Significance
We generated intestinal organoids from stem cells from a patient with infantile inflammatory bowel disease harboring a homozygous loss-of-function variant in the IL10RB gene, leaving the patient’s cells unable to respond to interleukin-22. Using both human stem cell and murine models, we show that IL-22 primes intestinal epithelial cells to control Salmonella infection more efficiently and that this control is abated in the patient organoids. This control is restored by introduction of a functional copy of the IL10RB gene into the patient’s cells. This work demonstrates the utility of stem cell-derived intestinal organoids as a tool for studying the effect of defined mutations on pathogen control, showing that organoids can provide an invaluable resource for pathogenesis research.
Keywords: Salmonella, interleukin-22, intestinal organoids
Abstract
Intestinal epithelial cells (IECs) play a key role in regulating immune responses and controlling infection. However, the direct role of IECs in restricting pathogens remains incompletely understood. Here, we provide evidence that IL-22 primed intestinal organoids derived from healthy human induced pluripotent stem cells (hIPSCs) to restrict Salmonella enterica serovar Typhimurium SL1344 infection. A combination of transcriptomics, bacterial invasion assays, and imaging suggests that IL-22–induced antimicrobial activity is driven by increased phagolysosomal fusion in IL-22–pretreated cells. The antimicrobial phenotype was absent in hIPSCs derived from a patient harboring a homozygous mutation in the IL10RB gene that inactivates the IL-22 receptor but was restored by genetically complementing the IL10RB deficiency. This study highlights a mechanism through which the IL-22 pathway facilitates the human intestinal epithelium to control microbial infection.
Recent advances in stem cell biology have enabled the production of human induced pluripotent stem cells (hIPSCs) from different donors to become more routine (1, 2). hIPSCs can be generated from healthy individuals or from patients harboring mutations of clinical interest, facilitating studies on phenotypic–genotypic associations (3, 4). Inflammatory bowel disease (IBD) can develop in infants due to, among other causes, loss-of-function mutations in IL10, IL10RA, and IL10RB (5, 6). In addition to IBD with severe perianal disease, patients with IL-10–signaling defects develop chronic folliculitis and arthritis (7). The development of IBD involves complex interactions between host genetics that impact intestinal barrier function and intestinal immunity, the environment, and the intestinal microbiota (8).
IL10R2 encoded by the IL10RB gene is a component of the heterodimeric IL-10 receptor (IL10R) as well as a component of the receptor for IL-22 (IL22R). Due to the common IL10R2 receptor chain that can pair with several R1 subunits, patients with loss-of-function mutations in IL10RB are nonresponsive to the cytokines IL-10, IL-22, IL-26, IL-28A, IL-28B, and IL-29 (5). IL10R2 is expressed constitutively across many cell types in the body, whereas IL22R1 is expressed mainly on nonhematopoietic cells lining barrier sites, including the intestine (9–11). A key role for IL-22 is maintenance of the gastrointestinal barrier. Mice deficient in IL-22 or the IL-22 receptor Il22ra1 can be susceptible to severe disseminated bacterial infection after chemically induced colitis or Citrobacter rodentium challenge (12, 13).
Intestinal organoids are beginning to provide a reductionist system to dissect the cell-intrinsic role of intestinal epithelial cells (IECs) in host defense. Murine intestinal organoids exposed to IL-22 displayed evidence of an inflammatory response, enhanced antimicrobial activity, and wound healing (12). Here, we explore the role of IL-22 in priming IEC responses to the enteric pathogen Salmonella enterica serovar Typhimurium, using hIPSC-derived intestinal organoids (iHOs), including those generated from a patient with infantile IBD due to an IL10RB defect, as a model system. Our results provide insights into an IL-22–driven bacterial defense mechanism involving enhanced phagolysosomal fusion.
Results
iHOs Derived from a Patient with Infantile IBD Lack Expression of IL10RB.
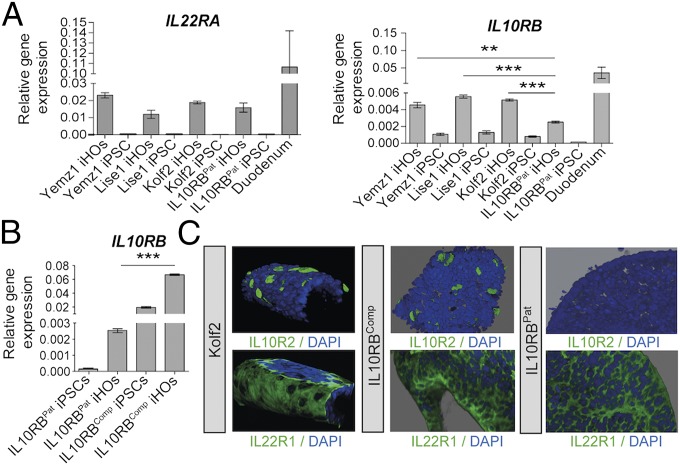
To explore whether iHOs provide a suitable system to study the IL-22 pathway, the expression patterns of mRNAs encoding IL22R1 and IL10R2, the receptor subunits for IL-22, were examined in iHOs derived from hIPSCs using our established protocol (Fig. 1) (14). iHOs were differentiated from the independently derived hIPSC lines Yemz1, Lise1, and Kolf2 as well as from hIPSCs derived from an individual with infantile IBD who harbored a homozygous splice site mutation at the boundary between intron and exon 3 in the IL10RB gene (hereafter “IL10RBPat”) (7). Six weeks after the generation of iHOs from each hIPSC line, the resulting iHOs were phenotyped for expression of markers of mature intestinal epithelium (Fig. 1 and SI Appendix, Fig. S1). mRNA was detected for IL22RA and IL10RB in iHOs from all lines, but significantly lower mRNA levels of IL10RB were detected in the IL10RBPat iHOs than in healthy control iHO lines (Fig. 1A). However, in contrast to mapped RNA-sequencing (RNA-seq) reads from Kolf2 iHOs, no reads mapped to exon 3 in the corresponding IL10RBPat iHO samples (SI Appendix, Fig. S2A). To generate a complemented isogenic control line for IL10RBPat (hereafter “IL10RBComp”), TALEN-based engineering was used to introduce a functional IL10RB gene into the adeno-associated virus site 1 (AAVS1 site) in the IL10RBPat genome (15, 16). After transfection and 7-d puromycin selection, RT-qPCR was performed on surviving colonies with IL10RB-specific primers to detect mRNA expression. One clone was selected for differentiation; the quantity and morphology of hIPSC colonies for this clone were indistinguishable from control hIPSCs. RNA was prepared from differentiated IL10RBPat and IL10RBComp iHOs, which displayed phenotypes similar to healthy control iHOs in culture (SI Appendix, Fig. S3), and from their hIPSC progenitors, and levels of IL10RB mRNA were assayed by RT-qPCR (Fig. 1B). IL10RB gene expression was significantly higher in IL10RBComp iHOs than in IL10RBPat iHOs. At the protein level, IL22R1 and IL10R2 expression was observed on the basal surface of Kolf2 iHOs but not IL10RBPat iHOs using immunostaining (Fig. 1C). The IL22R1 protein was clearly visible dispersed across the basal surface of the iHO epithelia. In contrast, the IL10R2 protein was more obvious as clusters associated with individual cells, often colocalizing with markers for enteroendocrine cells, a feature observed in both iHOs (SI Appendix, Fig. S2C) and primary intestinal tissue (SI Appendix, Fig. S4). As expression of receptors was basal, in subsequent assays IL-22 was delivered to the basal surface of iHOs. Although expression of IL10RB was consistent in healthy control hIPSC lines (Fig. 1A), IL10RB expression was routinely higher in IL10RBComp hIPSCs, probably due to the use of a different promoter (Fig. 1B). However, expression of IL10R2 protein was detected in IL10R2Comp iHOs at comparable levels to expression in Kolf2 iHOs (Fig. 1C). In control human ileal and colonic tissue similar patterns of marker staining were observed to iHOs (SI Appendix, Fig. S4). To confirm the lack of surface expression of IL10R2 in IL10RBPat iHOs, flow cytometry was performed. In Kolf2 iHOs 6% of IECs were positive for IL10R2 expression, confirming the diffuse expression observed via immunostaining, whereas IL10RBPat iHOs were entirely negative (SI Appendix, Fig. S2B).
Fig. 1.
Analysis of expression of IL-22R subunits IL22R1 and IL10R2 demonstrated that iHOs generated from healthy control hIPSCs express the receptors for IL-22, IL22R1, and IL10R2, in contrast to iHOs generated from a patient with infantile IBD, which lack the IL10R2 subunit. To generate IL10RBComp, an isogenic control line for patient iPSCs (IL10RBPat), TALEN-mediated gene integration was used to integrate a copy of the IL10RB gene into the AAVS1 site in IL10RBPat iPSCs. (A) mRNA levels determined by RT-qPCR for IL22RA and IL10RB in three healthy control iHO lines (Yemz1, Lise1, and Kolf2), IL10RBPat iHOs, and control duodenal tissue. IL10RB is significantly up-regulated in control iHOs in comparison with IL10RBPat (P = 0.0005, P < 0.0001, and P < 0.0001, respectively; Student’s t tests). (B) RT-qPCR analysis shows significant up-regulation of IL10RB expression in IL10RBComp iHOs in comparison with IL10RBPat iHOs (P < 0.0001; unpaired, two-tailed Student’s t test). RT-qPCR was performed with TaqMan gene-expression assays and analyzed via the comparative cycle threshold (CT) method with GAPDH as an endogenous control. Data are presented from four technical replicates, with assays repeated at least three times from independent iHO batches. **P < 0.001; ***P < 0.0001. (C) Z-stacked immunostaining for IL-22 receptors IL10R2 or IL22R1 (green) and DAPI (blue) on healthy control Kolf2, IL10RBComp, and IL10RBPat iHOs showing localization of IL22R1 and IL10R2 on the basal IEC surface, with IL10R2 not detected in IL10RBPat iHOs. (Original magnification: 20×.)
IL10R2 Is Necessary for iHO Responses to IL-22.
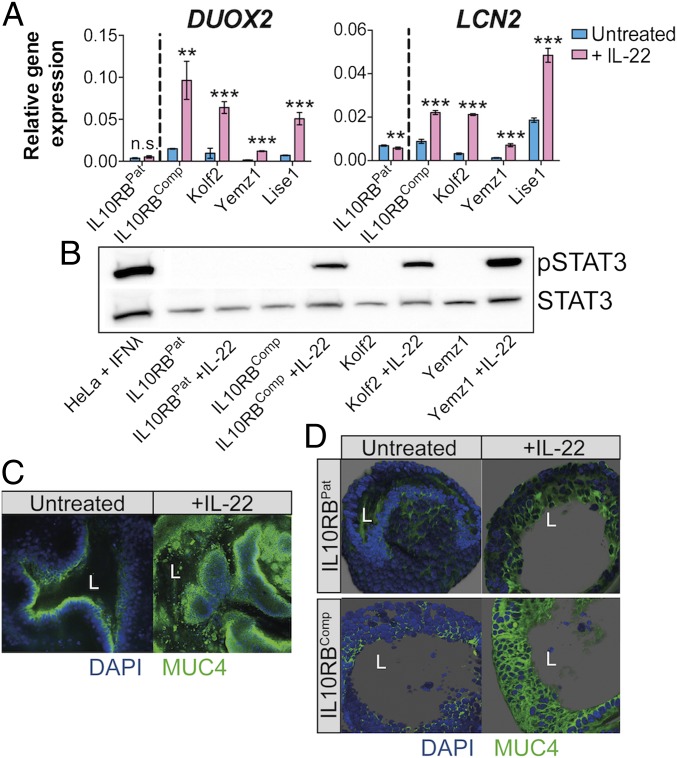
To probe for transcriptional changes in IL-22–prestimulated iHOs, a global transcriptomic analysis was performed in Kolf2 iHOs that were left untreated or were treated with IL-22 for 18 h (SI Appendix, Fig. S5 A and B). Principal component analysis (SI Appendix, Fig. S5A) showed that iHOs treated with IL-22 and untreated iHOs formed distinct groups. A heatmap for the 30 most highly up-regulated IL-22–responsive genes (SI Appendix, Fig. S5B and Datasets S1 and S2) included the antimicrobial proteins REGIIIα and REGIβ and the mucin-encoding gene MUC1, which are potentially expressed by Paneth and goblet cells, respectively. Several IFN-regulated genes associated with antiviral and antimycobacterial defenses, including IFITM1, IFITM2, and IFITM3, were relatively up-regulated (17). Components involved in the maturation and processing of the epithelial oxidase complex DUOX2, DUOXA2, and DUOXA1 were also up-regulated (18), along with DMBT1, which can restrict Salmonella entry into cultured IECs (19). When treated with IL-22 for 18 h, Yemz1, Lise1, and Kolf2 iHOs significantly up-regulated DUOX2 and LCN2 mRNA (Fig. 2A), previously demonstrated to be regulated by IL-22 in other model systems (12, 20). Levels of DUOX2 and LCN2 mRNA did not change significantly in IL10RBPat iHOs after IL-22 treatment; however, the IL-22 response was restored in IL10RBComp iHOs (Fig. 2A). At earlier time points of 3 and 6 h we did not observe up-regulation of DUOX2 and LCN2 (SI Appendix, Fig. S5D). STAT3 activation was detected by Western blotting in whole-cell lysates from healthy controls (Kolf2 and Yemz1 iHOs) and IL10RBComp iHOs but not the IL10RBPat iHOs after 30-min treatment with IL-22 (Fig. 2B). To visualize changes in the barrier phenotype of iHOs treated with IL-22, we performed immunostaining with antibodies for Mucin 4 after IL-22 treatment (Fig. 2C), showing enhanced Mucin 4 production. Mucin 4 production was also enhanced in IL10RBComp iHOs but not in IL10RBPat iHOs (Fig. 2D). We have shown here that, after IL-22 treatment, iHOs up-regulated various antimicrobials and components of the intestinal barrier defense. Hence, we performed RNA-seq and subsequent gene-expression analyses for mRNA in Kolf2 iHOs treated with IL-22 alone or with S. enterica Typhimurium SL1344 added to the medium for 3 h. S. enterica Typhimurium exacerbated the expression of numerous genes in iHOs (SI Appendix, Fig. S5C and Datasets S3 and S4), which was confirmed using RT-qPCR for LCN2, DUOX2, and CXCL2 (SI Appendix, Fig. S5E). Therefore, we hypothesized that during infection IL-22–pretreated iHOs might be less susceptible to invasion by S. enterica Typhimurium SL1344 due to the induction of barrier and host defense mechanisms by IL-22 treatment.
Fig. 2.
Expression of IL10RB was required for iHO responses to IL-22. iHOs were treated with 100 ng/mL recombinant human IL-22 (rhIL-22) added to the iHO medium. (A) In iHO lines with a functional copy of IL10RB, transcripts for IL-22–regulated genes lipocalin 2 (LCN2) and dual oxidase 2 (DUOX2) are significantly up-regulated after the addition of 100 ng/mL rhIL-22 for 18 h, in comparison with unstimulated iHOs (**P < 0.001; ***P < 0.0001; unpaired, two-tailed, Student’s t tests; n.s., not significant). Data are presented from four technical replicates; assays were repeated with at least three biological replicates. RT-qPCR was performed with TaqMan gene-expression assays and analyzed via the comparative CT method with GAPDH as an endogenous control. (B) Phospho-STAT3 level was detected by Western blot after stimulation of healthy control (Yemz1 and Kolf2), IL10RBPat, and IL10RBComp iHOs for 30 min with rhIL-22 and preparation of whole-cell extracts. To verify equal protein loading, the blot was stripped and reprobed with STAT3 antibody. Lysate from HeLa cells stimulated with IFNλ were used as a positive control. (C) Kolf2 iHOs challenged with IL-22 for 18 h or left untreated were examined for Mucin 4 (MUC4; green) and DAPI (blue) by immunofluorescence. (D) Z-stacked immunostaining for Mucin 4 in IL10RBPat and IL10RBComp iHOs challenged with IL-22 for 18 h or were left untreated. (Original magnification: 20×; L = iHO lumen.)
IL-22 Induces a Protective Phenotype Against S. enterica Serovar Typhimurium Infection in iHOs.
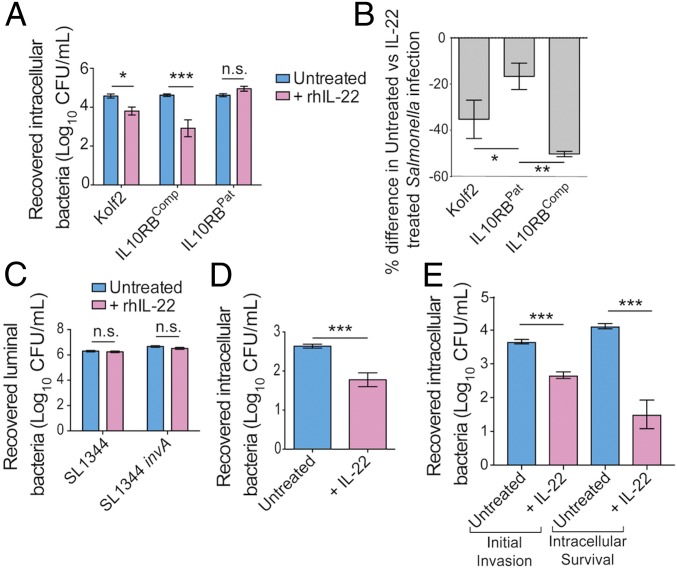
To establish whether IL-22 had an effect on the early interaction of S. enterica Typhimurium with iHOs, we pretreated iHOs with IL-22 for 18 h and then performed microinjections with S. enterica Typhimurium SL1344 (Fig. 3). Interestingly, modified gentamicin protection assays measuring bacterial intracellular survival demonstrated that in Kolf2 and IL10RBComp iHOs significantly less S. enterica Typhimurium was recovered from IL-22–pretreated iHOs than from untreated iHOs, in contrast to IL10RBPat iHOs, in which no significant difference was observed between pretreated and untreated iHOs (Fig. 3A). We then performed similar assays in Kolf2, IL10RBComp, and IL10RBPat iHOs using a fluorescently tagged S. enterica Typhimurium SL1344 (TIMERbac-Salmonella) (Fig. 3B), which can yield bright fluorescence in the green/orange channel, depending on replication state (21). The percentage differences in Salmonella counts between untreated and IL-22–pretreated iHOs in the IL10RBPat samples were significantly lower than the corresponding differences between untreated and IL-22–pretreated iHOs in the Kolf2 and IL10RBComp iHOs. These protection assays were designed to measure differences in levels of intracellular bacteria. In addition, we performed luminal killing assays using S. enterica Typhimurium SL1344 and an S. Typhimurium SL1344 invA mutant, which we have previously shown to be ∼30 times less invasive in the iHO system (Fig. 3C) (14). In these assays bacteria were harvested directly from the iHO luminal cavity. There was no significant difference in the counts recovered from untreated or IL-22–pretreated iHOs, suggesting that luminal killing did not explain the phenotypic differences observed in the invasion assays. We also measured initial invasion by performing protection assays with intracellular bacteria harvested after 30 min (Fig. 3E). Here, we saw a significant reduction in intracellular S. enterica Typhimurium counts in Kolf2 iHOs prestimulated with IL-22, suggesting that the initial invasion was inhibited by IL-22. However, assays were also performed to assess longer-term intracellular survival by incubating infected iHOs for a further 90 min post infection (Fig. 3E). Here, a greater difference was observed in bacterial counts recovered from IL-22–pretreated iHOs compared with untreated iHOs.
Fig. 3.
Pretreatment with rhIL-22 of iHOs that express both IL22R1 and IL10R2 restricts S. enterica Typhimurium SL1344 invasion into IECs. For protection assays iHOs were treated with 100 ng/mL IL-22 18 h before infection or were left untreated. After injection of S. enterica Typhimurium SL1344 into the luminal cavity, iHOs were incubated for 90 min unless otherwise stated. Bacteria were recovered either from the luminal cavity or from IECs. The data presented show the mean from three technical replicates from the combined total of 25 iHOs per replicate, ± SEM, unless otherwise stated. For significance testing unpaired, two-tailed, Mann–Whitney U tests were used for all assays. *P < 0.05; **P < 0.001; ***P < 0.0001; n.s., not significant. (A) Gentamicin protection assays in iHOs show that pretreatment with IL-22 results in significantly less invasion after microinjection of S. enterica Typhimurium SL1344 in healthy control lines with functional IL10R2 (Kolf2: P = 0.0012; IL10R2Comp: P < 0.0001), but this phenotype is not observed in IL10RBPat iHOs (P = 0.2). (B) Percentage difference in TIMERbac Salmonella-infected cells assayed by flow cytometry recovered from untreated or IL-22–treated iHOs (40 iHOs per condition), with gating on live cells. There were significant differences between Kolf2 samples (P = 0.0214) or IL10RBComp samples and IL10RBPat samples (P = 0.0002; Kruskal–Wallis test with Dunn’s multiple comparison test). (C) Log numbers of cfu/mL recovered from lumens of iHOs after microinjection of untreated or IL-22–pretreated Kolf2 iHOs with SL1344 or an invasion-deficient strain, S. enterica Typhimurium SL1344 invA. There was no significant difference in numbers recovered. (D) Protection against S. enterica Typhimurium infection after IL-22 pretreatment is observed in human primary duodenal organoids. Data presented show the mean from three biological replicates with 10 organoids injected per replicate, ± SEM (P < 0.0001). (E) Modified gentamicin protection assays in Kolf2 iHOs show that pretreatment with IL-22 results in significantly less initial invasion (30-min incubation) after microinjection of S. enterica Typhimurium SL1344 (P < 0.0001) and also significantly less intracellular survival, when infected iHO IECs were incubated for an additional 90 min after gentamicin treatment and before IEC lysis (P < 0.0001).
IL-22 Induces a Protective Phenotype in Primary Mouse and Human Organoids.
To ensure that IL-22–mediated restriction was not an artifact of the iHO system, we repeated gentamicin protection assays in primary organoid systems (Fig. 3D and SI Appendix, Fig. S6C). In primary human duodenal organoids pretreated with IL-22 we recovered significantly fewer cfu/mL in protection assays (Fig. 3D) and confirmed the up-regulation of IL-22–responsive genes assayed by RT-qPCR (SI Appendix, Fig. S6 A and B).To confirm that IL-22–mediated protection was driven by signaling via IL22R1, murine intestinal organoids (iMOs) were generated from mucosal tissue harvested from wild-type and Il22ra1−/− mice, and protection assays were performed (SI Appendix, Fig. S6C). In iMOs derived from Il22ra1−/− mice there was no significant difference in the bacterial numbers recovered upon IL-22 treatment.
The Protective Phenotype Induced by IL-22 Pretreatment of iHOs Is Driven by Enhanced Phagolysosomal Fusion.
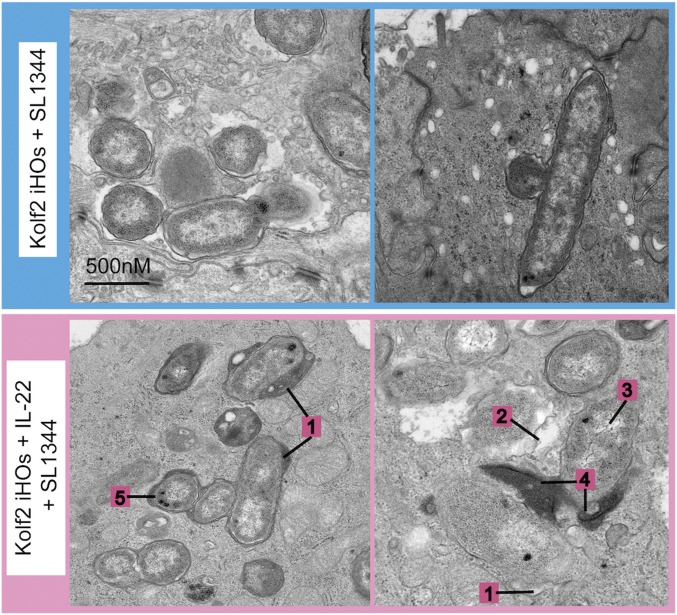
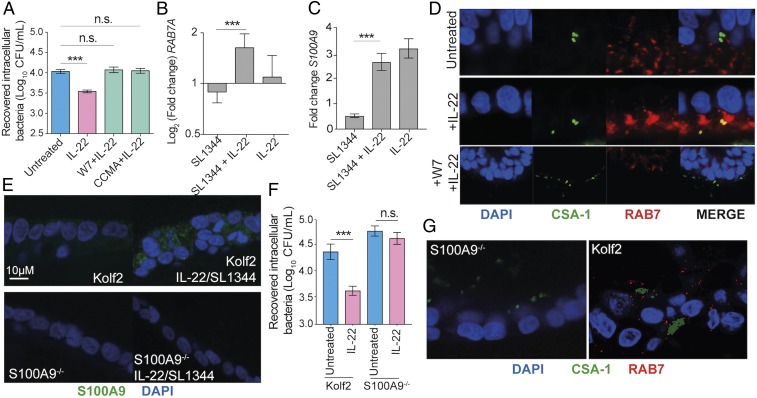
Our protection assays demonstrated that intracellular survival in IL-22–pretreated iHOs was significantly reduced, suggesting differences in the intracellular environment after induction of IL-22 signaling (Fig. 3). Therefore, transmission electron microscopy (TEM) was used to image intracellular bacterial populations at high resolution (Fig. 4 and SI Appendix, Fig. S7 A and B). Interestingly, a distinct difference in bacterial populations was observed within IL-22–treated compared with untreated iHOs. In IL-22–pretreated iHOs, bacteria were visibly more degraded based on the scoring system presented in SI Appendix, Fig. S7B. Histopathological scores from 100-μM sections of 1 mm of iHO mucosa demonstrated that there were more degraded bacteria in Kolf2 iHOs pretreated with IL-22 than in untreated iHOs (49 in pretreated iHOs vs. 11 in untreated iHOs). Moreover, more phagolysosomes were observed in IL-22–pretreated iHOs. To test this observation quantitatively, we used blinded scoring and collected counts for bacterial localization according to the feature scoring system presented in SI Appendix, Fig. S7A. In IL-22–responsive iHOs, the proportion of phagolysosomes to total Salmonella-containing vacuoles (SCVs) was significantly higher in iHOs pretreated with IL-22 before infection than in untreated iHOs (SI Appendix, Table S1). In IL10RBPat iHOs this phenotype was lost. IL-22 can restrict the growth of Mycobacterium tuberculosis intracellularly in macrophages by enhancing phagolysosomal fusion (22) and could be blocked by treatment with the phagolysosomal fusion inhibitor W7. W7 is a specific inhibitor of Ca2+/calmodulin interactions with binding partners, which have been shown to be part of a signaling pathway necessary for converting phagosomes into phagolysosomes (23). We added W7 for 6 h to iHOs pretreated with IL-22 for 18 h before infection with S. enterica Typhimurium SL1344. This revealed that, in the presence of W7, intracellular bacterial counts recovered were similar to those recovered from untreated iHOs (Fig. 5A). This phenotype was then replicated using concanamycin A, a vacuolar ATPase inhibitor that is also known to inhibit phagolysosomal fusion and acidification of phagosomes (Fig. 5A) (24, 25). To confirm that W7 and concanamycin A were not impacting bacterial invasion by restricting any other pathways, invasion assays with S. enterica Typhimurium in Kolf2 iHOs were repeated, and iHOs were treated with either W7 or concanamycin A (SI Appendix, Fig. S7C) or were left untreated; no significant difference in recovered intracellular bacteria was observed. W7 and CCMA assays were repeated in IL10RBPat and IL10RBComp iHOs, demonstrating that in IL10RBComp blocking of phagolysosomal fusion with W7 also inhibited IL-22–mediated intracellular S. enterica Typhimurium restriction (SI Appendix, Fig. S7E). It is well established that the Rab family GTPase RAB7 marks late endosomes and late phagosomes (26). Therefore, we performed RT-qPCR and immunostaining of infected iHOs with RAB7-specific primers or antibodies. In Kolf2 iHOs a significant difference in fold change of RAB7A was observed between iHOs pretreated with IL-22 and those left untreated before infection (Fig. 5B). We also observed strong RAB7 staining in S. enterica Typhimurium-infected iHOs pretreated with IL-22, with visible colocalization between CSA-1, which stains Salmonella, and RAB7 (Fig. 5D). This phenotype was lost in IL10RBPat iHOs and was restored in IL10RBComp iHOs (SI Appendix, Fig. S7D). In addition, less RAB7 expression via immunostaining was observed in iHOs in which W7 was added before microinjection of S. enterica Typhimurium SL1344 (Fig. 5D, Bottom).
Fig. 4.
IL-22 protection was mediated through enhanced lysosomal fusion with SCVs in IL-22–treated iHOs. TEM images of S. enterica Typhimurium SL1344 internalized into IECs 90 min after injection into the lumen of Kolf2 iHOs pretreated for 18 h with 100 ng/mL IL-22 (Lower) or left untreated (Upper), showing healthy bacteria in untreated organoids and degraded bacteria, often associated with lysosomes, in IL-22–pretreated iHOs. Representative images for each treatment condition were selected from 30 iHOs injected and processed per condition. Numbers indicate characterizations of bacterial cell damage/stress used for scoring: widening of periplasmic space (1); membrane damage and ragged appearance (2); decrease in cytosol density (3); direct contact with lysosomes (4); and presence of volutin granules (5).
Fig. 5.
IL-22–induced calgranulin B enhanced iHO colonization resistance to S. enterica Typhimurium infection. (A) Direct blocking of phagolysosomal fusion with phagolysosomal inhibitors restricts IL-22–mediated protection in iHOs. Kolf2 iHOs were pretreated with 100 ng/mL IL-22 for 18 h and then were treated with the phagolysosomal inhibitors W7 at 50 μM for 6 h or concanamycin A (CCMA) at 100 nM for 4 h or were left untreated, after which gentamicin protection assays were performed. There was no significant difference in intracellular recovered bacteria from W7- or concanamycin A-treated iHOs and untreated iHOs (W7, P = 0.7015; CCMA P = 0.0631, IL-22, ***P < 0.0001; unpaired, two-tailed Mann–Whitney U tests). Data presented show the mean from four biological replicates with three technical replicates per assay, ± SEM. (B and C) RT-qPCR was performed with a TaqMan gene-expression assay specific for RAB7A (B) or S100A9 (C) and was analyzed via the comparative CT method with GAPDH as an endogenous control. Data presented show the mean fold change between untreated samples and infected samples from three biological replicates, ± SEM. There was a significant difference in RAB7A and S100A9 expression in iHOs pretreated with IL-22 in comparison with iHOs left untreated before infection (RAB7A: P = 0.0001; S100A9: P < 0.0001; unpaired, two-tailed Student’s t tests). (D) Kolf2 iHOs challenged with IL-22 for 18 h or left untreated were microinjected with S. enterica Typhimurium SL1344 and examined for RAB7 (red), CSA-1 (green), and DAPI (blue) by immunofluorescence with colocalization between RAB7 and CSA-1 (yellow) visible in IL-22–pretreated samples. (Original magnification: 63×.) (E) Expression of S100A9 (green) by IL-22/S. enterica Typhimurium-treated Kolf2 and S100A9−/− iHOs was examined using immunofluorescence. (F) Protection assays in S100A9−/− iHOs show that pretreatment with IL-22 results in no significant difference in recovered intracellular bacterial after microinjection of S. enterica Typhimurium SL1344, in contrast to Kolf2 iHOs. (G) S100A9−/− or Kolf2 iHOs treated with IL-22 for 18 h and then microinjected with S. enterica Typhimurium SL1344 were examined for RAB7 (red), CSA-1 (green), and DAPI (blue) by immunofluorescence; RAB7 was not visible in S100A9−/− samples. (Original magnification: 40×.) n.s., not significant; ***P < 0.0001.
Calgranulin B Enhances IL-22–Induced Phagolysosomal Fusion.
Observations by Dhiman et al. (27) suggested that increased expression of calgranulin A (S100A8) was essential for IL-22–dependent mycobacterial growth inhibition through enhancement of phagolysosomal fusion. No differential expression of S100A8 was observed in our RNA-seq experiments; however, increased expression of calgranulin B (S100A9) was observed after exposure of iHOs to IL-22 and S. enterica Typhimurium (Dataset S3), and was confirmed via RT-qPCR (Fig. 5C) and immunostaining (SI Appendix, Fig. S7G). S100A9 is induced by IL-6 signaling in colonic cells via the interaction of activated STAT3 with the S100A9 promoter (28). Therefore, we reasoned that the IL-22 pathway might be triggering similar induction of S100A9 through STAT3 activation and thus decided to assess the role of S100A9 in IL-22–mediated phagolysosomal fusion. To this end, a biallelic mutation in S100A9 was generated in Kolf2 hIPSCs using CRISPR/Cas9 engineering. In iHOs derived from S100A9−/− hIPSCs, which were morphologically similar to other iHO lines (SI Appendix, Fig. S3), no expression of S100A9 protein via immunostaining was detected after stimulation with IL-22 and S. enterica Typhimurium SL1344 (Fig. 5E). Gentamicin protection assays in S100A9−/− iHOs demonstrated no significant difference in intracellular S. enterica Typhimurium recovered from IL-22–pretreated iHOs in comparison with untreated iHOs (Fig. 5F), suggesting that IL-22–induced S100A9 directly inhibits S. enterica Typhimurium colonization. To ensure that this phenotype was not due to lack of IL-22 response in S100A9−/− iHOs, mRNA levels of the IL-22–regulated genes LCN2 and DUOX2 were assessed after IL-22 treatment, revealing significant increases in the expression of both genes (SI Appendix, Fig. S7F). We then performed immunostaining for RAB7 in Kolf2 or S100A9−/− iHOs treated with IL-22 and infected with S. enterica Typhimurium (Fig. 5G), observing a lack of RAB7 in S100A9−/− iHOs.
Discussion
We have shown that iHOs generated from healthy control hIPSCs expressed IL-22 receptors on their basal surface and were responsive to IL-22. Conversely, IL-22 response was lost in iHOs generated from a patient with a loss-of-function mutation in IL10RB. Pretreatment of responsive iHOs with IL-22 appeared to drive a “barrier phenotype” as IL-22–primed iHOs were less susceptible to colonization by S. enterica Typhimurium, and prolonged incubation enhanced differences in bacterial survival. Through detailed imaging studies we attributed the IL-22–dependent protective phenotype to increased intracellular fusion of SCVs with lysosomes. Since fusion with the lysosome ends in degradation, some pathogens that enter the cell through the endocytic or phagocytic pathway are adapted to avoid this fusion (29). S. enterica Typhimurium has been shown to be capable of avoiding phagolysosomal fusion by modulating networks of Rab proteins that are recruited to the phagosome (30). To our knowledge, there have been no published reports showing that IL-22 prompts increased lysosomal fusion as a mechanism for restricting S. enterica Typhimurium growth in IECs. IL-22 can restrict the growth of M. tuberculosis intracellularly in macrophages by enhancing phagosomal fusion (22), and calgranulin A (S100A8) is required for this process (27). In iHOs we found that calgranulin B (S100A9) was required for IL-22–mediated intracellular killing. Further studies will be required to establish the functional relevance of this pathway in an intact immune system and to establish the specific interactions among S100A9, RAB7, and the phagolysome. Although our data suggest that IECs may be less susceptible to S. enterica Typhimurium colonization when the IL-22 pathway is functional, in vivo Salmonella has been observed to cross the epithelial barrier via M cells and direct capture from the lumen by phagocytes (31). This might help explain why Il22−/− mice are not more susceptible to S. enterica Typhimurium infection in vivo, although this lack of susceptibility could also be attributable to depletion of the protective microbiota by IL-22–mediated antimicrobials (20). Here, our data showed differences after preincubation with IL-22 that might be more relevant to later stages of infection. However, in several models S. enterica Typhimurium has been shown to take longer than expected to breach the intestinal epithelium (32), and IL-22 expression, along with genes induced by IL-22, are highly expressed during the initial phases of infection (33, 34), suggesting that this pathway may also have an impact on earlier interactions in vivo.
It would be tempting to speculate that the phenotype of infantile severe colitis in IL10R2-deficient patients may be at least partially attributable to reduced barrier function at the epithelial tissues. In addition to their well-characterized defective response to the antiinflammatory cytokine IL-10 in immune cells (35), patients with IL10RB defects might also exhibit reduced barrier function (7). However, the clinical presentation in patients with loss of function mutations in IL-10, IL10RA, and IL10RB is similarly severe, and IL10RB mutations have not been associated with enhanced enteric infection, suggesting that this pathway in the epithelium may be redundant in vivo. Further understanding of the epithelial defects in patients with IL-10 signaling defects is becoming increasingly important, given that hematopoietic transplantation is a curative therapeutic option in patients with IL-10/IL10R deficiency (7, 36), and it will be important to investigate whether there are differences in the posttransplant intestinal infection susceptibility between patients with IL-10 and IL10RA defects on the one side and patients with IL10RB defects on the other.
Materials and Methods
Generation and Culture of hIPSCs and Directed Differentiation to iHOs.
The healthy control hIPSC lines Yemz1, Lise1, and Kolf2, were acquired through the Human Induced Pluripotent Stem Cells Initiative Consortium (HipSci; www.hipsci.org), through which they were also characterized (37). Consent was obtained for the use of cell lines for the HIPSCI project from healthy volunteers. A favorable ethical opinion was granted by the National Research Ethics Service (NRES) Research Ethics Committee Yorkshire and The Humber – Leeds West, reference number 15/YH/0391. IL10RBPat hIPSCs were generated at the Wellcome Trust Sanger Institute (WTSI) as part of the Oxford IBD cohort study/COLORS in IBD (Colitis of Early Onset—Rare Diseases Within IBD Disease Phenotypes) project with appropriate ethical approvals [REC: 09/H1204/30; North Staffs Local Research Ethics Committee/NRES Committee West Midlands – The Black Country (IBD in Oxford)]. hIPSC lines were differentiated into endoderm, hindgut, and then iHOs using a previously published protocol (14, 38). For production of primary organoids, mucosal tissue was harvested and seeded according to protocols described by Sato and coworkers (39) and Fordham et al. (40). Control human intestinal samples were collected from the ascending colon and ileum of children under 16 y of age undergoing routine diagnostic endoscopy, following ethical approval (REC-12/EE/0482; NRES Committee East of England–Hertfordshire) and informed consent. All other methods are described in SI Appendix, SI Materials and Methods.
Data Availability.
The RNA-seq data have been deposited in the European Nucleotide Archive (accession no. ERP024278).
Supplementary Material
Acknowledgments
We thank Yoonha Choi of the Wellcome Trust Sanger Institute (WTSI) for help with the design of the functional IL10RB-targeting vector; the WTSI Core Scientific Operations team for conducting Illumina transcriptome sequencing; the WTSI Pathogen Informatics RNA-sequencing pipeline for mapping RNA-seq data; the WTSI High-Throughput Gene-Editing pipeline for generating S100A9−/− iPSCs; Dirk Bumann of the University of Basel for donating the TIMERbac Salmonella strain; and the Oxford IBD Cohort investigators, in particular Simon Travis and Huei-Ting, for help and discussions. This work was supported by The Wellcome Trust. F.P. and H.H.U. are supported by the Leona M. and Harry B. Helmsley Charitable Trust, the Crohn’s and Colitis Foundation of America, and the National Institute for Health Research Biomedical Research Centre, Oxford. R.E.W.H. holds a Canada Research Chair and is a University of British Columbia Killam Professor. The COLORS in IBD project is supported by the European Society for Paediatric Gastroenterology Hepatology and Nutrition.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The RNA-sequencing data have been deposited in the European Nucleotide Archive (accession no. ERP024278).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1811866115/-/DCSupplemental.
References
- 1.Villa-Diaz LG, Ross AM, Lahann J, Krebsbach PH. Concise review: The evolution of human pluripotent stem cell culture: From feeder cells to synthetic coatings. Stem Cells. 2013;31:1–7. doi: 10.1002/stem.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McKernan R, Watt FM. What is the point of large-scale collections of human induced pluripotent stem cells? Nat Biotechnol. 2013;31:875–877. doi: 10.1038/nbt.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.An MC, et al. Genetic correction of Huntington’s disease phenotypes in induced pluripotent stem cells. Cell Stem Cell. 2012;11:253–263. doi: 10.1016/j.stem.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bellin M, et al. Isogenic human pluripotent stem cell pairs reveal the role of a KCNH2 mutation in long-QT syndrome. EMBO J. 2013;32:3161–3175. doi: 10.1038/emboj.2013.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glocker EO, et al. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N Engl J Med. 2009;361:2033–2045. doi: 10.1056/NEJMoa0907206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glocker EO, et al. Infant colitis–It’s in the genes. Lancet. 2010;376:1272. doi: 10.1016/S0140-6736(10)61008-2. [DOI] [PubMed] [Google Scholar]
- 7.Engelhardt KR, et al. Clinical outcome in IL-10- and IL-10 receptor-deficient patients with or without hematopoietic stem cell transplantation. J Allergy Clin Immunol. 2013;131:825–830. doi: 10.1016/j.jaci.2012.09.025. [DOI] [PubMed] [Google Scholar]
- 8.Maloy KJ, Powrie F. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature. 2011;474:298–306. doi: 10.1038/nature10208. [DOI] [PubMed] [Google Scholar]
- 9.Sabat R, Ouyang W, Wolk K. Therapeutic opportunities of the IL-22-IL-22R1 system. Nat Rev Drug Discov. 2014;13:21–38. doi: 10.1038/nrd4176. [DOI] [PubMed] [Google Scholar]
- 10.Liang SC, et al. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J Exp Med. 2006;203:2271–2279. doi: 10.1084/jem.20061308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie MH, et al. Interleukin (IL)-22, a novel human cytokine that signals through the interferon receptor-related proteins CRF2-4 and IL-22R. J Biol Chem. 2000;275:31335–31339. doi: 10.1074/jbc.M005304200. [DOI] [PubMed] [Google Scholar]
- 12.Pham TA, et al. Sanger Mouse Genetics Project Epithelial IL-22RA1-mediated fucosylation promotes intestinal colonization resistance to an opportunistic pathogen. Cell Host Microbe. 2014;16:504–516. doi: 10.1016/j.chom.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng Y, et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14:282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 14.Forbester JL, et al. Interaction of Salmonella enterica serovar Typhimurium with intestinal organoids derived from human induced pluripotent stem cells. Infect Immun. 2015;83:2926–2934. doi: 10.1128/IAI.00161-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sadelain M, Papapetrou EP, Bushman FD. Safe harbours for the integration of new DNA in the human genome. Nat Rev Cancer. 2011;12:51–58. doi: 10.1038/nrc3179. [DOI] [PubMed] [Google Scholar]
- 16.Chang CJ, Bouhassira EE. Zinc-finger nuclease-mediated correction of α-thalassemia in iPS cells. Blood. 2012;120:3906–3914. doi: 10.1182/blood-2012-03-420703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ranjbar S, Haridas V, Jasenosky LD, Falvo JV, Goldfeld AE. A role for IFITM proteins in restriction of Mycobacterium tuberculosis infection. Cell Rep. 2015;13:874–883. doi: 10.1016/j.celrep.2015.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grasberger H, El-Zaatari M, Dang DT, Merchant JL. Dual oxidases control release of hydrogen peroxide by the gastric epithelium to prevent Helicobacter felis infection and inflammation in mice. Gastroenterology. 2013;145:1045–1054. doi: 10.1053/j.gastro.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosenstiel P, et al. Regulation of DMBT1 via NOD2 and TLR4 in intestinal epithelial cells modulates bacterial recognition and invasion. J Immunol. 2007;178:8203–8211. doi: 10.4049/jimmunol.178.12.8203. [DOI] [PubMed] [Google Scholar]
- 20.Behnsen J, et al. The cytokine IL-22 promotes pathogen colonization by suppressing related commensal bacteria. Immunity. 2014;40:262–273. doi: 10.1016/j.immuni.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Claudi B, et al. Phenotypic variation of Salmonella in host tissues delays eradication by antimicrobial chemotherapy. Cell. 2014;158:722–733. doi: 10.1016/j.cell.2014.06.045. [DOI] [PubMed] [Google Scholar]
- 22.Dhiman R, et al. IL-22 produced by human NK cells inhibits growth of Mycobacterium tuberculosis by enhancing phagolysosomal fusion. J Immunol. 2009;183:6639–6645. doi: 10.4049/jimmunol.0902587. [DOI] [PubMed] [Google Scholar]
- 23.Vergne I, Chua J, Deretic V. Tuberculosis toxin blocking phagosome maturation inhibits a novel Ca2+/calmodulin-PI3K hVPS34 cascade. J Exp Med. 2003;198:653–659. doi: 10.1084/jem.20030527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dröse S, Altendorf K. Bafilomycins and concanamycins as inhibitors of V-ATPases and P-ATPases. J Exp Biol. 1997;200:1–8. doi: 10.1242/jeb.200.1.1. [DOI] [PubMed] [Google Scholar]
- 25.Yates RM, Hermetter A, Russell DG. The kinetics of phagosome maturation as a function of phagosome/lysosome fusion and acquisition of hydrolytic activity. Traffic. 2005;6:413–420. doi: 10.1111/j.1600-0854.2005.00284.x. [DOI] [PubMed] [Google Scholar]
- 26.Mottola G. The complexity of Rab5 to Rab7 transition guarantees specificity of pathogen subversion mechanisms. Front Cell Infect Microbiol. 2014;4:180. doi: 10.3389/fcimb.2014.00180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dhiman R, et al. Interleukin 22 inhibits intracellular growth of Mycobacterium tuberculosis by enhancing calgranulin A expression. J Infect Dis. 2014;209:578–587. doi: 10.1093/infdis/jit495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee MJ, et al. Interleukin-6 induces S100A9 expression in colonic epithelial cells through STAT3 activation in experimental ulcerative colitis. PLoS One. 2012;7:e38801. doi: 10.1371/journal.pone.0038801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luzio JP, Pryor PR, Bright NA. Lysosomes: Fusion and function. Nat Rev Mol Cell Biol. 2007;8:622–632. doi: 10.1038/nrm2217. [DOI] [PubMed] [Google Scholar]
- 30.Smith AC, et al. A network of Rab GTPases controls phagosome maturation and is modulated by Salmonella enterica serovar Typhimurium. J Cell Biol. 2007;176:263–268. doi: 10.1083/jcb.200611056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Velge P, et al. Multiplicity of Salmonella entry mechanisms, a new paradigm for Salmonella pathogenesis. MicrobiologyOpen. 2012;1:243–258. doi: 10.1002/mbo3.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mayuzumi H, Inagaki-Ohara K, Uyttenhove C, Okamoto Y, Matsuzaki G. Interleukin-17A is required to suppress invasion of Salmonella enterica serovar Typhimurium to enteric mucosa. Immunology. 2010;131:377–385. doi: 10.1111/j.1365-2567.2010.03310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raffatellu M, et al. Simian immunodeficiency virus-induced mucosal interleukin-17 deficiency promotes Salmonella dissemination from the gut. Nat Med. 2008;14:421–428. doi: 10.1038/nm1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Godinez I, et al. T cells help to amplify inflammatory responses induced by Salmonella enterica serotype Typhimurium in the intestinal mucosa. Infect Immun. 2008;76:2008–2017. doi: 10.1128/IAI.01691-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moran CJ, et al. IL-10R polymorphisms are associated with very-early-onset ulcerative colitis. Inflamm Bowel Dis. 2013;19:115–123. doi: 10.1002/ibd.22974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murugan D, et al. Very early onset inflammatory bowel disease associated with aberrant trafficking of IL-10R1 and cure by T cell replete haploidentical bone marrow transplantation. J Clin Immunol. 2014;34:331–339. doi: 10.1007/s10875-014-9992-8. [DOI] [PubMed] [Google Scholar]
- 37.Leha A, et al. HipSci Consortium A high-content platform to characterise human induced pluripotent stem cell lines. Methods. 2016;96:85–96. doi: 10.1016/j.ymeth.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Forbester JL, Hannan N, Vallier L, Dougan G. Derivation of intestinal organoids from human induced pluripotent stem cells for use as an infection system. Methods Mol Biol. 2016 doi: 10.1007/7651_2016_7. [DOI] [PubMed] [Google Scholar]
- 39.Jung P, et al. Isolation and in vitro expansion of human colonic stem cells. Nat Med. 2011;17:1225–1227. doi: 10.1038/nm.2470. [DOI] [PubMed] [Google Scholar]
- 40.Fordham RP, et al. Transplantation of expanded fetal intestinal progenitors contributes to colon regeneration after injury. Cell Stem Cell. 2013;13:734–744. doi: 10.1016/j.stem.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data have been deposited in the European Nucleotide Archive (accession no. ERP024278).