Abstract
O-Mannose glycans, a family of highly heterogeneous complex glycans account up to 30% of total O-glycans in brain. Previous synthesis and functional studies only focused on the Core M3 O-mannose glycans of α-dystroglycan which are a causative factor for various muscular diseases. In this study, a highly efficient chemoenzymatic strategy was developed that enabled the first collective synthesis of 63 Core M1 and Core M2 O-mannose glycans. This chemoenzymatic strategy features the gram-scale chemical synthesis of 5 judiciously designed core structures, and the diversity-oriented modification of the core structures with 3 enzyme modules to provide 58 complex O-mannose glycans in a linear sequence that does not exceed 4 steps. Of note, 55 of these O-mannose glycans are synthesized for the first time. Using the printed O-mannose glycan array, the human brain protein CD33, a key factor in the modulation of Alzheimer’s disease was identified as strongly binding to sialylated Core M1 and Core M2.
Keywords: O-mannose glycan, chemoenzymatic synthesis, glycosylation, dystroglycan, glycosyltransferase
α-Dystroglycan (α-DG) is a highly glycosylated membrane-associated protein with more than half of the mass contributed by glycosylation. In addition to N-linked and conventional mucin-type O-linked N−acetylgalactosamine (O-GalNAc) glycans, the O-mannose glycans have been identified as the third most common glycosylation modification on α-DG. The O-mannose glycans on α-DG are known to play essential roles in muscle structure and functions. Defects in O-mannosylation lead to hypoglycosylation of α-DG, which are a causative factor for multiple forms of congenital muscular dystrophy (CMD).1 The O-mannose glycans of α-DG are also demonstrated to be involved in mesenchymal stem cell differentiation, lymphocytic choriomeningitis virus (LCMV) and Lassa fever virus (LFV) infection, and cancer metastasis.2
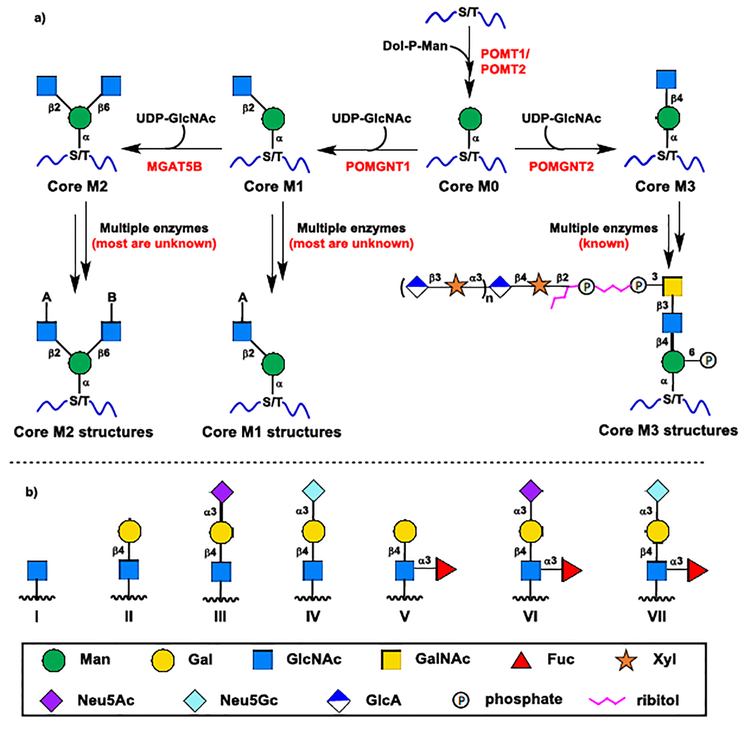
The O-mannose glycan structures of α-DG are highly heterogeneous with various branching structures linked to three O-mannose Cores (Core M1–3 in Scheme 1).1 The Core M3 can be further elaborated to a glycosaminoglycan-like polysaccharide−containing structure (Scheme 1).1,3 The Core M3 structures have been exclusively identified on α-DG and have been confirmed as the only O-mannose Core structures capable of directly binding to extracellular matrix (ECM) components.1 The Core M1 and Core M2 structures are far more abundant and diverse than the Core M3 structures on α-DG, and these highly heterogeneous and complex structures have also been characterized in many other brain glycoproteins.1,4 However, the biosynthetic pathway and the functional roles of the branching structures remain poorly understood.1 The Lewis x (Lex, V), sialyl Lewis x (sLex, VI and VII) with one of the two sialic acid forms N−acetylneuraminic acid (Neu5Ac) and N−glycolylneuraminic acid (Neu5Gc), and their substructures I–IV are the most common branching structures for Core M1 and Core M2 glycans (GlcNAc-A or GlcNAc-B in Scheme 1b).1,4 The sulfated I–VII and a sulfated trisaccharide human natural killer-1 epitope (HNK-1) have also been identified as the branching structures for Core M1 and Core M2.1,4 So far, only over 20 Core M1 and Core M2 structures have been well characterized. However, according to the known branching structures I–VII and their high abundance (up to 30% of total O-glycans) in brain glycoproteins, there are at least 56 putative non-sulfated Core M1 and Core M2 structures with appendant branching structures I–VII potentially existing in various O-mannosylated proteins, including 7 Core M1 structures (Scheme 4b), 7 symmetrical Core M2 structures (Scheme 4c) and 42 asymmetrical Core M2 structures (Scheme 5)
Scheme 1.
a) Biosynthetic pathway of α-dystroglycan O-mannose glycans; b) Seven common non-sulfated branching structures of Core M1 and Core M2
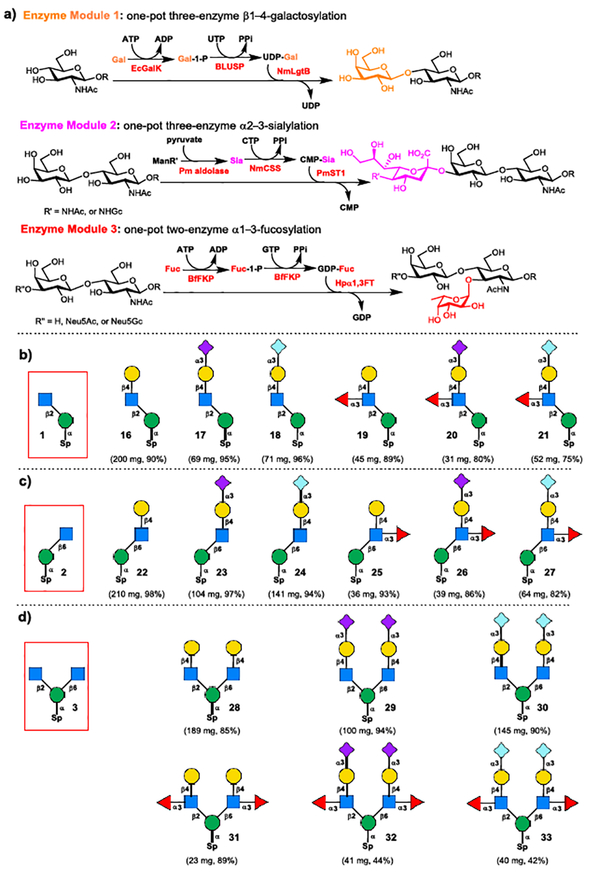
Scheme 4.
a) Three enzyme modules for enzymatic diversification; b) Enzymatic assembly of Core M1 O-mannose glycans 16−21 from 1; c) Enzymatic assembly of C6-branched Core M1 O-mannose glycan isomers 22−27 from 2; d) Enzymatic assembly of symmetrical Core M2 O-mannose glycans 28−33 from 3.
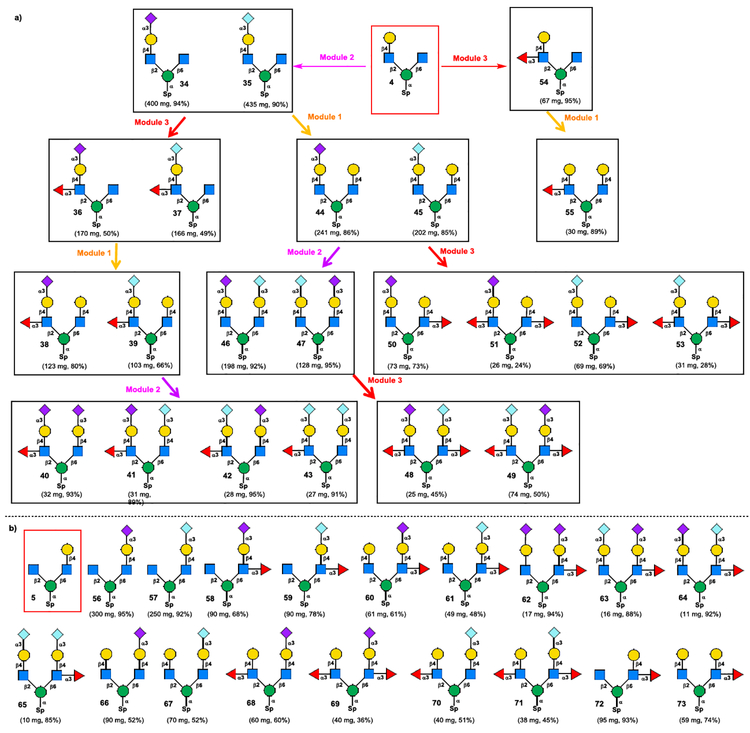
Scheme 5.
a) Enzymatic modular assembly of C2 “heavy chain”-branched asymmetrical Core M2 glycans 34−55 from tetrasaccharide 4 (50 and 51 were derived from 44 in one-pot, 52 and 53 were derived from 45 in one-pot); b) Enzymatic modular assembly of C6 “heavy chain”-branched asymmetrical Core M2 glycans 56−73 from tetrasaccharide 5 (68 and 69 were derived from 66 in one-pot, 70 and 71 were derived from 67 in one-pot).
To decipher the puzzling biosynthetic pathway, and disclose the functional roles of these highly heterogeneous O-mannose glycans in many brain glycoproteins, a collection of all putative Core M1 and Core M2 structures in sufficient amounts is highly desirable. Unfortunately, the collective synthesis of these complex glycans, especially the asymmetrical Core M2 structures is very challenging. Previous chemical or chemoenzymatic synthesis mainly focused on the simple Core M1 glycans with branching structures I–III5, and the phosphorylated Core M3 trisaccharide6. Except for a symmetrical Core M2 trisaccharide core structure7, all other Core M2 symmetrical and asymmetrical structures have not been synthesized. We describe herein the first collective synthesis of 63 putative nonsulfated Core M1 and Core M2 structures of O-mannose glycans.
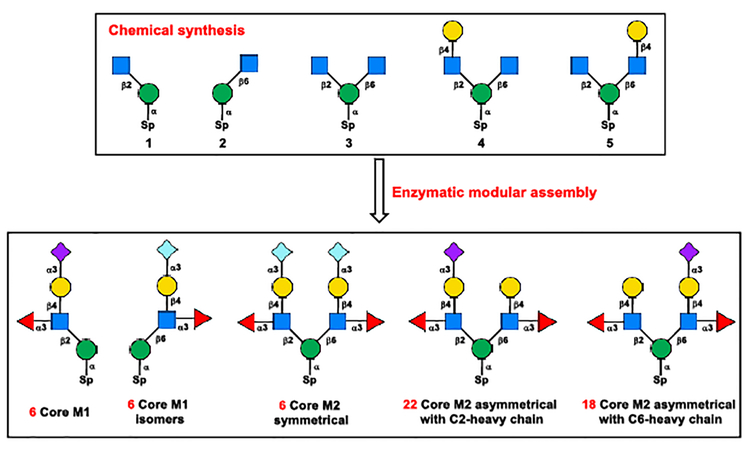
The overall chemoenzymatic synthetic plan is depicted in Scheme 2. To overcome the enzyme limitation, five chemically synthesized core structures 1–5 were designed as the key intermediates for further enzymatic diversification. The core structures 1 and 2 were designed for the enzymatic synthesis of linear Core M1 and C6-branched Core M1 isomers, respectively. The C6-branched Core M1 structures have not been identified from nature, while these structures can be used as controls and standards for future functional and enzymology studies. Trisaccharide 3 was designed to target the symmetrical Core M2 structures by synchronous enzymatic extension at both C2- and C6-branched GlcNAc, and the asymmetrical tetrasaccharides 4 and 5 were designed for the selective enzymatic extension to generate all remaining 40 putative asymmetrical Core M2 structures.
Scheme 2.
Synthetic plan.
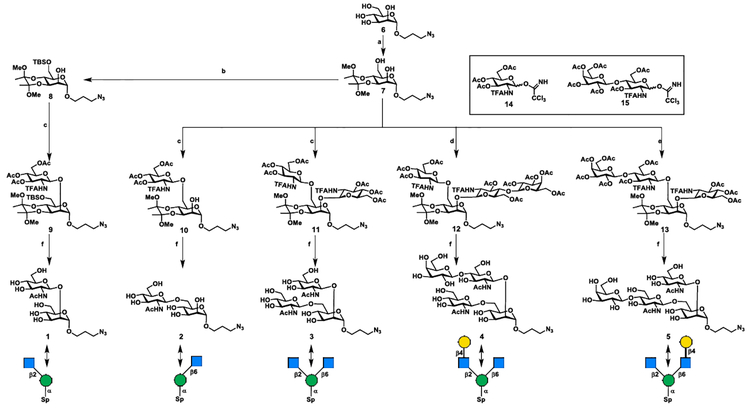
Chemical synthesis of core structures 1–5.
To acquire core structures 1–5 at gram scales for further enzymatic diversification, a concise chemical approach was designed using 2,6-diol 7, a known monosaccharide imidate 148, and a disaccharide imidate 159 as the key building blocks (Scheme 3).
Scheme 3.
Chemical assembly of core structures 1−5. (a) 2,3-butanedione, trimethyl orthoformate, (±)-camphorsulfonic acid, 110 °C, 12 h, 58%; (b) TBSCl, imidazole, DMF, rt, 1 h, 84%; (c) 14, TMSOTf, 4 Å MS, −50 °C to −20 °C, 83% for 9, 86% for 10, 85% for 11; (d) 14 (1.0 equiv), TMSOTf, 4 Å MS, −50 °C, 1 h, then 15 (1.5 equiv), −20 °C, 1 h, 65% for 2 steps; (e) 15 (1.0 equiv), TMSOTf, 4 Å MS, −50 °C, 1 h, then 14 (1.5 equiv), −20 °C, 1 h, 74% for 2 steps; (f) 1) TFA/H2O (9:1, v/v); 2) NaOMe/MeOH, then 1.0 M LiOH; 3) Ac2O/MeOH, 1 (1.8 g) 87% for 3 steps; 2 (1.88 g) 90% for 3 steps; 3 (1.13 g) 84% for 3 steps; 4 (1.58 g) 91% for 3 steps; 5 (1.41 g) 90% for 3 steps.
It was envisaged that the 2,6-diol 7 could serve as the only acceptor required for the synthesis of core structures 2–5 via regio- and stereoselective glycosylation (Scheme 3). The regioselective glycosylation at C6-OH or double glycosylation at 2,6-diol of 7 with imidate 14 provided C6-branched disaccharide 10 (81%) or C2 and C6-branched trisaccharide 11 (85%), respectively. Taking advantage of the difference in reactivity between the primary C6-OH and the secondary C2-OH of 7, a one-pot sequential glycosylation approach was applied for the synthesis of asymmetrical tetrasaccharide isomers 12 and 13 by simply changing the glycosylation sequence of donor 14 and 15. This operationally simple one-pot double glycosylation approach provided tetrasaccharides 12 (65% for two steps) and 13 (75% for two steps) in good yields. For the synthesis of fully protected Core M1 disaccharide 9, the 2,6-diol 7 was selectively protected with tert−butyldimethylsiyl (TBS) at C6-OH. The resulting mannoside 8 was glycosylated with imidate 14, yielding the desired disaccharide 9 in 70% yield for two steps. Next, a unified 3-step procedure was applied for the global deprotection of 9–13 to provide core structure 1–5. In this manner, the gram-scale synthesis of all 5 designed core structures 1–5 was achieved, which makes it possible to explore the enzymatic diversification in the next stage.
Enzymatic diversification of core structures 1–5 for the collective synthesis of Core M1 and Core M2 O-mannose glycans 16–71.
Previous, we developed three enzyme modules to target three different glycosidic linkages presenting in branching structures II–VII (Scheme 1) for the synthesis of serine-appended Core M1 O-mannose glycans.8 By taking advantage of the promiscuous substrate specificity of bacterial enzymes, we anticipate these enzyme modules could also be applied for the synthesis of sterically hindered Core M2 structures. As shown in Scheme 4a, the enzyme module 1 is a one-pot three-enzyme system that was designed for β1–4-galactosylation to form the Galβ1–4GlcNAc (LacNAc) moiety.10 The enzyme module 2 is another one-pot three-enzyme system which is designed for the α2–3-sialylation to form both Neu5Acα2–3Gal and Neu5Gcα2–3Gal units.11 The enzyme module 3 is a one-pot two-enzyme system which is applied for the α1–3-fucosylation to form the Fucα1–3GlcNAc linkage.12
Enzymatic diversification of core structures 1–5 for the synthesis of diverse Core M1 and Core M2 structures was commenced with the Core M1 structures 16–21. The synthesis of Core M1 structures 16–21 was achieved through stepwise enzymatic extension of disaccharide 1 following the general procedures of three enzyme modules (see Scheme S1 and Experimental section in Supporting Information for details). Accordingly, the disaccharide 1 was firstly converted to LacNAc-terminated trisaccharide 16 (200 mg, 90%) using enzyme module 1, then the purified trisaccharide 16 was used as the acceptor by enzyme module 2 for the parallel synthesis Neu5Acα2–3-linked sialoside 17 and Neu5Gcα2–3-linked sialoside 18 by using ManNAc or ManNGc as the sialic acid precursor, respectively. The isolated sialosides 17 (69 mg, 95%), 18 (71 mg, 96%), and trisaccharide 16 were elaborated parallelly using enzyme module 3 to yield Lex−containing structure 19 (45 mg, 89%), sLex−containing Core M1 structures 20 (31 mg, 80%) and 21 (52 mg, 75%).
The same stepwise assembly procedure was applied to the synthesis of C6-branched Core M1 isomers 22–27 from disaccharide 2, and symmetrical Core M2 O-mannose glycans 28–33 from trisaccharide 3. All these compounds were excellent substrates for 3 enzyme modules affording C6-branched Core M1 isomers 22–27 (Scheme 4c) and symmetrical Core M2 O-mannose glycans 28–33 (Scheme 4d) in good yields (see Scheme S2–S3 and Experimental section in Supporting Information for details).
The asymmetric Core M2 glycans were divided into two subgroups in our synthetic plan according to the number of monosaccharide units on C2- and C6-branched structures. As shown in Scheme 5a, the first group with C2-branched “heavy chain” as the C2-branch has greater or equal number of sugar units to C6-branched structures. The C2 “heavy chain”-branched Core M2 asymmetric structures 34–55 can be derived from chemically prepared asymmetric tetrasaccharide 4 (Scheme 5a). In contrast, the C6 “heavy chain”-branched Core M2 asymmetric structures 56–73 can be assembled from tetrasaccharide 5 (Scheme 5b). The overall synthetic scheme for the assembly of C2 “heavy chain”-branched Core M2 asymmetric structures 34–49 was depicted in Scheme 5a. Starting from tetrasaccharide 4, the sequential glycosylation using three enzyme modules (Enzyme modules 1–3 in Scheme 4a) afford 22 diverse complex Core M2 asymmetric structures 34–55 in 25–435 mg scales in a linear sequence that does not exceeded 4 steps (see Experimental section in Supporting Information for details). Following the similar enzymatic assembly strategy, the C6 “heavy chain”-branched Core M2 asymmetric structures 56–73 were also obtained in good yields (Scheme 5b, see Scheme S4 and Experimental section in Supporting Information for details).
Recognition of sialylated O-mannose glycans by human CD33-Fc (Siglec-3).
Despite the fact O-mannose glycans account for up to one-third of all O-linked glycans in the brain, and more than 50 O-mannosylated proteins have been identified, α-DG is the only O-mannosylated protein that has been intensively studied.1 Most recently, the biosynthetic pathway of Core M3 O-mannose glycans and their functional roles in various congenital muscular dystrophies have been well established.1 In contrast to Core M3 O-mannose glycans which exclusively presented on α-DG, the diverse and complex Core M1 and Core M2 O-mannose glycans have been identified on a number of brain proteins. However, the biosynthetic pathway of Core M1 and Core M2 O-mannose glycans have not been fully elucidated, and their biological functions or cognate receptors in the brain tissues are still unknown. The high prevalence of O-mannose glycans in brain encouraged us to screen their potential binding receptors with the printed microarray of our systematically synthesized Core M1 and Core M2 glycans. Among three tested brain proteins, only the recombinant human Fc-fusion CD33 (hSiglec-3)13 showed strong binding to all the sialylated O-mannose glycans. The response is very specific for sialic acids, and no signal of binding was displayed by the non-sialylated glycans (Figure S1). Human L1 cell adhesion molecule (L1CAM/CD171) and human myelin-associated glycoprotein (MAG/Siglec-4a) were also screened with O-mannose glycan array, however, no significant binding were exhibited for these two human brain proteins.
In summary, an efficient chemoenzymatic approach has been developed for the diversity-oriented assembly of complex Core M1 and Core M2 O-mannose glycans. Five core structures 1–5 designed to target different O-mannose glycan subgroups were obtained at gram-scale through concise chemical synthesis. The enzymatic diversification of 5 readily accessible core structures 1–5 were performed with 4 simple monosaccharides (Gal, ManNAc, ManNGc and Fuc) as donor precursors and 3 enzyme modules through diversity-oriented parallel enzymatic glycosylation. These provided 58 Core M1 and Core M2 structures in a linear sequence that does not exceeded 4 steps. All 8 recombinant enzymes in 3 enzyme modules are cloned from bacteria, have broad substrate specificities, and can be expressed at large scales in a conventional E. coli expression system, thus making the described synthesis of complex O-mannose glycans readily accessible. The 63 diverse chemoenzymatically synthesized O-mannose glycans, 55 of them are synthesized for the first time, constitute the most comprehensive O-mannose glycan library to date. These systematically synthesized O-mannose glycans enable to explore their unknown functions in brain. Using the printed O-mannose glycan array, the human brain protein CD33, a key factor in the modulation of Alzheimer’s disease, was identified as strongly binding to sialylated Core M1 and Core M2. This unique binding profile observed here indicates the O-mannose glycans may play roles in the neurodegenerative disease, and also provides important information on design O-mannose glycan-based probes to target the sterically rigid binding pocket of CD33.
Supplementary Material
Acknowledgements
We thank Prof. Xi Chen at the University of California at Davis for sharing plasmids and providing helpful discussions, and Sandra Diaz at UCSD for technical help. This project was financially supported by National Natural Science Foundation of China (Grant Nos. 21372145, 21672128 and 21702123), State Key Laboratory of Microbial Technology (M2016–06), Department of Science and Technology of Shandong Province (2016GGH4502, 2016GSF121002) and NIH grant R01GM032373.
Footnotes
Experimental Section
Experimental Details are available in Supporting information.
References
- [1].For recent reviews: Sheikh MO, Halmo SM, Wells L, Glycobiology 2017, 27, 806; Endo T, J. Biochem 2015, 157, 1; Praissman JL, Wells L, Biochemistry 2014, 53, 3066.
- [2].a) Ragni E, Lommel M, Moro M, Crosti M, Lavazza C, Parazzi V, r S, Strahl S, Lazzari L, Cell. Mol. Life Sci 2016, 73, 445–458; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Cao W, Henry MD, Borrow P, Yamada H, Elder JH, Ravkov EV, Nichol ST, Compans RW, Campbell KP, Oldstone MB, Science 1998, 282, 2079; [DOI] [PubMed] [Google Scholar]; c) Jae LT, Raaben M, Riemersma M, Beusekom E, Blomen VA, Velds A, Kerkhoven RM, Carette JE, Topaloglu H, Meinecke P, Science 2013, 340, 479; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Imperiali M, Thoma C, Pavoni E, Brancaccio A, Callewaert N, Oxenius A, J. Virol 2005, 79, 14297; [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Esser AK, Miller MR, Huang Q, Meier MM, Beltran-Valero de Berna D, Stipp CS, Campbell KP, Lynch CF, Smith BJ, Cohen MB, Henry MD, J. Biol. Chem 2013, 288, 2132; [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Bao X, Kobayashi M, Hatakeyama S, Angata K, Gullberg D, Nakayama J, Fukuda MN, Fukuda M, Proc. Natl. Acad. Sci. U. S. A 2009, 106, 12109; [DOI] [PMC free article] [PubMed] [Google Scholar]; g) Muschler J, Levy D, Boudreau R, Henry M, Campbell K, Bissell MJ, Cancer Res 2002, 62, 7102; [PubMed] [Google Scholar]; h) Dobson CM, Hempel SJ, Stalnaker SH, Stuart R, Wells L, Cell. Mol. Life Sci 2013, 70, 2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].a) Praissman JL, Willer T, Sheikh MO, Toi A, Chitayat D, Lin Y-Y, Lee H, Stalnaker SH, Wang S, Prabhakar PK, Nelson SF, Stemple DL, Moore SA, Moremen KW, Campbell KP, Wells L, eLife 2016, 5, e14473; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kanagawa M, Kobayashi K, Tajiri M, Manya H, Kuga A, Yamaguchi Y, Akasaka-Manya K, Furukawa J.-i., Mizuno M, Kawakami H, Shinohara Y, Wada Y, Endo T, Toda T, Cell Rep 2016, 14, 2209; [DOI] [PubMed] [Google Scholar]; c) Gerin I, Ury B, Breloy I, Seraphin C, Bolsée J, Halbout M, Graff J, Vertommen D, Muccioli GG, Seta N, Cuisset J-M, Dabaj I, Quijano-Roy S, Grahn A, Schaftingen EV, Bommer GT, Nat. Commun 2016, 7, 11534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].a) Vester-Christensen MB, Halim A, Joshi HJ, Steentoft C, Bennett EP, Levery SB, Vakhrushev SY, Clausen H, Proc. Natl. Acad. Sci. U. S. A 2013, 110, 21018; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Lommel M, Winterhalter PR, Willer T, Dahlhoff M, Schneider MR, Bartels MF, Renner-Mueller, Ruppert T, Wolf E, Strahl S, Proc. Natl. Acad. Sci. U. S. A 2013, 110, 21024; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Chai W, Yuen CT, Kogelberg H, Carruthers RA, Margolis RU, Feizi T, Lawson AM, FEBS J 1999, 263, 879; [DOI] [PubMed] [Google Scholar]; d) Dwyer CA, Katoh T, Tiemeyer M, Matthews RT, J. Biol. Chem 2015, 290, 10256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].a) Yu J, Westerlind U, ChemBioChem 2014, 15, 939; [DOI] [PubMed] [Google Scholar]; b) Sardzik R, Green AP, Laurent N, Both P, Fontana C, Voglmeir J, Weissenborn MJ, Haddoub R, Grassi P, Haslam SM, Widmalm G, Flitsch SL, J. Am. Chem. Soc 2012, 134, 4521; [DOI] [PubMed] [Google Scholar]; c) Seifert J, Ogawa T, Kurono S, Ito Y, Glycoconj. J 2000, 17, 407; [DOI] [PubMed] [Google Scholar]; d) Seifert J, Ogawa T, Ito Y, Tetrahedron Lett 1999, 40, 6803; [Google Scholar]; e) Matsuo I, Isomura M, Ajfsaka K, Tetrahedron Lett 1999, 40, 5050. [Google Scholar]
- [6].Mo KF, Fang T, Stalnaker SH, Kirby PS, Liu M, Wells L, Pierce M, Live DH, Boons GJ, J. Am. Chem. Soc 2011, 133, 14418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Yu J, Grant OC, Pett C, Stahl S, Woods RJ, Westerlind U, Chem. Eur. J 2017, 23, 3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zhang Y, Meng C, Jin L, Chen X, Wang F, Cao H, Chem. Commun 2015, 51, 11654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Yao W, Yan J, Chen X, Wang F, Cao H, Carbohydr. Res 2015, 401, 5. [DOI] [PubMed] [Google Scholar]
- [10].a) Chen X, Fang J, Zhang J, Liu Z, Shao J, Kowal P, Andreana P, Wang PG, J. Am. Chem. Soc 2001, 123, 2081; [DOI] [PubMed] [Google Scholar]; b) Muthana MM, Qu J, Li Y, Zhang L, Yu H, Ding L, Malekan H, Chen X, Chem. Commun 2012, 48, 2728; [DOI] [PubMed] [Google Scholar]; c) Lau K, Thon V, Yu H, Ding L, Chen Y, Muthana MM, Wong D, Huang R, Chen X, Chem. Commun 2010, 46, 6066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].a) Li Y, Yu H, Cao H, Lau K, Muthana S, Tiwari VK, Son B, Chen X, Appl. Microbiol. Biotechnol 2008, 79, 963; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Yu H, Yu H, Karpel R, Chen X, Bioorg. Med. Chem 2004, 12, 6427; [DOI] [PubMed] [Google Scholar]; c) Sugiarto G, Lau K, Qu J, Li Y, Lim S, Mu S, Ames JB, Fisher AJ, Chen X, ACS Chem. Boil 2012, 7, 1232; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Yu H, Chokhawala H, Karpel R, Yu H, Wu BY, Zhang JB, Zhang YX, Jia Q, Chen X, J. Am. Chem. Soc 2005, 127, 17618. [DOI] [PubMed] [Google Scholar]
- [12].a) Zhao G, Guan W, Cai L, Wang PG, Nat. Protoc 2010, 5, 636; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Sugiarto G, Lau K, Yu H, Vuong S, Thon V, Li Y, Huang S, Chen X, Glycobiology 2011, 21, 387; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Wang W, Hu T, Frantom PA, Zheng T, Gerwe B, del Amo DS, Garret S, Seidel RD III, Wu P, Proc. Natl. Acad. Sci. U. S. A 2009, 106, 16096; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Sun HY, Lin SW, Ko TP, Pan JF, Liu CL, Lin CN, Wang AH, Lin CH, J. Biol. Chem 2007, 282, 9973; [DOI] [PubMed] [Google Scholar]; e) Lin SW, Yuan TM, Li JR, Lin CH, Biochemistry 2006, 45, 8108. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.