Significance
X-ray crystallography studies of soluble ectodomains of influenza hemagglutinins (HA) have previously revealed details of their two functions in virus infection: receptor binding and membrane fusion. We have now used cryo-EM to determine the structures of full-length hemagglutinin solubilized in detergent, alone, and in complex with the Fab of an infectivity neutralizing antibody. We observe the structure of the region that anchors HA in the virus membrane as a bundle of three α-helices that are joined to the ectodomain by flexible linkages. The Fab binds near the linkages and restricts their flexibility. This may indicate that the flexibility is required for HA-mediated membrane fusion and that interference with it represents a mechanism for neutralizing virus infectivity.
Keywords: influenza, hemagglutinin, cryo-EM, membrane protein, membrane fusion
Abstract
Viruses with membranes fuse them with cellular membranes, to transfer their genomes into cells at the beginning of infection. For Influenza virus, the membrane glycoprotein involved in fusion is the hemagglutinin (HA), the 3D structure of which is known from X-ray crystallographic studies. The soluble ectodomain fragments used in these studies lacked the “membrane anchor” portion of the molecule. Since this region has a role in membrane fusion, we have determined its structure by analyzing the intact, full-length molecule in a detergent micelle, using cryo-EM. We have also compared the structures of full-length HA−detergent micelles with full-length HA−Fab complex detergent micelles, to describe an infectivity-neutralizing monoclonal Fab that binds near the ectodomain membrane anchor junction. We determine a high-resolution HA structure which compares favorably in detail with the structure of the ectodomain seen by X-ray crystallography; we detect, clearly, all five carbohydrate side chains of HA; and we find that the ectodomain is joined to the membrane anchor by flexible, eight-residue-long, linkers. The linkers extend into the detergent micelle to join a central triple-helical structure that is a major component of the membrane anchor.
The hemagglutinin (HA) membrane glycoprotein of influenza virus is a trimer of identical subunits that are formed of two disulfide-linked polypeptides: membrane-distal HA1 and the smaller, membrane-proximal, HA2.
HA has two functions in virus infection: receptor binding and membrane fusion (1). Viruses are bound by HAs to cell-surface sialic acid receptors and are taken into cells by endocytosis (2). At low pH in endosomes, about pH 5.5, depending on the virus strain, fusion of virus and endosomal membranes is activated in a process that involves extensive changes in HA conformation (3, 4). Antibodies that block receptor binding and membrane fusion neutralize virus infectivity and provide the immune pressure that selects the antigenic variants responsible for recurrent outbreaks of influenza (5–10). Many strain-specific antibodies bind in or near the HA1, membrane-distal, receptor-binding site. By contrast, a number of cross-reactive anti-HA antibodies have been described that bind to the membrane proximal regions of HA and block membrane fusion (11). The human monoclonal antibody that we have studied here (FISW84) binds to H1 influenza viruses. Our analysis indicates that it binds near the junction between the ectodomain and the membrane anchor.
The 3D structures of the ectodomain of HA, the nature of its receptor-binding site, and structural details of the changes in its conformation required for membrane fusion have been determined by analysis of soluble fragments of the glycoprotein using X-ray crystallography (4, 12–20). Suitable fragments can be obtained by proteolytic removal of the membrane-associated, membrane anchor region of virus HA or by selective expression of the predicted ectodomain. A structural description of the intact molecule has not been reported, but a number of studies, including site-specific mutation and structural analyses (18, 21), have indicated that the membrane anchor region is required for HA-mediated membrane fusion, possibly in the final stages of the fusion process.
The images obtained by EM of negative-stained full-length HA indicate a longer but very similar structure to the soluble ectodomain of HA determined by X-ray crystallography (12, 22). To obtain information on its structure, we have examined full-length HA in detergent micelles, by cryo-EM.
Studies, by X-ray crystallography, cryo-EM, and different spectroscopic procedures, of single-span, membrane-anchored proteins from cellular membrane proteins and viruses, have indicated predominantly α-helical components of their membrane anchors. Examples of the small number of cellular membrane proteins that have been examined include synaptobrevin, one of the components of SNARE complexes involved in neurotransmitter exocytosis (23), and Glycophorin A, the major surface glycoprotein of erythrocytes (24). Most of the data on the helical components of membrane-associated regions of virus proteins in the prefusion conformation come from cryo-EM studies of virus particles. For example, the membrane-associated regions of the glycoproteins of the alphaviruses VEEV, Chikungunya, and Sindbis (25–27) have been observed as well as those of the glycoproteins of the flaviviruses, Dengue and Zika (28, 29), and the paramyxovirus F glycoprotein (30–32)
Images of Foamy Virus, a retrovirus, indicate a membrane-associated region that contains six helices (33). HIV glycoprotein preparations containing the membrane-associated region have also been reported by cryo-EM (34), and NMR analyses of peptide analogs of the HIV gp41 transmembrane region indicate either a continuous trimeric α-helical component or a continuous monomeric α-helix (35–37). For the Ebola virus glycoprotein, also, NMR and EPR spectroscopy data suggest a membrane-associated monomeric helix (38).
Whether any of these structures is equivalent to the prefusion or the postfusion forms of the transmembrane regions is unclear.
In our studies of full-length HA, we find that the trimeric ectodomain is linked to the membrane anchor by short, flexible chains that extend into the detergent micelle to join a bundle of three α-helices. We describe linker flexibility in relation to observed variation in micelle orientation relative to the ectodomain. We consider its potential significance in the process of membrane fusion, in B cell recognition of membrane-proximal, cross-reactive antibody binding sites, and in mechanisms of infectivity neutralization by the H1-subtype-specific, human monoclonal antibody, FISW84, that binds near the ectodomain−membrane anchor junction.
Results and Discussion
Overall Structure of the HA and HA−FISW84 Fab Complex.
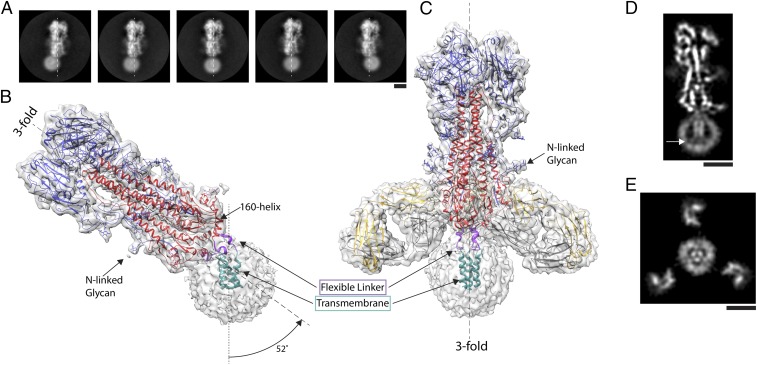
We used cryo-EM to visualize HA from A/duck/Alberta/35/76(H1N1) virus, solubilized in detergent and purified by affinity and size exclusion chromatography. The 2D class averages were generated for HA alone (Fig. 1A and SI Appendix, Fig. S1A) and also in complex with the Fab of monoclonal antibody FISW84 (SI Appendix, Fig. S1B). Both samples reveal a well-defined HA ectodomain flexibly linked to a detergent micelle containing the transmembrane domain of the HA (Fig. 1 B and C). Data analysis, using about 300,000 particles, yielded high-resolution structures of the ectodomain of the full-length HA which are very similar to an H1 HA structure determined by X-ray crystallography [Protein Data Bank (PDB) ID code: 1RUY; RMSD: 1.2 Å]. This observation confirms the conclusion from CD spectroscopy (39) that, once the HA trimer has assembled, the membrane anchor does not influence the structure of the ectodomain. The ectodomains have a global resolution of 3.3 Å, with better local resolution in core regions (SI Appendix, Fig. S2). There is well-defined density for amino acid side chains in most of the structure and also in the variable domain of the Fab, as well as for the five predicted Asn-linked carbohydrate side chains, extending up to 15 Å from the HA surface (Fig. 1 B and C and SI Appendix, Fig. S3).
Fig. 1.
The structure of full-length hemagglutinin from A/duck/Alberta/35/76 H1N1 determined by cryo-EM. (A) Image classes of similarly oriented HAs illustrating the flexibility of the detergent micelle containing the transmembrane domain. The threefold axis of symmetry of the ectodomain trimer is marked in each frame by a dashed line. (B and C) Two structures of HA with density suitable for model building of the transmembrane domain: (B) the transmembrane domain in a detergent micelle exhibits a tilt of 52° from the threefold axis of the ectodomain, and (C) a complex of HA with the Fab of the human monoclonal antibody FISW84 where classification led to identification of a subset of molecules in which the transmembrane region follows the threefold symmetry axis of the ectodomain. In both B and C, examples of well-defined density for N-linked glycans are indicated. Both structures indicate a linker region (purple) between the base of the ectodomain (HA1 shown in blue, HA2 shown in red), defined by the 160 helix, and the transmembrane region (cyan) formed of a bundle of three α-helices. (D and E) Slices of map of HA−Fab complex shown in C. At the C termini of the transmembrane α-helices, they begin to splay apart, and extend toward the surface of the micelle, indicated by arrow in D. Also, in E, the micelle is seen to adopt the trimeric shape of the α-helical bundle, with Fab fragments visible in the periphery. (Scale bars: 5 nm.)
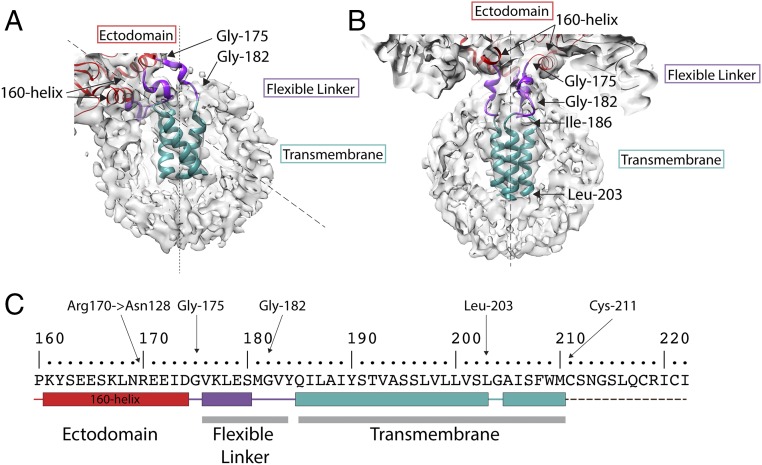
The HA ectodomain ends in the 160 helix (162 to 175) (Figs. 1B and 2 A and B). Following a conserved glycine (175), there are five residues of α-helix (176 to 180) and four residues of extended chain (181 to 184), that, together, constitute the flexible linker (Fig. 2). This connects the ectodomain to a trimeric helical bundle (186 to 203) which represents the major component of the membrane anchor. In different structures calculated for HA, the membrane anchor detergent micelles are positioned at angles from 0° up to 25° (n = 10) off the threefold axis of the ectodomain, and, in one structure, the micelle is at 52° (Figs. 1B and 2A and SI Appendix, Fig. S4). In the case of the HA−Fab complex, the range is, in most cases (n = 12), restricted to less than 20°, with one structure at 25° with respect to the ectodomain threefold axis. These different orientations are accommodated by rearrangements within and between the three flexible linkers and transmembrane domains, while the ectodomain remains unaltered.
Fig. 2.
The structure of the membrane-associated region. Detailed views of (A) tilted and (B) straight micelles, as shown in Fig. 1 B and C respectively. The flexible linker region (purple) runs between Gly-175, at the C terminus of the 160 helix, and Gly-182 and extends to the N termini of the α-helices of a trimeric α-helical bundle, residues 186 to 203 (cyan). (C) The amino acid sequence of the transmembrane domain. The sequence shown begins at the 160 helix to the C terminus of HA2. Color-coded block diagrams indicate the positions of these structural elements in the sequence.
In the full-length HA−Fab complex, the FISW84 Fab binds at the junction of the ectodomain and the membrane-associated region. This antibody neutralizes a range of H1N1 viruses but not other subtypes (SI Appendix, Fig. S5 A and C). The interactions of this antibody with HA mainly involve the HCDR2 and LCDR3 loops interacting with the membrane-proximal five-stranded β-sheet of the ectodomain (Fig. 1C). The Fab links across three of the strands of the β-sheet and likely restricts the conformational changes in HA at low pH that are required for membrane fusion as indicated by virus-mediated hemolysis assays (SI Appendix, Fig. S5B).
The Transmembrane Domain.
To visualize the transmembrane and linker structures, we performed a 3D classification using images from which most of the ectodomain was computationally subtracted (SI Appendix, Fig. S6). This procedure enabled us to resolve a structure of the membrane proximal parts of the HA at sufficient resolution for de novo model building. In the case of some particles of the full-length HA−Fab complex, the linker and transmembrane domain obey the same threefold symmetry as the ectodomain (Figs. 1C and 2B). We obtained a structure with a similar orientation for the transmembrane domain, but at somewhat lower resolution, from particles of HA alone.
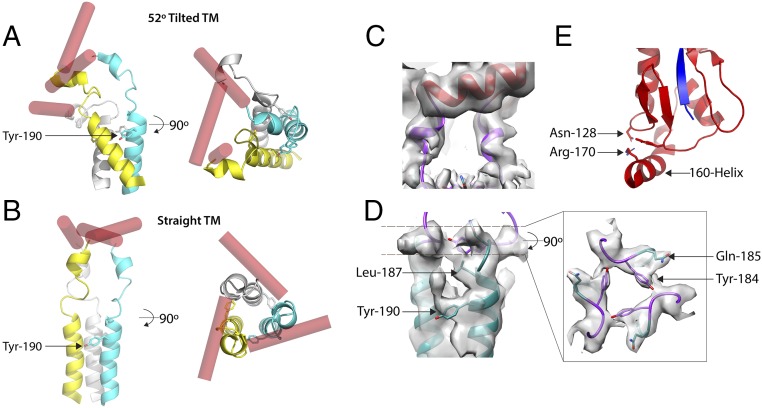
Examination of the structure indicates that the 160 helix (162 to 175), at the base of the ectodomain, adopts an almost horizontal position (as viewed in Fig. 2B), with its helical axis lying in a plane orthogonal to the molecular threefold axis. In addition to packing interactions, the position of this helix is stabilized by potential hydrogen bonds between conserved Arg-170, at the C terminus of the helix, and Asn-128, thus providing a stable attachment point for the proceeding flexible linker (Fig. 3).
Fig. 3.
A comparison of the overall structures of tilted and straight transmembrane domains. Schematic diagrams of (A) 52° tilted and (B) straight transmembrane domains to indicate that the linker regions undergo extensive rearrangements in the tilted domain by comparison with the straight, and that each linker adopts a different structure. The individual subunits of the trimer are colored differently as blue, yellow, and silver, while the 160 helices are in red. The α-helices of the α-helical bundle in the tilted domain are also extensively rearranged. (C and D) Details of the map (gray) used to build the structure of the linker region of the straight transmembrane domain. C shows the short α-helix of the linker region (purple) residues 176 to 180. (D) The N-terminal region of the bundled α-helices (cyan) and its junction with the linker region (purple) and conserved residues Gln-185 and Tyr-184 at the linker−α-helix junction. (E) The stabilizing interaction of the 160-helix between Asn-128 and Arg-170.
Two conserved glycine residues enable the polypeptide chain to change direction; firstly at residue 175, immediately after the 160 helix, where the chain turns toward the micelle and, secondly, at residue 182, where the chain turns again at the surface of the micelle (Figs. 2 A and B and 3C) to form a four-residue extension. Located at the C terminus of the linkage are conserved Tyr-184 and Gln-185, for both of which there is side chain density (Fig. 3D). At this point, the linker connects to the α-helices of a triple-stranded α-helical bundle beginning with conserved Ile-186 and Leu-187 and extending to Leu-203. There is a prominent linkage between the chains of the trimeric helix, at Tyr-190, the hydroxyl of which forms potential interactions with the main chain, or side chains of Ser-191 or Thr-192 of the neighboring chain. From Gly-204, the chains splay apart and become less ordered (Fig. 1D). The dimensions of the remaining density could accommodate α-helices extending to the first sites of palmitoylation, Cys-211, which we assume is the C terminus of the transmembrane region. Additional density on the surface of the micelle may be the location of the 11-residue cytoplasmic region, but this is not interpretable further. Also, viewed down the threefold axis of symmetry, it is evident that the three helices of the membrane anchor impose a triangular shape on the detergent micelle (Fig. 1E).
Membrane Anchor Orientation.
In addition to the subclass of membrane-associated regions from the HA−Fab complex where the ectodomain threefold axis aligns with the transmembrane region, we observed a number of different subclasses where the micelle axis was rotated by varying amounts from the ectodomain axis (Fig. 1A and SI Appendix, Fig. S4). In contrast to the HA−Fab complex, where the angle of tilt extends up to 20°, for HA alone, the tilt angle extends, in several cases, to 25° and, in one case, up to 52°. The ability to distinguish, and average, these subclasses demonstrates that there are a number of discrete, preferred orientations for the membrane anchor-bound micelles. The structure at 52° may be stabilized by interactions between the linker and the 160 helix that are possible at this extreme angle.
The changes in structure required to achieve these different orientations seem to be afforded by the unique properties of glycine residues located just after the 160 helix (Gly-175) and after the five-residue linker helix (Gly-182) (Fig. 2 A and B). Both of these residues, and also Gly-204, are conserved and, as glycine, can adopt dihedral angles not permitted for other residues.
In addition to variations in chain orientation in the linker region, the helices of the helical bundle in the transmembrane region, while retaining their individual secondary structures, rotate relative to each other in the tilted HAs (Figs. 2 A and B and 3 A and B). Nevertheless, density links between transmembrane helices consistent with interactions involving Tyr-190 suggest that this conserved residue is involved in maintaining the integrity of the helical bundle at different tilt angles. The observations of independent movement of the α-helices of the HA membrane anchor are similar to the sorts of changes seen in other membrane proteins such as channels (40) or in functions such as signal transduction across cellular membranes (41). By contrast with these flexibilities, the ectodomain retains the same well-defined structure irrespective of the orientation of the transmembrane domain.
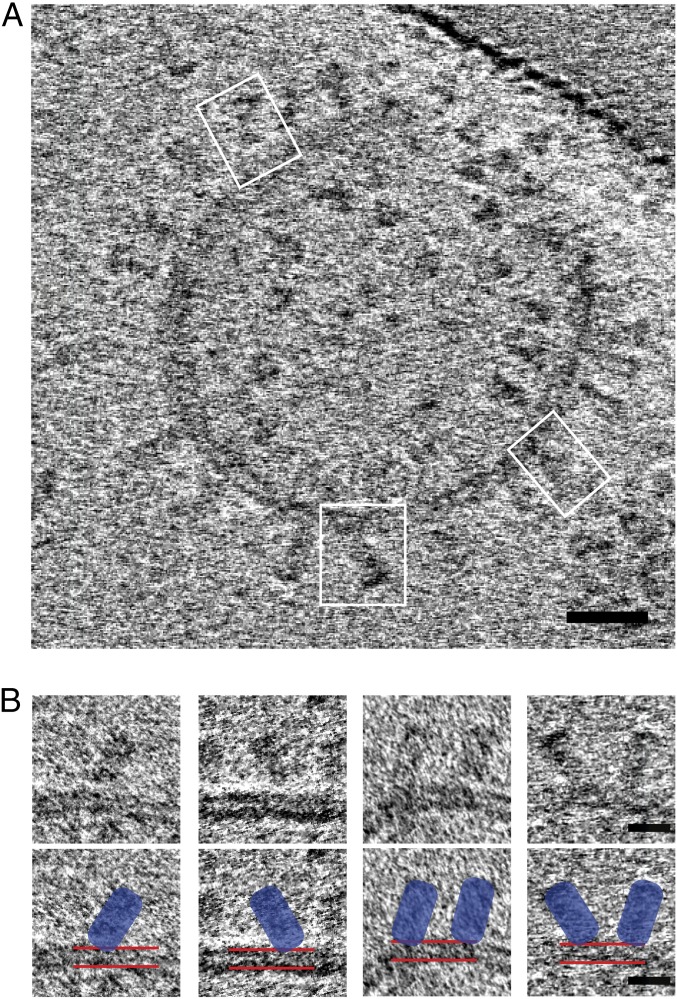
The lipid composition of virus membranes is determined by the composition of the cell membrane at which the viruses are assembled and released by budding. For influenza viruses, the membranes appear to be enriched in cholesterol and sphingolipid (42). The LMNG/MNA-C12 detergent micelle used here, although different from the native environment for the transmembrane region of HA, was selected for its ability to preserve the structure of membrane proteins (43, 44). We have also shown that HAs reconstituted in liposomes with cell membrane lipid composition and bilayer thickness similar to the thickness of the micelle are able to assume a range of orientations with respect to the membrane similar to what we have seen in micelles (Fig. 4).
Fig. 4.
HA reconstituted in liposomes. (A) Cryotomogram section showing cross-section of a liposome with examples of HAs tilted with respect to the lipid bilayer (white boxes). (Scale bar: 20 nm.) (B) Gallery of subtomograms of tilted HA in liposomes. Images in the second row are identical to those above but indicate HAs (blue) and liposome bilayer (red lines). (Scale bar: 10 nm.)
Flexibility for Membrane Fusion.
The flexibility of the linkage between the membrane-associated region and the ectodomain may facilitate the pH-dependent changes in HA conformation required for membrane fusion that reposition the activated HA ectodomain in the narrowing space between virus and cellular membranes as they are brought into contact for fusion (45). Consistent with this notion we have observed by cryo-EM, influenza viruses displaying an HA fusion intermediate with ectodomain components oriented nearly parallel to the membranes of fusing liposomes (46).
In this study, FISW84 antibody binding is seen to limit the range of orientations of the ectodomain with respect to the transmembrane domain and may neutralize virus infectivity by sterically restricting changes in orientation required for fusion. This would represent a previously unrecognized mechanism of infectivity neutralization. The membrane-proximal binding site of FISW84 on HA is topologically similar to those of cross-reactive antibodies that recognize the Membrane Proximal External Region of gp41 of the HIV membrane fusion glycoprotein (47), and it may be that the ability of such antibodies to neutralize virus infectivity involves similar principles. It is also possible that flexibility at the junction of the ectodomain and the transmembrane domain facilitates antibody recognition of HA expressed at the surfaces of infected cells.
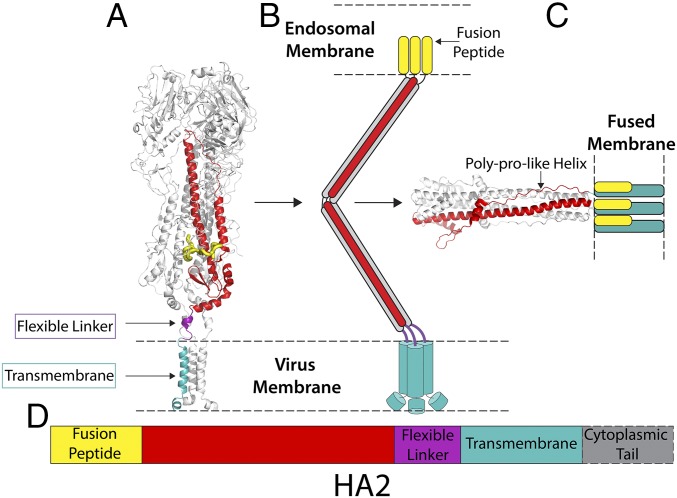
The crystal structure of HA2 in the fusion pH conformation indicates that the N-terminal and C-terminal regions are extensively refolded compared with the neutral pH structure reported here (Fig. 5). The C-terminal chain, preceding the linker and membrane anchor, adopts an extended polyproline type II helix that is packed into the outward-facing grooves between the α-helices of the central trimeric coiled coil (18) (Fig. 5). In this low-pH conformation, residues near the N and C termini of HA2 interact to form an N cap that would position the N-terminal fusion peptide and C-terminal membrane anchor regions closely together, although these regions are absent from the structure (Fig. 5). In contrast to the parainfluenza virus fusion glycoprotein, for example, the N-cap structure of low-pH HA indicates that it is unlikely there are continuous α-helices at either the N or C terminus of HA2, consistent with our observation of a flexible linkage to the transmembrane anchor. The flexibility of the linkage region may also be important to facilitate the substantial refolding event at low pH in which the C-terminal polyproline region assumes its eventual position on the exterior of the coiled coil (Fig. 5). An understanding of this final part of the process will require determination of the structure of HA2, containing both the fusion peptide and the membrane anchor, in the fusion pH conformation.
Fig. 5.
Structural snapshots toward membrane fusion. (A) The structure of straight full-length HA from cryo-EM (this study). (B) Schematic representation of possible fusion pH intermediate (46). (C) The structure of fusion pH HA2 (18), with modeled membrane anchor and fusion peptide colocalized in a fused membrane. (D) A color-coded diagram showing the location of the different regions of HA2 in its primary structure.
Materials and Methods
Protein Preparation.
Full-length HA protein was isolated from purified virus. The virus, A/duck/Alberta/35/76(H1N1), was inoculated in 11- to 12-d-old embryonated hens’ eggs and incubated for 48 h. The virus was purified from the allantoic fluid by sucrose gradient centrifugation. To isolate HA alone, virus glycoproteins were extracted from the viral membrane with 1% Lauryldimethylamine N-oxide (LDAO) (all detergents obtained from Anatrace) and purified by anion exchange, and exchanging the detergent to 0.01% Lauryl Maltose Neopentyl Glycol (LMNG). Neuraminidase was removed by affinity chromatography using an oseltamivir−biotin conjugate (48) immobilized on a streptavidin Sepharose column. The HA was further purified by gel filtration using a Superose 6 Increase 10/300 column in the buffer: 25 mM Tris pH 8, 150 mM NaCl, 1 mM TCEP, 0.01% LMNG. For the HA−Fab complex, HA was purified by extraction with LDAO, transferring the HA to 0.05% Dodecyl Maltoside (DDM) during the anion exchange step. The HA was purified by the same steps as above, but, in DDM and as a final step, the protein was incubated with a molar excess of the FISW84 Fab fragment and 0.1% of the detergent MNA-C12. The complex was purified from excess detergent and free Fab by Superose 6 gel filtration in the buffer: 25 mM Tris pH 8, 150 mM NaCl, 0.002% MNA-C12.
Grid Preparation and Data Collection.
Grids of full-length HA were plunge-frozen using an Mk III Vitrobot (FEI) at a protein concentration of ∼2 mg/mL, supplemented with 0.1% octyl-β-glucoside (OG) to reduce orientational bias. Then 4 μL of sample was applied to glow-discharged 300-mesh copper R2/4 quantifoil grids and blotted for 5 s before plunging into liquid ethane. Grids of HA−Fab complex were frozen using an Mk IV Vitrobot with protein at a concentration of ∼1.5 mg/mL also supplemented with 0.1% OG. Then 4 uL of sample was applied to glow-discharged 200-mesh copper R2/2 quantifoil grids and blotted for 3 s before plunge-freezing. Both specimens were imaged using a Titan Krios electron microscope operating at 300 kV. Micrographs were recorded in counting mode using a Gatan K2 Summit detector mounted at the end of a Gatan GIF Quantum energy filter operating in zero-loss mode with a slit width of 20 eV. Exposures were 8 s, with a total dose of 43.7 e/Å2 fractionated into 20 frames with a calibrated pixel size of 1.08 Å. Images were recorded with a defocus of 1 µm to 4 µm.
Image Processing.
Movie frames were corrected for motion using MotionCor2 (49), and contrast transfer function was estimated using CTFfind4 (50). The full-length HA dataset consisted of 10,861 movies, of which 10,348 were used. The HA+FISW84 dataset consisted of 17,329 movies, of which 17,250 were used. Particles were picked from the micrographs using RELION template-based particle picking (51). This yielded 1,095,719 particles for the full HA dataset and 1,385,993 particles for the +Fab dataset. Particles were initially screened using two rounds of reference-free 2D classification followed by particle sorting. This reduced the particle numbers to 744,665 and 853,524. An initial model was generated from ∼20,000 randomly selected particles for both samples using the ab initio reconstruction program in CryoSPARC (52). This initial model was used for a first round of RELION 3D classification. This left 531,849 and 473,081 particles in the well-defined classes.
High-resolution ectodomain structures of HA and HA+Fab were generated by 3D classification. The detergent micelle and transmembrane density were removed by masking of the initial model; 365,887 and 291,146 particles were used for the final refinements of the HA ectodomain and HA ectodomain+Fab structures. All final refinements were carried out using the Homogeneous Refinement program of CryoSPARC using particles extracted in a 512 × 512 pixel box.
Structures which include the transmembrane were generated from the particle stacks from the initial round of 3D classification. The HA+Fab class with the straight transmembrane was generated by RELION refinement of all particles, imposing C3 symmetry followed by signal subtraction (53). Subtraction masks were designed to leave density for the detergent micelle, linker region, and a small amount of the base of the HA ectodomain. These subtracted particles were subjected to a number of rounds of 3D classification, imposing C3 symmetry using the particle orientations determined in the refinement. In the first two rounds of classification, classes were chosen which had well-defined density in the transmembrane region. The unsubtracted 83,024 particles were refined, imposing C3 symmetry yielding a map of 3.7-Å resolution and were used to build the linking regions (residues 175 to 184). Subclassification led to a smaller class with better-defined density in the transmembrane domain. This class consisted of 23,286 particles of which the unsubtracted particles refined to 4.1-Å resolution when imposing C3 symmetry.
Maps with transmembrane domains not aligned with the ectodomain were generated for both the HA and HA+Fab. Signal-subtracted particles were classified using the alignments from C1 RELION refinements of each dataset. Each dataset was classified into 20 classes. Particles from classes with well-defined transmembrane regions were refined, leading to 10 maps of HA and 12 maps of HA+FISW84. One class from the full-length HA dataset had a transmembrane domain with defined secondary structure with an angle of tilt of 52°. This class was made up of 41,043 particles and refined to a map with a global resolution of 4.2 Å.
Refined half-maps were sharpened automatically (54) and global resolution was corrected by high-resolution noise substitution (55) using the RELION postprocessing tool. Local resolution was estimated using blocres, implemented in CryoSPARC (56).
Angle Measurement.
Angles of transmembrane tilt were measured by aligning C1 maps by the HA ectodomain using chimera, reducing the location of the transmembrane to a 120° angular region about the ectodomain trimer. Rotation and tilt were measured relative to the vertical axis.
Model Building.
High-resolution models of the ectodomain and ectodomain+Fab were constructed using a previously determined structure of the closely related A/swine/Iowa/15/30(H1N1) (PDB ID code: 1RUY) (19) and a crystal structure of the FISW84 (PDB ID code: 6HKG). The sequence was mutated to that of A/duck/Alberta/35/76(H1N1). This model was manually adjusted in Coot (57) and real-space−refined using PHENIX (58), followed by refinement in REFMAC (59) with half-map validation. Ectodomain and ectodomain+Fab structures were used as the basis for building transmembrane domains. All sequence numbers described are in H3 numbering.
Models of tilted transmembrane regions were generated by rigid-body fitting the transmembrane domain of the straight transmembrane class and manually adjusting the position of the linker helices before a single round of real-space refinement in PHENIX to correct structure geometry.
Supplementary Material
Acknowledgments
We acknowledge Phil Walker and Andrew Purkiss of the Structural Biology Science Technology Platform for computational support and the Electron Microscopy science technology platform for the use of screening microscopes. This work was funded by the Francis Crick Institute which receives its core funding from Cancer Research UK (FC001078 and FC001143), the UK Medical Research Council (FC001078 and FC001143), and the Wellcome Trust (FC001078 and FC001143). U.N. was also funded by a Marie Curie Action Intra-European Fellowship (Grant 629829).
Footnotes
Conflict of interest statement: A.L. is the scientific founder of Humabs BioMed SA. A.L. holds shares in Humabs BioMed. D.C. is an employee of Humabs Biomed. N.L.K. was employed by MedImmune, LLC when work was executed and may currently hold AstraZeneca stock or stock options.
Data deposition: Maps and models have been deposited in the Electron Microscopy Data Bank, http://www.ebi.ac.uk/pdbe/emdb/ (accession nos. EMD-0234, EMD-0235, EMD-0236, and EMD-0237). Models have been deposited in the Protein Data Bank, https://www.ebi.ac.uk/pdbe/ (PDB ID codes 6HJN, 6HJP, 6HJQ, 6HJR, and 6HKG).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1810927115/-/DCSupplemental.
References
- 1.Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: The influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 2.Cossart P, Helenius A. Endocytosis of viruses and bacteria. Cold Spring Harb Perspect Biol. 2014;6:a016972. doi: 10.1101/cshperspect.a016972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Skehel JJ, et al. Changes in the conformation of influenza virus hemagglutinin at the pH optimum of virus-mediated membrane fusion. Proc Natl Acad Sci USA. 1982;79:968–972. doi: 10.1073/pnas.79.4.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bullough PA, Hughson FM, Skehel JJ, Wiley DC. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 5.Gerhard W, Yewdell J, Frankel ME, Webster R. Antigenic structure of influenza virus haemagglutinin defined by hybridoma antibodies. Nature. 1981;290:713–717. doi: 10.1038/290713a0. [DOI] [PubMed] [Google Scholar]
- 6.Laver WG, Air GM, Webster RG. Mechanism of antigenic drift in influenza virus. Amino acid sequence changes in an antigenically active region of Hong Kong (H3N2) influenza virus hemagglutinin. J Mol Biol. 1981;145:339–361. doi: 10.1016/0022-2836(81)90209-6. [DOI] [PubMed] [Google Scholar]
- 7.Sleigh MJ, Both GW, Underwood PA, Bender VJ. Antigenic drift in the hemagglutinin of the Hong Kong influenza subtype: Correlation of amino acid changes with alterations in viral antigenicity. J Virol. 1981;37:845–853. doi: 10.1128/jvi.37.3.845-853.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wiley DC, Wilson IA, Skehel JJ. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature. 1981;289:373–378. doi: 10.1038/289373a0. [DOI] [PubMed] [Google Scholar]
- 9.Knossow M, Daniels RS, Douglas AR, Skehel JJ, Wiley DC. Three-dimensional structure of an antigenic mutant of the influenza virus haemagglutinin. Nature. 1984;311:678–680. doi: 10.1038/311678a0. [DOI] [PubMed] [Google Scholar]
- 10.Knossow M, Skehel JJ. Variation and infectivity neutralization in influenza. Immunology. 2006;119:1–7. doi: 10.1111/j.1365-2567.2006.02421.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corti D, Lanzavecchia A. Broadly neutralizing antiviral antibodies. Annu Rev Immunol. 2013;31:705–742. doi: 10.1146/annurev-immunol-032712-095916. [DOI] [PubMed] [Google Scholar]
- 12.Wilson IA, Skehel JJ, Wiley DC. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature. 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 13.Weis W, et al. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature. 1988;333:426–431. doi: 10.1038/333426a0. [DOI] [PubMed] [Google Scholar]
- 14.Sauter NK, et al. Crystallographic detection of a second ligand binding site in influenza virus hemagglutinin. Proc Natl Acad Sci USA. 1992;89:324–328. doi: 10.1073/pnas.89.1.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bizebard T, et al. Structure of influenza virus haemagglutinin complexed with a neutralizing antibody. Nature. 1995;376:92–94. doi: 10.1038/376092a0. [DOI] [PubMed] [Google Scholar]
- 16.Eisen MB, Sabesan S, Skehel JJ, Wiley DC. Binding of the influenza A virus to cell-surface receptors: Structures of five hemagglutinin-sialyloligosaccharide complexes determined by X-ray crystallography. Virology. 1997;232:19–31. doi: 10.1006/viro.1997.8526. [DOI] [PubMed] [Google Scholar]
- 17.Chen J, et al. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell. 1998;95:409–417. doi: 10.1016/s0092-8674(00)81771-7. [DOI] [PubMed] [Google Scholar]
- 18.Chen J, Skehel JJ, Wiley DC. N- and C-terminal residues combine in the fusion-pH influenza hemagglutinin HA(2) subunit to form an N cap that terminates the triple-stranded coiled coil. Proc Natl Acad Sci USA. 1999;96:8967–8972. doi: 10.1073/pnas.96.16.8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gamblin SJ, et al. The structure and receptor binding properties of the 1918 influenza hemagglutinin. Science. 2004;303:1838–1842. doi: 10.1126/science.1093155. [DOI] [PubMed] [Google Scholar]
- 20.Stevens J, et al. Structure of the uncleaved human H1 hemagglutinin from the extinct 1918 influenza virus. Science. 2004;303:1866–1870. doi: 10.1126/science.1093373. [DOI] [PubMed] [Google Scholar]
- 21.Melikyan GB, Markosyan RM, Roth MG, Cohen FS. A point mutation in the transmembrane domain of the hemagglutinin of influenza virus stabilizes a hemifusion intermediate that can transit to fusion. Mol Biol Cell. 2000;11:3765–3775. doi: 10.1091/mbc.11.11.3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruigrok RW, et al. Electron microscopy of the low pH structure of influenza virus haemagglutinin. EMBO J. 1986;5:41–49. doi: 10.1002/j.1460-2075.1986.tb04175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stein A, Weber G, Wahl MC, Jahn R. Helical extension of the neuronal SNARE complex into the membrane. Nature. 2009;460:525–528. doi: 10.1038/nature08156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trenker R, Call ME, Call MJ. Crystal structure of the Glycophorin A transmembrane dimer in lipidic cubic phase. J Am Chem Soc. 2015;137:15676–15679. doi: 10.1021/jacs.5b11354. [DOI] [PubMed] [Google Scholar]
- 25.Zhang R, et al. 4.4 Å cryo-EM structure of an enveloped alphavirus Venezuelan equine encephalitis virus. EMBO J. 2011;30:3854–3863. doi: 10.1038/emboj.2011.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang J, et al. Molecular links between the E2 envelope glycoprotein and nucleocapsid core in Sindbis virus. J Mol Biol. 2011;414:442–459. doi: 10.1016/j.jmb.2011.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Porta J, et al. Structural studies of Chikungunya virus-like particles complexed with human antibodies: Neutralization and cell-to-cell transmission. J Virol. 2015;90:1169–1177. doi: 10.1128/JVI.02364-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X, et al. Cryo-EM structure of the mature dengue virus at 3.5-Å resolution. Nat Struct Mol Biol. 2013;20:105–110. doi: 10.1038/nsmb.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sirohi D, et al. The 3.8 Å resolution cryo-EM structure of Zika virus. Science. 2016;352:467–470. doi: 10.1126/science.aaf5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baker KA, Dutch RE, Lamb RA, Jardetzky TS. Structural basis for paramyxovirus-mediated membrane fusion. Mol Cell. 1999;3:309–319. doi: 10.1016/s1097-2765(00)80458-x. [DOI] [PubMed] [Google Scholar]
- 31.Bissonnette MLZ, Donald JE, DeGrado WF, Jardetzky TS, Lamb RA. Functional analysis of the transmembrane domain in paramyxovirus F protein-mediated membrane fusion. J Mol Biol. 2009;386:14–36. doi: 10.1016/j.jmb.2008.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee M, et al. Conformation and trimer association of the transmembrane domain of the parainfluenza virus fusion protein in lipid bilayers from solid-state NMR: Insights into the sequence determinants of trimer structure and fusion activity. J Mol Biol. 2018;430:695–709. doi: 10.1016/j.jmb.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Effantin G, et al. Cryo-electron microscopy structure of the native prototype foamy virus glycoprotein and virus architecture. PLoS Pathog. 2016;12:e1005721. doi: 10.1371/journal.ppat.1005721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee JH, Ozorowski G, Ward AB. Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science. 2016;351:1043–1048. doi: 10.1126/science.aad2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roche J, Louis JM, Grishaev A, Ying J, Bax A. Dissociation of the trimeric gp41 ectodomain at the lipid-water interface suggests an active role in HIV-1 Env-mediated membrane fusion. Proc Natl Acad Sci USA. 2014;111:3425–3430. doi: 10.1073/pnas.1401397111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dev J, et al. Structural basis for membrane anchoring of HIV-1 envelope spike. Science. 2016;353:172–175. doi: 10.1126/science.aaf7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chiliveri SC, Louis JM, Ghirlando R, Baber JL, Bax A. Tilted, uninterrupted, monomeric HIV-1 gp41 transmembrane helix from residual dipolar couplings. J Am Chem Soc. 2018;140:34–37. doi: 10.1021/jacs.7b10245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee J, et al. Structure of the Ebola virus envelope protein MPER/TM domain and its interaction with the fusion loop explains their fusion activity. Proc Natl Acad Sci USA. 2017;114:E7987–E7996. doi: 10.1073/pnas.1708052114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Flanagan MT, Skehel JJ. 1977. The conformation of influenza virus haemagglutinin, FEB Letts 80:57−60. [DOI] [PubMed]
- 40.Hite RK, et al. Cryo-electron microscopy structure of the Slo2.2 Na(+)-activated K(+) channel. Nature. 2015;527:198–203. doi: 10.1038/nature14958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sansom MS, Weinstein H. Hinges, swivels and switches: The role of prolines in signalling via transmembrane α-helices. Trends Pharmacol Sci. 2000;21:445–451. doi: 10.1016/s0165-6147(00)01553-4. [DOI] [PubMed] [Google Scholar]
- 42.Rossman JS, Lamb RA. Influenza virus assembly and budding. Virology. 2011;411:229–236. doi: 10.1016/j.virol.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chae PS, et al. Maltose-neopentyl glycol (MNG) amphiphiles for solubilization, stabilization and crystallization of membrane proteins. Nat Methods. 2010;7:1003–1008. doi: 10.1038/nmeth.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hussain H, et al. Accessible mannitol-based amphiphiles (MNAs) for membrane protein solubilisation and stabilisation. Chemistry. 2016;22:7068–7073. doi: 10.1002/chem.201600533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weissenhorn W, et al. Structural basis for membrane fusion by enveloped viruses. Mol Membr Biol. 1999;16:3–9. doi: 10.1080/096876899294706. [DOI] [PubMed] [Google Scholar]
- 46.Calder LJ, Rosenthal PB. Cryomicroscopy provides structural snapshots of influenza virus membrane fusion. Nat Struct Mol Biol. 2016;23:853–858. doi: 10.1038/nsmb.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cerutti N, Loredo-Varela JL, Caillat C, Weissenhorn W. Antigp41 membrane proximal external region antibodies and the art of using the membrane for neutralization. Curr Opin HIV AIDS. 2017;12:250–256. doi: 10.1097/COH.0000000000000364. [DOI] [PubMed] [Google Scholar]
- 48.Streicher H, et al. A phospha-oseltamivir-biotin conjugate as a strong and selective adhesive for the influenza virus. Bioorg Med Chem Lett. 2014;24:1805–1807. doi: 10.1016/j.bmcl.2014.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng SQ, et al. MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scheres SHW. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA. cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 53.Bai XC, Rajendra E, Yang G, Shi Y, Scheres SH. Sampling the conformational space of the catalytic subunit of human γ-secretase. eLife. 2015;4:e11182. doi: 10.7554/eLife.11182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 55.Chen S, et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy. 2013;135:24–35. doi: 10.1016/j.ultramic.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cardone G, Heymann JB, Steven AC. One number does not fit all: Mapping local variations in resolution in cryo-EM reconstructions. J Struct Biol. 2013;184:226–236. doi: 10.1016/j.jsb.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Emsley P, Lohkamp B, Scott WG, Cowtan K, Cr IU. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.