Significance
Dysregulated expression of master transcription factor c-MYC has been shown to promote proliferation and cell survival programs in cancer cells to mediate resistance to anticancer therapies and promote metastasis. Development of pharmacological agents to inhibit c-MYC as an anticancer therapy is a longstanding but elusive goal in the cancer field. Our study provides a potential widely applicable pharmacological strategy for targeting c-MYC–driven oncogenic activity by inhibiting cIAP1 E3 ubiquitin ligase activity as a treatment for cancers. Furthermore, we demonstrate the pharmacological interference in the dynamic interaction of an E3 ubiquitin ligase with its E2s as a strategy for inhibiting ubiquitination reactions.
Keywords: cIAP1, apoptosis, MYC, MAD1, ubiquitination
Abstract
Protooncogene c-MYC, a master transcription factor, is a major driver of human tumorigenesis. Development of pharmacological agents for inhibiting c-MYC as an anticancer therapy has been a longstanding but elusive goal in the cancer field. E3 ubiquitin ligase cIAP1 has been shown to mediate the activation of c-MYC by destabilizing MAD1, a key antagonist of c-MYC. Here we developed a high-throughput assay for cIAP1 ubiquitination and identified D19, a small-molecule inhibitor of E3 ligase activity of cIAP1. We show that D19 binds to the RING domain of cIAP1 and inhibits the E3 ligase activity of cIAP1 by interfering with the dynamics of its interaction with E2. Blocking cIAP1 with D19 antagonizes c-MYC by stabilizing MAD1 protein in cells. Furthermore, we show that D19 and an improved analog (D19-14) promote c-MYC degradation and inhibit the oncogenic function of c-MYC in cells and xenograft animal models. In contrast, we show that activating E3 ubiquitin ligase activity of cIAP1 by Smac mimetics destabilizes MAD1, the antagonist of MYC, and increases the protein levels of c-MYC. Our study provides an interesting example using chemical biological approaches for determining distinct biological consequences from inhibiting vs. activating an E3 ubiquitin ligase and suggests a potential broad therapeutic strategy for targeting c-MYC in cancer treatment by pharmacologically modulating cIAP1 E3 ligase activity.
Cellular inhibitors of apoptosis protein-1/2 (cIAP1/2) are E3 ubiquitin ligases characterized by the presence of three tandem Baculoviral IAP Repeat (BIR) domains (BIR1–3) at the N-terminal part and a C-terminal RING domain that binds with E2-ubiquitin thioester to activate the discharge of ubiquitin cargo to substrate proteins (1). Since cIAP1/2 are frequently overexpressed in a variety of human cancers and can inhibit apoptosis by blocking the caspase activation, considerable efforts have been made to develop IAP antagonists as cancer therapies. Small-molecule mimetics of the second mitochondria-derived activator of caspases, or Smac, bind the BIR2–3 domains of cIAP1/2, activating their E3 ubiquitin ligase activity by triggering dimerization. Thus, Smac mimetics activate the E3 ubiquitin ligase activity of cIAP1/2, which eventually leads to their degradation (2–5). Several Smac mimetics have been evaluated in human clinical trials; however, none has been able to demonstrate efficacy as a monotherapy for the treatment of cancer (6).
Protooncogene c-MYC, a master transcription factor, is a major driver of human tumorigenesis (7). Deregulated MYC alleles, including mutations, rearrangements, or amplification, are frequently observed in many human cancers (8). Elevated c-MYC in cancer cells can transmit a broad transcriptional response to differentially regulate a multitude of cellular processes such as proliferation, metabolism, and cell survival programs, leading to tumorigenesis and metastasis (9, 10). Development of pharmacological agents to inhibit c-MYC as an anticancer therapy has been a longstanding but elusive goal in the cancer field.
The regulation of MYC by the MYC/MAX/MAD network of bHLH/LZip transcription factors is critical in the control of gene-specific transcriptional activation or repression (11, 12). While MYC/MAX heterodimers form a transcriptional activator complex, MAD/MAX heterodimers form a transcriptional repressor complex. The turnover of MAD1 (encoded by the gene MXD1) is regulated by the ubiquitin–proteasome system (UPS) mediated by the E3 ubiquitin ligase activity of cIAP1 (13, 14). The levels of MAD1 are tightly regulated during cell proliferation and differentiation, and overexpression of MAD1 has been shown to suppress c-MYC/Ras-mediated oncogenic transformations in vivo (15–17). MAD1 antagonizes the function of c-MYC by recruiting MAX and the mSin3 repressor complex to MYC-responsive elements and directly competing with the MYC/MAX dimer for the access to binding sites on the key downstream promoters (15, 18, 19).
Here we show that enhancing the E3 ligase activity of cIAP1 by Smac mimetics stabilizes c-MYC. We developed a high-throughput screen for cIAP1 E3 ubiquitin ligase activity and identified a small-molecule inhibitor of cIAP1, D19. We show that D19 binds directly to the RING domain of cIAP1 to limit E3 ubiquitin ligase activity. D19 suppresses c-MYC oncogenic function and cancer cell proliferation by stabilizing MAD1 protein and promoting the degradation of c-MYC. Moreover, we show that D19 can reduce c-MYC levels in cells treated with Smac mimetics to inhibit cancer cell growth. Finally, we developed D19-14, an improved analog of D19, and demonstrated its efficacy in blocking cancer growth and reducing c-MYC in an in vivo cancer xenograft model. Our study provides a potential strategy for targeting c-MYC–driven oncogenic activity, a longstanding elusive goal in cancer treatment.
Results
Smac Mimetics Stabilize c-MYC Through Promoting E3 Ubiquitin Ligase Activity of cIAP1.
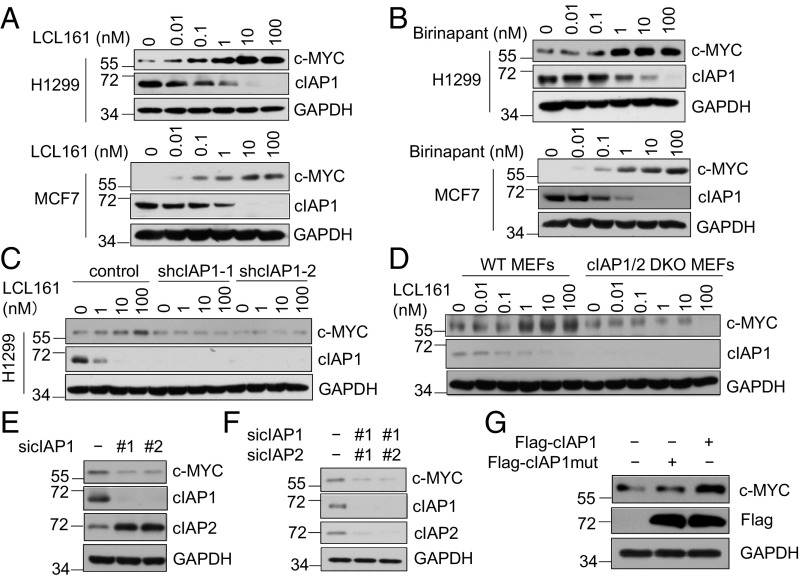
Since cIAP1 has been implicated in promoting the turnover of MAD1, which antagonizes c-MYC (14), we investigated the effect of two Smac mimetics, LCL161 (20) and Birinapant (21), on the protein levels of c-MYC in multiple cancer cell lines that do not die in the presence of Smac. As expected, the treatment with Smac mimetics promoted cIAP1 E3 ubiquitin ligase activity, which led to the degradation of cIAP1 (2–5). Interestingly, the treatment with Smac mimetics dose-dependently increased the levels of c-MYC protein, but not mRNA (Fig. 1 A and B and SI Appendix, Fig. S1 A and B). The effect of Smac mimetics on the levels of c-MYC was dependent on cIAP1 as Smac mimetics could no longer increase the levels of c-MYC in cells with cIAP1 knocked-down or knocked-out (Fig. 1 C and D and SI Appendix, Fig. S1C). We compared the effect of cIAP1 and cIAP2 on the levels of c-MYC individually. We found that knockdown of cIAP1 alone or knockout of both cIAP1/2 was sufficient to reduce levels of c-MYC (Fig. 1 E and F), whereas knockdown of cIAP2 alone had no effect on c-MYC levels (SI Appendix, Fig. S1D). Thus, cIAP1, but not cIAP2, positively regulates the levels of c-MYC.
Fig. 1.
Smac mimetics stabilize c-MYC by promoting the E3 ubiquitin ligase activity of cIAP1. (A and B) The effects of Smac mimetics on protein levels of cIAP1 and c-MYC. H1299 and MCF7 cells were treated with LCL161 (A) or Birinapant (B) at the indicated concentrations for 4 h. (C and D) The up-regulation of c-MYC protein levels by Smac mimetics is mediated by cIAP1. H1299 cIAP1 knockdown cells (C) or cIAP1/2 double-knockout MEFs (D) were treated with LCL161 at the indicated concentrations for 4 h. (E and F) H1299 cells expressing nontarget siRNA, cIAP1 siRNAs, or cIAP1/2 siRNAs were harvested for Western blotting using the indicated antibodies. (G) HEK293T cells were transfected with Flag-cIAP1 or Flag-cIAP1 H588A. After 48 h, the cells were harvested for Western blotting using the indicated antibodies.
Since Smac mimetics stimulate the E3 ligase activity of cIAP1 to promote its autoubiquitination and degradation (2–5), we next characterized the role of cIAP1 catalytic activity in regulating the abundance of c-MYC protein in cells overexpressing wild-type (WT) or H588A mutant cIAP1, which contains a mutation in the RING domain rendering it unable to maintain normal E3 ligase activity (22). Overexpression of WT cIAP1 led to increased levels of c-MYC protein whereas overexpression of catalytically inactive H588A cIAP1 did not (Fig. 1G). This finding suggests that promoting E3 ubiquitin ligase activity of cIAP1 can increase the levels of c-MYC.
MAD1 Promotes c-MYC Ubiquitination and Degradation.
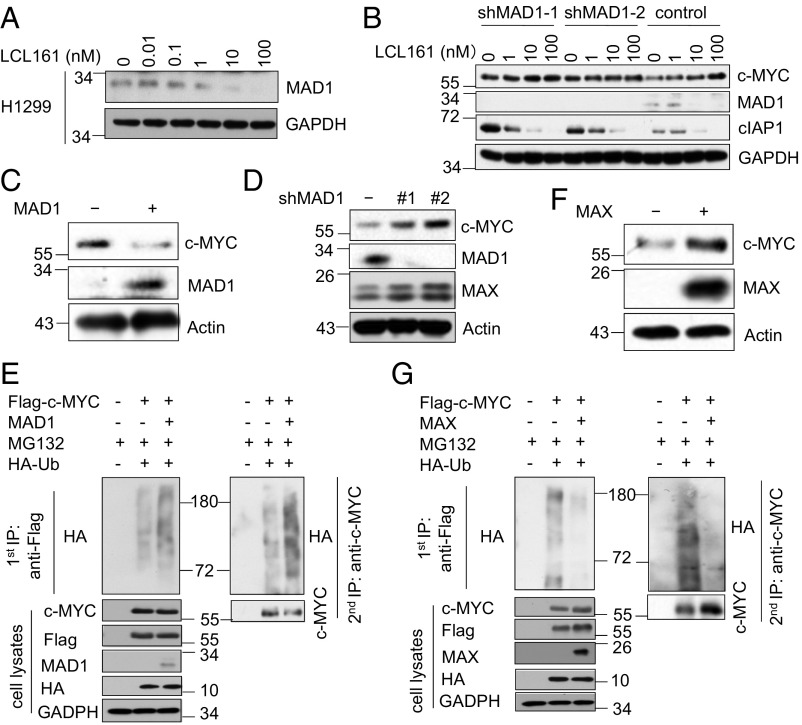
As MAD1, a key antagonist of c-MYC function, is degraded by cIAP1-mediated K48 ubiquitination (14), we then examined if the increased levels of c-MYC protein in cells treated with Smac mimetics were dependent on MAD1. We found that the treatment with LCL161 dose-dependently decreased the levels of MAD1 protein in H1299 cells (Fig. 2A). Moreover, in cells with MAD1 knockdown, LCL161 treatment could no longer increase the levels of c-MYC, suggesting that MAD1 is involved in mediating Smac mimetics-induced c-MYC up-regulation (Fig. 2B). We then asked if increasing the levels of MAD1 might regulate the stability of c-MYC. We found that overexpression of MAD1 reduced the level of c-MYC protein (Fig. 2C). Conversely, MAD1 knockdown increased the levels of c-MYC (Fig. 2D). Consistently, c-MYC was more extensively modified by polyubiquitination when coexpressed with MAD1 than when expressed alone in 293T cells (Fig. 2E).
Fig. 2.
MAD1 promotes c-MYC ubiquitination and degradation. (A) The effects of Smac mimetics on protein levels of MAD1. H1299 cells were treated with LCL161 at the indicated concentrations for 4 h. (B) The up-regulation of c-MYC protein levels by Smac mimetics is dependent on MAD1. H1299 cells stably expressing control shRNA or MAD1 shRNAs were treated with LCL161 at the indicated concentrations for 4 h. (C) H1299 cells stably expressing empty or MAD1 vector were harvested for Western blotting using the indicated antibodies. (D) The cell lysates from H1299 cells stably expressing control shRNA or MAD1 shRNAs were analyzed by Western blotting using the indicated antibodies. (E) The ubiquitination of c-MYC is increased following MAD1 overexpression. HEK293T cells were transfected with Flag–c-MYC, HA-Ub, and control or MAD1 vector for 48 h, followed by treatment with MG132 (10 μM) for 2 h. The anti-Flag immunoprecipitates were denatured by boiling in buffer with 1% SDS and re-IPed with anti–c-MYC antibody. The immunoprecipitates and total cell lysates were immunoblotted with the indicated antibodies. (F) H1299 cells stably expressing empty or MAX vector were harvested for Western blotting using the indicated antibodies. (G) The ubiquitination of c-MYC is decreased by MAX overexpression. HEK293T cells were transfected with Flag–c-MYC, HA-Ub, and control or MAX vector for 48 h, followed by treatment with MG132 (10 μM) for 2 h. The anti-Flag immunoprecipitates were denatured by boiling in buffer with 1% SDS and re-IPed with anti–c-MYC antibody. The immunoprecipitates and cell lysates were immunoblotted with the indicated antibodies.
Because MAD1 competes with c-MYC for dimerization with MAX (15, 23), we hypothesized that MAX/c-MYC dimerization might protect c-MYC from proteasomal degradation. Indeed, overexpression of MAX increased the levels of c-MYC protein (Fig. 2F). Meanwhile, the polyubiquitination levels of c-MYC were reduced by coexpression with MAX, suggesting that dimerization of c-MYC with MAX may provide the resistance to proteasomal degradation by reducing its ubiquitination (Fig. 2G). These results suggest that Smac mimetics alter the balance of MYC/MAX and MYC/MAD heterodimers by promoting the E3 ubiquitin ligase activity of cIAP1.
Identification of a Small-Molecule D19 as an Inhibitor of cIAP1 E3 Ligase.
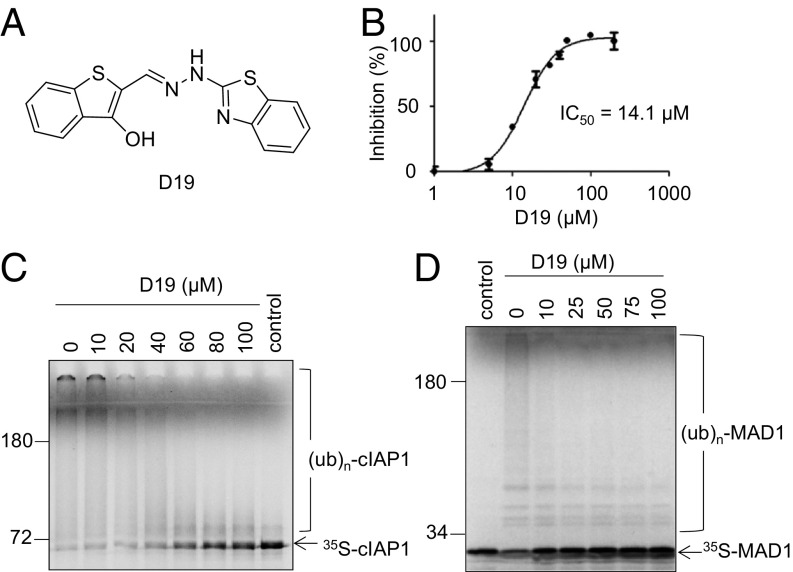
To directly test the possibility of inhibiting cIAP1 as a strategy to antagonize c-MYC, we developed a colorimetric assay using malachite green to quantitatively measure the levels of cIAP1 ubiquitination in vitro. This assay uses ATP consumption by E1 as a surrogate marker for E3 ubiquitin ligase activity. The pyrophosphate produced by E1-mediated ATP hydrolysis is hydrolyzed into phosphate by a pyrophosphatase that can form a colorimetric complex with malachite green (SI Appendix, Fig. S2 A–C). Using this assay, we screened a chemical library of ∼50,000 compounds and identified a compound named D19 that inhibited cIAP1 autoubiquitination with IC50 of 14.1 μM (Fig. 3 A and B). The activity of D19 was further confirmed by using radioactively labeled cIAP1, where treatment with D19 led to increases in unmodified cIAP1 protein and decreases in polyubiquitinated species (Fig. 3C). The autoubiquitination of cIAP2, the closest homolog of cIAP1, was also inhibited by D19 (SI Appendix, Fig. S2D). Importantly, consistent with the effect of cIAP1 in mediating ubiquitination of MAD1 (14), D19 also inhibited the ubiquitination of MAD1 by cIAP1 (Fig. 3D). However, D19 had no effect on autoubiquitination of BRCA1/BARD1 (SI Appendix, Fig. S2E), suggesting that D19 exhibits selectivity toward cIAP1/2 ubiquitin ligase activity.
Fig. 3.
Identification of a small-molecule D19 as an inhibitor of cIAP1 E3 ligase. (A) The chemical structure of D19. (B) D19 inhibits cIAP1 autoubiquitination with an IC50 of 14.1 μM. The autoubiquitination assay of c-IAP1 was performed in the presence of the indicated concentrations of D19 for 2 h and then monitored by pyrophosphate production. Values are the mean of three independent experiments. (C) D19 inhibits cIAP1 autoubiquitination in a dose-dependent manner. In vitro cIAP1 autoubiquitination assay utilizing 35S-labeled cIAP1 in the presence of DMSO or indicated concentrations of D19 for 1 h. (D) The ubiquitination of MAD1 by cIAP1 is inhibited by D19 in a dose-dependent manner. 35S-labeled MAD1 was subjected to an in vitro ubiquitination assay with purified GST-cIAP1 for 1 h in the presence of DMSO or indicated concentrations of D19.
cIAP1 has been shown to cooperate with distinct E2 enzymes including UbcH5 and Ubc13/Uev1a to facilitate K11-, K48-, and K63-polyubiquitination chain formation (14, 24, 25). To explore if the effect of D19 was dependent on any specific E2, we profiled cIAP1 autoubiquitination in the presence of various E2s. We found that UbcH5a/b/c, UbcH6, and Ubc13/Uev1a were able to drive effective autoubiquitination of cIAP1 (SI Appendix, Fig. S2F). Consistently, treatment with D19 inhibited cIAP1 autoubiquitination with all of the E2s that can mediate its activity (SI Appendix, Fig. S2 G and H).
D19 Changes the Dynamics of cIAP1 and E2 Interaction.
To elucidate the molecular basis for the inhibition of cIAP1 ligase activity by D19, we first investigated whether the initial E1 activation and the concomitant step of E2-ubiquitin thioester formation were adversely affected by D19. We found that D19 had no effect on the formation of the UbcH5b-ubiquitin thioester catalyzed by E1 (SI Appendix, Fig. S3A). We next assessed the role of cIAP1’s caspase-activating recruitment domain (CARD) as it is involved in promoting autoinhibition by preventing RING dimerization and binding to E2 (26). Autoubiquitination of the ΔBIR1/2ΔCARD cIAP1 mutant was also significantly impaired by D19 (SI Appendix, Fig. S3B), indicating that the inhibitory effect of D19 was independent of the BIR1/2 or CARD.
The activation of cIAP1 involves dimerization (2). However, we found that the treatment of D19 could not promote the dimerization of Flag-tagged cIAP1 with GST-tagged cIAP1 (SI Appendix, Fig. S3C). In comparison, the addition of LCL161 promoted the dimerization of cIAP1 as reported (2) (SI Appendix, Fig. S3D). Thus, the mechanism of D19 is distinct from that of Smac mimetics.
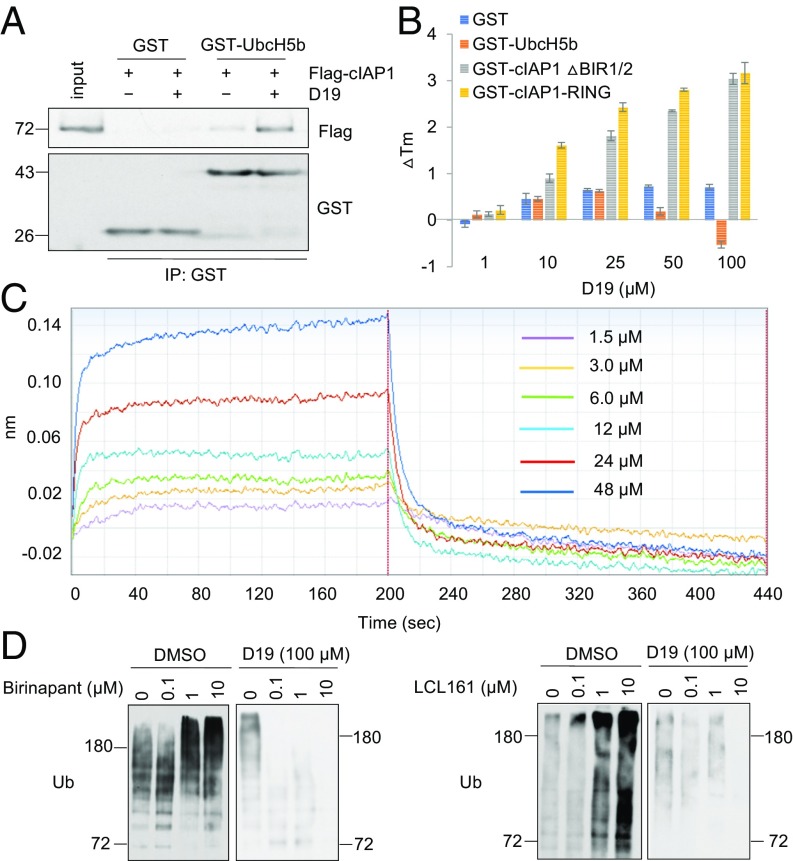
The catalytic transfer of ubiquitin from E2 to substrate involves the rapid assembly and disassembly of the E2–E3 complex, which is critical for progressive ubiquitination reactions (27). We next asked if D19 could affect the association of cIAP1 with E2. Under normal conditions, purified Flag-cIAP1 protein bound weakly to GST-UbcH5b. Interestingly, this interaction was significantly stabilized in the presence of D19 (Fig. 4A). In contrast, an inactive analog of D19, D19-Cl, which is unable to inhibit c-IAP1 autoubiquitination (SI Appendix, Fig. S3 E and F), had no impact on cIAP1/UbcH5b binding (SI Appendix, Fig. S3G). Moreover, we found that D19 did not inhibit cIAP1-Ub conjugate formation when preloaded E2-Ub was incubated with cIAP1 (SI Appendix, Fig. S3H). Taken together, we conclude that D19 may impair cIAP1 ligase activity by stabilizing the dynamic interaction of cIAP1 and the E2 complex to prevent successive rounds of cIAP1 and E2-Ub interaction in ubiquitination reaction.
Fig. 4.
D19 changes the dynamics of cIAP1 and E2 interaction. (A) D19 enhances binding of cIAP1 and E2. Flag-tagged cIAP1 was coprecipitated with recombinant GST-tagged UbcH5b or GST alone immobilized on glutathione agarose in the presence or absence of D19 (100 μM). (B) D19 binds to cIAP1. Purified GST, GST-UbcH5b, GST-cIAP1ΔBIR1/2, or GST-cIAP1-RING was subjected to a DSF assay in the presence of indicated concentrations of D19. The thermal shift (ΔTm) was calculated and plotted. The data represent the mean ± SD of three independent experiments. (C) D19 directly binds to the RING domain of cIAP1. A Biolayer Interferometry assay was performed with the indicated concentrations of D19. (D) D19 inhibits cIAP1 autoubiquitination stimulated by Smac mimetics. A GST-cIAP1 autoubiquitination assay was performed for 30 min with the indicated concentrations of Birinapant or LCL161 with or without D19 (100 μM).
To identify the protein-targeting mechanism of D19, we performed a differential scanning fluorimetry (DSF) assay to measure the protein melting temperature (Tm) change induced by D19. The presence of D19 dose-dependently increased the Tm of GST-tagged ΔBIR1/2-cIAP1 and RING protein, while it had no obvious effect on the Tm of GST alone or UbcH5b (Fig. 4B). Notably, D19, but not D19-Cl, increased the Tm of GST-RING domain protein (SI Appendix, Fig. S3I), suggesting that D19 may directly bind to RING domain. Furthermore, using a BioLayer Interferometry assay, we found a clear dose-dependent interaction of D19 with the RING domain protein immobilized on the biosensor tip (Fig. 4C). Taken together, these results suggest that D19 inhibits cIAP1 ubiquitination activity by binding to the RING domain to interfere its dynamic interaction with E2.
Since Smac mimetics are known to promote the E3 ubiquitin ligase activity of cIAP1 (4, 5), we next characterized the effect of D19 on ubiquitination of cIAP1 with Smac. We found that D19 was highly effective in blocking cIAP1 autoubiquitination induced by Smac mimetics (Fig. 4D). Thus, the ability of D19 to block the dynamics of the E2–E3 complex is dominant over the activity of Smac mimetics in promoting cIAP1 E3 ligase activity.
D19 Promotes the Proteasomal Degradation of c-MYC.
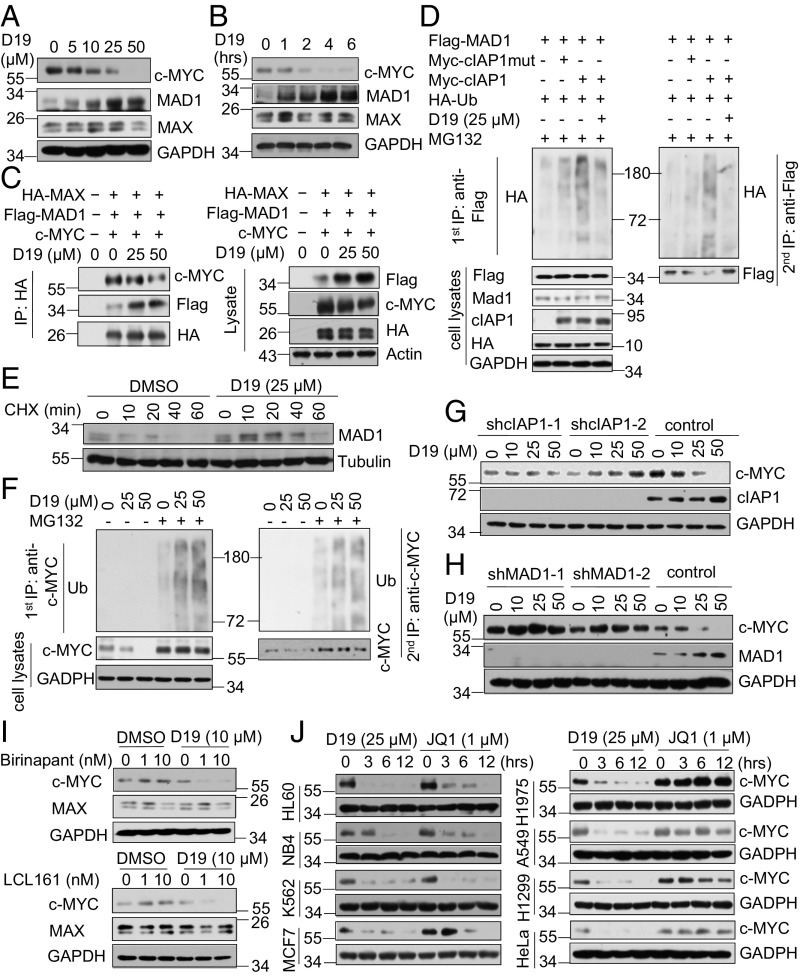
In contrast to Smac mimetics, which promoted increases in the levels of c-MYC (Fig. 1 A and B), we found that treatment with D19 led to a dose- and time-dependent reduction of c-MYC and an increase of MAD1 protein levels with no significant effect on their mRNA levels (Fig. 5 A and B and SI Appendix, Fig. S4A). As a result, D19 treatment reduced the prevalence of bound dimeric c-MYC/MAX while increasing that of MAD1/MAX (Fig. 5C). Consistent with the role of cIAP1 as an E3 ubiquitin ligase of MAD1 (14), we found that treatment with D19 inhibited the ubiquitination of MAD1 by cIAP1 (Fig. 5D). In contrast, the inactive analog D19-Cl had no effect on the levels of MAD1 protein in cells, confirming that D19 acts directly on the ubiquitination functions of cIAP1 (SI Appendix, Fig. S4B). In addition, D19 had no effect on p53 ubiquitination by MDM2 or on total protein ubiquitination (SI Appendix, Fig. S4 C and D). Treatment with D19 slowed the decay of MAD1 protein levels in the presence of the protein synthesis inhibitor cycloheximide (CHX) (Fig. 5E). The effect of D19 was cIAP1-dependent as D19 was unable to increase MAD1 protein in cIAP1 knockdown H1299 cells or cIAP1/2 double-knockout mouse embryonic fibroblasts (MEFs) (SI Appendix, Fig. S4 E and F). These data suggest that D19 is a selective inhibitor of cIAP1 ubiquitination activity in vivo.
Fig. 5.
D19 promotes the proteasomal degradation of c-MYC. (A and B) D19 decreases c-MYC and increases MAD1 protein levels. H1299 cells were treated with D19 at the indicated concentrations for 4 h (A) and time points at 25 μM (B). (C) D19 decreases the binding of c-MYC/MAX while increasing that of MAD1/MAX. HEK293T cells were transfected with HA-MAX, Flag-MAD1, and c-MYC. Forty-eight hours later, the cells were treated with D19 at the indicated concentration for 4 h. Anti-HA immunoprecipitates and cell lysates were immunoblotted with the indicated antibodies. (D) D19 inhibits MAD1 ubiquitination by cIAP1 in cells. HEK293T cells were transfected with Flag-MAD1, HA-Ub, Myc-cIAP1, or Myc-cIAP1-H588A for 24 h. Cells were subsequently treated with D19 (25 μM) or DMSO for 12 h and then with MG132 (10 μM) for 4 h. The anti-Flag immunoprecipitates were denatured by boiling in buffer with 1% SDS and re-IPed with anti-FLAG antibody. The immunoprecipitates and cell lysates were immunoblotted with the indicated antibodies. (E) D19 inhibits MAD1 degradation. H1299 cells were preincubated with DMSO or D19 (25 μM) for 2 h, followed by treatment with CHX (100 ng/mL) for the indicated time periods. (F) D19 promotes the ubiquitination and proteasomal degradation of c-MYC protein. H1299 cells were preincubated with MG132 (20 μM) for 2 h and then treated with D19 at the indicated concentrations for 4 h. The anti–c-MYC immunoprecipitates were denatured by boiling in buffer with 1% SDS and re-IPed with anti–c-MYC antibody. The immunoprecipitates and cell lysates were immunoblotted with indicated antibodies. (G and H) The down-regulation of c-MYC protein levels by D19 is mediated by cIAP1 and MAD1. H1299 cells stably expressing control shRNA and either cIAP1 shRNAs (G) or MAD1 shRNAs (H) were treated with D19 at the indicated concentrations for 4 h. (I) D19 abolishes the up-regulation of c-MYC protein levels by Smac mimetics. H1299 cells were preincubated with D19 for 1 h, followed by treatment with LCL161 or Birinapant at the indicated concentrations for 4 h. (J) A comparison of D19 versus JQ1 in cells. The indicated cell lines were treated with D19 (25 μM) or JQ1 (1 μM) for the indicated time periods, and protein levels of c-MYC were measured by Western blot.
We subsequently investigated the mechanism by which D19 regulates c-MYC protein levels. Preincubation with MG132 before D19 treatment blocked the down-regulation of c-MYC (Fig. 5F). Correspondingly, the polyubiquitination of endogenous c-MYC, barely detectable under control conditions even in the presence of MG132 due to its rapid degradation, was markedly enhanced by D19 treatment in the presence of MG132 (Fig. 5F), suggesting that D19 promotes the ubiquitination and proteasomal degradation of c-MYC protein. The accelerated turnover of c-MYC by D19 treatment was mediated by the cIAP1/MAD1 axis because D19 failed to decrease c-MYC protein levels in cells lacking either cIAP1 or MAD1 (Fig. 5 G and H).
As we found that the effects of D19 are dominant over those of Smac mimetics, we next examined if D19 could abolish the up-regulation of c-MYC protein driven by Smac mimetics. Interestingly, treatment with D19 blocked increases in the levels of c-MYC protein in cells treated by Smac mimetics (Fig. 5I). Thus, inhibition of cIAP1 E3-ubiquitinating activity by D19 can abrogate the activation of c-MYC induced by Smac mimetics.
We next compared D19 with JQ1, a BET bromodomain inhibitor, which has been shown to reduce the levels of c-MYC by repressing transcription (28, 29). We profiled the activities of D19 and JQ1 on c-MYC expression in eight cancer cell lines. We found that, while JQ1 treatment reduced levels of c-MYC in only four of eight cell lines, D19 showed efficacy in all eight cancer cell lines tested (Fig. 5J). Thus, targeting cIAP1 may provide a broad strategy for inhibiting c-MYC across a variety of tumor types.
Developing cIAP1 E3 Ligase Inhibitors to Antagonize c-MYC in Cancer Treatment.
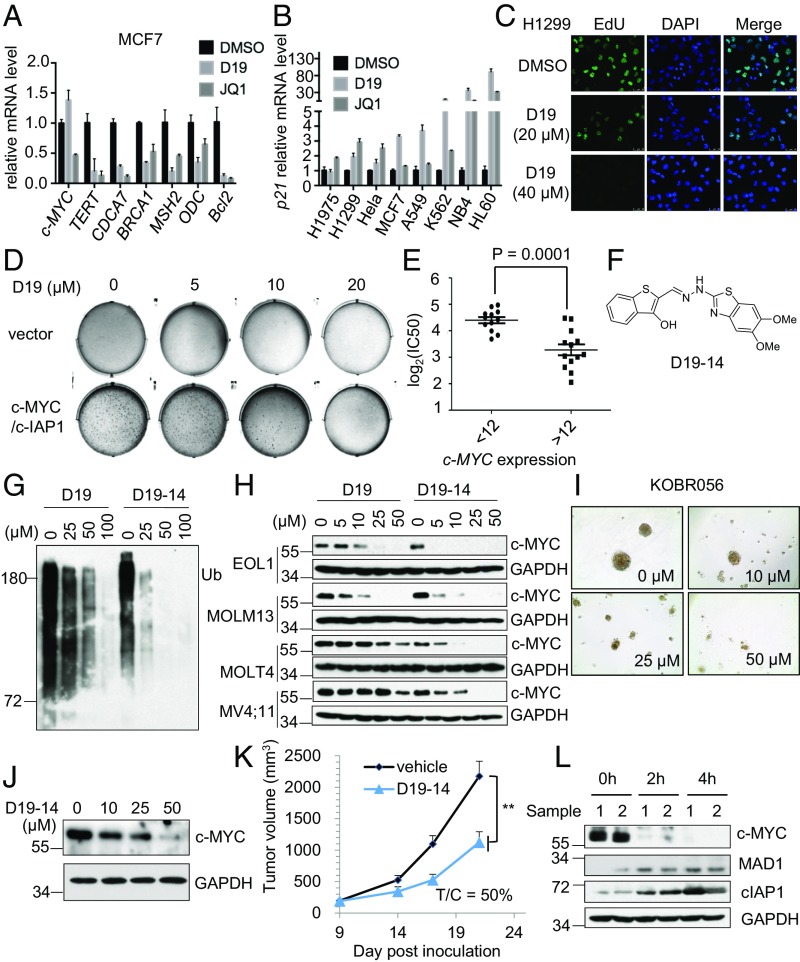
We next evaluated inhibition of cIAP1 E3 ligase activity with D19 as a potential anti–c-MYC strategy in cancer treatment. We first asked if D19 could antagonize the downstream oncogenic functions of c-MYC in cancer cells. Transcription of ODC, CDCA7, BRCA1, and MSH2 is promoted by c-MYC while p21 is repressed (30–33). We found that treating cells with either D19 or JQ1 diminished the mRNA levels of ODC, CDCA7, BRCA1, and MSH2, although D19 did not change MYC transcription (Fig. 6A). Interestingly, the mRNA levels of p21 were profoundly elevated by D19 treatment in multiple cell lines (Fig. 6B). Consistent with induction of p21, treatment with D19 led to inhibition of DNA synthesis and substantial G1 arrest (Fig. 6C and SI Appendix, Fig. S5 A and B).
Fig. 6.
The cIAP1 inhibitor D19-14 antagonizes c-MYC in a cancer xenograft model. (A) D19 inhibits the transcription of c-MYC target genes. MCF7 cells were treated with D19 (40 μM) or JQ1 (2.0 μM) for 24 h. The mRNA levels of c-MYC target genes were analyzed by quantitative RT-PCR. GAPDH mRNA was used as an internal control, and the data were normalized to the DMSO control. The data represent the mean ± SD of three independent experiments. (B) D19 up-regulates the mRNA level of p21.The indicated cell lines were treated with D19 (40 μM) or JQ1 (2.0 μM) for 24 h. p21 mRNA levels were analyzed by quantitative RT-PCR. The data represent the mean ± SD of three independent experiments. (C) D19 inhibits DNA synthesis. EdU incorporation was measured using imaging analysis in H1299 cells after D19 treatment for 24 h. (D) D19 inhibits c-MYC/cIAP1–induced transformation. The colony formation of NIH 3T3 cells stably expressing empty vector or cIAP1/c-MYC with DMSO or D19 treatment at the indicated concentrations for 2 wk. (E) The correlation of c-MYC expression and the IC50 of D19 in 25 cancer cell lines. Column scatterplot with mean ± SD of three independent experiments (P = 0.0001). (F) The chemical structure of D19-14. (G) D19-14 is a more effective inhibitor of cIAP1 autoubiquitination than D19. Autoubiquitination assay of cIAP1 in the presence of the indicated concentrations of D19 or D19-14. (H) D19-14 more potently decreases the c-MYC protein level than D19. The indicated cell lines were treated with D19 or D19-14 for 4 h. (I) D19-14 inhibits PDTO proliferation. The PDTO KOBR056 was treated with D19-14 at the indicated concentrations for 6 d. (J) D19-14 decreases c-MYC protein levels in PDTO. The PDTO KOBR056 was treated with D19-14 at the indicated concentrations for 6 h. (K) Growth curve of s.c. EOL1 xenograft tumors in mice treated with D19-14 (50 mg/kg) by i.p. injection once daily (eight mice per group; mean ± SEM). Tumor volume was calculated using the formula: tumor volume = [length × width × width]/2. T/C: the average tumor size of treatment group versus control group. Data represent means ± SD; **P ≤ 0.001, two-tailed unpaired Student’s t test. (L) The mice bearing EOL1 xenograft tumors were removed at the indicated times following the last dose of D19-14 in vivo and lysed immediately. The protein levels of c-MYC in tumor lysates were analyzed by Western blotting.
Consistent with the ability of cIAP1 to synergistically cooperate with c-MYC to induce tumorigenesis (14, 34), coexpression of cIAP1 and c-MYC readily transformed NIH 3T3 cells, leading to the rapid formation of massive colonies in soft agar, which were effectively eliminated by the treatment of D19 (Fig. 6D). The formation of foci in MCF7 cells was also dose-dependently attenuated by D19 treatment (SI Appendix, Fig. S5C). Moreover, treatment with D19 resulted in pronounced cellular senescence as determined by β-galactosidase staining in treated H1299 cells (SI Appendix, Fig. S5D).
We next examined if the antitumor activity of D19 is dependent on c-MYC expression. We profiled the sensitivity of 25 cancer cell lines treated with D19 (SI Appendix, Table S1). The expression levels of c-MYC in these 25 cell lines obtained from the Cancer Cell Line Encyclopedia (CCLE) were compared with the logarithm of their respective IC50 values. We found that cell lines with high c-MYC expression were significantly more sensitive to D19 than those with low c-MYC expression (Fig. 6E), suggesting that the antitumor activity of D19 is dependent on c-MYC overexpression.
Our data suggest that inhibition of cIAP1 E3 ligase activity is a potential strategy to target c-MYC–dependent cancers. We next tested this hypothesis in animal models. Since the pharmacokinetic properties of D19 limited its application for in vivo treatment, we conducted medicinal chemistry optimization of D19 and identified an improved D19 analog, known as D19-14 (Fig. 6F), with a significantly increased ability to inhibit cIAP1 autoubiquitination and reduce protein levels of c-MYC in vitro compared with D19 (Fig. 6 G and H). Consistently, D19-14 was more potent in inhibiting the proliferation of cancer cells than D19 (SI Appendix, Fig. S5E). Patient-derived tumor organoids (PDTOs) have shown the advantage of mimicking the biological characteristics of the human tumor both phenotypically and genetically (35). We therefore tested D19-14 in three PDTOs from breast cancer patients. D19-14 effectively inhibited PTDO proliferation (Fig. 6I and SI Appendix, Fig. S5F) and decreased c-MYC protein levels (Fig. 6J).
In addition, D19-14 exhibited acceptable pharmacokinetics (PK) properties in vivo [area under curve (AUC) 4,075 nM⋅h and T1/2 4.9 h] and improved bioavailability in tumors (SI Appendix, Fig. S5 G and H). Pharmacodynamic studies indicated that D19-14 effectively decreased the levels of c-MYC protein in tumors (SI Appendix, Fig. S5I). We further evaluated the antitumor activity of D19-14 in a human xenograft model of acute myeloid leukemia using EOL1 cells with a high c-MYC mRNA level (CCLE data). Randomized cohorts of mice with established tumors were treated with D19-14 (50 mg/kg) or vehicle. Daily i.p. administration of D19-14 was well tolerated by mice (SI Appendix, Fig. S5J). The tumor growth was remarkably reduced by 12 d of D19-14 treatment (Fig. 6K and SI Appendix, Fig. S5K). Consistently, treatment with D19-14 led to a clear decrease in the protein levels of c-MYC in tumors compared with that of the vehicle-treated group when examined following the final day of treatment (Fig. 6L). Thus, inhibiting E3 ubiquitin ligase activity of cIAP1 reduces c-MYC levels and cancer growth in vivo.
Discussion
In this article, we describe a study using chemical biological approaches to differentiate the distinct biological consequences of inhibiting E3 ubiquitin ligase activity of cIAP1 by D19 vs. activating the E3 ubiquitin ligase activity of cIAP1 by Smac mimetics. Since the turnover of MAD1 is highly regulated by cIAP1-mediated ubiquitination and proteasomal turnover, rapid activation of cIAP1 E3 activity before cIAP1’s degradation in the presence of Smac mimetics is sufficient to promote the turnover of MAD1 to lead to stabilization of c-MYC, whereas stabilizing the interaction of cIAP1 with its E2 by D19 and D19-14 block successive rounds of ubiquitination reaction, which leads to stabilization of MAD1 and inhibition of c-MYC. Our study provides an interesting example of using chemical biological approaches to determine distinct biological consequences from inhibiting vs. activating an E3 ubiquitin ligase. D19-14 may serve as a lead compound for further development to improve the potency and pharmacokinetic properties as a treatment for cancers with c-MYC dysregulation.
Our study explored the possibility of targeting IAP proteins using pharmacological strategy to block the oncogenic activity of c-MYC. We show that inhibition of cIAP1 by D19 and its analog D19-14 can reduce the levels of c-MYC protein in a broad range of cancer cell lines and in xenograft tumor models in vivo. While ubiquitination modification is known to be critical in controlling key biochemical reactions regulating diverse cellular processes, developing small-molecule inhibitors for E3 ubiquitin ligases has been challenging to date. We show that D19 inhibits cIAP1 ubiquitination activity by binding to the RING domain to interfere with its dynamic interaction with E2. The mechanism by which the RING domain promotes ubiquitin transfer is not fully understood because the RING-binding site on the E2s is distant from the active site. It has been proposed that the RING domain brings the E2-Ub conjugate into close proximity with the substrate (36, 37). Recharging utilized E2 requires the residues within the H1 α-helix to be exposed to engage with E1; however, these residues are normally buried at the E2–E3 interface (36, 38). Therefore, the interactions between RING domains and E2s are generally transient with modest affinity, facilitating E2 “hit-and-run” from E3 before participation in the next cycle of E2-Ub conjugation (39). Meanwhile, E3 is released from E2 and accepts a new E2-Ub molecule for ubiquitin transfer. Thus, the ubiquitin modification of proteins relies on the rapid assembly and disassembly of the E2–E3 complex, which allows the catalytic transfer of ubiquitin from E2 to substrate and the initiation of subsequent rounds of ubiquitination (27). In this regard, the enhanced interaction between cIAP1 and UbcH5b driven by D19 treatment may disturb the dynamics of the ubiquitin transfer process and result in the termination of ubiquitination. Our data suggest that stabilization of the weak interactions between ubiquitin and ubiquitination enzymes by small molecules may provide a useful strategy to selectively inhibit E3 ubiquitin ligase activities. Thus, D19 provides a prototypic example of stabilizing E2/RING–E3 interaction as an approach to block E3 ubiquitin ligase activity. This strategy may be extended to develop small-molecule inhibitors for other RING finger-E3 ubiquitin ligases, the largest ubiquitin ligase family in the human genome that includes many important E3s such as the anaphase-promoting complex.
While a subset of cancer cell lines can die by TNFα-mediated apoptosis when treated with Smac mimetics, many cancer cells are resistant to Smac mimetics (14, 40, 41). Our study uncovered the ability of Smacs to induce the protein levels of c-MYC in these resistance cancer cell lines. We found that the protein levels of c-MYC were elevated following treatment with Smac mimetics in such Smac nonresponsive cell lines, suggesting that increased levels of c-MYC may provide an underlying mechanism for this resistance. We confirm that Smac mimetics function as cIAP1 agonists to promote c-IAP1 E3 ligase activity that triggers ubiquitination and degradation of its substrates, such as MAD1, with close proximity and high affinity. The ubiquitination of MAD1 by cIAP1 is important for the control of MAD1 protein levels in cancer cells; therefore, activating E3 ligase activity of cIAP1 by the treatment with Smac mimetics can boost ubiquitination and degradation of MAD1, resulting in the stabilization of c-MYC as MAD1 is no longer able to antagonize c-MYC’s oncogenic functions. Compared with Smac mimetics, the endogenous mitochondrial protein Smac is released into cytosol, but does not enter into the nucleus (42), which may explain why cellular Smac protein is not able to stabilize c-MYC and instead triggers apoptosis.
The MYC/MAX/MAD network model proposed by Eisenman and colleagues centers on MAX forming transactivating complexes when associated with MYC or repressive complexes when bound to MAD (15, 19). The MYC/MAX/MAD transcription network is tightly controlled through UPS during the cell cycle, proliferation, and differentiation. Both c-MYC and MAD1 are short-lived proteins that dynamically respond to, or guide, cell-fate determination following external stimuli (15). However, how exactly protein levels of c-MYC and MAD1 are intrinsically balanced and regulated during cell-fate transitions is not fully understood. We found that MAD1 promotes c-MYC ubiquitination and degradation, revealing a mechanism of c-MYC/MAX/MAD network regulation during cell-fate transition. We propose inhibiting cIAP1-mediated ubiquitination of MAD1 as an anti-MYC strategy for the treatment of cancers.
Materials and Methods
Constructs and Reagents.
The cDNAs coding Flag–c-MYC, c-MYC, HA-MAX, MAX, Flag-cIAP1, cIAP1, and Flag-cIAP1-H588A were cloned into pl-MCS vector. The cDNAs coding Flag-p53, MDM2, Flag-MAD1, MAD1, Myc-cIAP1-H588A, and Myc-cIAP1 were cloned into the pMSCV vector (#68469; Addgene). The cDNAs coding BRCA1, BARD1, UbcH5b, cIAP1, cIAP1ΔBIR1/2, cIAP1ΔBIR1/2ΔCARD, and cIAP1-RING were subcloned into pGEX-4T-1 vectors (28-9545-49; GE Healthcare). The cDNAs coding E1 were subcloned into the pMAL vector. Other plasmids were prepared as described previously (14).
The shRNAs targeting human MAD1 and cIAP1 were obtained from Sigma-Aldrich. All shRNA primers (SI Appendix, Table S2) were annealed and cloned into linearized pLKO.1-puro. All plasmids were verified by DNA sequencing. The sequences of the siRNA oligonucleotides are shown in SI Appendix, Table S3.
MG132 (S2916), Birinapant (S7015), and JQ1 (S7110) were purchased from Selleck Chem. LCL161 (SX-171009) was purchased from Medsyin. Cycloheximide (C104450) was purchased from Sigma-Aldrich.
Statistical Analysis.
All data are presented as the mean ± SEM or SD from at least three independent determinations, and statistical analyses were done using the software Graphpad Prism version 6.0 (GraphPad Software). Differences of means were tested for statistical significance with the two-tailed Student’s t test. The animal protocol (IACUC#IRCBC-2017-003) was approved by The Institutional Animal Care and Use Committee of Interdisciplinary Research Center on Biology and Chemistry, Shanghai Institute of Organic Chemistry, Chinese Academy of Sciences.
Supplementary Material
Acknowledgments
We thank Lauren Mifflin (Harvard Medical School) for thoughtful discussion and critical reading of the manuscript and K2 Oncology Co., Ltd. for the PTDO samples. This work was supported in part by the Chinese Academy of Sciences (J.Z. and J.Y.) and by the Ludwig Center at Harvard (J.Y.). This work was supported by the National Key R&D Program of China (Grant 2016YFA0501900), the Natural Science Foundation of China (Grant 21532002), and the Shanghai Municipal Natural Science Foundation (Grant 14ZR1448600).
Footnotes
Conflict of interest statement: J.Y. and Eileen White are coauthors of a 2016 paper on guidelines for autophagy.
See Commentary on page 9821.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1807711115/-/DCSupplemental.
References
- 1.Salvesen GS, Duckett CS. IAP proteins: Blocking the road to death’s door. Nat Rev Mol Cell Biol. 2002;3:401–410. doi: 10.1038/nrm830. [DOI] [PubMed] [Google Scholar]
- 2.Dueber EC, et al. Antagonists induce a conformational change in cIAP1 that promotes autoubiquitination. Science. 2011;334:376–380. doi: 10.1126/science.1207862. [DOI] [PubMed] [Google Scholar]
- 3.Vince JE, et al. IAP antagonists target cIAP1 to induce TNFalpha-dependent apoptosis. Cell. 2007;131:682–693. doi: 10.1016/j.cell.2007.10.037. [DOI] [PubMed] [Google Scholar]
- 4.Varfolomeev E, et al. IAP antagonists induce autoubiquitination of c-IAPs, NF-kappaB activation, and TNFalpha-dependent apoptosis. Cell. 2007;131:669–681. doi: 10.1016/j.cell.2007.10.030. [DOI] [PubMed] [Google Scholar]
- 5.Feltham R, et al. Smac mimetics activate the E3 ligase activity of cIAP1 protein by promoting RING domain dimerization. J Biol Chem. 2011;286:17015–17028. doi: 10.1074/jbc.M111.222919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fulda S. Smac mimetics to therapeutically target IAP proteins in cancer. Int Rev Cell Mol Biol. 2017;330:157–169. doi: 10.1016/bs.ircmb.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 7.Dang CV, et al. Function of the c-Myc oncogenic transcription factor. Exp Cell Res. 1999;253:63–77. doi: 10.1006/excr.1999.4686. [DOI] [PubMed] [Google Scholar]
- 8.Stine ZE, Walton ZE, Altman BJ, Hsieh AL, Dang CV. MYC, metabolism, and cancer. Cancer Discov. 2015;5:1024–1039. doi: 10.1158/2159-8290.CD-15-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsieh AL, Walton ZE, Altman BJ, Stine ZE, Dang CV. MYC and metabolism on the path to cancer. Semin Cell Dev Biol. 2015;43:11–21. doi: 10.1016/j.semcdb.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grandori C, Cowley SM, James LP, Eisenman RN. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol. 2000;16:653–699. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- 12.Eilers M, Eisenman RN. Myc’s broad reach. Genes Dev. 2008;22:2755–2766. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu J, Blenis J, Yuan J. Activation of PI3K/Akt and MAPK pathways regulates Myc-mediated transcription by phosphorylating and promoting the degradation of Mad1. Proc Natl Acad Sci USA. 2008;105:6584–6589. doi: 10.1073/pnas.0802785105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu L, et al. c-IAP1 cooperates with Myc by acting as a ubiquitin ligase for Mad1. Mol Cell. 2007;28:914–922. doi: 10.1016/j.molcel.2007.10.027. [DOI] [PubMed] [Google Scholar]
- 15.Conacci-Sorrell M, McFerrin L, Eisenman RN. An overview of MYC and its interactome. Cold Spring Harb Perspect Med. 2014;4:a014357. doi: 10.1101/cshperspect.a014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cerni C, et al. Repression of in vivo growth of Myc/Ras transformed tumor cells by Mad1. Oncogene. 2002;21:447–459. doi: 10.1038/sj.onc.1205107. [DOI] [PubMed] [Google Scholar]
- 17.Larsson LG, Pettersson M, Oberg F, Nilsson K, Lüscher B. Expression of mad, mxi1, max and c-myc during induced differentiation of hematopoietic cells: Opposite regulation of mad and c-myc. Oncogene. 1994;9:1247–1252. [PubMed] [Google Scholar]
- 18.Rottmann S, Lüscher B. The Mad side of the Max network: Antagonizing the function of Myc and more. Curr Top Microbiol Immunol. 2006;302:63–122. doi: 10.1007/3-540-32952-8_4. [DOI] [PubMed] [Google Scholar]
- 19.Ayer DE, Kretzner L, Eisenman RN. Mad: A heterodimeric partner for Max that antagonizes Myc transcriptional activity. Cell. 1993;72:211–222. doi: 10.1016/0092-8674(93)90661-9. [DOI] [PubMed] [Google Scholar]
- 20.Weisberg E, et al. Smac mimetics: Implications for enhancement of targeted therapies in leukemia. Leukemia. 2010;24:2100–2109. doi: 10.1038/leu.2010.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Condon SM, et al. Birinapant, a smac-mimetic with improved tolerability for the treatment of solid tumors and hematological malignancies. J Med Chem. 2014;57:3666–3677. doi: 10.1021/jm500176w. [DOI] [PubMed] [Google Scholar]
- 22.Li X, Yang Y, Ashwell JD. TNF-RII and c-IAP1 mediate ubiquitination and degradation of TRAF2. Nature. 2002;416:345–347. doi: 10.1038/416345a. [DOI] [PubMed] [Google Scholar]
- 23.Lüscher B. MAD1 and its life as a MYC antagonist: An update. Eur J Cell Biol. 2012;91:506–514. doi: 10.1016/j.ejcb.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 24.Bertrand MJ, et al. cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol Cell. 2008;30:689–700. doi: 10.1016/j.molcel.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 25.Dynek JN, et al. c-IAP1 and UbcH5 promote K11-linked polyubiquitination of RIP1 in TNF signalling. EMBO J. 2010;29:4198–4209. doi: 10.1038/emboj.2010.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lopez J, et al. CARD-mediated autoinhibition of cIAP1’s E3 ligase activity suppresses cell proliferation and migration. Mol Cell. 2011;42:569–583. doi: 10.1016/j.molcel.2011.04.008. [DOI] [PubMed] [Google Scholar]
- 27.Kleiger G, Saha A, Lewis S, Kuhlman B, Deshaies RJ. Rapid E2-E3 assembly and disassembly enable processive ubiquitylation of cullin-RING ubiquitin ligase substrates. Cell. 2009;139:957–968. doi: 10.1016/j.cell.2009.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Delmore JE, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Filippakopoulos P, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dang CV. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol. 1999;19:1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Osthus RC, et al. The Myc target gene JPO1/CDCA7 is frequently overexpressed in human tumors and has limited transforming activity in vivo. Cancer Res. 2005;65:5620–5627. doi: 10.1158/0008-5472.CAN-05-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Menssen A, Hermeking H. Characterization of the c-MYC-regulated transcriptome by SAGE: Identification and analysis of c-MYC target genes. Proc Natl Acad Sci USA. 2002;99:6274–6279. doi: 10.1073/pnas.082005599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoffman B, Liebermann DA. Apoptotic signaling by c-MYC. Oncogene. 2008;27:6462–6472. doi: 10.1038/onc.2008.312. [DOI] [PubMed] [Google Scholar]
- 34.Zender L, et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Drost J, Clevers H. Organoids in cancer research. Nat Rev Cancer. 2018;18:407–418. doi: 10.1038/s41568-018-0007-6. [DOI] [PubMed] [Google Scholar]
- 36.Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102:533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
- 37.Mace PD, et al. Structures of the cIAP2 RING domain reveal conformational changes associated with ubiquitin-conjugating enzyme (E2) recruitment. J Biol Chem. 2008;283:31633–31640. doi: 10.1074/jbc.M804753200. [DOI] [PubMed] [Google Scholar]
- 38.Huang DT, et al. Structural basis for recruitment of Ubc12 by an E2 binding domain in NEDD8’s E1. Mol Cell. 2005;17:341–350. doi: 10.1016/j.molcel.2004.12.020. [DOI] [PubMed] [Google Scholar]
- 39.Christensen DE, Brzovic PS, Klevit RE. E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat Struct Mol Biol. 2007;14:941–948. doi: 10.1038/nsmb1295. [DOI] [PubMed] [Google Scholar]
- 40.Petersen SL, et al. Autocrine TNFalpha signaling renders human cancer cells susceptible to Smac-mimetic-induced apoptosis. Cancer Cell. 2007;12:445–456. doi: 10.1016/j.ccr.2007.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petersen SL, Peyton M, Minna JD, Wang X. Overcoming cancer cell resistance to Smac mimetic induced apoptosis by modulating cIAP-2 expression. Proc Natl Acad Sci USA. 2010;107:11936–11941. doi: 10.1073/pnas.1005667107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Du C, Fang M, Li Y, Li L, Wang X. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell. 2000;102:33–42. doi: 10.1016/s0092-8674(00)00008-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.