Significance
Chromosomal rearrangements are early and essential events in the formation of many tumors. Two distinct end-joining pathways, classic and alternative nonhomologous end joining, can mediate rearrangement formation. Previous studies have shown that genetic factors mediating rearrangements differ significantly between mouse and human cells. Here we show that poly(ADP)ribose polymerase 3 (Parp3) uniquely promotes chromosomal rearrangements in both species. Using next-generation sequencing of rearrangement junctions, we investigated the mechanistic contribution of Parp3 and a closely related enzyme, Parp1, that is also known to promote rearrangements in murine cells. We find differences in the phenotypes of rearrangements in cells lacking Parp3, Parp1, or both, suggesting that these enzymes promote rearrangements through distinct mechanisms and providing insight into this essential mechanism of tumorigenesis.
Keywords: Parp3, rearrangements, nonhomologous end joining
Abstract
Chromosomal rearrangements, including translocations, are early and essential events in the formation of many tumors. Previous studies that defined the genetic requirements for rearrangement formation have identified differences between murine and human cells, most notably in the role of classic and alternative nonhomologous end-joining (NHEJ) factors. We reported that poly(ADP)ribose polymerase 3 (PARP3) promotes chromosomal rearrangements induced by endonucleases in multiple human cell types. We show here that in contrast to classic (c-NHEJ) factors, Parp3 also promotes rearrangements in murine cells, including translocations in murine embryonic stem cells (mESCs), class–switch recombination in primary B cells, and inversions in tail fibroblasts that generate Eml4–Alk fusions. In mESCs, Parp3-deficient cells had shorter deletion lengths at translocation junctions. This was corroborated using next-generation sequencing of Eml4–Alk junctions in tail fibroblasts and is consistent with a role for Parp3 in promoting the processing of DNA double-strand breaks. We confirmed a previous report that Parp1 also promotes rearrangement formation. In contrast with Parp3, rearrangement junctions in the absence of Parp1 had longer deletion lengths, suggesting that Parp1 may suppress double-strand break processing. Together, these data indicate that Parp3 and Parp1 promote rearrangements with distinct phenotypes.
Chromosomal rearrangements are critical events in the pathogenesis of malignant and nonmalignant disorders (1). These aberrant events drive malignant transformation and congenital disorders, including deafness, schizophrenia, and infertility. Many efforts to elucidate the genetic basis of rearrangement formation have relied upon experiments in mouse cells. Studies in murine embryonic stem cells (mESCs) identified a cohort of genetic factors that promote or suppress rearrangements (2–6). In aggregate, these studies suggest that chromosomal rearrangements form by a noncanonical or alternative nonhomologous end-joining pathway (alt-NHEJ). However, a subsequent report demonstrated that in human cells the rearrangement between endonuclease-induced double-strand breaks (DSBs) depends on classic NHEJ (c-NHEJ) (7). The disparate results suggest that the genetic basis for rearrangements differs significantly between human and murine cells.
We recently reported that PARP3, a member of the ADP-ribose polymerase family of enzymes, promotes chromosomal rearrangement formation in human cells (8). We showed that PARP3 regulates G quadruplex (G4) DNA in response to DNA damage. Chemical stabilization of G4 DNA in PARP3−/− cells led to widespread DSBs. This suggested a model in which PARP3 suppresses G4 DNA, which allows processing of DSB ends to intermediates that participate in rearrangements in human cells. Here we investigated the role of Parp3 within murine cells using a range of cell types and approaches to quantify frequency and characterize rearrangement junction phenotypes. In this study we have examined large rearrangements, including interchromosomal translocations between chromosomes 17 and 14, ∼100- to 200-kb intrachromosomal rearrangements in switch regions, and ∼10-Mb intrachromosomal inversions on chromosome 17.
Results
Parp3 Promotes Targeted Translocations in mESCs.
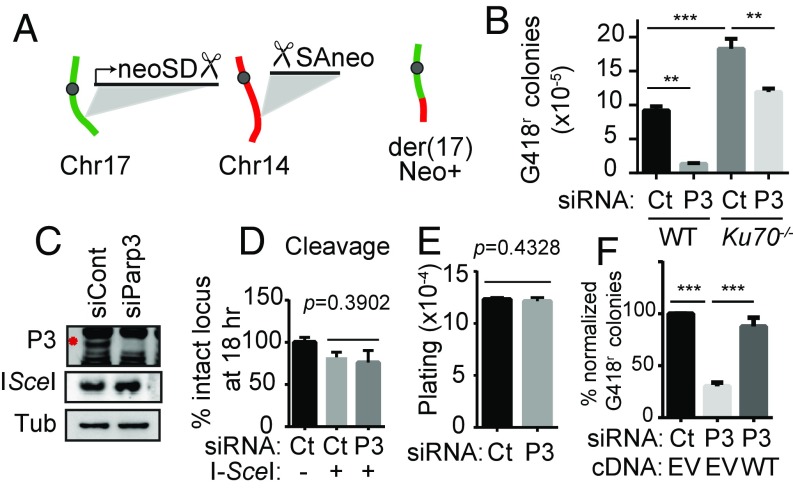
First, we tested the effects of Parp3-depletion mESCs harboring the pCr15 reporter (2). Introduction of the I-SceI endonuclease into these cells leads to concurrent DSBs on chromosomes 14 and 17. Translocation between these DSBs generates a functional neomycin-resistance gene on derivative chromosome 17. Knockdown of Parp3 reduced the frequency of targeted translocations between chromosomes 14 and 17 induced by I-SceI by ∼80% compared with a control siRNA (Fig. 1 A–C). Targeted DSBs that are repaired by inversion, large deletions that extend into the neomycin resistance gene-coding sequence, or other rare chromosomal events are not detected in this assay. The siParp3-dependent reduction in targeted translocations is approximately the same extent of reduction previously reported after knockdown of the DSB end-resection factor CtIP (5). PARP3 depletion did not affect I-SceI protein expression, cleavage by I-SceI, or colony-plating efficiency (Fig. 1 C–E). PARP3 expression after suppression with a UTR-directed siRNA rescued the siRNA effect on rearrangement frequency (Fig. 1F). Cells lacking the c-NHEJ factor Ku70 had approximately twofold higher rearrangement frequency than wild-type pCr15 cells (Fig. 1B and ref. 2). PARP3 knockdown also reduced rearrangement frequency in Ku70−/− pCr15 cells (Fig. 1B), indicating that PARP3 promotes rearrangements both in the presence and absence of Ku70.
Fig. 1.
Parp3 promotes translocations in mESCs. (A) Schematic of the targeted translocation assay (2). Scissors indicate the I-SceI site. (B) Absolute translocation frequency in wild-type and Ku70−/− pCr15 mESCs with siRNA against Parp3 (P3) or nontargeting control (Ct). (C) Immunoblots in pCr15 cells. The red asterisk indicates PARP3. Tub, tubulin. (D and E) Cleavage by I-SceI measured by qPCR across the I-SceI site (D) and plating efficiency of mESCs (E). (F) Translocation frequency with 5′ UTR siParp3 and expression of PARP3 transcript with empty vector (EV) or PARP3 (WT) pCAGGS. **P < 0.01, ***P < 0.001; ANOVA with Tukey’s correction (B and F) or an unpaired Student’s t test (D and E).
We examined the characteristics of the rearrangement junctions to elucidate Parp3-dependent mechanisms involved in rearrangement formation. A central challenge of this analysis is to interpret Parp3-dependent changes in rearrangement junctions in the context of a Parp3-dependent reduction in overall rearrangement frequency (Fig. 1B). Therefore, throughout our analysis, we have referred to junction phenotypes in “residual” events. Among the residual translocations in Parp3-depleted cells, there was a notable reduction in junctions with longer deletions (i.e., >200 bp) (SI Appendix, Fig. S1 A and B). This resulted in a statistically significant reduction in the mean length of deletions at rearrangement junctions in both wild-type pCr15 cells and Ku70−/− pCr15 cells (SI Appendix, Table S1 and Dataset S1). This suggests that Parp3 plays a Ku70-independent role in promoting long deletions at rearrangement junctions.
We compared the phenotypes reported from previous studies using pCr15 cells (2, 4, 5). In these studies, knockout of the c-NHEJ factors Ku70 or Xrcc4 increased the proportion of junctions with >200-bp deletions in residual translocations (SI Appendix, Fig. S1C and Dataset S1). In contrast, Parp3 knockdown led to a reduction in junctions with >200-bp deletions, similar to CtIP knockdown (SI Appendix, Fig. S1C). We did not find any significant effects from Parp3 knockdown on insertions or microhomology (i.e., 1- to 10-bp stretches of homology) at translocation junctions (SI Appendix, Fig. S1 D and E and Table S1).
Parp3 Promotes Class-Switch Recombination.
To explore the in vivo phenotypes of Parp3 deficiency, we used CRISPR/Cas9 mutagenesis to establish Parp3−/− mice. We deleted a 492-bp region containing two of the three Parp3 catalytic residues in mESCs (SI Appendix, Fig. S1 F–H). Cells with the deletion exhibited complete loss of Parp3 expression (SI Appendix, Fig. S1 I and J), and Parp3−/− mice were established from these mESCs in the 129s background. As previously reported in a separate knockout mouse (9), Parp3−/− mice were born in expected Mendelian ratios and had no apparent gross phenotypes.
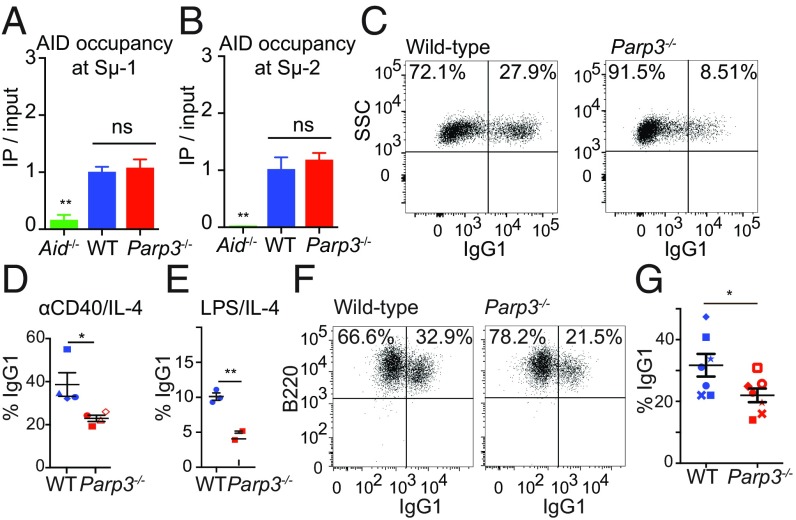
Based on our observation that Parp3 promotes targeted rearrangements in mESCs, we hypothesized that Parp3 would also promote class-switch recombination (CSR), a physiological form of long-range, intrachromosomal rearrangement across ∼100–200 kb. Several factors that mediate interchromosomal rearrangements also play roles in CSR, and aberrant CSR is known to contribute to pathological rearrangements (10). In particular, 53bp1 plays a role in long-range rearrangements in multiple contexts in both murine and human cells (8, 11, 12). However, a recent study reported that Parp3 loss increased CSR frequency (13) by increasing activation-induced deaminase (AID) occupancy at switch regions. Therefore, we first sought to establish whether Parp3 influences AID occupancy at switch regions Sμ-1 and Sμ-2 in our Parp3−/− mouse. We performed ChIP for AID in wild-type and Parp3−/− primary B cells and qPCR for Sμ-1 and Sμ-2. As expected, ChIP-qPCR demonstrated marked enrichment of Sμ-1 and Sμ-2 sequences in wild-type B cells compared with B cells from Aid−/− mice. Unlike the previous report, we did not find any significant differences in AID occupancy at Sμ-1 and at Sμ-2 in Parp3-deficient cells (Fig. 2 A and B).
Fig. 2.
Parp3 promotes CSR in murine B cells. (A and B) AID occupancy at Sμ-1 (A) and Sμ-2 (B) measured by ChIP. (C–E) Ex vivo CSR to IgG1 in primary B cells following stimulation with αCD40/IL-4 (C and D) or LPS/IL-4 (E). Data are shown as mean ± SE. (F and G) Ex vivo CSR to IgG1 in primary B cells from Rag2-deficient blastocyst complementation with WT or Parp3−/−-deficient mESCs. Two different Parp3−/− clones were tested. Data are shown as mean ± SE with a representative experiment from n = 7. P values were calculated by unpaired Student’s t test (A and B) or one-way ANOVA with Tukey’s correction (D and G). *P < 0.05, **P < 0.01, ***P < 0.001. ns, not significant.
The absence of an effect on recruitment within our model allows us to directly investigate the contribution of Parp3 to recombination downstream of AID recruitment. We confirmed that Parp3−/− mice have no statistically significant defects in the early stages of B cell maturation that precede CSR (SI Appendix, Fig. S2). Parp3−/− B cells underwent CSR to IgG1 ∼40–50% less efficiently than wild-type cells upon in vitro stimulation with either αCD40/IL-4 or LPS/IL-4 (Fig. 2 C–E). Despite reduced CSR, Parp3−/− B cells had similar or increased AID expression, Sμ-1 and Sμ-2 transcript levels, and in vitro proliferation following stimulation compared with wild-type B cells (SI Appendix, Fig. S3 A–D). Analysis of switch-junction sequences showed no significant Parp3-dependent changes in insertions or microhomology (SI Appendix, Fig. S3E).
To confirm these findings with a second approach, we performed Rag2-deficient blastocyst complementation (14) to reconstitute the lymphoid compartment with two different Cas9-generated Parp3−/− mESC clones. After in vitro stimulation with αCD40/IL-4, Parp3−/− B cells isolated from complemented Rag2−/− mice had ∼35% reduced CSR to IgG1 compared with wild-type B cells (Fig. 2 F and G). As a third approach, we used the CH12F3 murine B cell line, which undergoes CSR from IgM to IgA upon stimulation with αCD40, IL-4, and TGF-β (15). Compared with wild-type cells, shRNA-mediated knockdown of Parp3 reduced CSR to IgA by ∼50% compared with cells with control shRNA (SI Appendix, Fig. S3 F and G). This reduction was similar in effect to shRNA-mediated knockdown of 53bp1, a factor known to promote CSR (11). When we used a retrovirus to express PARP3 in Parp3-depleted cells, we found that CSR from IgM to IgA was restored to the level of the control cells (SI Appendix, Fig. S3 H and I).
Loss of Parp3 Reduces Eml4–Alk Inversions.
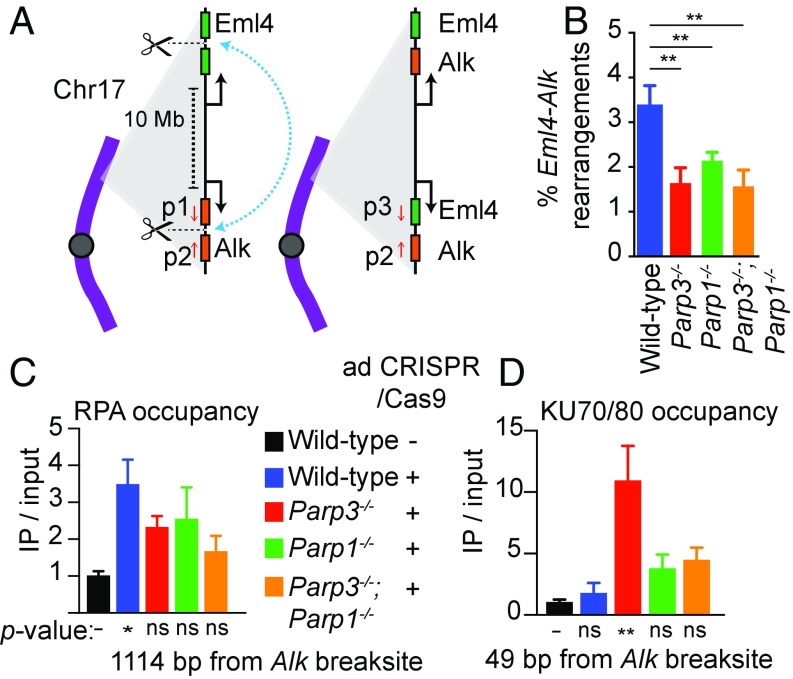
To study a phenotype of long-range rearrangement junctions in murine cells in more depth, we examined inversions between the Eml4 and Alk loci located 10 Mb apart on mouse chromosome 17 (Fig. 3A). Previous studies have suggested that intrachromosomal rearrangements of this scale share mechanistic features with interchromosomal rearrangements (16, 17). We chose Eml4–Alk because it is a known driver of non-small cell lung cancer in both mice and humans (18). In addition, the relatively high frequency of rearrangements in this system (18) coupled to high-throughput analysis of amplicon deep sequencing allowed us to compare Eml4–Alk rearrangement junctions (i.e., distal repair events) with non-rearrangement repair events at the Alk locus (i.e., proximal repair events). The murine Alk locus is predicted to contain abundant G4 DNA structures (SI Appendix, Fig. S3J). Thus, we hypothesized that Parp3 deletion would affect chromosomal rearrangements involving this locus.
Fig. 3.
High-throughput analysis of amplicon deep sequencing. (A) Schematic of Eml4–Alk rearrangements on mouse chromosome 17. Scissors indicate Cas9-mediated DSB. p1, p2, and p3 are PCR primers amplifying proximal repair (p1 and p2) or rearrangements (p2 and p3). (B) Frequency of Eml4–Alk rearrangements in murine tail fibroblasts measured by droplet digital PCR 5 d after adenovirus-mediated expression of CRISPR/Cas9. (C and D) ChIP to measure RPA (C) and KU70/KU80 (D) occupancy at the Alk locus measured 24 h after adenovirus-mediated expression of CAS9 with gRNA targeting Alk or dsRed as a control. ChIP data are shown as the mean ± SE of n = 3 biological replicates. *P < 0.05; **P < 0.01; one-way ANOVA with Dunnett’s correction. ns, not significant.
In cultures of primary murine tail fibroblasts, we used an established adenoviral approach to express CRISPR/Cas9 with gRNA targeting intron 13 of Eml4 and intron 19 of Alk (Fig. 3A) (18, 19). We performed droplet digital PCR to measure the frequencies of rearrangements and observed that rearrangement frequency was reduced ∼50% in Parp3−/− tail fibroblasts (Fig. 3B) without an appreciable difference in adenoviral transduction efficiency (SI Appendix, Fig. S3K) or CAS9 cutting efficiency (SI Appendix, Fig. S4A).
Loss of Parp3 Reduces Replication Protein A and Increases KU70/KU80 Deposition at Targeted DSBs.
To examine the mechanistic roles of Parp3 and Parp1 in DSB repair at this locus, we used ChIP to measure occupancy of repair factors at the CRISPR/Cas9-mediated DSBs. Replication protein A (RPA) is a heterotrimeric complex that binds to ssDNA intermediates during homologous recombination (HR) (20), one of two predominant DSB repair pathways. RPA was significantly increased in wild-type tail fibroblasts upon DSB induction at Alk (Fig. 3C) with a similar pattern observed at Eml4 (SI Appendix, Fig. S4B). In contrast, RPA deposition at both DSBs was reduced in Parp3−/−, Parp1−/−, and Parp3−/−;Parp1−/− cells (Fig. 3C and SI Appendix, Fig. S4B). These findings are consistent with our studies in human cells (8) and additionally suggest that Parp3 and Parp1 have overlapping roles in promoting RPA deposition with no significant additive effect observed from loss of both enzymes.
The KU70/KU80 complex binds DSB ends during NHEJ to protect the broken DNA and recruit downstream repair factors (20). In Parp3−/− cells, KU70/KU80 binding was significantly increased at sites close to both DSB ends (Fig. 3D and SI Appendix, Fig. S4C). In wild-type, Parp1−/−, and Parp3−/−;Parp3−/− cells the increases in KU70/KU80 were more modest (Fig. 3D and SI Appendix, Fig. S4C). Taken together, these results suggest that KU70/KU80 occupancy at DSBs is higher in the absence of Parp3 than in the other genotypes and that loss of Parp1 in the Parp3−/− cells largely reverses this effect.
Differences in Repair Junction Phenotypes in Parp3−/− and Parp1−/− Cells.
High-throughput analysis of amplicon deep sequencing can facilitate the rapid interrogation of repair phenotypes (21–23). We used this approach to address an aspect of the long-standing question of PARP enzyme redundancy in the context of DNA repair. Parp1 and Parp3 share ∼60% homology in their catalytic and WGR domains (24) and modify overlapping and distinct targets in DNA repair (9, 25). We have observed that while both enzymes promote RPA deposition at targeted DSBs (Fig. 3C and ref. 8), Parp3 uniquely suppresses KU70/KU80 accumulation (Fig. 3D). Both enzymes promote chromosomal rearrangements in murine cells (Fig. 1B and ref. 6). Together, these findings suggest that they may act in a partially overlapping mechanism. To examine the relationship between Parp3 and Parp1 in rearrangement formation, we first compared Eml4–Alk rearrangement frequencies. Parp1−/− tail fibroblasts exhibited ∼50% reduced rearrangement frequency (Fig. 3B), similar to Parp3−/− tail fibroblasts. Tail fibroblasts lacking both Parp3−/− and Parp1−/− also had ∼50% reduced rearrangement frequency (Fig. 3B), suggesting the enzymes are epistatic with respect to this phenotype.
Next, we used PCR in the linear range (SI Appendix, Fig. S4D) to amplify ∼250-bp regions surrounding either the Eml4–Alk rearrangement junction or the Alk CRISPR/Cas9 cut site (Fig. 3A). The amplicons were deep sequenced in multiplexed, paired-end MiSeq reactions. To validate our assay, we designed five different synthetic sequences or “spike-in controls” that mimic distinct repair outcomes at the Alk locus (SI Appendix, Table S2). We created mixtures of defined quantities of these spike-in controls and performed amplicon deep sequencing in triplicate. The comparison between the number of input molecules and the readcounts resulted in a Spearman correlation coefficient (rs) of 0.982 (SI Appendix, Fig. S4E), indicating that the deep sequencing approach is highly quantitative across at least five orders of magnitude.
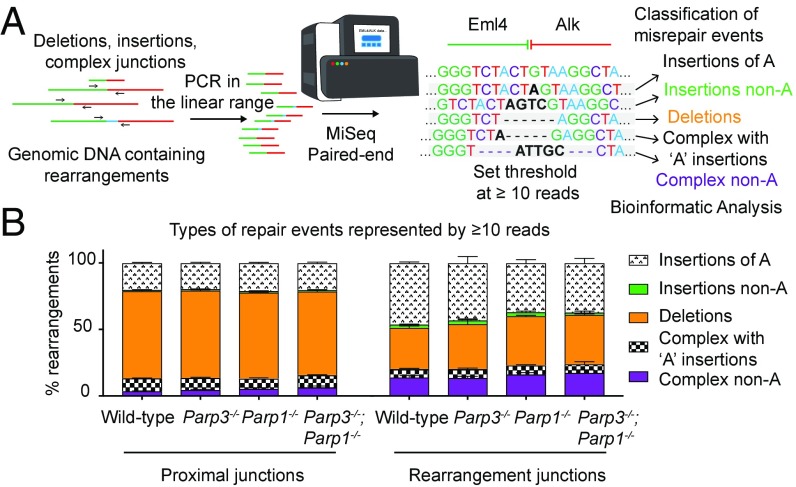
Similar to recent studies, our analysis cannot distinguish uncut loci from error-free repair (21, 22). Therefore, we considered only sequences with insertions or deletions to be unambiguous products of end joining. We used HiFiBR (26) to categorize all misrepaired sequences represented by ≥10 reads as deletions, insertions, or complex repair events (deletion combined with insertion) (Fig. 4A). We set our threshold at ≥10 reads for a unique junction to reduce PCR artifacts and increase the likelihood that repair events would be reproducibly observed across samples. We also sequenced control amplicons from cells without CRISPR/Cas9 cutting. “Misrepaired” sequences detected in the uncut controls were present at very low levels (0.09–0.001%) in the cells with CRISPR/Cas9 cutting (SI Appendix, Table S3). Approximately 80% of these sequences were excluded from our analysis because they were represented by <10 reads. However, a handful of these sequences may be real repair events that also appear as sequencing artifacts. Therefore, we have not excluded repair sequences based solely on their appearance in the uncut control dataset. Three biological replicates were performed for each of the four genotypes for both Eml4–Alk rearrangement and proximal repair at the Alk locus. We determined the relative frequencies of repair events by dividing by the total number of reads (for rearrangements) or the total number of misrepaired reads (proximal repair) (Fig. 4B and SI Appendix, Fig. S4F). We further analyzed all misrepaired sequences represented by one or more reads rather than the threshold of ≥10 reads, and this analysis yielded very similar distributions (SI Appendix, Fig. S5A).
Fig. 4.
Amplicon deep sequencing of Eml4–Alk repair junctions. (A) Schematic of sequencing and analysis pipeline for repair amplicons. PCR in the linear range generates amplicons from proximal repair or rearrangements. After MiSeq paired-end sequencing, differences from the reference sequence are classified as deletions, insertions, or complex repair events. Insertions or complex repair events containing a single A nucleotide insertion are designated separately. (B) Distributions of types of repair events represented by misrepair sequences with ≥10 reads.
We first noted that an unexpectedly high proportion of both proximal repair junctions and rearrangements were classified as insertions (∼25% and ∼50%, respectively) (Fig. 4B). Of these insertions, the large majority consisted of a single “A” base pair (Fig. 4B and SI Appendix, Table S4), a common repair outcome for CRISPR/Cas9 DSBs (27, 28). Among the complex junctions, ∼35–75% consisted of 1-bp insertions of A concurrent with deletions (Fig. 4B). We validated our experimental strategy using Sanger sequencing of repair products, and 95% of the misrepaired events were also seen with amplicon deep sequencing, including a high proportion of single A insertions (SI Appendix, Table S5). For the purposes of our initial analysis, we retained these single A insertion events. We compared the distributions of repair events found at rearrangement junctions versus proximal junctions. To do so, we constructed a multinomial logistic regression model with a random effect for repeated measures using type of junction as the dependent variable (SI Appendix, Table S6). Due to the large number of reads in the analysis (SI Appendix, Fig. S4F), most comparisons were significant (P < 0.0001), so we considered only those with an odds ratio (OR) ≥1.25 or ≤0.75 to be of interest. Rearrangements in each genetic background (wild-type, Parp3−/−, Parp1−/−, and Parp3−/−;Parp1−/−) were less likely to have deletions and more likely to have insertions relative to proximal repair in the same genetic background (P < 0.0001; ORs 4.93, 4.05, 3.13, and 3.05, respectively) (Fig. 4B and SI Appendix, Table S6). As the great majority of these insertions are likely CRISPR/Cas9-mediated, we evaluated the dataset neglecting all events containing insertions of single A nucleotides and found similar patterns for all genotypes (P < 0.0001; ORs 6.03, 4.72, 4.47, and 2.61 respectively) (SI Appendix, Fig. S5B and Table S7).
Using the complete dataset, we compared distributions of rearrangement repair events between genotypes. Rearrangements in Parp3−/− or Parp1−/− cells were more likely to have deletions than those in wild-type cells (ORs = 1.25 and 1.48, respectively) (Fig. 4B and SI Appendix, Table S6). Cells lacking both enzymes exhibited a pattern similar to either single knockout (OR = 1.52) (Fig. 4B and SI Appendix, Table S6). In addition, rearrangements in both Parp1−/− and Parp3−/−;Parp1−/− cells were less likely to have insertions than those in wild-type cells (ORs = 0.7 and 0.64, respectively) (SI Appendix, Table S6). In comparisons across genotypes for proximal repair, the ORs were <1.25 or >0.75, indicating that the differences between distributions of repair events during proximal repair of the Alk locus were less dramatic than in Eml4–Alk rearrangements (Fig. 4B). In the analysis of the dataset without single A insertions, rearrangements in Parp3−/−;Parp1−/− cells remained less likely to have insertions than wild-type cells (OR = 0.47) (SI Appendix, Fig. S5B and Table S7). However, in this dataset, the Parp3- and Parp1-dependent effects were extinguished (SI Appendix, Fig. S5B and Table S7), suggesting that they were CRISPR/Cas9-mediated effects. Taken together, these results indicate that the combined loss of Parp3 and Parp1 decreases insertions during rearrangement formation.
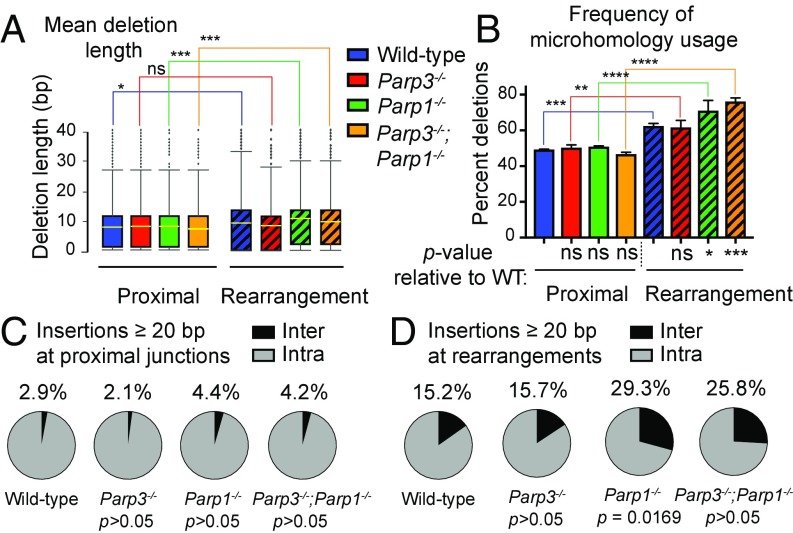
Next, we examined mean lengths of deletions, microhomologies, and insertions (SI Appendix, Table S8) in the residual rearrangement junctions. In wild-type cells, we observed that the mean deletion length was significantly longer in rearrangement junctions than in proximal junctions (Fig. 5A and SI Appendix, Table S8), consistent with the finding that rearrangements in murine cells involve more extensive processing of DNA breaks (2, 29). We observed the same pattern in Parp1−/− and Parp3−/−;Parp1−/− cells (Fig. 5A and SI Appendix, Table S8). In contrast, in Parp3−/− cells, the mean deletion lengths for rearrangements and proximal repair were nearly the same (Fig. 5A and SI Appendix, Table S8).
Fig. 5.
Repair phenotypes in murine tail fibroblasts. (A) Distribution of deletion lengths in proximal repair and at rearrangements. Boxes show the quartile range; whiskers show minimum to 99th percentiles; diamonds are outliers, yellow lines are the mean deletion length. (B) Percent of deletions that exhibit microhomology. P values below the x axis compare each genotype to the wild-type value within proximal or rearrangements. (C and D) Proportion of inter- and intrachromosomal templated insertions ≥20 bp in length in proximal repair (C) and at rearrangements (D). *P < 0.05, **P < 0.01, ***P < 0.001; two-way ANOVA with Tukey’s correction (A and B) or two-sided χ2 (C and D). ns, not significant.
Based on our previous report (8), we hypothesized that Parp3 promotes the processing of DNA DSBs during rearrangement in murine cells, which results in repair that involves deletions of the end sequence. As expected, rearrangements in Parp3−/− cells had a shorter mean deletion length than those in wild-type cells. In contrast, rearrangements in Parp1−/− cells had longer mean deletion length than rearrangements in wild-type cells. These data suggest opposing trends, although neither difference achieved statistical significance (P = 0.1565 and P = 0.0525, respectively) (Fig. 5A and SI Appendix, Table S8). In support of these trends, rearrangements in Parp1−/− cells had significantly longer deletions than those in Parp3−/− cells (P = 0.0001) (Fig. 5A). In addition, rearrangements in Parp3−/−;Parp1−/− cells had significantly shorter deletions than those in Parp1−/− cells (P = 0.0202) (Fig. 5A). Taken together, these data suggest that during rearrangement formation, Parp3 promotes and Parp1 suppresses longer deletions in murine fibroblasts. Within the proximal repair junctions, we did not find any statistically significant differences in mean deletion lengths between genotypes (Fig. 5A and SI Appendix, Table S8).
During DSB repair, deletions can uncover microhomologies that may be preferentially utilized to facilitate repair by alt-NHEJ (20). Reports in the literature indicate that rearrangements in murine cells occur predominantly by alt-NHEJ (2–6). A comparison of the flanking sequences indicated comparable opportunities for microhomology usage during proximal repair or during Eml4–Alk rearrangement (SI Appendix, Fig. S5C). However, rearrangements exhibited significantly increased frequencies of microhomology usage compared with proximal repair in wild-type, Parp3−/−, Parp1−/−, and Parp3−/−;Parp1−/− cells (Fig. 5B). The mean length of microhomology was also significantly increased at rearrangements compared with proximal repair in all four genotypes (SI Appendix, Fig. S5D).
Among rearrangements, there were no significant Parp3-dependent differences in either the proportion of repair events with microhomology or the mean length of microhomology (Fig. 5B and SI Appendix, Fig. S5D), consistent with our findings in pCr15 cells and Parp3−/− B cells (SI Appendix, Figs. S1D and S3E). In contrast, Parp1−/− and Parp3−/−;Parp1−/− cells had significantly increased frequencies of microhomology usage at rearrangements compared with wild-type cells (Fig. 5B). In addition, rearrangements in Parp3−/−;Parp1−/− cells exhibited significantly longer mean microhomology usage than rearrangements in wild-type cells (SI Appendix, Fig. S5D). No significant differences in microhomology were observed in proximal repair (Fig. 5B and SI Appendix, Fig. S5D).
To evaluate mean insertion lengths during DSB repair, we included both simple insertions and insertions occurring with a deletion (complex events). Loss of Parp1 led to a significant increase in mean insertion length (SI Appendix, Fig. S5E and Table S8) in both proximal repair and rearrangements. Mean insertion lengths were also increased in double-knockout cells, although the increase in proximal repair did not achieve statistical significance (SI Appendix, Fig. S5E and Table S8). No additional Parp3-dependent effect was observed, indicating that the increases in insertion length depend on Parp1. Because CRISPR/Cas9 can lead to a predominance of single-nucleotide insertions during repair (27, 28), we also evaluated insertion lengths without these events. When we excluded single A insertions, the overall mean insertion lengths were increased, but the Parp1-dependent increase in insertion length remained statistically significant only in proximal repair. (SI Appendix, Fig. S5F).
Given that each insertion is a unique event, we reasoned that some insertions in our dataset would be represented by fewer than 10 reads. Therefore, to understand the origins of inserted sequences in our system, we combined the three biological replicates for each condition and considered all insertions (both simple insertions and insertions in complex junctions) represented by ≥1 read. We aligned every insertion of ≥20 bp with the genomic sequence and normalized its representation by the number of reads for that sequence. None of the sequences that we interrogated showed significant alignment to the adenoviral genome (30) or Cas9 sequence. Overall, ∼99.8% of insertions >20 bp aligned with the mouse genome. Upon manual curation of the remaining 0.2%, we observed single insertions containing a sequence from multiple genomic loci; we excluded these events from further analysis.
We divided all aligned insertions from both rearrangement and proximal junctions into sequences that originated from chromosome 17 or nonchromosome-17 locations. We noted that insertions from nonchromosome-17 locations (i.e., interchromosomal insertions) were distributed throughout the genome without any appreciable chromosomal bias (SI Appendix, Fig. S5 G and H). For all genotypes, the fraction of insertions that involved a nonchromosome-17 sequence was higher in rearrangements than at proximal junctions (Fig. 5 C and D). Both Parp1−/− and Parp1−/−;Parp3−/− cells had approximately twofold increases in the proportion of nonchromosome-17 sequence insertions compared with wild-type cells (P = 0.0169 for rearrangements) (Fig. 5 C and D), suggesting that Parp1 uniquely suppresses interchromosomal insertions. In conclusion, our data indicate that while loss of either Parp3 or Parp1 leads to a reduction in chromosomal rearrangements, rearrangement junctions in Parp3−/− and Parp1−/− cells exhibit different phenotypes (SI Appendix, Fig. S6).
Discussion
In this study, we confirm that Parp3 promotes chromosomal rearrangements in murine cells including I-SceI–mediated translocations in mESCs, class-switch recombination in primary B cells, and inversions of the Eml4–Alk locus in tail fibroblasts (Figs. 1–3). To date, most factors known to mediate chromosomal rearrangements have species-specific roles (2–5, 7); Parp3 is an exception to this paradigm. The observation that Parp3 promotes rearrangements in both murine and human cells suggests that Parp3 either acts in both pathways or performs a function upstream of both pathways. Supporting the latter, we previously reported that PARP3 suppresses G4 DNA to facilitate DSB repair in human cells (8). In murine cells, the Alk locus is predicted to contain abundant G4 DNA structures (SI Appendix, Fig. S3J), raising the possibility that Parp3 could act upstream of both NHEJ pathways by suppressing G4 DNA in mouse cells as well.
Ig switch regions form abundant G4 structures that may be involved in recruitment and oligomerization of AID (31). In our Parp3 knockout, we do not observe the Parp3-dependent suppression of AID recruitment found by Robert et al. (13). We noted that while our Parp3−/− cells (both the germ-line–knockout mouse and the cells used for Rag-deficient blastocyst complementation) are in the 129 background (32), Robert et al. (13) established their Parp3−/− mice in a B6;129 mixed background. As differences have been noted in antibody class switching between strains of inbred mice (33), this could explain the observed differences.
One interesting question in our data arises from the observation that both Parp3−/− and Parp1−/− cells exhibit reduced deposition of RPA at DSBs (Fig. 3 and SI Appendix, Fig. S4B), suggesting that both enzymes promote processing of DSBs to ssDNA. Parp3−/− cells had shorter deletions at Eml4–Alk rearrangements, which is consistent with reduced RPA deposition. However, Parp1−/− cells unexpectedly had longer deletions (Fig. 5A and SI Appendix, Table S8), leading us to ask how reduced RPA deposition can be reconciled with increased deletion length. One possibility is that RPA is an imperfect measure of ssDNA. Reduced RPA deposition could also result from defective RPA loading onto ssDNA without changes in resection. A second possibility stems from the observation that Parp1−/− cells have defects in single-strand break repair (SSBR) (34). Faulty SSBR in Parp1−/− cells could result in the longer deletions observed at rearrangements if single-strand breaks near the targeted DSB lead to the loss of additional sequences from the ends.
Parp3 can interact with and PARylate the KU70/KU80 heterodimer (35, 36). A previous report found that PARP3 depletion impaired the recruitment of ectopically expressed KU80 to sites of laser-induced DNA damage (35), and our data show that the Parp3-dependent effects on rearrangement frequency are Ku-independent (Fig. 1B). The ChIP results in Fig. 3D suggest that one possible role for the Parp3 interaction may be to promote the removal of the Ku heterodimer from the DSBs as the repair progresses. Alternatively, KU70/KU80 has been found to interact with G4 DNA structures (37, 38), and we find that G4 DNA accumulates in the absence of Parp3 (8).
Analysis of amplicon deep sequencing to profile repair outcomes is a potentially powerful technique with several important caveats. While the goal of this analysis is to evaluate the frequency of individual repair events, both cellular proliferation and PCR may confound the results. Cellular proliferation is an inherent challenge in cell-based assays owing to the time required to achieve adequate CRISPR/Cas9 expression. One possible strategy to address this issue has been to rapidly induce nuclear localization of an endonuclease using a small molecule such as estrogen (39). In our hands, however, this strategy has been leaky, resulting in constitutive, low-level cutting that has so far outweighed its benefit. To address the challenge of possible bias introduced by PCR amplification, we created a library of synthetic repair events, mixed them in defined ratios, and repeated our amplicon deep-sequencing protocol. We observed a strong correlation between the input and the number of reads for each synthetic repair event (SI Appendix, Fig. S4E and Table S2). These results suggest that our PCR-based amplification led to minimal bias, so that the readcounts closely reflected the representation in the original population.
In conclusion, we find that Parp3 promotes rearrangements in a locus- and species-independent manner. We harnessed the power of high-throughput analysis of deeply sequenced amplicons to find that Parp3-deficient cells have decreased deletion lengths at rearrangement junctions. Although the closely related enzyme Parp1 also promotes rearrangement formation, we uncovered phenotypic differences at residual junctions that support differences in function at DSBs. Our results demonstrate the promising applications of this experimental technique to efficiently evaluate a large number of variables for DNA-repair outcomes.
Materials and Methods
Five days after infection with CRISPR/Cas9 adenovirus, genomic DNA (gDNA) was collected from each population using the Qiagen QIAamp kit. For defined mixtures of spike-in control repair products, 20-bp barcodes (SI Appendix, Table S2) flanked on each side by 95 and 145 bp of the Alk locus sequence were obtained as gBlocks Gene Fragments (Integrated DNA Technologies). Eight PCR reactions were made, each containing 5, 0.5, 0.05, 0.005, and 0.0005 fg of spikes 1, 2, 3, 4, and 5, respectively, and 100 ng of 293T gDNA. PCR in the linear range was performed using Bioneer AccuPrime with 100 ng of gDNA template or a spike-in control mixture for each reaction. PCR conditions were 3 min at 95 °C followed by cycles of 15 s at 95 °C, 15 s at 55 °C, and 15 s at 72 °C. Twenty-seven cycles and 33 cycles were performed for the proximal repair at the Alk locus and the Eml4–Alk rearrangement, respectively. Eight PCR products were pooled for each condition and were purified using the Qiagen PCR purification kit. Purified products were visualized on an agarose gel to ensure the presence of a single band and were sequenced (SI Appendix, Supporting Information Materials and Methods).
Supplementary Material
Acknowledgments
We thank members of the Price and D.M.W. laboratories for thoughtful comments. This work was supported by American Cancer Society and Alex Lemonade Stand fellowships (both to T.A.D.), the Claudia Adams Barr Fund for Basic Cancer Research (T.A.D. and D.M.W.), a National Science Foundation Graduate Research Fellowship under Grant DGE1144152 (to J.V.L.), and NIH/National Cancer Institute Grants R01 CA151898 and R01 CA172387 (both to D.M.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1801591115/-/DCSupplemental.
References
- 1.Kong F, et al. dbCRID: A database of chromosomal rearrangements in human diseases. Nucleic Acids Res. 2011;39:D895–D900. doi: 10.1093/nar/gkq1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weinstock DM, Brunet E, Jasin M. Formation of NHEJ-derived reciprocal chromosomal translocations does not require Ku70. Nat Cell Biol. 2007;9:978–981. doi: 10.1038/ncb1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simsek D, et al. DNA ligase III promotes alternative nonhomologous end-joining during chromosomal translocation formation. PLoS Genet. 2011;7:e1002080. doi: 10.1371/journal.pgen.1002080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simsek D, Jasin M. Alternative end-joining is suppressed by the canonical NHEJ component Xrcc4-ligase IV during chromosomal translocation formation. Nat Struct Mol Biol. 2010;17:410–416. doi: 10.1038/nsmb.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y, Jasin M. An essential role for CtIP in chromosomal translocation formation through an alternative end-joining pathway. Nat Struct Mol Biol. 2011;18:80–84. doi: 10.1038/nsmb.1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wray J, et al. PARP1 is required for chromosomal translocations. Blood. 2013;121:4359–4365. doi: 10.1182/blood-2012-10-460527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghezraoui H, et al. Chromosomal translocations in human cells are generated by canonical nonhomologous end-joining. Mol Cell. 2014;55:829–842. doi: 10.1016/j.molcel.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Day TA, et al. PARP3 is a promoter of chromosomal rearrangements and limits G4 DNA. Nat Commun. 2017;8:15110. doi: 10.1038/ncomms15110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boehler C, Dantzer F. PARP-3, a DNA-dependent PARP with emerging roles in double-strand break repair and mitotic progression. Cell Cycle. 2011;10:1023–1024. doi: 10.4161/cc.10.7.15169. [DOI] [PubMed] [Google Scholar]
- 10.Casellas R, et al. Mutations, kataegis and translocations in B cells: Understanding AID promiscuous activity. Nat Rev Immunol. 2016;16:164–176. doi: 10.1038/nri.2016.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ward IM, et al. 53BP1 is required for class switch recombination. J Cell Biol. 2004;165:459–464. doi: 10.1083/jcb.200403021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jankovic M, et al. 53BP1 alters the landscape of DNA rearrangements and suppresses AID-induced B cell lymphoma. Mol Cell. 2013;49:623–631. doi: 10.1016/j.molcel.2012.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robert I, et al. Parp3 negatively regulates immunoglobulin class switch recombination. PLoS Genet. 2015;11:e1005240. doi: 10.1371/journal.pgen.1005240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen J, Lansford R, Stewart V, Young F, Alt FW. RAG-2-deficient blastocyst complementation: An assay of gene function in lymphocyte development. Proc Natl Acad Sci USA. 1993;90:4528–4532. doi: 10.1073/pnas.90.10.4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakamura M, et al. High frequency class switching of an IgM+ B lymphoma clone CH12F3 to IgA+ cells. Int Immunol. 1996;8:193–201. doi: 10.1093/intimm/8.2.193. [DOI] [PubMed] [Google Scholar]
- 16.Boeva V, et al. Breakpoint features of genomic rearrangements in neuroblastoma with unbalanced translocations and chromothripsis. PLoS One. 2013;8:e72182. doi: 10.1371/journal.pone.0072182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, et al. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell. 2012;148:908–921. doi: 10.1016/j.cell.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blasco RB, et al. Simple and rapid in vivo generation of chromosomal rearrangements using CRISPR/Cas9 technology. Cell Rep. 2014;9:1219–1227. doi: 10.1016/j.celrep.2014.10.051. [DOI] [PubMed] [Google Scholar]
- 19.Maddalo D, et al. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature. 2014;516:423–427. doi: 10.1038/nature13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang HHY, Pannunzio NR, Adachi N, Lieber MR. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat Rev Mol Cell Biol. 2017;18:495–506. doi: 10.1038/nrm.2017.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wyatt DW, et al. Essential roles for polymerase θ-mediated end joining in the repair of chromosome breaks. Mol Cell. 2016;63:662–673. doi: 10.1016/j.molcel.2016.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khodaverdian VY, et al. Secondary structure forming sequences drive SD-MMEJ repair of DNA double-strand breaks. Nucleic Acids Res. 2017;45:12848–12861. doi: 10.1093/nar/gkx1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soong CP, et al. Development of a novel method to create double-strand break repair fingerprints using next-generation sequencing. DNA Repair (Amst) 2015;26:44–53. doi: 10.1016/j.dnarep.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 24.Hanzlikova H, Gittens W, Krejcikova K, Zeng Z, Caldecott KW. Overlapping roles for PARP1 and PARP2 in the recruitment of endogenous XRCC1 and PNKP into oxidized chromatin. Nucleic Acids Res. 2017;45:2546–2557. doi: 10.1093/nar/gkw1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gibson BA, et al. Chemical genetic discovery of PARP targets reveals a role for PARP-1 in transcription elongation. Science. 2016;353:45–50. doi: 10.1126/science.aaf7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown AJ, et al. High-throughput analysis of DNA break-induced chromosome rearrangements by amplicon sequencing. Methods Enzymol. 2018;601:111–144. doi: 10.1016/bs.mie.2017.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Overbeek M, et al. DNA repair profiling reveals nonrandom outcomes at Cas9-mediated breaks. Mol Cell. 2016;63:633–646. doi: 10.1016/j.molcel.2016.06.037. [DOI] [PubMed] [Google Scholar]
- 28.Lemos BR, et al. CRISPR/Cas9 cleavages in budding yeast reveal templated insertions and strand-specific insertion/deletion profiles. Proc Natl Acad Sci USA. 2018;115:E2040–E2047. doi: 10.1073/pnas.1716855115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robbiani DF, et al. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell. 2008;135:1028–1038. doi: 10.1016/j.cell.2008.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dehghan S, et al. Complete genome sequence of human adenovirus prototype 17. J Virol. 2011;85:11540–11541. doi: 10.1128/JVI.06051-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qiao Q, et al. AID recognizes structured DNA for class switch recombination. Mol Cell. 2017;67:361–373.e4. doi: 10.1016/j.molcel.2017.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deng C, Wynshaw-Boris A, Zhou F, Kuo A, Leder P. Fibroblast growth factor receptor 3 is a negative regulator of bone growth. Cell. 1996;84:911–921. doi: 10.1016/s0092-8674(00)81069-7. [DOI] [PubMed] [Google Scholar]
- 33.Kaminski DA, Stavnezer J. Antibody class switching differs among SJL, C57BL/6 and 129 mice. Int Immunol. 2007;19:545–556. doi: 10.1093/intimm/dxm020. [DOI] [PubMed] [Google Scholar]
- 34.Ray Chaudhuri A, Nussenzweig A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat Rev Mol Cell Biol. 2017;18:610–621. doi: 10.1038/nrm.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beck C, et al. PARP3 affects the relative contribution of homologous recombination and nonhomologous end-joining pathways. Nucleic Acids Res. 2014;42:5616–5632. doi: 10.1093/nar/gku174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rouleau M, et al. PARP-3 associates with polycomb group bodies and with components of the DNA damage repair machinery. J Cell Biochem. 2007;100:385–401. doi: 10.1002/jcb.21051. [DOI] [PubMed] [Google Scholar]
- 37.Zhang T, Zhang H, Wang Y, McGown LB. Capture and identification of proteins that bind to a GGA-rich sequence from the ERBB2 gene promoter region. Anal Bioanal Chem. 2012;404:1867–1876. doi: 10.1007/s00216-012-6322-y. [DOI] [PubMed] [Google Scholar]
- 38.Xodo L, Paramasivam M, Membrino A, Cogoi S. Protein hnRNPA1 binds to a critical G-rich element of KRAS and unwinds G-quadruplex structures: Implications in transcription. Nucleic Acids Symp Ser (Oxf) 2008:159–160. doi: 10.1093/nass/nrn081. [DOI] [PubMed] [Google Scholar]
- 39.Aymard F, et al. Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat Struct Mol Biol. 2014;21:366–374. doi: 10.1038/nsmb.2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.