Significance
Inhibitors of dengue virus, a mosquito-borne pathogen, are urgently needed. Virally encoded NS2B/3, a two-subunit serine proteinase, is responsible for many cleavages within the viral polyprotein. We demonstrate that two cleavage sites flanking NS3 and one that is internal are self-processing. This strict intramolecular cleavage dictates that, if uncleaved precursors can inhibit viral growth, this phenotype will be trans-dominant. Indeed, a mutation that abrogates one of the self-processing sites caused trans-dominant inhibition of wild-type virus. This site is also the most susceptible to a dengue proteinase inhibitor, providing a compelling rationale for its efficacy. We suggest that explicit targeting of specific intramolecular cleavage sites will be an effective antiviral strategy that can suppress multiple variants in an intracellular quasispecies.
Keywords: dengue virus, antiviral agents, polyproteins, trans-dominant inhibition
Abstract
Many positive-strand RNA viruses translate their genomes as single polyproteins that are processed by host and viral proteinases to generate all viral protein products. Among these is dengue virus, which encodes the serine proteinase NS2B/3 responsible for seven different cleavages in the polyprotein. NS2B/3 has been the subject of many directed screens to find chemical inhibitors, of which the compound ARDP0006 is among the most effective at inhibiting viral growth. We show that at least three cleavages in the dengue polyprotein are exclusively intramolecular. By definition, such a cis-acting defect cannot be rescued in trans. This creates the possibility that a drug-susceptible or inhibited proteinase can be genetically dominant, inhibiting the outgrowth of drug-resistant virus via precursor accumulation. Indeed, an NS3-G459L variant that is incapable of cleavage at the internal NS3 junction dominantly inhibited negative-strand RNA synthesis of wild-type virus present in the same cell. This internal NS3 cleavage site is the junction most inhibited by ARDP0006, making it likely that the accumulation of toxic precursors, not inhibition of proteolytic activity per se, explains the antiviral efficacy of this compound in restraining viral growth. We argue that intramolecularly cleaving proteinases are promising drug targets for viruses that encode polyproteins. The most effective inhibitors will specifically target cleavage sites required for processing precursors that exert trans-dominant inhibition.
Dengue virus is a mosquito-borne pathogen that causes an estimated 390 million annual cases globally (1). Infection can lead to dengue fever, dengue hemorrhagic fever, or dengue shock syndrome, and no vaccines or antiviral compounds are currently approved for use. The development of preventative and therapeutic treatments for this and other neglected or emerging diseases is a priority for global healthcare organizations.
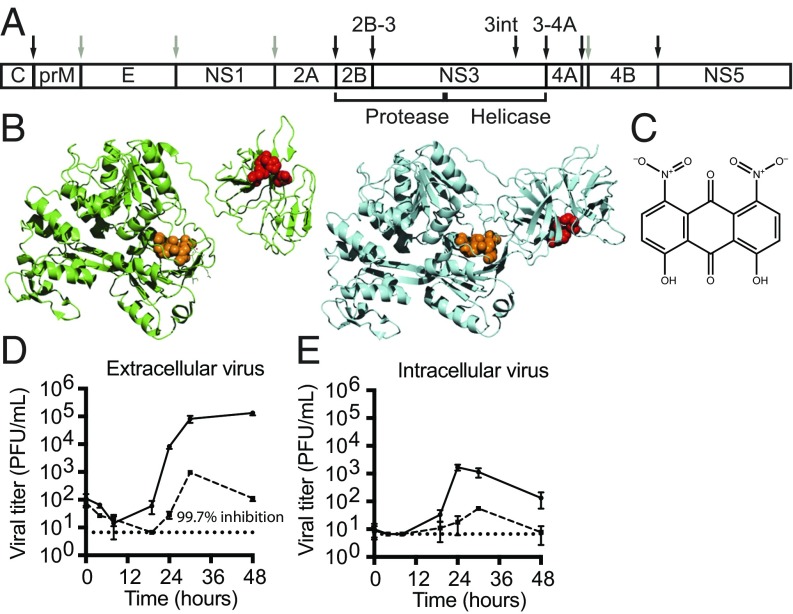
Positive-strand RNA viruses encode polyproteins that must be cleaved during and after translation. A virus-encoded serine proteinase (NS3) and its essential cofactor (NS2B) are responsible for virally mediated cleavages among all members of the Flavivirus genus including West Nile (WNV), yellow fever, Japanese encephalitis, Zika, and dengue viruses. When first synthesized, NS2B/3 is embedded within the dengue polyprotein, and is responsible for seven of the processing events within it (2) (Fig. 1A). In WNV, cleavage between NS2B and NS3 has been demonstrated to be exclusively intramolecular (3). For dengue virus, this cleavage is dilution-insensitive (4), suggesting that strict intramolecular processing at this junction is common among flaviviruses. In addition to the proteolytic events that originally defined the viral proteins, there is an internal NS3 cleavage (NS3int). The significance of this cleavage is not yet known, though it has been reported to occur in mammalian but not insect cells (5, 6).
Fig. 1.
Inhibition of dengue virus by ARDP0006. (A) Dengue virus polyprotein organization showing cleavages made by NS2B/3 (black arrows) and host proteases (gray arrows). (B) Two high-resolution crystal structures of full-length NS2B/3 display multiple conformations of the helicase and protease domains relative to each other (Protein Data Bank ID codes 2VBC and 2WHX, green and teal, respectively). The proteinase active site is shown in red with the internal NS3 cleavage site in orange. (C) Chemical structure of NS2B/3 inhibitor ARDP0006. (D and E) Effect of 25 μM ARDP0006 (dashed lines) on extracellular and intracellular dengue virus (multiplicity of infection, 0.1 PFU per cell) during growth in BHK-21 cells. Inhibition of viral growth is comparable to that observed previously (11), with a reported IC50 of 4.2 μM. n = 4 in D and E.
NS3 makes an attractive drug target, as it has multiple essential functions. The N-terminal domain contains the proteinase active site, and the C-terminal domain exhibits RNA helicase, RNA 5′ triphosphatase (RTPase), and NTPase activities (7). In addition, mutations that alter conserved, nonenzymatic surface residues reduce viral fitness (8). Structures determined by X-ray crystallography have shown two different orientations (Fig. 1B) of the proteinase domain relative to the helicase domain of NS3 (9, 10), arguing that these domains might be flexible with respect to each other in solution as well.
An inhibitor of dengue NS2B/3 proteinase ARDP0006 (1,8-dinitro-4,5-dihydroanthraquinone; Fig. 1C) was previously identified in a high-throughput screen. The NS2B/3 proteinase tested was engineered to contain only the cofactor regions required for NS3 proteinase activity (NS2B residues 49 to 96) joined to residues 1 to 185 of NS3 via a flexible Gly4-Ser-Gly4 sequence. This minimal proteinase efficiently cleaves substrates supplied in trans but cannot self-process. Thus, this screen tested for inhibitors of intermolecular cleavage (11, 12). It is of note that, while the half-maximal inhibitory concentration (IC50) for intermolecular cleavage in solution was 432 μM (12), the IC50 during infection was 4.2 μM (11). The additional inhibitory effect observed in cells could be due to increased local concentrations or off-target effects. However, we envisaged an alternative mechanism in which accumulated proteinase precursors inhibit viral growth.
We set out to investigate mechanisms of viral inhibition by ARDP0006. We tested whether other polyprotein cleavages mediated by NS2B/3 were self-processing, and whether precursors that would accumulate in the absence of self-cleavage would be trans-dominant inhibitors of wild-type (WT) virus. Our findings support the hypothesis that inhibiting one particular self-processing event leads to trans-dominant inhibition of RNA replication. This provides insight into the previously unexplored role of dengue precursors in infection and suggests an approach to antiproteinase drug development by preferentially targeting specific intramolecular cleavages.
Results
Inhibition of Dengue Virus and Proteinase Activity by ARDP0006.
Despite extensive screening and chemical synthesis, ARDP0006 remains one of the most potent NS2B/3-targeted inhibitors of dengue viral growth, with a reported IC50 of 4.2 μM (13). Addition of 25 μM ARDP0006 reduced the yield of extracellular and intracellular dengue virus by 100-fold during a single 24-h infectious cycle (Fig. 1 D and E). Negligible cytotoxicity was observed at this concentration of ARDP0006 (SI Appendix, Fig. S1).
ARDP0006 was identified as an inhibitor of fluorogenic substrate cleavage by an NS2B/3 proteinase incapable of self-processing (11). We investigated whether this compound also inhibits NS2B/3 self-cleavage. To this end, a minimal self-cleaving proteinase termed NS2B/3pro was used (14). This construct contains the required cofactor domain of NS2B (amino acids 47 to 95), the last 10 residues of NS2B to reconstruct the cleavage site (amino acids 122 to 131), and the protease domain of NS3 (amino acids 1 to 185), but lacks the transmembrane domains.
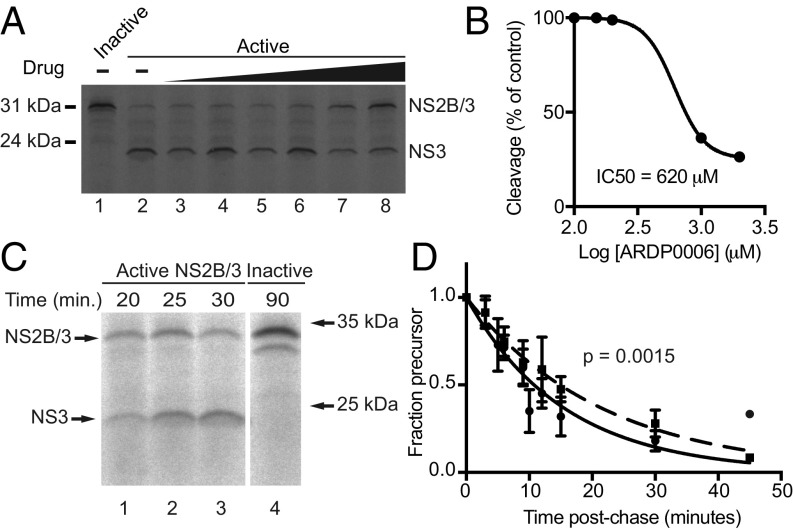
To study NS2B/3pro cleavage, conditions under which the kinetic analysis could be performed immediately after synthesis were needed. To this end, rabbit reticulocyte lysate (RRL) was used to produce NS2B/3pro radioactively labeled with [35S]methionine. A proteinase active-site mutation, NS3-S135A, caused this protein to be produced as a precursor only (Fig. 2A, lane 1), and provides a marker for intact NS2B/3pro. We first wanted to establish whether ARDP0006 inhibits NS2B/3 cleavage at the IC50 of intermolecular proteinase activity (432 μM) or of viral inhibition (4.2 μM). The active version was efficiently translated and cleaved into products after a 90-min incubation (Fig. 2A, lane 2). ARDP0006 inhibited this cleavage, as seen by the greater proportion of labeled proteinase precursor with increasing inhibitor concentration (Fig. 2A, lanes 3 to 8). However, the IC50 for NS2B/3pro self-cleavage was high, comparable in magnitude to the Ki for cleavage of small substrates in vitro (Fig. 2B).
Fig. 2.
Inhibition of NS2B/3pro in vitro by ARDP0006. An in vitro assay was developed to measure inhibition of intramolecular cleavage at NS2B-3 in the dengue minimal proteinase. (A and B) Minimal proteinase was produced in RRL in the presence of increasing concentrations of ARDP0006 (50 μM to 2 mM), and remaining precursor was plotted to determine the IC50 of self-cleavage in vitro (n = 1). (C and D) A pulse–chase assay was developed to monitor the effect of the inhibitor on reaction rate. Peak precursor abundance in RRL occurred at 25 to 30 min (C, lanes 1 to 3), and inactive NS3-S135A protein was robustly translated (C, lane 4) but no cleavage products were observed. The 32-kDa species seen in lane 4 is frequently observed background in this system. After labeling and translation, addition of 100 μM ARDP0006 resulted in modest but statistically significant inhibition of cleavage (D, dashed line) relative to mock treated reactions (D, solid line). Line fitting was performed using GraphPad Prism v.7 using a log[inhibitor] vs. response variable-slope equation for determining the apparent IC50 (B) and single-phase exponential decay functions to fit the data in D. The P value in D was determined by comparing the fit of individual and shared models using the extra sum-of-squares F test. n = 3 (dashed line) or 5 (solid line).
One explanation for the observation that ARDP0006 is a more efficacious inhibitor of viral growth than of NS2B/3 proteinase activity is that, although it was identified as an inhibitor of NS2B/3, it inhibits the virus through an off-target effect. Given that identification of drug targets is often facilitated by the identification of drug-resistant variants, we attempted to select ARDP0006-resistant dengue virus in cultured cells (SI Appendix). From two independent selection pools, there were no individual or shared mutations in NS2B/3, but the mutation A21V in nearby NS4A did arise. This valine substitution is extremely rare in natural dengue isolates, occurring in only 4 of 3,871 sequences deposited in the National Center for Biotechnology Information (NCBI) Virus Variation database (15). Reconstruction of the A21V mutation in isolation showed that it did not confer specific resistance to ARDP0006 but increased the rate of viral growth in both the presence and absence of inhibitor (SI Appendix, Fig. S2). We conclude that there is a high barrier to resistance against ARDP0006 but that rapidly growing variants can emerge, possibly due to off-target effects of the compound.
Recently, another NS2B/3-targeted molecule, termed NSC135618, was shown to have potent antiflaviviral activity (16). We tested this compound’s activity using the assays described for ARDP0006. We determined viral and intramolecular cleavage IC50 values of 1.7 and 490 μM, respectively (SI Appendix, Fig. S3), which mirrored the discrepancy in these values found for ARDP0006. Thus, both NS2B/3 proteinase inhibitors are 100-fold more effective at inhibiting viral growth than the enzymatic activity of the viral proteinase, making it more likely that both compounds indeed inhibit dengue viral growth through inhibition of the protease, but by potentially complex mechanisms.
To monitor the kinetics and efficiency of NS2B/3pro cleavage, pulse–chase experiments were performed. For the minimal proteinase, a translation time course revealed that precursor and product could be readily quantified at 25 to 30 min (Fig. 2C). Pulse–chase assays were performed in which protein was translated with radioactively labeled methionine for 30 min, followed by addition of excess unlabeled methionine and either ARDP0006 or its DMSO solvent. The solid line in Fig. 2D quantifies the degradation of labeled NS2B/3pro precursor. The amount of inhibition observed was statistically significant, though slight, as expected from the IC50 of ∼620 μM (Fig. 2D and Table 1). A concentration of 100 μM was used because it is between the IC50 for virus and proteinase inhibition and to provide a benchmark for subsequent examinations of individual cleavage sites.
Table 1.
Processing rates of proteinase constructs
| Construct | Treatment | Rate (SE), min−1 | t1/2, min |
| NS2B/3pro | DMSO | 0.064 (0.0037) | 10.8 |
| ARDP0006 | 0.047 (0.0033) | 14.9** | |
| NS2B/3/4A | DMSO | 1.0 (0.057) | 40 |
| G459L S1L | ARDP0006 | 0.66 (0.034) | 66*** |
| NS2B/3/4A | DMSO | 0.59 (0.11) | 70 |
| S1L | ARDP0006 | <10−16 | >1015*** |
| NS2B/3/4A | DMSO | 2.2 (0.16) | 19 |
| G459L | ARDP0006 | 1.7 (0.15) | 25** |
Constructs were expressed in reticulocyte lysate reactions in the absence or presence of 100 μM ARDP0006, and precursor loss was normalized to the amount of precursor at time 0. Rates were calculated by fitting a single-exponential curve constrained to K > 0 and plateau = 0. t1/2, half-life. Asterisks indicate statistical significance for rates in the presence of ARDP0006 vs. DMSO treatment for each construct (**P < 0.01, ***P < 0.001). The bold type represents the conditions in which the compound was added.
Three NS2B/3 Cleavages Are Exclusively Intramolecular.
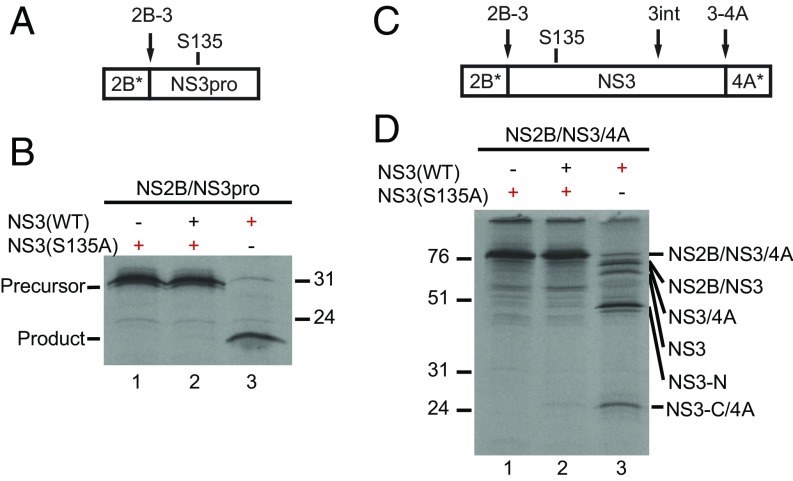
In WNV, the NS2B-3 junction is processed exclusively intramolecularly (3). We hypothesized that this would be the case for dengue virus as well and that, given the flexibility of the proteinase domain, other NS3 cleavages might also be self-processed. To test this, mixing experiments were performed with catalytically active and inactive versions of NS2B/3pro. After 120 min of incubation, S135A-NS2B/3pro remained uncleaved (Fig. 3B, lane 1). As before, cleavage of labeled, active NS2B/3pro was efficient and essentially complete by 120 min (Fig. 3B, lane 3). However, when [35S]methionine-labeled catalytically inactive proteinase was mixed with unlabeled, active proteinase, no specific cleavage products were observed (Fig. 3B, lane 2). Therefore, cleavage of the dengue NS2B-3 junction occurs exclusively intramolecularly.
Fig. 3.
Intramolecular cleavage of NS3-proximal polyprotein junctions. (A) Schematic of the NS2B/3 minimal proteinase. The black arrow indicates the cleavage site; S135 indicates catalytic serine mutated to create inactive proteinase. (B) Active (WT) and inactive (S135A) minimal proteinases were translated in RRL reactions separately or together as indicated for 120 min in the presence (red symbols) or absence (black symbols) of [35S]methionine and visualized by SDS/PAGE. (C) Schematic of NS2B/3/4A proteinase constructs including full-length NS3 and residues 1 to 49 of NS4A. Labeled as in A. (D) Active (WT) and inactive (S135A) proteinases were translated and mixed as in B. Asterisks in A and C indicate protein truncations described in the text.
Two other cleavage junctions occur within or flanking NS3: NS3int and NS3-4A. To test whether these could be cleaved intermolecularly, we created elongated versions of NS2B/3pro that included the helicase domain of NS3 and the first 49 residues of NS4A (Fig. 3C). Inactive, labeled NS2B/3/4A alone gave rise to intact precursor (Fig. 3D, lane 1). When active, unlabeled proteinase was added, no additional bands or observable disappearance of the precursor was revealed (Fig. 3D, lane 2). This is in contrast to active, labeled proteinase, which was converted from a single precursor to the expected products (Fig. 3D, lane 3), demonstrating that all three NS3-proximal cleavages (NS2B-3, NS3int, and NS3-4A) are strictly intramolecular. We reasoned that one or more of these could be specifically targeted by ARDP0006.
Inhibition of NS2B-3, NS3int, and NS3-4A Cleavages by ARDP0006.
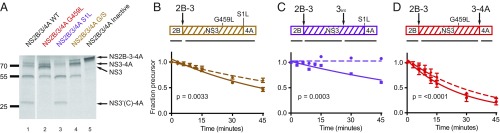
To monitor the effect of ARDP0006 inhibition at these sites, it was useful to simplify the cleavage patterns (Fig. 4A). To create a protein at which only the NS2B-3 site was cleaved, mutations were made that destroyed the NS3int and NS3-4A cleavage junctions (Fig. 4A). Cleavage was modestly but significantly inhibited by 100 μM ARDP0006 (Fig. 4B and Table 1).
Fig. 4.
NS3-proximal cleavages are sensitive to ARDP0006 inhibition. (A) Reticulocyte lysate reactions programmed with plasmids encoding NS2B/3/4A that contain the following mutations: NS3-S135A (inactive; black), NS3-G459L/NS4A-S1L (both cleavage sites; orange), NS4A-S1L (NS3-4 junction; purple), or NS3-G459L (NS3int junction; red) were incubated for 90 min in the presence of [35S]methionine, and products were visualized by SDS/PAGE to confirm cleavage patterns. (B–D) Degradation kinetics of the gene products from A were monitored in the absence (solid lines) and presence (dashed lines) of 100 μM ARDP0006. Aliquots were taken at the indicated times postchase with unlabeled methionine and drug. Proteolysis was modeled as a single-exponential decay reaction with baseline constrained to 0. Kinetic parameters in the presence of inhibitor were statistically significantly different for all constructs (B–D; n = 4, P = 0.0033; n = 2, P = 0.0003; n = 4, P < 0.0001; respectively) as determined by the extra sum-of-squares F test.
The S1L mutant is capable of cleavage at NS2B-3 and NS3int. In this case, processing was significantly and dramatically inhibited by 100 μM ARDP0006 (Fig. 4C and Table 1). We conclude that cleavage of the internal NS3 junctions, when NS3 is still coupled to NS4A, is preferentially susceptible to ARDP0006 and may trap NS3 in an inflexible conformation. The failure to cleave the NS2B-3 junction in the presence of the S1L mutation argues that these cleavages are not independent.
The G459L mutant is not cleavable at the internal NS site, but can be cleaved at the NS2B-3 and NS3-4A junctions. ARDP0006 slowed processing of this construct to approximately the same degree as for the NS2B/3pro alone (Figs. 2D and 4 B and D and Table 1). We conclude that the lack of independence of the N-terminal, internal, and C-terminal reaction sites may cause the temporal order of cleavage to differ in differently mutated polyproteins. Importantly, the NS3int cleavage site is highly susceptible to inhibition, with sensitivity to ARDP0006 comparable to that of the virus itself.
NS3-G459L Is a Dominant Inhibitor of WT Viral Growth.
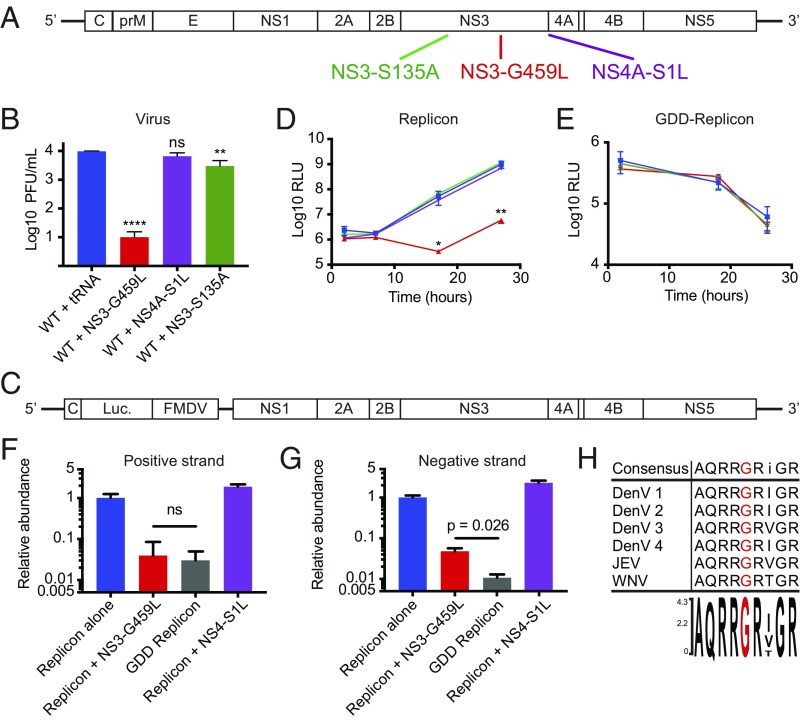
To determine whether dengue precursors are inhibitory to viral growth, we monitored growth of WT virus in the presence of variant genomes with either particular defects in proteinase cleavage sites or general inhibition of proteinase activity. All mutations shown in Fig. 5A were lethal to the virus that contained them (SI Appendix, Table S1). We tested whether any of these mutant genomes was capable of dominantly inhibiting WT virus, as would be predicted if a mutation caused an inhibitory precursor to accumulate upon translation of input RNA.
Fig. 5.
Internal NS3 cleavage mutant genome is a trans-dominant inhibitor. (A) Diagram of genomic viral RNA is shown with the location of internal cleavage site NS3-G459L, NS3-4A NS4A-S1L, and protease-dead NS3-S135A mutations shown in red, purple, and green, respectively. (B) WT vRNA was cotransfected with equal masses of yeast tRNA, NS3-G459L mutant vRNA, NS4A-S1L mutant vRNA, or NS3-S135A mutant vRNA and viral yields were determined 48 h posttransfection (n = 4). (C) Dengue luciferase replicon construct, lacking structural proteins. (D) Replicon RNA (1 μg per well) was cotransfected in 24-well plates with equal masses of WT, NS3-G459L, NS4A-S1L, or NS3-S135A vRNAs and luciferase signal was measured at the indicated time points (n = 4). RLU, relative light unit. (E) Inactive polymerase replicon RNA (1 μg per well) was cotransfected in 24-well plates with equal masses of WT, NS3-G459L, or NS3-S135A vRNAs and luciferase signal was measured at the indicated time points (n = 4). (F) Positive-strand replicon RNA was detected by strand-specific RT-qPCR. (G) Negative-strand replicon RNA was detected by strand-specific RT-qPCR (n = 2). (H) Multiple sequence alignment of dengue virus serotypes 1 to 4, JEV, and WNV virus, accession nos. ACF49259, P29990, AAA99437, AAX48017, P32886, and P14335, respectively. Protein sequences were obtained from the NCBI and aligned by Clustal Omega using Lasergene MegAlign Pro. ns, not significant. *P < 0.05, **P < 0.01, ****P < 0.0001.
Upon cotransfection with equal amounts of WT and NS4-S1L viral RNA (vRNA), growth of WT virus was unperturbed (Fig. 5B). However, cotransfection of WT and NS3-G459L vRNA reduced viral growth by 100- to 1,000-fold (Fig. 5B). This mutation should allow the accumulation of intact NS3, possibly still tethered to flanking sequences. To determine whether inhibiting all cleavages by NS2B/3 also caused trans-dominant inhibition of WT virus, we tested the effect of the proteinase active-site mutation NS3-S135A. Upon cotransfection of WT and NS3-S135A vRNA, we observed significant but relatively modest inhibition of WT virus (Fig. 5B). Therefore, we hypothesize that it is the accumulation of specific precursors, not general proteinase inhibition, which is responsible for this dominant effect.
To determine the intracellular step that was dominantly inhibited by the NS3-G459L genome, we tested the effect of mutant vRNAs on a cotransfected WT replicon (Fig. 5C). This replicon encodes Renilla luciferase and dengue nonstructural proteins but lacks the dengue structural proteins (17). The luciferase signal therefore only reflects translation and RNA replication of transfected RNA. Viral genomes that were WT or contained the NS4-S1L mutation had no observable effect on replicon luciferase signal, but viral RNA that contained the NS3-G459L mutation was strongly inhibitory (Fig. 5D). This inhibition could result from inhibition of replicon translation, RNA amplification, or both. To determine whether the NS3-G459L genome exerted trans-dominant inhibition of translation, cotransfection of replicon that contained a polymerase active-site mutation (GDD) with WT and mutant genomic viral RNAs was performed (Fig. 5E). In this experiment, all luciferase signal derived from initial translation events, and diminished with time in the absence of new RNA synthesis. Luciferase production from the GDD replicon was unaffected by the presence of NS3-G459L or NS3-S135A vRNA (Fig. 5E). Therefore, the NS3-G459L mutation has no effect on early viral protein synthesis.
To identify whether the dominant NS3-G459L mutant genome inhibited negative-strand synthesis (and therefore positive-strand synthesis as well) or just positive-strand synthesis, we performed strand-specific RT-qPCR to measure both strands of WT and GDD replicons (Fig. 5 F and G). In all conditions, the amount of RNA measured was normalized to the values observed for the WT replicon alone. The amount of positive-strand replicon RNA detected when cotransfected with the NS3-G459L dominant mutant was indistinguishable from that detected for the GDD replicon alone, indicating that positive-strand synthesis was strongly inhibited (Fig. 5F). By contrast, the amount of negative-strand replicon RNA was significantly greater than that for the GDD replicon alone (Fig. 5G), indicating that the NS3-G459L dominant mutant allows negative-strand synthesis and therefore specifically inhibits positive-strand synthesis.
NS3 has multiple enzymatic functions in addition to proteinase activity. NS3-G459 lies within a region of NS3 known to be important for RNA helicase, ATPase, and RTPase activities. This region is highly conserved, and G459 itself has 100% identity between all four serotypes of dengue virus as well as Japanese encephalitis virus (JEV) and WNV (Fig. 5H). To determine whether the trans-dominance of the NS3-G459L mutation resulted from disrupting nonproteinase activities, we tested the effect of four different mutations known to destroy some combination of these other activities (7, 18, 19). Dominant inhibition did not correlate in a simple way with mutations in any of these functions (SI Appendix, Fig. S4). For example, mutant genomes NS3-K199A and NS3-K396A, with disrupted helicase functions, did not dominantly inhibit the growth of cotransfected WT RNA, but the helicase-dead NS3-R376A genome was a potent trans-dominant inhibitor (SI Appendix, Fig. S4B). We investigated whether this trans-dominance occurred by a mechanism similar to that of the NS3-G459L genome, perhaps via an unanticipated effect on proteinase activity. Unlike the G459L mutation, however, the NS3-R376A mutant displayed normal protein processing (SI Appendix, Fig. S4C) and showed no trans-dominant inhibition of replicon RNA synthesis (SI Appendix, Fig. S4D). Therefore, trans-dominant inhibition by the NS3-R376A genome is likely an effect distinct from that of G459L and later in the dengue infectious cycle. Interestingly, the NS3-R376A mutant protein has an increased affinity for double-stranded RNA (7), making it possible that such a gain-of-function effect contributes to its trans-dominance. Thus, none of the helicase-defective viruses phenocopies the trans-dominant effect of the NS3-G459L mutation. It is likely that there are many allele-specific gain-of-function defects that remain to be explored. Here, we show that inhibiting cleavage of the internal NS3 cleavage site specifically, either pharmacologically or genetically, leads to potent and, in the case of the G459L mutation, trans-dominant inhibition of viral RNA replication.
Discussion
The NS3 proteins of dengue and other flaviviruses have several enzymatic activities mediated by distinct domains. In addition, NS3 interacts with host proteins such as STING and MAVS to disrupt innate immune signaling. We have shown that three different NS2B/3-mediated cleavages occur exclusively intramolecularly, requiring conformational flexibility of the proteinase domain. We found that abrogating one of these cleavages leads to accumulation of a precursor that trans-dominantly inhibits all viruses in the same cell. These findings both identify a mechanism of viral inhibition and provide a path for suppressing the outgrowth of drug-resistant variants.
Rapid emergence of drug resistance is the major obstacle to development of effective antivirals for all RNA viruses, including dengue. Low polymerase fidelity and fast generation time result in high population diversity, including potentially preexisting resistant variants for any given drug. Typical strategies for inhibiting viral growth also apply strong selection pressure, allowing rapid selection in a population of viruses infecting a host. Multidrug therapy decreases the probability of generating a virus that contains mutations conferring resistance to all components of an antiviral mixture. Another approach to suppress drug resistance is to accept the inevitable generation of drug-resistant variants but to choose a drug target that prevents their outgrowth. For example, targeting oligomeric viral proteins can reduce the selection of drug-resistant variants due to the mixing of proteins from both drug-resistant and drug-susceptible genomes (20). Capsids are obvious targets for this antiviral paradigm; however, it may be possible to create other destructive oligomers of highly interactive proteins.
Viral polyprotein processing is exquisitely timed for optimal viral growth. Incorrectly processed viral precursors interfere with viral growth and pathogenesis. In poliovirus, mutations that abrogate cleavage at the VP1/2A junction are trans-dominant, probably because an uncleaved precursor containing the VP1 and capsid proteins destroys virus assembly (21). In hepatitis C, increasing the rate of NS4B-5 precursor cleavage by manipulating the recognition site inhibits viral RNA replication (22). Additionally, Sindbis virus exhibits exquisite temporal regulation of proteolysis. Uncleaved P123 polyprotein is essential for minus-strand synthesis but P123 cleavage is required for efficient plus-strand synthesis, and the relative abundance of these protein species is regulated by variations in substrate specificity (23, 24).
NS2B-3 Cleavage.
The N-terminal domain of NS2B protein is membrane-associated and its C-terminal sequence acts as a cofactor for the NS3 proteinase domain. NS2B remains bound to NS3 after junction cleavage, and the specific proteinase activity is greatly increased (25). Additionally, the hydrophobic regions of NS2B should tether this holoenzyme to the intracellular membranes on which viral replication takes place (26). Dengue virus NS2B-3 cleavage has been shown to be exclusively intramolecular, even though it can be reconstituted biochemically in trans (27). Given that NS2B-3 cleavage is rapid and increases the activity of the proteinase (4, 28), it is likely to be rate-limiting in protein processing. Therefore, we were surprised that NS2B-3 cleavage was not more dramatically inhibited by ARDP0006.
NS3-4A Cleavage.
NS4A contains several membrane-associated sequences that induce membrane curvature and rearrangements. We have observed that cleavage at the NS3-4 junction of dengue virus is strictly intramolecular. This has also been reported for the related pestivirus, classical swine fever virus (29). When the NS3-4 junction is not processed, there is no observable trans-dominant effect, suggesting that the resultant precursors, which necessarily contain NS4A, do not interfere with viral growth. Like at the NS2B-3 junction, ARDP0006 only modestly inhibited cleavage between NS3 and NS4A. Genomes encoding noncleavable NS3/4A sites exhibit no inhibitory effects on other viral genomes in the same cell.
NS3 Internal Cleavage.
Internal cleavage of NS3 has been observed in dengue and many closely related viruses (3, 6, 29, 30), but its significance has remained cryptic. This cleavage generates an NS2B/3 proteinase freed from its NS4A membrane tether. Mutations that abrogate this cleavage site are lethal, which may be due to either lack of cleavage or disruption of helicase activities. We have shown that this junction is processed in a strictly intramolecular manner. Of the three intramolecularly processed sites within and flanking NS3, this cleavage junction is the most strongly inhibited by ARDP0006 (Fig. 4 and Table 1). When the internal NS3 junction is inhibited by ARDP0006, cleavage at the NS2B-3 junction is also inhibited. This suggests allosteric effects upon inhibition of the NS3 internal cleavage that prevent access to the NS2B-3 and possibly other NS3 junctions. Most importantly, viral genomes that abrogate this self-processing event trans-dominantly inhibit positive-strand RNA synthesis of WT genomes present in the same cell, reminiscent of the effect of precursors on Sindbis virus replication. We hypothesize that the antiviral effect of ARDP0006 is due to its specific inhibition of the internal cleavage in NS3, which is also abrogated by the G459L mutation, and that in both cases this leads to the accumulation of trans-dominant NS3 precursors tethered to flanking sequences.
We propose that an effective inhibitory strategy for viruses with self-cleaving polyprotein precursors will be to first identify the cleavages that, when not made, give rise to trans-dominant inhibitors. Upon inhibition of that cleavage event, the accumulation of inhibitory precursors will block the replication of susceptible viruses and, most likely, any drug-resistant genomes as they arise in infected cells.
Methods
Plasmids.
Dengue proteinase expression plasmid pGEX-NS2B-3prot, in which the transmembrane domain in NS2B was deleted, was constructed as previously described (14). NS2B/3/4A plasmids were generated from this plasmid by NsiI/BamHI digestion and T4 DNA ligase-mediated (New England Biolabs) insertion of NS3-4A amplicons (nucleotides 7473 to 13699), adding a BamHI site to the 3′ end. Infectious clone cDNA for dengue virus 2 strain 16681 was used as a parent plasmid for all constructs (31). All mutations were introduced using QuikChange Site-Directed Mutagenesis Kits (Agilent Technologies).
Viruses and Cell Culture.
Dengue virus strain 16681 was used in this study. See SI Appendix for detailed methods. BHK-21 cells were grown at 37 °C with 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% bovine serum, 1 U/mL penicillin/streptomycin, and 10 mM Hepes. For plaque assays, viruses were allowed to adsorb to BHK-21 cells for 1 h at 37 °C, overlaid with 1 mL of DMEM plus 0.375% (wt/vol) sodium bicarbonate and 0.8% Aquacide II, grown for 7 d, fixed (5% formaldehyde), and stained (0.1% crystal violet).
ARDP0006 Treatments.
1,8-Dihydroxy-4,5-dinitroanthraquinone (Sigma-Aldrich) was dissolved in DMSO and added to cell-culture media 30 min before infection. Extracellular virus was collected from cellular supernatant. Intracellular virus was collected in DMEM by washing cells (0.5 M NaCl, 10 mM Tris), freeze–thawing (three times), and centrifuging (5 min, 3,200 × g).
Proteinase Cleavage Assays.
Dengue protein constructs were expressed in RRL (TNT coupled T7; Promega BioSystems) programmed with 1 μg DNA/50 μL and labeled with 20 μCi of l-[35S]methionine (EasyTag; PerkinElmer). Except where noted, radioactive pulse–chase reactions were incubated at 30 °C for 30 min before adding l-methionine (1 mM final) and DMSO (2% final). For treated samples, ARDP0006 was dissolved in DMSO. Aliquots were immediately diluted with Laemmli sample buffer (4 volumes, 1× final), denatured (60 °C, 10 min), and separated by SDS/PAGE. Gels were dried under vacuum (80 °C, 120 min) and exposed to a low-energy phosphor storage screen (Molecular Probes). Protein quantification was performed using ImageQuant TL 8.1 software (GE Healthcare Life Sciences). For intermolecular proteinase cleavage assays, NS2B/3 and NS2B/3/4A constructs were expressed either with or without l-[35S]methionine as above. Labeled reactions were chased with excess unlabeled l-methionine, mixed with unlabeled translation reactions (1:1), and incubated for 90 min. Reactions were stopped and visualized as above.
Cotransfections.
Cotransfections were performed on BHK-21 cells in 24-well plates with 1 μg of each RNA added per well (Lipofectamine 3000; Thermo Fisher Scientific). Transfection solution was removed after 2 h at 37 °C, and cells were washed twice and incubated for 48 h.
Luciferase Assays.
An efficient dengue replicon system has been previously described (32). This replicon is derived from strain 16681 (17) and expresses Renilla luciferase (gift of Jan Carette, Stanford University School of Medicine). Replicon transfections were performed as described above. Cells were collected and assayed according to the Renilla Luciferase Assay System protocol (Promega BioSystems) using a luminometer set for 10-s signal integration (GloMax; Promega BioSystems).
Strand-Specific RT-qPCR.
Viral strand-specific RT-qPCR was carried out based on previously described strategies (33, 34). Total RNA from BHK-21 cells was collected 26 h after transfection. Reverse-transcription reactions were performed with SuperScript II RT (Thermo Fisher Scientific) and 200 ng of RNA primed with replicon-specific primers tagF Pos or tagF Neg, yielding cDNA for positive- or negative-strand replicon sequences. These cDNAs were diluted 25-fold with water, and quantitative PCR was performed with primers TAG and Rev Pos or Rev Neg using Platinum SYBR Green (Thermo Fisher Scientific) on an Applied Biosystems 7300 Real-Time PCR System (Thermo Fisher Scientific). Primer sequences are in SI Appendix, Table S2.
Supplementary Material
Acknowledgments
We thank Drs. Scott Crowder, Gabriele Fuchs, Caleb Marceau, and Poornima Paramaswaram for contributing to initial stages of this project, and Jan Carette for reagents and advice. We appreciate critical reading by Jasmine S. Moshiri and Peter Sarnow. This work was supported by an NIH Director’s Pioneer Award (to K.K.), National Institute of General Medical Sciences of the NIH Awards T32GM007276 and U19AI109662, and funding from the Stanford SPARK program. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. P.-Y.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1805195115/-/DCSupplemental.
References
- 1.Bhatt S, et al. The global distribution and burden of dengue. Nature. 2013;496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perera R, Kuhn RJ. Structural proteomics of dengue virus. Curr Opin Microbiol. 2008;11:369–377. doi: 10.1016/j.mib.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bera AK, Kuhn RJ, Smith JL. Functional characterization of cis and trans activity of the flavivirus NS2B-NS3 protease. J Biol Chem. 2007;282:12883–12892. doi: 10.1074/jbc.M611318200. [DOI] [PubMed] [Google Scholar]
- 4.Preugschat F, Yao CW, Strauss JH. In vitro processing of dengue virus type 2 nonstructural proteins NS2A, NS2B, and NS3. J Virol. 1990;64:4364–4374. doi: 10.1128/jvi.64.9.4364-4374.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arias CF, Preugschat F, Strauss JH. Dengue 2 virus NS2B and NS3 form a stable complex that can cleave NS3 within the helicase domain. Virology. 1993;193:888–899. doi: 10.1006/viro.1993.1198. [DOI] [PubMed] [Google Scholar]
- 6.Teo KF, Wright PJ. Internal proteolysis of the NS3 protein specified by dengue virus 2. J Gen Virol. 1997;78:337–341. doi: 10.1099/0022-1317-78-2-337. [DOI] [PubMed] [Google Scholar]
- 7.Sampath A, et al. Structure-based mutational analysis of the NS3 helicase from dengue virus. J Virol. 2006;80:6686–6690. doi: 10.1128/JVI.02215-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gebhard LG, et al. A proline-rich N-terminal region of the dengue virus NS3 is crucial for infectious particle production. J Virol. 2016;90:5451–5461. doi: 10.1128/JVI.00206-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo D, et al. Flexibility between the protease and helicase domains of the dengue virus NS3 protein conferred by the linker region and its functional implications. J Biol Chem. 2010;285:18817–18827. doi: 10.1074/jbc.M109.090936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo D, et al. Crystal structure of the NS3 protease-helicase from dengue virus. J Virol. 2008;82:173–183. doi: 10.1128/JVI.01788-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tomlinson SM, et al. Structure-based discovery of dengue virus protease inhibitors. Antiviral Res. 2009;82:110–114. doi: 10.1016/j.antiviral.2009.02.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomlinson SM, Watowich SJ. Anthracene-based inhibitors of dengue virus NS2B-NS3 protease. Antiviral Res. 2011;89:127–135. doi: 10.1016/j.antiviral.2010.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chu JJH, et al. Antiviral activities of 15 dengue NS2B-NS3 protease inhibitors using a human cell-based viral quantification assay. Antiviral Res. 2015;118:68–74. doi: 10.1016/j.antiviral.2015.03.010. [DOI] [PubMed] [Google Scholar]
- 14.Clum S, Ebner KE, Padmanabhan R. Cotranslational membrane insertion of the serine proteinase precursor NS2B-NS3(Pro) of dengue virus type 2 is required for efficient in vitro processing and is mediated through the hydrophobic regions of NS2B. J Biol Chem. 1997;272:30715–30723. doi: 10.1074/jbc.272.49.30715. [DOI] [PubMed] [Google Scholar]
- 15.Hatcher EL, et al. Virus Variation Resource—Improved response to emergent viral outbreaks. Nucleic Acids Res. 2017;45:D482–D490. doi: 10.1093/nar/gkw1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brecher M, et al. A conformational switch high-throughput screening assay and allosteric inhibition of the flavivirus NS2B-NS3 protease. PLoS Pathog. 2017;13:e1006411. doi: 10.1371/journal.ppat.1006411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marceau CD, et al. Genetic dissection of Flaviviridae host factors through genome-scale CRISPR screens. Nature. 2016;535:159–163. doi: 10.1038/nature18631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chiang P-Y, Wu H-N. The role of surface basic amino acids of dengue virus NS3 helicase in viral RNA replication and enzyme activities. FEBS Lett. 2016;590:2307–2320. doi: 10.1002/1873-3468.12232. [DOI] [PubMed] [Google Scholar]
- 19.Matusan AE, Pryor MJ, Davidson AD, Wright PJ. Mutagenesis of the dengue virus type 2 NS3 protein within and outside helicase motifs: Effects on enzyme activity and virus replication. J Virol. 2001;75:9633–9643. doi: 10.1128/JVI.75.20.9633-9643.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanner EJ, et al. Dominant drug targets suppress the emergence of antiviral resistance. eLife. 2014;3:e03830. doi: 10.7554/eLife.03830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crowder S, Kirkegaard K. Trans-dominant inhibition of RNA viral replication can slow growth of drug-resistant viruses. Nat Genet. 2005;37:701–709. doi: 10.1038/ng1583. [DOI] [PubMed] [Google Scholar]
- 22.Herod MR, Jones DM, McLauchlan J, McCormick CJ. Increasing rate of cleavage at boundary between non-structural proteins 4B and 5A inhibits replication of hepatitis C virus. J Biol Chem. 2012;287:568–580. doi: 10.1074/jbc.M111.311407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lemm JA, Rümenapf T, Strauss EG, Strauss JH, Rice CM. Polypeptide requirements for assembly of functional Sindbis virus replication complexes: A model for the temporal regulation of minus- and plus-strand RNA synthesis. EMBO J. 1994;13:2925–2934. doi: 10.1002/j.1460-2075.1994.tb06587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Groot RJ, Hardy WR, Shirako Y, Strauss JH. Cleavage-site preferences of Sindbis virus polyproteins containing the non-structural proteinase. Evidence for temporal regulation of polyprotein processing in vivo. EMBO J. 1990;9:2631–2638. doi: 10.1002/j.1460-2075.1990.tb07445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yusof R, Clum S, Wetzel M, Murthy HM, Padmanabhan R. Purified NS2B/NS3 serine protease of dengue virus type 2 exhibits cofactor NS2B dependence for cleavage of substrates with dibasic amino acids in vitro. J Biol Chem. 2000;275:9963–9969. doi: 10.1074/jbc.275.14.9963. [DOI] [PubMed] [Google Scholar]
- 26.Li Y, Li Q, Wong YL, Liew LSY, Kang C. Membrane topology of NS2B of dengue virus revealed by NMR spectroscopy. Biochim Biophys Acta. 2015;1848:2244–2252. doi: 10.1016/j.bbamem.2015.06.010. [DOI] [PubMed] [Google Scholar]
- 27.Zhang L, Mohan PM, Padmanabhan R. Processing and localization of dengue virus type 2 polyprotein precursor NS3-NS4A-NS4B-NS5. J Virol. 1992;66:7549–7554. doi: 10.1128/jvi.66.12.7549-7554.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leung D, et al. Activity of recombinant dengue 2 virus NS3 protease in the presence of a truncated NS2B co-factor, small peptide substrates, and inhibitors. J Biol Chem. 2001;276:45762–45771. doi: 10.1074/jbc.M107360200. [DOI] [PubMed] [Google Scholar]
- 29.Lamp B, Riedel C, Wentz E, Tortorici M-A, Rümenapf T. Autocatalytic cleavage within classical swine fever virus NS3 leads to a functional separation of protease and helicase. J Virol. 2013;87:11872–11883. doi: 10.1128/JVI.00754-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang L, Padmanabhan R. Role of protein conformation in the processing of dengue virus type 2 nonstructural polyprotein precursor. Gene. 1993;129:197–205. doi: 10.1016/0378-1119(93)90269-9. [DOI] [PubMed] [Google Scholar]
- 31.Kinney RM, et al. Construction of infectious cDNA clones for dengue 2 virus: Strain 16681 and its attenuated vaccine derivative, strain PDK-53. Virology. 1997;230:300–308. doi: 10.1006/viro.1997.8500. [DOI] [PubMed] [Google Scholar]
- 32.Alvarez DE, De Lella Ezcurra AL, Fucito S, Gamarnik AV. Role of RNA structures present at the 3′UTR of dengue virus on translation, RNA synthesis, and viral replication. Virology. 2005;339:200–212. doi: 10.1016/j.virol.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 33.Vashist S, Urena L, Goodfellow I. Development of a strand specific real-time RT-qPCR assay for the detection and quantitation of murine norovirus RNA. J Virol Methods. 2012;184:69–76. doi: 10.1016/j.jviromet.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peyrefitte CN, Pastorino B, Bessaud M, Tolou HJ, Couissinier-Paris P. Evidence for in vitro falsely-primed cDNAs that prevent specific detection of virus negative strand RNAs in dengue-infected cells: Improvement by tagged RT-PCR. J Virol Methods. 2003;113:19–28. doi: 10.1016/s0166-0934(03)00218-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.