptional machinery and alter the transcription level of viral and cellular genes. In the early stages of leukemogenesis, the HTLV-1 protein Tax activates transcription of viral genes by forming a complex with CREB and the KIX domain of CBP/p300 (13). Tax is strongly immunogenic and is down-regulated after infection. By competing with Tax for binding to the MLL site on KIX, HBZ functions to repress viral transcription and help the virus evade detection by the host immune system (17). In addition, by directly competing with c-Jun, MLL, E2A, and other transcription factors that utilize the MLL surface of KIX, HBZ is expected to disrupt key hematopoietic transcriptional programs (16). In contrast, HBZ and c-Myb bind synergistically to KIX, mutually enhancing each other’s binding affinity. Through its interaction with CBP/p300, c-Myb regulates differentiation and proliferation of hematopoietic stem and progenitor cells and is implicated in development and growth of leukemia and other cancers (21, 34–38). By strongly enhancing binding of c-Myb to KIX, HBZ would be expected to up-regulate c-Myb transcriptional programs and thereby contribute to aberrant T cell proliferation and progression of leukemia. Although the role of HBZ in HTLV-1 pathogenesis remains a subject of intense investigation, our data establish the HBZ and c-Myb activation domains as highly synergistic KIX domain ligands that reprogram CBP/p300-mediated transcription and deregulate hematopoiesis.
Materials and Methods
A full description of the materials and methods is provided in SI Appendix.
Protein Preparation.
Peptides corresponding to the HTLV-1 HBZ activation domain were coexpressed with KIX in Escherichia coli as His6-tagged GB1 or SUMO fusion proteins. A 32-residue peptide spanning the mouse c-Myb transactivation domain (Myb32, residues 284–315) was expressed in E. coli as a His6-SUMO fusion protein. Fusion proteins were cleaved after purification with TEV protease or Ulp1 protease and further purified by reverse-phase HPLC. The KIX domain of mouse CBP (residues 586–683) was expressed and purified as described (39).
Crystallization and Structure Determination of HBZ AD:KIX:Myb Ternary Complexes.
Crystals were cryoprotected and flash-cooled in liquid nitrogen. Diffraction data were collected at Stanford Synchrotron Radiation Lightsource (SSRL) beamline 9–2 and processed with HKL-2000 (40). Molecular replacement was performed using Phaser (41) with KIX:c-Myb25 chains from the solution structure ensemble of KIX:c-Myb25:MLL (6), where all 20 NMR structures were used as ensemble input.
NMR Spectroscopy.
15N-labeled and 13C, 15N-labeled proteins were prepared in E. coli using isotope-enriched M9 minimal media. NMR spectra were acquired on Bruker spectrometers, and data were processed and analyzed using NMRPipe (42) and NMRView (43).
ITC.
ITC measurements were performed using a MicroCal iTC200 instrument.
CD.
CD spectra were measured on an Aviv62DS spectropolarimeter.
Supplementary Material
Acknowledgments
We thank Gerard Kroon for expert assistance with NMR experiments and insightful discussions, Peter Haberz for acquiring preliminary data, and Jeanne Matteson for assistance with ITC. This work was supported by Grant CA214054 from the National Institutes of Health and the Skaggs Institute for Chemical Biology. Use of the SSRL, SLAC National Accelerator Laboratory, is supported by the Department of Energy (DOE), Office of Science, Office of Basic Energy Sciences under Contract DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including Grant P41GM103393).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 6DMX and 6DNQ) and Biological Magnetic Resonance Data Bank (accession no. 27517).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1810397115/-/DCSupplemental.
References
- 1.Motlagh HN, Wrabl JO, Li J, Hilser VJ. The ensemble nature of allostery. Nature. 2014;508:331–339. doi: 10.1038/nature13001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nam Y, Sliz P, Song L, Aster JC, Blacklow SC. Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes. Cell. 2006;124:973–983. doi: 10.1016/j.cell.2005.12.037. [DOI] [PubMed] [Google Scholar]
- 3.Goto NK, Zor T, Martinez-Yamout M, Dyson HJ, Wright PE. Cooperativity in transcription factor binding to the coactivator CREB-binding protein (CBP). The mixed lineage leukemia protein (MLL) activation domain binds to an allosteric site on the KIX domain. J Biol Chem. 2002;277:43168–43174. doi: 10.1074/jbc.M207660200. [DOI] [PubMed] [Google Scholar]
- 4.Berlow RB, Dyson HJ, Wright PE. Hypersensitive termination of the hypoxic response by a disordered protein switch. Nature. 2017;543:447–451. doi: 10.1038/nature21705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wright PE, Dyson HJ. Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol. 2015;16:18–29. doi: 10.1038/nrm3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Guzman RN, Goto NK, Dyson HJ, Wright PE. Structural basis for cooperative transcription factor binding to the CBP coactivator. J Mol Biol. 2006;355:1005–1013. doi: 10.1016/j.jmb.2005.09.059. [DOI] [PubMed] [Google Scholar]
- 7.Kimbrel EA, et al. Systematic in vivo structure-function analysis of p300 in hematopoiesis. Blood. 2009;114:4804–4812. doi: 10.1182/blood-2009-04-217794. [DOI] [PubMed] [Google Scholar]
- 8.Kasper LH, et al. Genetic interaction between mutations in c-Myb and the KIX domains of CBP and p300 affects multiple blood cell lineages and influences both gene activation and repression. PLoS One. 2013;8:e82684. doi: 10.1371/journal.pone.0082684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsuoka M, Jeang KT. Human T-cell leukemia virus type 1 (HTLV-1) and leukemic transformation: Viral infectivity, Tax, HBZ and therapy. Oncogene. 2011;30:1379–1389. doi: 10.1038/onc.2010.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arnold J, et al. Enhancement of infectivity and persistence in vivo by HBZ, a natural antisense coded protein of HTLV-1. Blood. 2006;107:3976–3982. doi: 10.1182/blood-2005-11-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoshida M. Discovery of HTLV-1, the first human retrovirus, its unique regulatory mechanisms, and insights into pathogenesis. Oncogene. 2005;24:5931–5937. doi: 10.1038/sj.onc.1208981. [DOI] [PubMed] [Google Scholar]
- 12.Nicot C. Current views in HTLV-I-associated adult T-cell leukemia/lymphoma. Am J Hematol. 2005;78:232–239. doi: 10.1002/ajh.20307. [DOI] [PubMed] [Google Scholar]
- 13.Ramírez JA, Nyborg JK. Molecular characterization of HTLV-1 Tax interaction with the KIX domain of CBP/p300. J Mol Biol. 2007;372:958–969. doi: 10.1016/j.jmb.2007.06.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsuoka M, Jeang KT. Human T-cell leukaemia virus type 1 (HTLV-1) infectivity and cellular transformation. Nat Rev Cancer. 2007;7:270–280. doi: 10.1038/nrc2111. [DOI] [PubMed] [Google Scholar]
- 15.Sugata K, et al. HTLV-1 bZIP factor impairs cell-mediated immunity by suppressing production of Th1 cytokines. Blood. 2012;119:434–444. doi: 10.1182/blood-2011-05-357459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cook PR, Polakowski N, Lemasson I. HTLV-1 HBZ protein deregulates interactions between cellular factors and the KIX domain of p300/CBP. J Mol Biol. 2011;409:384–398. doi: 10.1016/j.jmb.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clerc I, et al. An interaction between the human T cell leukemia virus type 1 basic leucine zipper factor (HBZ) and the KIX domain of p300/CBP contributes to the down-regulation of tax-dependent viral transcription by HBZ. J Biol Chem. 2008;283:23903–23913. doi: 10.1074/jbc.M803116200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Radhakrishnan I, et al. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: A model for activator:coactivator interactions. Cell. 1997;91:741–752. doi: 10.1016/s0092-8674(00)80463-8. [DOI] [PubMed] [Google Scholar]
- 19.Zor T, De Guzman RN, Dyson HJ, Wright PE. Solution structure of the KIX domain of CBP bound to the transactivation domain of c-Myb. J Mol Biol. 2004;337:521–534. doi: 10.1016/j.jmb.2004.01.038. [DOI] [PubMed] [Google Scholar]
- 20.Shammas SL, Travis AJ, Clarke J. Allostery within a transcription coactivator is predominantly mediated through dissociation rate constants. Proc Natl Acad Sci USA. 2014;111:12055–12060. doi: 10.1073/pnas.1405815111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramsay RG, Gonda TJ. MYB function in normal and cancer cells. Nat Rev Cancer. 2008;8:523–534. doi: 10.1038/nrc2439. [DOI] [PubMed] [Google Scholar]
- 22.Arai M, Sugase K, Dyson HJ, Wright PE. Conformational propensities of intrinsically disordered proteins influence the mechanism of binding and folding. Proc Natl Acad Sci USA. 2015;112:9614–9619. doi: 10.1073/pnas.1512799112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van der Lee R, et al. Classification of intrinsically disordered regions and proteins. Chem Rev. 2014;114:6589–6631. doi: 10.1021/cr400525m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Csizmok V, Follis AV, Kriwacki RW, Forman-Kay JD. Dynamic protein interaction networks and new structural paradigms in signaling. Chem Rev. 2016;116:6424–6462. doi: 10.1021/acs.chemrev.5b00548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ernst P, Wang J, Huang M, Goodman RH, Korsmeyer SJ. MLL and CREB bind cooperatively to the nuclear coactivator CREB-binding protein. Mol Cell Biol. 2001;21:2249–2258. doi: 10.1128/MCB.21.7.2249-2258.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang N, Lodge JM, Fierke CA, Mapp AK. Dissecting allosteric effects of activator-coactivator complexes using a covalent small molecule ligand. Proc Natl Acad Sci USA. 2014;111:12061–12066. doi: 10.1073/pnas.1406033111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Law SM, Gagnon JK, Mapp AK, Brooks CL., 3rd Prepaying the entropic cost for allosteric regulation in KIX. Proc Natl Acad Sci USA. 2014;111:12067–12072. doi: 10.1073/pnas.1405831111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brüschweiler S, Konrat R, Tollinger M. Allosteric communication in the KIX domain proceeds through dynamic repacking of the hydrophobic core. ACS Chem Biol. 2013;8:1600–1610. doi: 10.1021/cb4002188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brüschweiler S, et al. Direct observation of the dynamic process underlying allosteric signal transmission. J Am Chem Soc. 2009;131:3063–3068. doi: 10.1021/ja809947w. [DOI] [PubMed] [Google Scholar]
- 30.Korkmaz EN, Nussinov R, Haliloğlu T. Conformational control of the binding of the transactivation domain of the MLL protein and c-Myb to the KIX domain of CREB. PLOS Comput Biol. 2012;8:e1002420. doi: 10.1371/journal.pcbi.1002420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palazzesi F, Barducci A, Tollinger M, Parrinello M. The allosteric communication pathways in KIX domain of CBP. Proc Natl Acad Sci USA. 2013;110:14237–14242. doi: 10.1073/pnas.1313548110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Satou Y, Yasunaga J, Yoshida M, Matsuoka M. HTLV-I basic leucine zipper factor gene mRNA supports proliferation of adult T cell leukemia cells. Proc Natl Acad Sci USA. 2006;103:720–725. doi: 10.1073/pnas.0507631103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma G, Yasunaga J, Matsuoka M. Multifaceted functions and roles of HBZ in HTLV-1 pathogenesis. Retrovirology. 2016;13:16. doi: 10.1186/s12977-016-0249-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sandberg ML, et al. c-Myb and p300 regulate hematopoietic stem cell proliferation and differentiation. Dev Cell. 2005;8:153–166. doi: 10.1016/j.devcel.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 35.Bies J, et al. Myeloid-specific inactivation of p15Ink4b results in monocytosis and predisposition to myeloid leukemia. Blood. 2010;116:979–987. doi: 10.1182/blood-2009-08-238360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin S, et al. c-Myb binds MLL through menin in human leukemia cells and is an important driver of MLL-associated leukemogenesis. J Clin Invest. 2010;120:593–606. doi: 10.1172/JCI38030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uttarkar S, et al. Targeting acute myeloid leukemia with a small molecule inhibitor of the Myb/p300 interaction. Blood. 2015;127:1173–1182. doi: 10.1182/blood-2015-09-668632. [DOI] [PubMed] [Google Scholar]
- 38.Uttarkar S, Frampton J, Klempnauer K-H. Targeting the transcription factor Myb by small-molecule inhibitors. Exp Hematol. 2017;47:31–35. doi: 10.1016/j.exphem.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 39.Sugase K, Dyson HJ, Wright PE. Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature. 2007;447:1021–1025. doi: 10.1038/nature05858. [DOI] [PubMed] [Google Scholar]
- 40.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 41.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 43.Johnson BA, Blevins RA. NMR view: A computer program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.