Abstract
Plant shoot stem cell pool is constantly maintained by a negative feedback loop through peptide-receptor mediated signaling pathway. CLAVATA3 (CLV3) encode a 96 amino-acid protein which is processed to 12-amino-acid or arabinosylated 13-amino-acid peptides, acting as a ligand signal to regulate stem cell homeostasis in the shoot apical meristem (SAM). Although arabinosylated 13-amino-acid CLV3 peptide (CLV3p) shows more significant binding affinity to its receptors and biological activities in the SAM, the physiological function of two mature forms of CLV3p remained an unresolved puzzle in the past decade due to the technical difficulties of arabinosylation modification in the peptide synthesis. Here, we analyzed the role of two mature CLV3 peptides with newly synthesized arabinosylated peptide. Beside shoot meristem phenotypes, arabinosylated CLV3p showed the conventional trait of CLV2-dependent root growth inhibition. Moreover, both 12-amino-acid and arabinosylated 13-amino-acid CLV3 peptides have analogous activities in shoot stem cell signaling. Notably, we demonstrated that non-arabinosylated 12-amino acid CLV3p can affect shoot stem cell signaling at the physiological level unlike previously suggested (Ohyama et al., 2009; Shinohara and Matsubayashi, 2013; Shinohara and Matsubayashi, 2015). Therefore, these results support the physiological role of the 12-amino-acid CLV3p in shoot stem cell signaling in the deficient condition of arabinosylated 13-amino-acid CLV3p in Arabidopsis thaliana.
Keywords: Arabidopsis thaliana, CLAVATA3, Peptide modification, Shoot apical meristem, Stem cell signaling
Introduction
A stable stem cell pool in the shoot apical meristem (SAM) is the self-renewable reservoir to generate new cells for leaf, stem and flower organogenesis, and is dynamically maintained by complex regulatory networks in plants. The CLAVATA-WUSHCEL (CLV-WUS) feedback signaling pathway, first discovered in Arabidopsis thaliana, appears to be conserved for stem cell control in the SAM of higher plants (Somssich et al., 2016; Yamaguchi et al., 2016). The CLAVATA3 peptide (CLV3p) produced in the stem cells of the shoot apex functions as a signaling ligand of the CLAVATA1 (CLV1) and related receptor kinases to restrict the level of WUS transcription factor in the organizing center (OC), which promotes CLV3 expression via cell-to-cell communication through plasmodesmata and the stem cell fate but represses differentiation (Fletcher et al., 1999; Yadav et al., 2011; Yadav et al., 2013; Lee, 2014; Somssich et al., 2016; Yamaguchi et al., 2016). The signaling components between CLV3-CLV1 and WUS remains enigmatic despite great efforts in the past two decades. Although loss-of-function and various genetic manipulations of CLV3 expression have facilitated the analysis of long-term phenotypes, exogenous application of physiological CLV3 peptides complementing clv3 mutants will aid in the discovery of primary and dynamic processes in peptide-receptor signaling (Somssich et al., 2016; Yamaguchi et al., 2016).
Mass spectrometry (MS) analyses have identified two forms of mature CLV3 peptides. The first mature form is the 12-amino-acid CLV3 peptide (MCLV3p) by in situ MALDI-TOF MS analyses from CLV3 overexpression calli with hydroxyproline residues in the 4th and 7th positions. MCLV3p binds to the ectodomain of CLV1 in vitro with the dissociation constant (Kd) values of 17.5-24 nM (Kondo et al., 2006; Ogawa et al., 2008; Ohyama et al., 2009). A secreted arabinosylated 13-amino-acid peptide [Ara3]CLV3p is also identified in liquid medium using nano-LC-MS/MS from CLV3 overexpression seedlings (Ohyama et al., 2009). Because the high affinity (Kd=1 nM) of the purified [Ara3]CLV3p binding to the ectodomain of CLV1 in vitro, it has been assumed that the glycopeptide is the physiological ligand in CLV3-CLV1 signaling (Ohyama et al., 2009). When comparing the biological effects of chemically synthesized peptides in clv3-1 with enlarged SAM, synthetic [Ara3]CLV3p at 30-100 nM leads to SAM size reduction but not the non-arabinosylated 13-amino-acid CLV3p with much lower binding affinity to CLV1 (Kd=280 nM) (Shinohara and Matsubayashi, 2013). However, the performance of chemically synthesized 12-amino-acid MCLV3p and 13-amino-acid [Ara3]CLV3p have not been carefully compared due to the difficulties of determining the stereoselective nature of glycosidic linkages in chemical synthesis of the pure [Ara3]CLV3 peptides (Shinohara and Matsubayashi, 2013). Inevitably, high concentrations of synthetic CLV3/EMBRYO SURROUNDING REGION-related (CLE) peptides in the range of 1-100 μM have been commonly applied in the measurement of SAM size in biossays in different plant species, leading to concerns about biological relevance of the findings (Somssich et al., 2016; Yamaguchi et al., 2016).
Three studies spanning the past decade have led to unresolved puzzles regarding the physiological function of arabinosylation in the Arabidopsis CLV3 peptides (Kondo et al., 2008; Song et al., 2012; Song et al., 2013; MacAlister et al., 2016). Alanine-scan analyses were conducted using synthetic MCLV3 peptide variants in bioassays and in vitro competitive binding with the CLV1 ectodomain (Kondo et al., 2008), or using in vivo complementation of the SAM size and WUS expression in clv3-2 transgenic plants expressing MCLV3 peptide variants from CLV3 transgenes (Song et al., 2012). These systematic analyses of peptide mutants in vitro and in vivo failed to support a critical biological role of the 7th hydroxyproline residue, where three arabinoses are covalently attached (Kondo et al., 2008; Ohyama et al., 2009; Song et al., 2012; Shinohara and Matsubayashi, 2013). Although recent findings on hydroxyproline O-arabinosyltransferases (HPATs), responsible for the arabinosylation of proteins or peptides, have confirmed the importance of peptide arabinosylation in tomato plants (Xu et al., 2015), the thorough genetic and phenotypic analyses of the single, double and triple Arabidopsis hpat1,2,3 mutants do not support a role of HPATs in stem cell signaling in the SAM (MacAlister et al., 2016).
In this study, we functionally analyzed the role of newly synthesized the arabinosylated 13-amino-acid CLV3p in shoot stem cell signaling. Our results revealed similar biological activities between MCLV3p and [Ara3]CLV3p in physiological conditions. These results support the idea of the redundant role of different in vivo CLV3 peptides in the Arabidopsis shoot stem cell signaling and development.
Results
Effect of Chemically Synthesized Arabinosylated CLV3 Peptide in Root Growth Inhibition
As the activity of MCLV3p and [Ara3]CLV3p at physiological concentrations in bioassays is a prerequisite for integrated analyses of dynamic CLV3 peptide signaling, we chemically synthesized [Ara3]CLV3p (Fig. 1A, B; see Materials and Methods) and MCLV3p. Because of the difficulty in the chemical synthesis of glycosylated CLV3 peptides, we tested whether newly synthesized [Ara3]CLV3p has a conventional property as a CLV3 signal. The biological activities of [Ara3]CLV3p and MCLV3p were first examined in parallel using the well-established and sensitive inhibitory assays in the root apical meristem (RAM) (Kondo et al., 2006; Kondo et al., 2008; Kondo et al., 2011). Both [Ara3]CLV3p and MCLV3p but not unarabinosylated 13-amino-acid CLV3p displayed similar and prominent root growth inhibition at 10-100 nM and this inhibition was almost saturated 1 μM or higher concentrations (Fig. 1C) (Kondo et al., 2006; Kondo et al., 2008; Lee et al., 2012). This result indicates that [Ara3]CLV3p has similar biological activity in addition to MCLV3p in the consumption of root meristems via the misspecification of cell type (Fiers et al., 2005). Because the primary root growth inhibition by [Ara3]CLV3p as well as previously reported MCLV3p was not observed in the clv2-1 mutant seedlings (Fig. 1D, E) (Lee et al., 2012), the result demonstrates that newly synthesized [Ara3]CLV3p also possesses a typical biological trait known as CLV2-dependent root growth inhibition (Fiers et al., 2005).
Fig. 1.
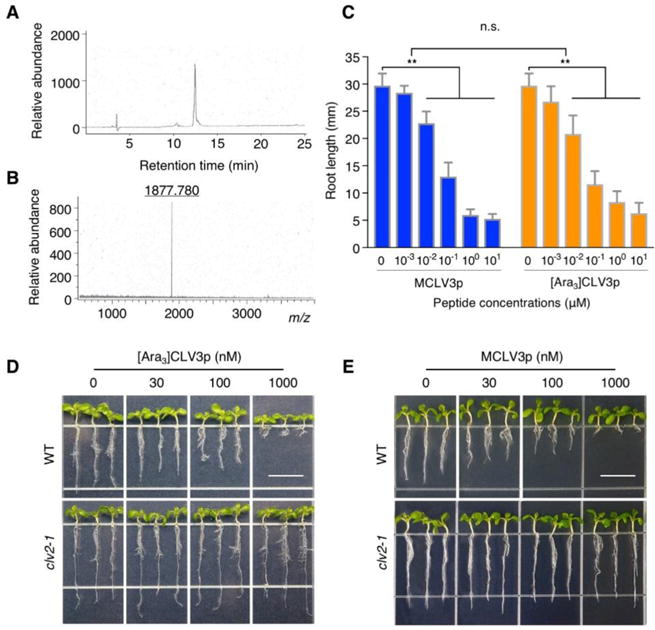
In vivo activity of chemically synthesized arabinosylated CLV3 peptides.
(A) HPLC purification of the chemically synthesized [Ara3]CLV3 peptide.
(B) Selected ion at m/z 1877.780 that corresponds to the 13-amino-acid [Ara3]CLV3 peptide in MS analysis.
(C) Similar root growth inhibition mediated by MCLV3p and [Ara3]CLV3p. Ler seedlings were treated with synthetic peptides (0, 1, 10, 100, 1000, 10,000 nM) for 14 days. Error bars represent SD from repeats (n = 10). ** indicates significant difference (two-way ANOVA test, p<0.001). n.s., not significant.
(D, E) CLV2-dependent root growth inhibition mediated by [Ara3]CLV3p and MCLV3p. WT Ler and clv2-1 mutant seedlings were treated without or with 30 nM, 100 nM and 1000 nM [Ara3]CLV3p or MCLV3p for 7 days. Scale bar = 10 mm.
Effect of chemically synthesized CLV3 peptides in Shoot Growth Arrest
We next determined the effect of both CLV3 peptides on stem cell signaling in the SAM by applying a physiological range of exogenous MCLV3p and [Ara3]CLV3p. The CLV-WUS negative feedback loop controls the homeostasis of stem cell population in the SAM, and it therefore affects the SAM size depending on the balance between cell division and differentiation. To investigate more sensitive effect of CLV3p-mediated SAM growth arrest, we used and measured the size of shoot meristems of clv3-2, which is a null mutant lacking the endogenous CLV3 peptide signal (Fletcher et al., 1999; Ohyama et al., 2009; Song et al., 2012; Shinohara and Matsubayashi, 2013; Xu et al., 2015). clv3-2 mutant seedlings were treated with each type of CLV3p with various concentrations for 7 days and then we measured the width and the height between two franking primordia in the SAM after sectioning paraffin-embedded tissues (Fig. S1). Surprisingly, unlike previous reports (Shinohara and Matsubayashi, 2013; Shinohara and Matsubayashi, 2015), both MCLV3p and [Ara3]CLV3p at 30-100 nM similarly reduced the enlarged SAM in clv3-2 to the size in WT (Fig. 2A, B). It is possible that different clv3 alleles and experimental conditions make different effects. Notably, these results demonstrate that MCLV3p can also reduce meristem size at physiologically low concentration in addition to [Ara3]CLV3p (Shinohara and Matsubayashi, 2013). Consistent with a key role of CLV3p signaling in controlling stem cell numbers in the SAM (Je et al., 2016; Mandel et al., 2016; Somssich et al., 2016; Yamaguchi et al., 2016), CLV3p-mediated reduction of the shoot meristem size was most significantly reflected in the height of the clv3-2 SAM rather than the width (Fig. 2A, B). In particular, we could observe the different penetrance between [Ara3]CLV3p and MCLV3p in causing the depletion of shoot stem cell population. About 18.2 % of clv3-2 seedlings treated with 100 nM [Ara3]CLV3p but not MCLV3p were manifested as a flat structure in the region of the SAM (Fig. S1 and Table 1). Consistent with this, the slightly enlarged shoot meristems in well-known CLV3 receptor mutants, such as clv1 or barely any meristem 1 (bam1), were also similarly reduced by the treatment of two kinds of CLV3 peptides (Fig. S2). Taken together, these results indicate that both MCLV3p and [Ara3]CLV3p have similar biological activities on SAM growth and development despite of more sensitive effect of [Ara3]CLV3p.
Fig. 2.
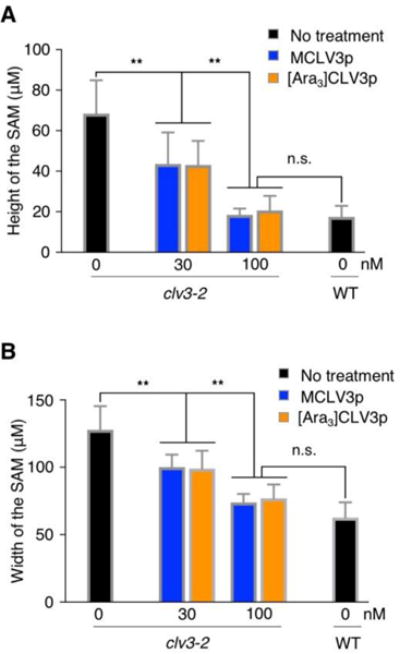
MCLV3p and [Ara3]CLV3p complement the SAM size in clv3-2
(A, B) The size of shoot meristems was quantified by measuring the height (A) or the width (B). Error bars represent SD from biological repeats (n = 11). ** indicates significant difference (two-way ANOVA test, p<0.001). n.s., not significant.
Table 1.
Shoot meristem growth arrested by different CLV3 peptides.
| Genotype | Peptide | Width (μm) | Height (μm) | n | |
|---|---|---|---|---|---|
| Ler | no treatment | 62.5 ± 13.0 | 17.4 ±6.8 | 11 | |
|
| |||||
| clv3-2 | no treatment | 127.8 ±19.5 | 68.3 ± 17.5 | 11 | |
|
| |||||
| clv3-2 | MCLV3p | 30 nM | 100.7 ±11.9 | 43.6 ±16.1 | 11 |
|
| |||||
| 100 nM | 72.3 ±9.9 | 18.6 ±4.3 | 11 | ||
|
| |||||
| [Ara3] CLV3p | 30 nM | 98.9 ±14.3 | 43.3 ± 12.4 | 11 | |
|
| |||||
| 100 nM | 78.0 ± 10.6 | 20.2 ±6.6 | 11* | ||
Some (18.2 %) of observed SAMs showed the flat structure.
Effect of chemically synthesized CLV3 peptides in the Regulation of Downstream Genes
To investigate molecular responses stimulated by MCLV3p and [Ara3]CLV3p, we also performed reverse transcription-quantitative polymerase chain reaction (RT-qPCR) using the SAM tissues from clv3-2 seedlings, which possess higher basal level of WUS expression by the deficient endogenous CLV3 peptide signal for repressing WUS expression in the CLV-WUS pathway (Lee, 2014; Somssich et al., 2016). When we applied 100 nM of two different batches of MCLV3p and [Ara3]CLV3p for 6 h, both chemically synthesized peptides equivalently decreased about 60 % of WUS expression in clv3-2 (Fig. 3A), which correlated well with the reduced height in the SAM size (Fig. 2A and Table 1). Interestingly, the reduced WUS level in the clv3-2 SAM caused by the treatment of two kinds of CLV3 peptides seemed to be quite similar to the endogenous level of WUS gene in the WT Ler SAM (Fig. 3A and Fig. S3A). Consistently, the treatment of 100 nM MCLV3p or [Ara3]CLV3p almost complemented the enlarged size of the clv3-2 SAM like that of the WT Ler SAM (Figs. 2, 3 and Fig. S3A). In addition, we detected similar reduction of other marker genes co-expressed with WUS in the SAM, such as SULFOTRANSFERASE 17 (SOT17) and ADENOSINE-5´-PHOSPHOSULFATE KINASE 2 (APK2) genes (Fig. 3B, C) (Yadave et al., 2009), which are known to be involved in the promotion of stem cell fate and proliferation in root meristems by glucose-TOR signaling (Xiong et al., 2013). It suggests the reciprocal promotion of stem cell proliferation in Arabidopsis shoot meristems. However, since the endogenous levels of SOT17 and APK2 genes in the WT Ler SAM were not correlated with the reduced expression caused by two kinds of CLV3 peptides in the clv3-2 SAM (Fig. 3B, C and Fig. S3B, C), it indicates that SOT17 and APK2 genes involved in stem cell proliferation seem to be indirectly affected by the CLV-WUS pathway.
Fig. 3.
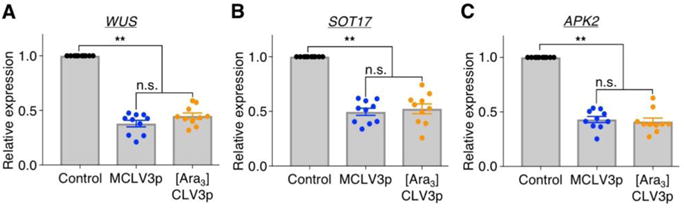
MCLV3p and [Ara3]CLV3p suppress marker genes in the SAM of clv3-2 mutant plants
(A-C) Seedlings (7 days) were treated with 100 nM CLV3 peptides for 6 h, and RNAs from the SAM tissues were analyzed by RT-qPCR. ACT2 (At3g18780) was used as an internal control to normalize the expression levels of WUS (At2g17950) (A), SOT17 (At1g18590) (B) and APK2 (At4g39940) (C). Error bars represent SEM (n = 10). ** indicates significant difference (one-way ANOVA test, p<0.001). n.s., not significant.
Discussion
Our findings with the chemically synthesized [Ara3]CLV3p and MCLV3p in this study provide compelling phenotypic and molecular evidence to support their analogous biological activities in various in vivo analyses (Figs. 1–3 and Table 1). These observations are consistent with prior in vitro and in vivo analyses supporting physiological functions of dual CLV3 peptides identified in calli and liquid medium of seedlings overexpression the Arabidopsis CLV3 transgene (Kondo et al., 2006; Kondo et al., 2008; Ohyama et al., 2009; Song et al., 2012; Song et al., 2013). Moreover, a recent study of the Arabidopsis loss-of-function mutants of three HPATs without overt SAM defect supports the likely physiological role of the 12-amino-acid MCLV3p in shoot stem cell signaling when the 13-amino-acid [Ara3]CVL3p cannot be generated (MacAlister et al., 2016). Indeed, since the arbinosylated modification of the 12-amino acid CLE2 peptide was almost disappeared even in the hpat3 single mutant (Ogawa-Ohnishi et al., 2013), the arabinosylation of CLV3p is not functionally indispensable in the Arabidopsis shoot stem cell signaling as previously suggested (Ohyama et al., 2009; Shinohara and Matsubayashi, 2013; Shinohara and Matsubayashi, 2015). Therefore, application of the synthetic 12-amino-acid MCLV3p, but not the arabinosylated 13-amino-acid CLV3p, in the clv3 null mutants will greatly simplify future research in uncovering the primary and dynamic signaling mechanisms in the CLV-WUS pathway in the SAM using the versatile Arabidopsis system (Kondo et al., 2006; Kondo et al., 2008; Song et al., 2012; Song et al., 2013).
Arabidopsis has been known to contain 32 CLE genes in the genome (Yamaguchi et al., 2016). Currently, three CLE peptides, including CLV3p, CLE2p and CLE9p, are purified with a glycosylated modification of three arabinose residues (Ohyama et al., 2009; Shinohara et al., 2012). Interestingly, only CLV3p has been revealed to have the 13-amino-acid length among Arabidopsis CLE peptides (Yamaguchi et al., 2016), suggesting that Arabidopsis has different proteolytic processing to produce different length of CLE peptides (Tabata and Sawa, 2014). In fact, it has been demonstrated that N-terminal flanking sequence of CLV3p affects its cleavage to produce two mature forms of 12-amino-acid and 13-amino-acid CLV3 peptides (Xu et al., 2013). Similar with our results, both arabinosylated/non-arabinosylated 12-amino-acid CLE9 peptides displayed similar activity in the binding affinity to the BARELY ANY MERISTEM 1 (BAM1) receptor (Shinohara et al., 2012), supporting an idea of the redundant role of two mature CLV3 peptides in Arabidopsis shoot stem cell signaling. Nevertheless, peptide arabinosylation could affect peptide conformation and receptor binding, and modulate peptide stability and trafficking in a context-dependent manner, as has been functionally proven for CLE and related peptides in other plant species (Ohyama et al., 2009; Shinohara and Matsubayashi, 2013; Xu et al., 2015; Somssich et al., 2016; Yamaguchi et al., 2016). Different environmental conditions may influence HPAT or other AT enzyme activity or expression at different developmental stages. It remained to be clarified whether peptides with or without arabinosylation will lead to differential signaling via distinct receptors in the Arabidopsis SAM signaling and development.
Materials and Methods
Plant materials and growth conditions
Landsberg erecta (Ler) ecotype was used as wild-type (WT) Arabidopsis plants in this study. The clv2-1 and clv3-2 are in Ler background. The mutation of clv2-1 causes the early stop codon at the 33rd residue and is a null allele (Jeong et al., 1995). The γ-ray-induced clv3-2 is a presumed null allele (Fletcher et al., 1999). For liquid culture of Arabidopsis seedlings, 10 seeds were germinated and grew in 6-well plates containing 1 ml of liquid medium (0.5 × MS and 0.5% sucrose, pH 5.8 adjusted with KOH). To quantify the primary root length of WT in various concentrations of peptides, seedlings were grown in liquid medium for two weeks (12-h light/12-h dark, 100 μmol m−2 s−1, 22°C±1) (Fig. 1C). To determine the root growth inhibition by [Ara3]CLV3p in clv2-1, seedlings were grown in liquid medium with various concentrations of peptides for seven days (16-h light/8-h dark, 100 μmol m−2 s−1, 22°C±1) (Fig. 1D). The SAM tissues from clv3-2 seedlings grown for seven days in liquid medium without or with 100 nM peptides under long days were used for RT-qPCR analysis of marker genes (Fig. 3).
CLV3p-mediated SAM arrest assay
Ten seeds of clv3-2 mutants were sowed in 6-well plates containing 1 ml of liquid medium without or with various concentrations of CLV3 peptides. Seedlings were grown at 22°C under the long day condition for seven days. The SAM tissues were harvested and fixed in 1X PBS/4% (w/v) paraformaldehyde/4% (v/v) DMSO at 4°C for overnight. Collected samples were dehydrated through ethanol series (30%, 40%, 50%, 60%, 70%, 85%, 95% for 1 h in each step) at 4°C, and were stained with 0.1% Eosin Y (Sigma) in 100% ethanol at 4°C for overnight. Ethanol was changed through histoclear series (75% ethanol:25% histoclear, 50% ethanol:50% histoclear, 25% ethanol:75 % histoclear for 30 min in each step and 100% histoclear twice for 1 h in each step) (National Diagnostics). Histoclear was then gradually replaced with melted paraffin (Leica) at 60°C. Replacement of freshly melted paraffin was performed once a day for four days. Paraffin embedded tissues were poured into the mold and adjusted in appropriate position. Section was carried out with a rotary microtome (Reichert-Jung) at 8 μm thickness. Sectioned ribbons were placed on poly-prep slides (Sigma) with pre-warm water and incubated on slide warmer (Chang Shin Science, Korea) at 42°C for overnight. For meristem staining, paraffins were removed by 100% histoclear twice for 10 min. Sectioned tissues were hydrated through reverse ethanol series (100%, 95%, 80%, 60%, 30% ethanol and water for 2 min in each step). Sections were stained with 0.1% Giemsa (Sigma) for 2 min and rinsed briefly with water. Stained sections were dehydrated through ethanol series (2 min in each step). For microscopic analysis, samples were dried and mounted with Cytoseal 60 (Richard-Allan Scientific) before observing and measuring the height and width of the SAM using a microscope (Olympus X22LED).
RT-qPCR analysis
Total RNA was isolated from SAM tissues with TRIzol reagent (Invitrogen). To harvest SAM tissues, other tissues including cotyledons, hypocotyls and roots were removed using fine forceps and single-edged blades. Harvested SAM tissues were immediately frozen by liquid nitrogen and ground in a microtube using a blue pestle powered by a motor drill. First strand cDNA was synthesized from 1 μg of total RNA with M-MLV reverse transcriptase (Promega). All RT-qPCR analyses were performed by Step One Plus™ real time PCR detection system with Power SYBR® green master mix (Applied Biosystem). ACT2 (At3g18780) was used as a control gene. Following primers were used: ACT2-F: 5′-TCCCT CAGCA CATTC CAGCA GAT-3′ and ACT2-R: 5′-AACGA TTCCT GGACC TGCCT CATC-3′ for ACT2 gene expression; WUS-F: 5′- TGCAA GCTCA GGTAC TGAAT G-3′ and WUS-R: 5′- ATGAT CCATG TTTGC CCATC-3′ for WUS (At2g17950) gene expression; SOT17-F: 5′-GGAAG AGGAG GGAAA TGTTG-3′ and SOT17-R: 5′-CAGCA GGACG ATCTT CTCTA TC-3′ for SOT17 (At1g18590) gene expression, APK2-F: 5′-TCCGT ACCGG AGAGA CAGAG-3′ and APK2-R: 5′-GGATC TCTCG ACTCG CACAC-3′ for APK2 (At4g39940) gene expression.
Synthesis of [Ara3]CLV3 peptides
The [Ara3]CLV3 peptide was synthesized by solid-phase peptide synthesis using a PS3 peptide synthesizer (Protein Technology). Starting with 0.1 mmole (0.166 g) of HMP (p-hydroxymethyl phenoxymethyl polystyrene) resin (1.66 mmole g−1) (CYW, unpublished), the synthesis was performed using a stepwise Fast Moc protocol. The amino acids were introduced using the prepacked cartridges (0.4 mmole each). The Fmoc-Hyp(Ara3) was introduced using the prepacked cartridges (0.2 mmole each) and stirring overnight. After synthesis, 0.412 g peptide resin was placed in a round-bottom flask containing a micro stirring bar. The cool mixture containing 0.375 g crystalline phenol, 0.125 mL EDT, 0.25 mL thioanisole, 0.25 mL water, and 5 mL TFA was put into the flask and stirred for 1 h at 0°C. The peptide was further lyophilized. Then the peptide was rinsed with 100 mL cold ether and filtered. The peptide was purified by HPLC using a C18 column (5 μm particle size, 250 × 10 mm, Supelco) with a gradient (9% to 11% buffer B in 24 min) using buffer A (0.1% TFA in water) and buffer B (0.1% TFA in acetonitrile) at a flow rate of 2.5 mL min−1 and monitored by absorbance at 214 nm. Mass spectra were determined using a Ultraflex II TOF/TOF (Bruker). All reagents and solvents were obtained commercially as reagent grade and used without further purification. The amino acid sequence of [Ara3]CLV3p is RTVHypSGHyp(3Ara)DPLHHH. Hyp indicates a hydroxyproline residue.
Statistical analysis
Statistical analysis was performed with one-way or two-way analysis of variation (ANOVA) with Prism software (version 7.0b) and significant differences were determined by Turkey’s multiple comparison test.
Supplementary Material
Fig. S1. CLV3 peptides complement clv3-2 in the SAM homeostasis.
Fig. S2. MCLV3p and [Ara3]CLV3p similarly reduce the SAM size in clv1 and bam1 mutants.
Fig. S3. Endogenous expression levels of WUS, SOT17 and APK2 genes in the SAM of clv3-2 and WT Ler seedlings.
Acknowledgments
Work in the lab of Molecular Plant Development and Signal Network is supported by funding from the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIP) (2014R1A1A1006288) and the Next Generation Biogreen 21 Program. CYW and HMY are supported by Genomic Research Center, Academia Sinica, Taiwan. JS is supported by grants from the NIH and the NSF, and WJC Special Project RDA-Korea. We are grateful to Dr. CH Wong for his guidance and support in the chemical synthesis of the [Ara3]CLV3 peptide.
Footnotes
No conflict of interest declared.
Author’s Contributions
HL and JS designed the research and wrote the manuscript. HJK and HL performed the experiments. HMY and CYW performed the chemical synthesis of CLV3 peptides.
References
- Fiers M, Golemiec E, Xu J, van der Geest L, Heidstra R, Stiekema W, Liu C-M. The 14-amino acid CLV3, CLE19, and CLE40 peptides trigger consumption of the root meristem in Arabidopsis through a CLVATA2-dependent pathway. Plant Cell. 2005;17:2542–2553. doi: 10.1105/tpc.105.034009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science. 1999;283:1911–1914. doi: 10.1126/science.283.5409.1911. [DOI] [PubMed] [Google Scholar]
- Je BI, Gruel J, Lee YK, Bommert P, Arevalo ED, Eveland AL, Wu Q, Goldshmidt A, Meeley R, Bartlett M, Komatsu M, Sakai H, Jönsson H, Jackson D. Signaling from maize organ primordia via FASCIATED EAR3 regulates stem cell proliferation and yield traits. Nature Genet. 2016;48:785–791. doi: 10.1038/ng.3567. [DOI] [PubMed] [Google Scholar]
- Jeong S, Trotochaud AE, Clark SE. The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell. 1995;11:1925–1933. doi: 10.1105/tpc.11.10.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Sawa S, Kinoshita A, Mizuno S, Kakimoto T, Fukuda H, Sakagami Y. A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science. 2006;313:845–848. doi: 10.1126/science.1128439. [DOI] [PubMed] [Google Scholar]
- Kondo T, Nakamura T, Yokomine K, Sakagami Y. Dual assay for MCLV3 activity reveals structure-activity relationship of CLE peptides. Biochem Biophys Res Commun. 2008;377:312–316. doi: 10.1016/j.bbrc.2008.09.139. [DOI] [PubMed] [Google Scholar]
- Kondo T, Yokomine K, Nakagawa A, Sakagami Y. Analogs of the CLV3 peptide: Synthesis and structure-activity relationships focused on proline residues. Plant Cell Physiol. 2011;52:30–36. doi: 10.1093/pcp/pcq146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. Molecular signaling networks in the shoot apical meristem. J Plant Biol. 2014;57:321–326. [Google Scholar]
- Lee H, Khatri A, Plotnikov JM, Zhang X-C, Sheen J. Complexity in differential peptide-receptor signaling: Response to Segonzac et al. and Mueller et al. Commentaries. Plant Cell. 2012;24:3177–3185. doi: 10.1105/tpc.112.099259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlister CA, Ortiz-Ramirez C, Becker JD, Feijo J, Lippman ZB. Hydroxyproline O-arabinosyltransferase mutants oppositely alter tip growth in Arabidopsis thaliana and Physcomitrella patens. Plant J. 2016;85:193–208. doi: 10.1111/tpj.13079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel T, Candela H, Landau U, Asis L, Zelinger E, Carles CC, Williams LE. Differential regulation of meristem size, morphology and organization by the ERECTA, CLAVATA and class III HD-ZIP pathway. Development. 2016;143:1612–1622. doi: 10.1242/dev.129973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Shinohara H, Sakagami Y, Matsubayashi Y. Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science. 2008;319:294. doi: 10.1126/science.1150083. [DOI] [PubMed] [Google Scholar]
- Ogawa-Ohnishi M, Matsushita W, Matsubayashi Y. Identification of three hydroxyproline O-arabinosyltransferases in Arabidopsis thaliana. Nature Chem Biol. 2013;9:726–730. doi: 10.1038/nchembio.1351. [DOI] [PubMed] [Google Scholar]
- Ohyama K, Shinohara H, Ogawa-Ohnishi M, Matsubayashi Y. A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nature Chem Biol. 2009;5:578–580. doi: 10.1038/nchembio.182. [DOI] [PubMed] [Google Scholar]
- Shinohara H, Matsubayashi Y. Chemical synthesis of Arabidopsis CLV3 glycopeptide reveals the impact of hydroxyproline arabinosylation on peptide conformation and activity. Plant Cell Phyiol. 2013;54:369–374. doi: 10.1093/pcp/pcs174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara H, Matsubayashi Y. Reevaluation of the CLV3-receptor interaction in the shoot apical meristem: dissection of the CLV3 signaling pathway from a direct ligand-binding point of view. Plant J. 2015;82:328–336. doi: 10.1111/tpj.12817. [DOI] [PubMed] [Google Scholar]
- Shinohara H, Moriyama Y, Ohyama K, Matsubayashi Y. Biochemical mapping of a ligand-binding domain within Arabidopsis BAM1 reveals diversified ligand recognition mechanisms of plant LRR-RKs. Plant J. 2012;70:845–854. doi: 10.1111/j.1365-313X.2012.04934.x. [DOI] [PubMed] [Google Scholar]
- Somssich M, Je BI, Simon R, Jackson D. CLAVATA-WUSCHEL signaling in the shoot meristem. Development. 2016;143:3238–3248. doi: 10.1242/dev.133645. [DOI] [PubMed] [Google Scholar]
- Song X-F, Yu D-L, Xu T-T, Ren S-C, Guo P, Liu C-M. Contributions of individual amino acid residues to the endogenous CLV3 function in shoot apical meristem maintenance in Arabidopsis. Mol Plant. 2012;5:515–523. doi: 10.1093/mp/ssr120. [DOI] [PubMed] [Google Scholar]
- Song X-F, Xu T-T, Ren S-C, Liu C-M. Individual amino acid residues in CLV3 peptide contribute to its stability in vitro. Plant Signal Behav. 2013;8:e25344. doi: 10.4161/psb.25344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata R, Sawa S. Maturation processes and structure of small secreted peptides in plants. Front Plant Sci. 2014;5:311. doi: 10.3389/fpls.2014.00311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, McCormack M, Li L, Hall Q, Xiang C, Sheen J. Glucose-TOR signalling reprograms the transcriptiome and activates meristems. Nature. 2013;496:181–186. doi: 10.1038/nature12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Liberatore KL, MacAlister CA, Huang Z, Chu Y-H, Jiang K, Brooks C, Ogawa-Ohnishi M, Xiong G, Pauly M, Van Eck J, Matsubayashi Y, van der Knaap E, Lippman ZB. A cascade of arabinosyltransferase controls shoot meristem size in tomato. Nature Genet. 2015;47:784–791. doi: 10.1038/ng.3309. [DOI] [PubMed] [Google Scholar]
- Xu T-T, Song X-F, Ren S-C, Liu C-M. The sequence flanking of the N-terminus of the CLV3 peptide is critical for its cleavage and activity in stem cell regulation in Aarabidopsis. BMC Plant Biol. 2013;13:225. doi: 10.1186/1471-2229-13-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav RK, Girke T, Pasala S, Xie M, Reddy GB. Gene expression map of the Arabidopsis shoot apical meristem stem cell niche. Proc Natl Acad Sci USA. 2009;106:4941–4946. doi: 10.1073/pnas.0900843106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav RK, Perales M, Gruel J, Girke T, Jönsson H, Reddy GV. WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes Dev. 2011;25:2025–2030. doi: 10.1101/gad.17258511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav RK, Perales M, Gruel J, Ohno C, Heisler M, Girke T, Jönsson H, Reddy GV. Plant stem cell mainternance involves direct transcriptional repression of differentiation program. Mol Sys Biol. 2013;9:654. doi: 10.1038/msb.2013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi YL, Ishida T, Sawa S. CLE peptides and their signaling pathways in plant development. J Exp Bot. 2016;67:4813–4826. doi: 10.1093/jxb/erw208. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. CLV3 peptides complement clv3-2 in the SAM homeostasis.
Fig. S2. MCLV3p and [Ara3]CLV3p similarly reduce the SAM size in clv1 and bam1 mutants.
Fig. S3. Endogenous expression levels of WUS, SOT17 and APK2 genes in the SAM of clv3-2 and WT Ler seedlings.


