Abstract
Background
Glioblastoma ranks among the most lethal cancers, with current therapies offering only palliation. Paracrine vascular endothelial growth factor (VEGF) signaling has been targeted using anti-angiogenic agents, whereas autocrine VEGF/VEGF receptor 2 (VEGFR2) signaling is poorly understood. Bevacizumab resistance of VEGFR2-expressing glioblastoma cells prompted interrogation of autocrine VEGF-C/VEGFR2 signaling in glioblastoma.
Methods
Autocrine VEGF-C/VEGFR2 signaling was functionally investigated using RNA interference and exogenous ligands in patient-derived xenograft lines and primary glioblastoma cell cultures in vitro and in vivo. VEGF-C expression and interaction with VEGFR2 in a matched pre- and post-bevacizumab treatment cohort were analyzed by immunohistochemistry and proximity ligation assay.
Results
VEGF-C was expressed by patient-derived xenograft glioblastoma lines, primary cells, and matched surgical specimens before and after bevacizumab treatment. VEGF-C activated autocrine VEGFR2 signaling to promote cell survival, whereas targeting VEGF-C expression reprogrammed cellular transcription to attenuate survival and cell cycle progression. Supporting potential translational significance, targeting VEGF-C impaired tumor growth in vivo, with superiority to bevacizumab treatment.
Conclusions
Our results demonstrate VEGF-C serves as both a paracrine and an autocrine pro-survival cytokine in glioblastoma, promoting tumor cell survival and tumorigenesis. VEGF-C permits sustained VEGFR2 activation and tumor growth, where its inhibition appears superior to bevacizumab therapy in improving tumor control.
Keywords: bevacizumab, glioblastoma, VEGF, VEGF-C, VEGFR2
Importance of the study
VEGF ligands stimulate tumor angiogenesis through effects on both endothelial cells and tumor cells, prompting the development of anti-angiogenic targeting strategies, including bevacizumab, which blocks VEGF-A ligand binding to VEGFR2. Unfortunately, bevacizumab generates only transient responses for glioblastoma patients with universal treatment failure in the long term. One potential mechanism of treatment failure could derive from compensatory expression of other VEGF ligands in glioblastoma cells with associated autocrine signaling through VEGFR2. Here, we demonstrate that the VEGF-C ligand maintains VEGFR2 activation in glioblastoma cells despite bevacizumab treatment. In patient-derived tumor specimens and cultures, VEGF-C was expressed and activated VEGFR2, unaffected by bevacizumab treatment. Targeting VEGF-C led to transcriptional reprogramming, inhibiting glioblastoma cell survival, cell cycle, and tumor-initiating capacity. Collectively, these results support autocrine VEGF-C/VEGFR2 signaling as important in glioblastoma growth and a potential mechanism to evade bevacizumab.
Glioblastoma ranks among the most lethal primary central nervous system tumors, with median survival of less than 15 months.1,2 Robust vascularization, often associated with the overexpression of vascular endothelial growth factor A (VEGF-A), is a hallmark of glioblastoma.3 The VEGF receptor 2 (VEGFR2) is a primary effector of VEGF-A signaling, expressed by both endothelial and tumor cells to stimulate paracrine and autocrine effects of VEGF signaling. Although VEGF-A/VEGFR2 signaling is most commonly associated with regulation of endothelial cells, autocrine VEGFR2 activation also promotes proliferation and self-renewal of glioblastoma stemlike cells.4–8 The collective pro-tumorigenic effects of VEGF signaling have led to the development of targeted therapeutics against VEGF-A/VEGFR2.9 Despite initial enthusiasm from preclinical and clinical studies, targeting VEGF-A using a humanized anti-VEGF antibody, bevacizumab (Avastin), failed to show improvement in overall survival for glioblastoma patients.10,11 Bevacizumab remains FDA approved for glioblastoma and continues to be widely used, despite the lack of survival benefit, supporting the importance of understanding treatment failure with bevacizumab. Glioblastoma evades bevacizumab through multiple mechanisms, revealing the complexity of pro-angiogenic signaling.12 These mechanisms, together with the lack of inhibitory effect of bevacizumab on tumor cells expressing VEGF-A/VEGFR2, suggest alternative ligand-mediated activation of VEGFR2 upon bevacizumab treatment.4,8,13
The VEGF ligand family comprises several ligands, including VEGF-C and VEGF-D, with overexpression associated with disease progression in various solid tumors.14–17 However, the roles of these ligands in glioblastoma is poorly defined. VEGF-C, a main driver of lymphatic vessel formation, serves critical roles during embryogenesis, tumorigenesis, and metastasis.18,19 VEGF-C undergoes extensive proteolytic processing, whereupon mature VEGF-C binds VEGFR3 and VEGFR2 primarily located on lymphatic and vascular endothelial cells, respectively.20 VEGF-C and its cognate receptors are expressed on tumor cells, including leukemic, skin, and gastric cells, facilitating tumor progression in part through autocrine signaling.21–23 In glioblastoma, VEGF-C is expressed by endothelial cells, tumor cells, and infiltrating macrophages.24–27 In contrast to VEGF-D, VEGF-C is overexpressed in glioblastoma compared with nonneoplastic brain,24–26,28 and high VEGF-C levels have been documented to inform poor prognosis.26–28 While most previous studies focused on the prognostic significance of VEGF-C and its role in endothelial stimulation,28,29 functional studies interrogating the role of VEGF-C signaling in glioblastoma tumor cells are still missing.
In the present study, we investigated the role of autocrine VEGF-C/VEGFR2 signaling in glioblastoma. We demonstrate that VEGF-C expressed by glioblastoma cells mediates their survival and cell cycle progression and promotes tumorigenicity, potentially contributing to bevacizumab resistance.
Methods
Cell Culture
Patient-derived glioblastoma xenograft lines CPH017 (p4), CPH036 (p6), CPH047 (p3m1), CPH048 (p6), and primary cell cultures (see Supplementary Table S1) were derived in the Hamerlik lab30; 1966 was provided by J. N. Rich (University of California San Diego)4; and IN326 and IN1123 were provided by I. Nakano (University of Alabama at Birmingham).31,32 Human dermal microvascular endothelial cells, human dermal lymphatic endothelial cells (HDLECs), and brain microvascular endothelial cells were purchased from Lonza, PromoCell, and 3H Biomedical, respectively. For cell culture conditions, see the Supplementary materials.
Inhibitors, Growth Factors, and Blocking Antibodies
We purchased SU1498 (Millipore), bevacizumab (Avastin, 25 mg/mL, Roche), recombinant VEGF-A-165aa (50 µg/mL, Miltenyi Biotec), and recombinant human mature VEGF-C (Thr103-Arg227~21 kDa; 10 µg/mL, R&D Systems).
SiRNA Transfection/shRNA Transduction
Cells were transfected using 75 pmol of either VEGF-C–small interfering (si)RNA (siVEGF-C-1: esiRNA pool/EHU013781, Sigma-Aldrich; siVEGF-C-2: single siRNA/HSS111277, ThermoFisher Scientific) or scrambled control siRNA (siCtrl: Stealth RNAi Negative Control Duplex Med CG, ThermoFisher Scientific). Lentiviral transduction was used for delivery of VEGF-C–short hairpin (sh)RNA (shVEGF-C-A: TRCN0000425238, shVEGF-C-B: TRCN000058507, Sigma-Aldrich) or nontargeting control shRNA (shCtrl: SHC007, Sigma Aldrich). For details, see the Supplementary materials.
Patient Surgical Biopsy Specimens
Paired formalin-fixed paraffin-embedded (FFPE) samples (n = 12 pathologically confirmed glioblastoma) collected pre- and postprogression on bevacizumab in combination with CPT-11 (irinotecan) were previously described.33 Moreover, blood-plasma samples were collected at time of surgical resection for glioblastoma of 7 other patients. Patient material was used according to the Declaration of Helsinki and Danish legislation under permission from the Danish Data Protection Agency (2006-41-6979) and the scientific ethical committee for Copenhagen and Frederiksberg (H-3-2009-136, H-2-2012-069).
Western Blot
Western blot (WB) analysis was performed as described previously.34 Semi-quantification of band intensities was performed using ImageJ software. Antibodies are listed in the Supplementary materials.
Quantitative Real-Time PCR
RNA was purified using the QIAshredder/RNeasy Mini kit (Qiagen). Synthesis of cDNA and quantitative real-time (qRT)-PCR reactions were performed using the Superscript III Platinum qRT-PCR kit with SYBR Green (ThermoFisher Scientific). Quantification of gene expression levels was done according to the comparative cycle threshold method and normalized to expression of reference human genes TOP1, EIF4A2, and CYC1. Primer sequences are listed in the Supplementary materials.
Cell Viability Assay
We plated 0.7–2 × 104 cells/well in 96-well plates. Viability was measured using MTT assay (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide) as previously described.34
Enzyme-Linked Immunosorbent Assay
Cells were plated at 1.25 × 106 cells/mL. Conditioned media were collected 48–72 hours later and subjected to Human VEGF-Quantikine enzyme-linked immunosorbent assay (ELISA) or Human VEGF-C-Quantikine ELISA (R&D Systems), following manufacturer’s instructions using a Synergy2 microplate reader (BioTek).
Proximity Ligation Assay
VEGF-C and VEGFR2 colocalization was analyzed using the Duolink In Situ Red Starter Kit Mouse/Rabbit (Sigma-Aldrich). Detailed method description can be found in the Supplementary materials.
Immunohistochemistry
Four-micrometer sections were deparaffinized, hydrated, blocked for endogenous peroxidase using H2O2, and pretreated in a microwave oven with a Tris/ethylene glycol tetra-acetic acid buffer (pH 9.0) before immunostaining. Staining was conducted on a Dako Cytomation autostainer using antibody for VEGF-C (Supplementary materials). Staining was evaluated using an Olympus BX51 microscope.
Gene Expression Analysis
Gene expression analysis was performed using the HumanGene 2.0 ST GeneChip array (Affymetrix), and differential expression analysis was conducted with P-value threshold of 0.05 and Bonferroni–Hochberg correction. Gene set enrichment analysis (GSEA) was performed by analyzing gene sets (with applied set size filters: min = 15, max = 500) from the Reactome pathway database. Detailed analysis description is found in the Supplementary materials.
Cell Cycle Analysis
Analysis of cell cycle profiles and mitotic index was done as previously described32,34 (see Supplementary materials), using the FACSVerse Cell Sorter (BD Biosciences) and FlowJo software.
In Vivo Tumor Formation
Intracranial glioblastoma xenografts were established by orthotopic injection of 50000–100000 viable cells (transfected with siVEGF-C/siCtrl or transduced with shVEGF-C/shCtrl) in 8- to 10-week-old Naval Medical Research Institute nude female mice (Taconic) as previously described.35 Selected mice were treated intraperitoneally with bevacizumab (5 mg/kg) or immunoglobulin (Ig)G every second day for 2 weeks from day 2 post cell injection. Mice were humanely euthanized when presenting neurological signs and/or 20% weight loss. Animal care and all experimental procedures were performed under the approval of the Danish Animal Welfare Council (2012-15-2934-00267/2012-15-2934-00636).
Statistical Analysis
Quantified in vitro data are presented as mean ± SD, ± SEM, or ± 95% CI, as indicated. For all in vitro experiments, n indicates the number of independent performed experiments, while for the in vivo data it represents the number of animals used. Statistical analysis was performed using GraphPad Prism (v7.02).
Results
VEGF-C Activated VEGFR2 in Bevacizumab-Treated Glioblastoma Cells
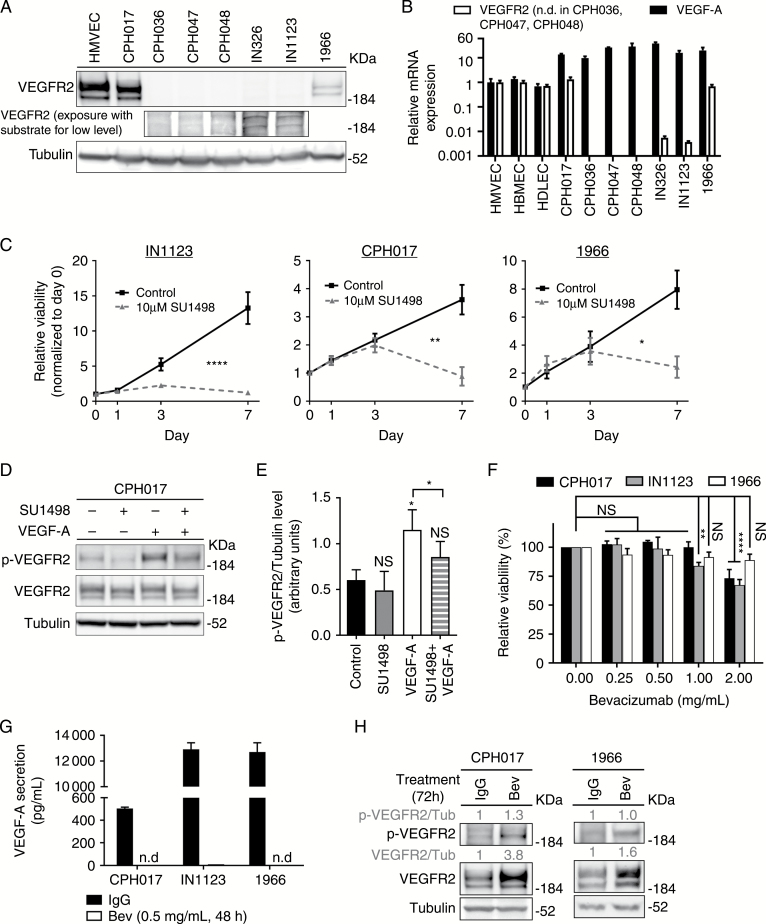
We previously demonstrated that glioblastoma cells express VEGFR2, stimulating tumor growth.4 To evaluate VEGFR2 expression in primary patient-derived glioblastoma models, we performed qRT-PCR and WB analysis. Human microvascular endothelial cells (HMVECs) and human brain microvascular endothelial cells (HBMECs) served as positive controls. VEGFR2 expression was heterogeneous, ranging from negative (CPH036, CPH047, CPH048) to moderate (IN326 and IN1123) to high (CPH017 and 1966). VEGFR2 levels in the high-expressing glioblastoma cultures were comparable to levels of HMVECs and HBMECs (Fig. 1A and B). Quantitative RT-PCR analysis confirmed VEGFA expression in all tested cultures with HMVECs as controls (Fig. 1B). We selected representative VEGFR2high (CPH017 and 1966) and VEGFR2moderate (IN1123) glioblastoma cells to interrogate the functionality of autocrine VEGFR2 signaling. Cells were treated with the exogenous VEGF-A165 ligand and/or the VEGFR2 tyrosine kinase inhibitor SU1498. CPH017, IN1123, and 1966 cells showed significant sensitivity to 10 µM SU1498 (Fig. 1C), whereas this concentration had no effect on VEGFR2negative CPH036 cells (Supplementary Figure S1). Pretreatment of CPH017 cells with SU1498 decreased VEGFR2 phosphorylation upon stimulation with VEGF-A165 (Fig. 1D and E), confirming the functionality of VEGFR2 in tumor cells. In contrast, although bevacizumab treatment of glioblastoma cells sequestered secreted VEGF-A, no significant impairment of glioblastoma cell viability was observed (Fig. 1F and G; Supplementary Figure S1). WB analysis of cells treated with bevacizumab indicated increased expression and activation of VEGFR2 (Fig. 1H). Collectively, these data demonstrate that tumor cell VEGFR2 remains activated in the absence of VEGF-A, suggesting that other VEGF ligands may stimulate VEGFR2 in the absence of VEGF-A.
Fig. 1.
VEGFR2 tyrosine kinase expressed by glioblastoma cells remains active under bevacizumab therapy. (A) Western blot (WB) for VEGFR2 and tubulin in a panel of patient-derived glioblastoma cultures. (B) Quantitative RT-PCR analysis of VEGF-A and VEGFR2 expression in glioblastoma cells standardized to expression in human microvascular endothelial cells (HMVECs). Data are presented as mean ± SD, n = 3. The y-axis is log10 transformed. (C) Viability of CPH017, IN1123, and 1966 cells treated with SU1498 (10 µM) or left unstimulated. Data are presented as mean ± SD, n = 3. Significance determined by 2-way ANOVA with Bonferroni post-test. (D) Representative WB of total and phosphorylated VEGFR2 and tubulin in CPH017 cells plated overnight in Neurobasal media without epidermal growth factor and basic fibroblast growth factor, then treated with either SU1498 (30 µM for 3 h), VEGF-A (40 ng/mL, 15 min), or the combination (30 µM SU1498 for 3 h followed by 40 ng/mL VEGF-A for 15 min). (E) Quantification of relative phosphorylated-VEGFR2/tubulin levels from WB analysis. Data are presented as mean ± SD, n = 3. Significance determined by one-way ANOVA with Dunnett’s post-test correction and ratio paired 2-sample t-test. (F) Viability of glioblastoma cells treated 7 days with bevacizumab (Bev) (0–2 mg/mL). Data are presented as mean ± SD, n = 3. Significance determined by 2-way ANOVA with Bonferroni post-test correction. (G) ELISA quantification of VEGF-A secretion by glioblastoma cells treated with Bev (0.5 mg/mL, 48 h) or IgG control. Data are presented as mean ± SD, n = 3. N.D. denotes that signal could not be detected. (H) Representative WB of total and phosphorylated VEGFR2 or tubulin in CPH017 and 1966 cells treated for 72 h with Bev (0.5 mg/mL) or IgG. *P ≤ 0.05; **P ≤ 0.01; ****P ≤ 0.0001; NS: nonsignificant.
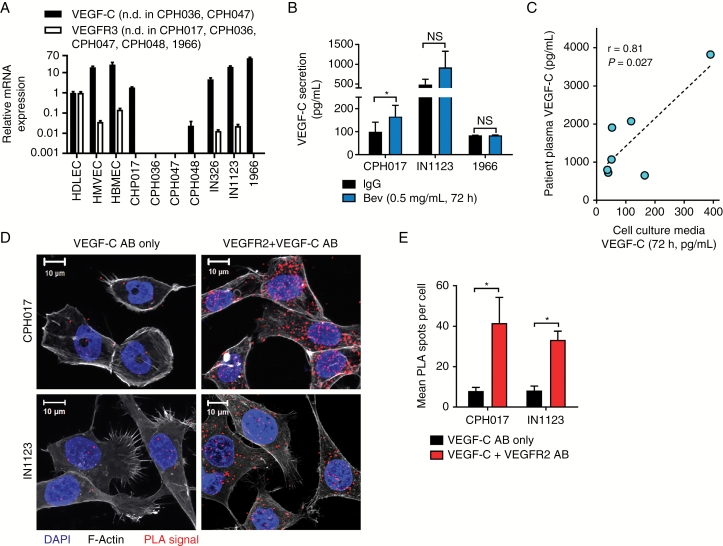
To determine the potential role of alternative VEGF ligands, we interrogated our patient-derived models for VEGF-C expression. As shown in Fig. 2A, 5 of 7 glioblastoma cultures expressed VEGF-C at levels comparable to control HDLECs, which were previously demonstrated to express both VEGF-C and VEGFR3.36 While VEGF-C expression in most cultures correlated with VEGFR2 expression, VEGFR3 was detected in only 2 cell cultures at very low levels (Fig. 2A). Bevacizumab treatment (0.5 mg/mL) did not alter VEGF-C secretion in 1966 cells and induced expression in CPH017 and IN1123 glioblastoma cells (Fig. 2B), supporting VEGF-C activation of VEGFR2 activation in the absence of VEGF-A. To rule out VEGF-C secretion as a cell culture artifact, we compared VEGF-C levels in matched collected plasma samples from a glioblastoma patient with conditioned media collected from in vitro primary cell cultures derived from the same patient (n = 7; Fig. 2C and Supplementary Table S1). A significant positive correlation indicated that findings for VEGF-C from in vitro cultures are representative of patient tumors.
Fig. 2.
VEGF-C is expressed under bevacizumab therapy and interacts with VEGFR2 in glioblastoma cells. (A) Quantitative RT-PCR analysis of VEGF-C and VEGFR3 expression in glioblastoma cells standardized to HDLECs. Data represented as mean ± SD; n = 2. The y-axis is log10 transformed. N.D. = not detected. (B) ELISA quantification of VEGF-C secretion by glioblastoma cells treated 72 h with bevacizumab (Bev; 0.5 mg/mL) or IgG. Data represented as mean ± SEM; n = 3. Significance determined by paired 2-sample t-test. *P ≤ 0.05. (C) Correlation of VEGF-C ELISA quantification in matched samples of blood-plasma and conditioned media (72 h) from tumor-derived cell cultures of 7 glioblastoma patients. Significance was determined by Spearman’s correlation analysis. (D) Representative confocal images showing PLA signal (red), F-actin (gray), and 4′,6′-diamidino-2-phenylindole (blue) of glioblastoma cells. Staining with only VEGF-C antibody (AB) was used as a negative control. (E) Quantification of PLA dots in glioblastoma cells. Data are represented as mean ± SEM; n > 40 cells.
VEGF-C Interacts with VEGFR2 In Vitro and Ex Vivo
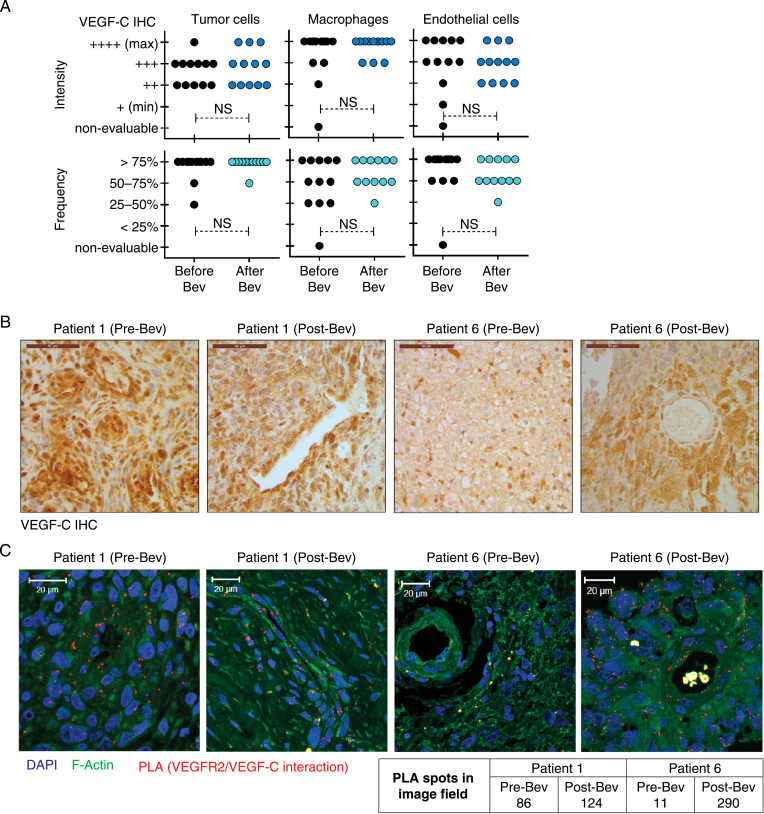
To verify that VEGF-C binds to VEGFR2 in glioblastoma cells, we performed a proximity ligation assay (PLA), which confirmed VEGF-C/VEGFR2 interaction. PLA spots, denoting interaction, were localized to both plasma membrane and cytosol (Fig. 2D, E and Supplementary Figure S2). VEGF-C and VEGFR2 colocalization was confirmed in FFPE glioblastoma surgical biopsy specimens collected pre and post bevacizumab treatment. The cohort (Supplementary Table S2) consisted of 12 pairs of parallel tumor samples from surgery before treatment with combined bevacizumab and irinotecan chemotherapy (Pre-Bev samples) and after treatment relapse (Post-Bev samples). Immunohistochemistry showed heterogeneous granular VEGF-C expression pattern in tumor cells, endothelial cells of both neoplastic and nonneoplastic vessels, and tumor-infiltrating microglia and macrophages (Fig. 3B, C and Supplementary Figure S3). Although heterogeneous, VEGF-C expression was found in all patient samples, and no systematic changes in VEGF-C expression were associated with bevacizumab treatment. PLA performed on FFPE specimens (6 selected patients of our cohort) confirmed the interaction between VEGF-C and VEGFR2 in samples collected before and after bevacizumab treatment (Fig. 2D and Supplementary Figure S4).
Fig. 3.
VEGF-C expression and interaction with VEGFR2 in glioblastoma cells is sustained upon bevacizumab therapy. (A) Immunohistochemical evaluation of VEGF-C in matched pre- and post-bevacizumab glioblastoma patient surgical biopsy specimens. Quantification of staining intensity and frequency in tumor cells, macrophages, and endothelial cells in each tumor sample is displayed. Statistical significance is determined by 2-sample t-test. NS: nonsignificant. (B) Representative images of 2 of 12 patients analyzed by VEGF-C immunohistochemistry. Scale bar, 50 μm. (C) Confocal images of PLA signal (red), F-actin (green), and 4′,6′-diamidino-2-phenylindole (blue) in FFPE pre- and post-bevacizumab patient glioblastoma specimens. Representative images with PLA spot quantification from tumor samples of 2 out of 6 analyzed patients are shown.
VEGF-C Signaling Regulates Glioblastoma Cell Survival
Our previous studies demonstrated that activation of VEGFR2 mediated by VEGF-A triggers Akt/extracellular signal-regulated kinase (ERK)/phospholipase C (PLC)‒gamma signaling in glioblastoma cells.37 Therefore, we measured the activation of these signaling pathways in response to exogenous VEGF-C. In gain-of-function studies, VEGF-C treatment induced activation of VEGFR2/PLC-gamma/ERK1/2 signaling in glioblastoma cells, in both the presence (Fig. 4A) and the absence of VEGF-A (Supplementary Figure S5A). To determine loss-of-function modulation of VEGF-C, we performed siRNA-mediated knockdown of VEGF-C using a nontargeting control siRNA (siCtrl) or 2 independent siRNAs against VEGF-C (siVEGF-C-1, siVEGF-C-2) in glioblastoma cells. Effective knockdown was validated by qRT-PCR and WB (Fig. 4B and C). VEGF-C loss led to reduced cell viability (Fig. 4F) and induced apoptosis (cleavage of caspase-3; Fig. 4G). In addition, we observed increased protein levels of signal transducer and activator of transcription 1 and interferon regulatory factor 1, mediators of interferon signaling with anti-proliferative effects in cancer following VEGF-C knockdown (Supplementary Figure S5B).38,39 Concordant with a potential adaptive role, VEGF-C knockdown resulted in increased VEGF-A secretion as assessed by qRT-PCR and ELISA (Fig. 4D and E; Supplementary Figure S6D). The differential regulation of VEGF-A and VEGF-C ligands prompted us to evaluate the combinatorial effect of VEGF-C knockdown and bevacizumab treatment, which reduced viability compared with VEGF-C knockdown alone, indicating additional proliferative effects of VEGF-C (Fig. 4H). In validation studies, transduction of glioblastoma cells with stable shRNA-mediated VEGF-C knockdown led to decreased PLC-gamma/ERK1/2 signaling and viability (Supplementary Figure S6).
Fig. 4.
VEGF-C regulates canonical VEGFR2 survival signaling. (A) WB analysis of downstream VEGFR2 signaling in CPH017 glioblastoma cells starved for 24 h followed by stimulation with VEGF-A (40 ng/mL) or VEGF-C (0.2 µg/mL). (B) Quantitative RT-PCR analysis of VEGF-C expression in glioblastoma cells 48 h following transfection with either siCtrl (nontargeting control) or siVEGF-C (VEGF-C targeting siRNA). Data are represented as mean ± SEM; n = 3 or 4. Significance determined with 1-sample t-tests setting the hypothetical value to 1. (C) WB for VEGF-C and tubulin in CPH017 cells 72 h following siCtrl or siVEGF-C transfection. (D) Quantitative RT-PCR analysis of VEGF-A expression in CPH017 and IN1123 cells 48 h following siCtrl or siVEGF-C transfection. Data are represented as mean ± SEM; n = 3 or 4. Significance determined with 1-sample t-tests setting the hypothetical value to 1. (E) ELISA quantification of VEGF-A in conditioned media from CPH017 glioblastoma cells transfected with siCtrl or siVEGF-C. Data are represented as mean ± SEM; n = 2. Significance determined by 1-way ANOVA with Dunnett’s post-test correction. (F) Viability of CPH017 and IN1123 glioblastoma cells transfected with siCtrl or siVEGF-C. Data are represented as mean ± SEM; n = 3–5. Significance determined by 2-way ANOVA with Bonferroni post-test correction. (G) WB of pro- and cleaved (Cl.) caspase-3 and tubulin in CPH017 and IN1123 glioblastoma cells 72 h following siCtrl or siVEGF-C transfection. (H) Viability of CPH017 and IN1123 glioblastoma cells 5 days after transfection with siCtrl or siVEGF-C alone and combined treatment with 0.5 mg/mL bevacizumab (Bev). Data are represented as mean ± SEM; n = 3. Significance determined by 1-way ANOVA with Dunnett’s post-test correction. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001.
VEGF-C Knockdown Impairs Cell Cycle Progression of Glioblastoma Cells
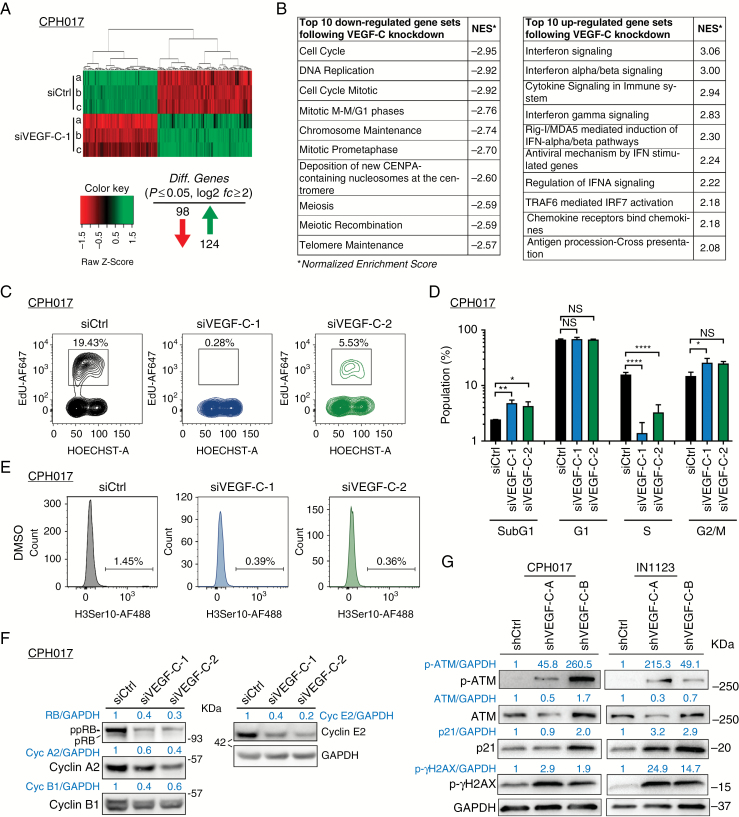
To better delineate autocrine signaling and identify the biological role of VEGF-C in glioblastoma cells, we interrogated gene expression changes in CPH017 cells following VEGF-C knockdown using Affymetrix GeneChip Human Gene 2.0 ST arrays. A total of 222 genes with absolute log2-fold change in expression above 2 (log2fc ≥ 2) were found to be significantly altered (P ≤ 0.05). Ninety-eight genes were downregulated and 124 genes upregulated (Fig. 5A; Supplementary Tables S3 and S4). Gene set enrichment analysis showed significant downregulation of pathways regulating the cell cycle, DNA replication, and mitosis (Fig. 4B and Supplementary Figure S7). Cyclin-family members, cell-division cycle regulators, and histone coding genes scored among the top downregulated candidates (Supplementary Table S3). Interferon/cytokine signaling associated with immunomodulatory responses ranked among the top upregulated GSEA pathways following VEGF-C knockdown (Fig. 5B and Supplementary Figure S8).
Fig. 5.
VEGF-C loss leads to transcriptional changes and cell cycle arrest in glioblastoma cells. (A) Heat map of significantly differentially expressed genes (log2fc ≥ 2 and adjusted P ≤ 0.05) 48 h after VEGF-C knockdown compared with siCtrl. (B) GSEA using a 479-gene set from the Reactome pathway database. Top 10 most significant downregulated and 10 most upregulated gene sets are shown with normalized enrichment score (NES). (C) Representative fluorescence activated cell sorting (FACS) plot showing cell cycle profile of CPH017 cells post transfection with siCtrl or siVEGF-C. (D) Quantification of sub-G1, G1, S, and G2/M cell cycle populations–based FACS analysis. Data are displayed as mean ± SEM; n = 3–4. The y-axis is log10 transformed. Statistical significance was determined by one-way ANOVA. *P ≤ 0.05; **P ≤ 0.01; ****P ≤ 0.0001; NS: nonsignificant. (E) Mitotic index (%) of CPH017 cells 72 h post siCtrl and siVEGF-C transduction. (F) WB analysis of phosphorylated and total retinoblastoma, cyclins A2/B1/E2, and glyceraldehyde 3-phosphate dehydrogenase after siCtrl and siVEGF-C transfection in glioblastoma cells. (G) WB for DNA damage markers phosphorylated and total ataxia telangiectasia mutated and gamma-H2AX in glioblastoma cells 72 h following shCtrl or shVEGF-C transduction.
To examine the effects of VEGF-C knockdown on the cell cycle, we performed a flow cytometry–based cell cycle analysis, including incorporation of EdU (5-ethynyl-2'-deoxyuridine; ethynyl-labeled (deoxy)uridine) to mark S phase cells and staining for phosphorylated histone 3Ser10 (p-H3Ser10) to mark mitotic cells. VEGF-C loss induced the apoptotic sub-G1 population from 2.4% for siCtrl to 4.8 and 4.3% for siVEGF-C-1 and siVEGF-C-2, respectively (Fig. 5C and D). The proliferative index, calculated as percentage of EdU-positive cells, decreased from 15% (siCtrl cells) to 1% (siVEGF-C-1) and 3% (siVEGF-C-2), respectively. The mitotic index, calculated as percentage of p-H3Ser10–positive cells, decreased from 1.45% (siCtrl) to 0.39% and 0.36% for siVEGF-C-1 and siVEGF-C-2, respectively (Fig. 5D). WB analysis confirmed reduced retinoblastoma hyperphosphorylation and cyclin E2/B1/A2 levels, supporting impaired G1-S phase transition in siVEGF-C cells (Fig. 5F). VEGF-C loss increased ataxia telangiectasia mutated and gamma-H2AX phosphorylation, indicating DNA damage accumulation, a prerequisite for cell cycle arrest (Fig. 5G).
Targeting of VEGF-C Reduces In Vivo Glioblastoma Tumor Growth
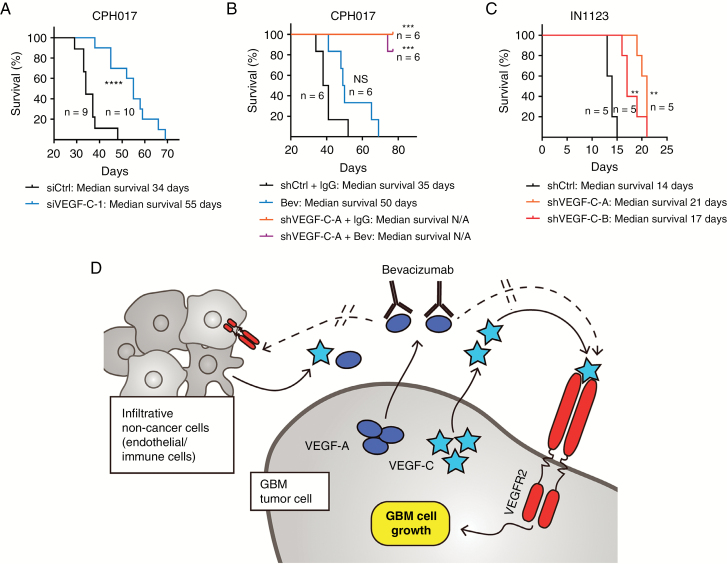
To assess the contribution of VEGF-C signaling to glioblastoma tumor growth, we performed in vivo survival studies using both siRNA and shRNA approaches validated in vitro, as noted above. VEGF-C knockdown extended the survival of immunocompromised mice bearing orthotopic xenografts of either CPH017 or IN1123 glioblastoma cells (Fig. 6). VEGF-C knockdown augmented survival more than did bevacizumab treatment (Fig. 6B). Although VEGF-C knockdown and bevacizumab treatment had combinatorial benefit in vitro, no additional benefit was detected in xenograft studies.
Fig. 6.
VEGF-C loss promotes DNA damage, reduces survival marker expression, and impairs in vivo glioblastoma growth. (A) Kaplan–Meier survival analysis of mice bearing orthotopically transplanted viable CPH017 glioblastoma cells transduced with either VEGF-C–siRNA or Ctrl–siRNA. Significance was determined by log-rank analysis; *P ≤ 0.05, **P ≤ 0.01, NS: nonsignificant. (B) Kaplan–Meier survival analysis of mice bearing orthotopically transplanted viable CPH017 glioblastoma cells. Group 1: transduced with shCtrl and treated with IgG control. Group 2: treated with bevacizumab alone (10 mg/kg; for details see “Materials and Methods” section). Group 3: transduced with shVEGF-C-A alone. Group 4: transduced with shVEGF-C-A and treated with bevacizumab (10 mg/kg; for details see “Materials and Methods” section). Significance was determined by log-rank analysis; *P ≤ 0.05, **P ≤ 0.01, NS: nonsignificant, N/A: not achieved. (C) Kaplan–Meier survival analysis of mice bearing orthotopically transplanted viable IN1123 glioblastoma cells transduced with shCtrl or shVEGF-C. Significance was determined by log-rank analysis; *P ≤ 0.05, **P ≤ 0.01, NS: nonsignificant. (D) Schematic illustration of model in which VEGF-C stimulates VEGFR2 in glioblastoma through both autocrine and paracrine pathways to promote survival, proliferation, and cell cycle progression and sustain VEGFR2 activity in the inhibition of VEGF-A signaling under bevacizumab therapy.
Discussion
Few options are available for glioblastoma patients suffering tumor recurrence or progression. Given the role of microvascular proliferation as a histologic hallmark of glioblastoma and detection of VEGF in glioblastoma surgical specimens, angiogenesis has been a prominent area of research in neuro-oncology. Bevacizumab treatment leads to a high response rate in early treatment, but tumor control is transient and no consistent impact on patient survival has been found.10,11 Despite the lack of survival benefit, bevacizumab was approved by the FDA and remains in wide use for tumor recurrence and treatment of radiation-induced necrosis. The molecular underpinnings for evasion of anti–VEGF-A therapy remain an area of active investigation.12,40
Our results demonstrate for the first time that autocrine VEGF-C/VEGFR2 signaling regulates glioblastoma cell viability, cell cycle, and in vivo tumor growth, underscoring the global requirement for VEGF signaling in glioblastoma. Despite VEGF-A sequestration by bevacizumab, VEGFR2 remains active on glioblastoma cells, suggesting an alternative pathway for receptor activation (Fig. 1H). Mahfouz and coworkers41 recently reported that bevacizumab induces VEGFR2 expression in gastrointestinal cancer. Collectively, our results support an important role of VEGF-C for glioblastoma growth, suggesting a novel anti-angiogenesis evasion mechanism, where glioblastoma cells secrete VEGF-C in response to bevacizumab treatment, facilitating sustained VEGFR2-mediated pro-survival signaling, tumor cell proliferation, and cell cycle progression (Fig. 6C). Quantitative RT-PCR and ELISA analyses confirmed VEGF-C expression in our collection of xenograft lines and primary glioblastoma cells. Model lines chosen for this study coexpressed VEGFR2 and VEGF-C but lacked or expressed very low VEGFR3 levels, indicating that VEGF-C may operate via VEGFR2 (Figures 1A, B and 2A). VEGF-C/VEGFR2 interacted in vitro and ex vivo with concordance between the VEGF-C protein levels and VEGF-C/VEGFR2 interaction (Figures 2C, D and 3B–D). VEGF-C expression and its interaction with VEGFR2 were sustained in glioblastomas after bevacizumab treatment (Fig. 3B–D and Supplementary Figure S3). Glioblastoma surgical specimens and patient-derived cultures display variable VEGF-C expression (Figures 2B and 3B, C), suggesting that targeting VEGF-C will require precise selection of patients. Intratumoral and intertumoral heterogeneity of VEGF-C expression may be driven by cellular environmental factors, like hypoxia, which induces VEGF-C expression in metastatic cancer cells.42 VEGF-C protein levels correlated with matched patient plasma samples and conditioned media collected from corresponding primary cultures (Fig. 3A), supporting patient-derived cultures as an accurate model to test VEGF-C biology in glioblastoma. Immunohistochemistry confirmed previously reported VEGF-C expression by tumor-infiltrating macrophages and other nontumor cell types (Supplementary Figure S3),25,43,44 suggesting that glioblastoma cells with low VEGF-C expression could be exposed to substantial amounts of VEGF-C ligand in vivo from other cell types.
Signaling downstream from VEGF-C includes canonical VEGFR2 signaling via PLC-gamma and ERK (Fig. 4A, Supplementary Figures S5 and S6). VEGFR1/2 signaling positively correlated with ERK activation,8 which is concordant with our data, where VEGF-C stimulation induced ERK activation in glioblastoma cells (Fig. 4A). Further, VEGF-C stimulated tumor viability and cell cycle progression (Figures 4–6 and Supplementary Figure S6), concordant with previous studies on autocrine VEGF-C signaling in other cancers.17
Bevacizumab treatment did not affect glioblastoma cell survival in vitro (Fig. 1F), with minimal change or increased secretion of VEGF-C in glioblastoma cultures after bevacizumab treatment (Fig. 2B). VEGF-A secretion was consistently increased after siRNA-mediated knockdown of VEGF-C (Fig. 4E). The mechanism(s) by which VEGF-C stimulates the expression of VEGF-A is not clear, but an increase in VEGF-C mRNA levels following bevacizumab therapy was previously demonstrated in glioblastoma and endothelial cells.29 Reciprocally, Zhao and coworkers27 reported VEGF-A induction of VEGF-C in retinal epithelial cells. As ERKs are ubiquitous intracellular signaling mediators of innumerous cellular functions, dysregulation of ERK, possibly in with PLC-gamma, may contribute to VEGF-A/VEGF-C compensatory expression in glioblastoma. Existence of this compensatory mechanism points to a global requirement for a VEGF signaling route in glioblastoma maintenance, yet the increase in VEGF-A levels did not compensate for the anti-proliferative and anti-tumorigenic effect of VEGF-C knockdown, suggesting non-overlapping effects of VEGF ligands. Combined VEGF-A (bevacizumab) and VEGF-C (siRNA) targeting in glioblastoma cells (Fig. 4H) was only modestly superior at reducing cell viability in comparison to inhibiting VEGF-C alone in vitro and had no added benefit in vivo, potentially due to dependency of the VEGF ligands on coreceptors, such as neuropilin-1 and -2,45 for VEGFR2 activation, thereby making coreceptor availability a key element in the impact of VEGF-A and VEGF-C ligand binding on downstream signaling.
In conclusion, we demonstrate that VEGF-C/VEGFR2 signaling promotes glioblastoma cell viability, cell cycle progression, and tumor growth in part through autocrine signaling. As VEGF-C expression and VEGF-C/VEGFR2 interaction remained stable under bevacizumab treatment, autocrine VEGF-C signaling may represent a novel escape mechanism employed by glioblastomas to counteract bevacizumab therapy.
Funding
This work was supported by the Danish Cancer Society, NovoNordisk, Bjarne Saxhofs, Arvid Nilssons, Torben & Alice Frimodts, King Christian IX’s, Dansk Kraeftforsknings, Fabrikant Ejner Willumsens Memorial, Sejer Perssons and Lis Klüwers, and an unrestricted grant from Roche A/S.
Supplementary Material
Acknowledgments
We thank P. Pedersen, C. B. Holst, and O. D. Laerum for technical/academic assistance and J. N. Rich for manuscript editing.
Conflict of interest statement. None declared.
References
- 1. Stupp R, Hegi ME, Mason WP, et al. ; European Organisation for Research and Treatment of Cancer Brain Tumour and Radiation Oncology Groups; National Cancer Institute of Canada Clinical Trials Group Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5):459–466. [DOI] [PubMed] [Google Scholar]
- 2. Ohgaki H. Epidemiology of brain tumors. Methods Mol Biol. 2009;472:323–342. [DOI] [PubMed] [Google Scholar]
- 3. Brem S, Cotran R, Folkman J. Tumor angiogenesis: a quantitative method for histologic grading. J Natl Cancer Inst. 1972;48(2):347–356. [PubMed] [Google Scholar]
- 4. Hamerlik P, Lathia JD, Rasmussen R, et al. Autocrine VEGF-VEGFR2-Neuropilin-1 signaling promotes glioma stem-like cell viability and tumor growth. J Exp Med. 2012;209(3):507–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Paul-Samojedny M, Pudełko A, Suchanek-Raif R, et al. Knockdown of the AKT3 (PKBγ), PI3KCA, and VEGFR2 genes by RNA interference suppresses glioblastoma multiforme T98G cells invasiveness in vitro. Tumour Biol. 2015;36(5):3263–3277. [DOI] [PubMed] [Google Scholar]
- 6. Xu C, Wu X, Zhu J. VEGF promotes proliferation of human glioblastoma multiforme stem-like cells through VEGF receptor 2. Scientific World Journal. 2013;2013:417413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kessler T, Sahm F, Blaes J, et al. Glioma cell VEGFR-2 confers resistance to chemotherapeutic and antiangiogenic treatments in PTEN-deficient glioblastoma. Oncotarget. 2015;6(31):31050–31068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Szabo E, Schneider H, Seystahl K, et al. Autocrine VEGFR1 and VEGFR2 signaling promotes survival in human glioblastoma models in vitro and in vivo. Neuro Oncol. 2016;18(9):1242–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chi AS, Sorensen AG, Jain RK, Batchelor TT. Angiogenesis as a therapeutic target in malignant gliomas. Oncologist. 2009;14(6):621–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gilbert MR, Dignam JJ, Armstrong TS, et al. A randomized trial of bevacizumab for newly diagnosed glioblastoma. N Engl J Med. 2014;370(8):699–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wick W, Gorlia T, Bendszus M, et al. Lomustine and bevacizumab in progressive glioblastoma. N Engl J Med. 2017;377(20):1954–1963. [DOI] [PubMed] [Google Scholar]
- 12. Tamura R, Tanaka T, Miyake K, Yoshida K, Sasaki H. Bevacizumab for malignant gliomas: current indications, mechanisms of action and resistance, and markers of response. Brain Tumor Pathol. 2017;34(2):62–77. [DOI] [PubMed] [Google Scholar]
- 13. Francescone R, Scully S, Bentley B, et al. Glioblastoma-derived tumor cells induce vasculogenic mimicry through Flk-1 protein activation. J Biol Chem. 2012;287(29):24821–24831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lin W, Jiang L, Chen Y, et al. Vascular endothelial growth factor-D promotes growth, lymphangiogenesis and lymphatic metastasis in gallbladder cancer. Cancer Lett. 2012;314(2):127–136. [DOI] [PubMed] [Google Scholar]
- 15. Yasuoka H, Kodama R, Hirokawa M, et al. Neuropilin-2 expression in papillary thyroid carcinoma: correlation with VEGF-D expression, lymph node metastasis, and VEGF-D-induced aggressive cancer cell phenotype. J Clin Endocrinol Metab. 2011;96(11):E1857–E1861. [DOI] [PubMed] [Google Scholar]
- 16. Tanaka M, Kitadai Y, Kodama M, et al. Potential role for vascular endothelial growth factor-D as an autocrine factor for human gastric carcinoma cells. Cancer Sci. 2010;101(10):2121–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang CA, Tsai SJ. The non-canonical role of vascular endothelial growth factor-C axis in cancer progression. Exp Biol Med (Maywood). 2015;240(6):718–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Karkkainen MJ, Haiko P, Sainio K, et al. Vascular endothelial growth factor C is required for sprouting of the first lymphatic vessels from embryonic veins. Nat Immunol. 2004;5(1):74–80. [DOI] [PubMed] [Google Scholar]
- 19. Chen JC, Chang YW, Hong CC, Yu YH, Su JL. The role of the VEGF-C/VEGFRs axis in tumor progression and therapy. Int J Mol Sci. 2012;14(1):88–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Joukov V, Sorsa T, Kumar V, et al. Proteolytic processing regulates receptor specificity and activity of VEGF-C. EMBO J. 1997;16(13):3898–3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cho HJ, Kim IK, Park SM, et al. VEGF-C mediates RhoGDI2-induced gastric cancer cell metastasis and cisplatin resistance. Int J Cancer. 2014;135(7):1553–1563. [DOI] [PubMed] [Google Scholar]
- 22. Yeh YW, Cheng CC, Yang ST, et al. Targeting the VEGF-C/VEGFR3 axis suppresses Slug-mediated cancer metastasis and stemness via inhibition of KRAS/YAP1 signaling. Oncotarget. 2017;8(3):5603–5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dias S, Choy M, Alitalo K, Rafii S. Vascular endothelial growth factor (VEGF)-C signaling through FLT-4 (VEGFR-3) mediates leukemic cell proliferation, survival, and resistance to chemotherapy. Blood. 2002;99(6):2179–2184. [DOI] [PubMed] [Google Scholar]
- 24. Grau SJ, Trillsch F, Herms J, et al. Expression of VEGFR3 in glioma endothelium correlates with tumor grade. J Neurooncol. 2007;82(2):141–150. [DOI] [PubMed] [Google Scholar]
- 25. Jenny B, Harrison JA, Baetens D, et al. Expression and localization of VEGF-C and VEGFR-3 in glioblastomas and haemangioblastomas. J Pathol. 2006;209(1):34–43. [DOI] [PubMed] [Google Scholar]
- 26. Xu Y, Zhong Z, Yuan J, et al. Collaborative overexpression of matrix metalloproteinase-1 and vascular endothelial growth factor-C predicts adverse prognosis in patients with gliomas. Cancer Epidemiol. 2013;37(5):697–702. [DOI] [PubMed] [Google Scholar]
- 27. Zhao H, Hou C, Hou A, Zhu D. Concurrent expression of VEGF-C and Neuropilin-2 is correlated with poor prognosis in glioblastoma. Tohoku J Exp Med. 2016;238(2):85–91. [DOI] [PubMed] [Google Scholar]
- 28. Carpenter RL, Paw I, Zhu H, et al. The gain-of-function GLI1 transcription factor TGLI1 enhances expression of VEGF-C and TEM7 to promote glioblastoma angiogenesis. Oncotarget. 2015;6(26):22653–22665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Grau S, Thorsteinsdottir J, von Baumgarten L, Winkler F, Tonn JC, Schichor C. Bevacizumab can induce reactivity to VEGF-C and -D in human brain and tumour derived endothelial cells. J Neurooncol. 2011;104(1):103–112. [DOI] [PubMed] [Google Scholar]
- 30. Stockhausen MT, Broholm H, Villingshøj M, et al. Maintenance of EGFR and EGFRvIII expressions in an in vivo and in vitro model of human glioblastoma multiforme. Exp Cell Res. 2011;317(11):1513–1526. [DOI] [PubMed] [Google Scholar]
- 31. Flavahan WA, Wu Q, Hitomi M, et al. Brain tumor initiating cells adapt to restricted nutrition through preferential glucose uptake. Nat Neurosci. 2013;16(10):1373–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rasmussen RD, Gajjar MK, Tuckova L, et al. BRCA1-regulated RRM2 expression protects glioblastoma cells from endogenous replication stress and promotes tumorigenicity. Nat Commun. 2016;7:13398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Poulsen HS, Grunnet K, Sorensen M, et al. Bevacizumab plus irinotecan in the treatment of patients with progressive recurrent malignant brain tumours. Acta Oncol. 2009;48(1):52–58. [DOI] [PubMed] [Google Scholar]
- 34. Staberg M, Michaelsen SR, Rasmussen RD, Villingshøj M, Poulsen HS, Hamerlik P. Inhibition of histone deacetylases sensitizes glioblastoma cells to lomustine. Cell Oncol (Dordr). 2017;40(1):21–32. [DOI] [PubMed] [Google Scholar]
- 35. Nedergaard MK, Michaelsen SR, Urup T, et al. 18F-FET microPET and microMRI for anti-VEGF and anti-PlGF response assessment in an orthotopic murine model of human glioblastoma. PLoS One. 2015;10(2):e0115315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu P, Zhou J, Zhu H, et al. VEGF-C promotes the development of esophageal cancer via regulating CNTN-1 expression. Cytokine. 2011;55(1):8–17. [DOI] [PubMed] [Google Scholar]
- 37. Knizetova P, Ehrmann J, Hlobilkova A, et al. Autocrine regulation of glioblastoma cell cycle progression, viability and radioresistance through the VEGF-VEGFR2 (KDR) interplay. Cell Cycle. 2008;7(16):2553–2561. [DOI] [PubMed] [Google Scholar]
- 38. Wu TH, Pabin CN, Qin Z, et al. Long-term suppression of tumor growth by TNF requires a Stat1- and IFN regulatory factor 1-dependent IFN-gamma pathway but not IL-12 or IL-18. J Immunol. 2004;172(5):3243–3251. [DOI] [PubMed] [Google Scholar]
- 39. Papageorgiou A, Dinney CP, McConkey DJ. Interferon-alpha induces TRAIL expression and cell death via an IRF-1-dependent mechanism in human bladder cancer cells. Cancer Biol Ther. 2007;6(6):872–879. [DOI] [PubMed] [Google Scholar]
- 40. Lu KV, Bergers G. Mechanisms of evasive resistance to anti-VEGF therapy in glioblastoma. CNS Oncol. 2013;2(1):49–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mahfouz N, Tahtouh R, Alaaeddine N, et al. Gastrointestinal cancer cells treatment with bevacizumab activates a VEGF autoregulatory mechanism involving telomerase catalytic subunit hTERT via PI3K-AKT, HIF-1α and VEGF receptors. PLoS One. 2017;12(6):e0179202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Morfoisse F, Kuchnio A, Frainay C, et al. Hypoxia induces VEGF-C expression in metastatic tumor cells via a HIF-1α-independent translation-mediated mechanism. Cell Rep. 2014;6(1):155–167. [DOI] [PubMed] [Google Scholar]
- 43. Ding M, Fu X, Tan H, Wang R, Chen Z, Ding S. The effect of vascular endothelial growth factor C expression in tumor-associated macrophages on lymphangiogenesis and lymphatic metastasis in breast cancer. Mol Med Rep. 2012;6(5):1023–1029. [DOI] [PubMed] [Google Scholar]
- 44. Špirić Z, Eri Ž, Erić M. Significance of vascular endothelial growth factor (VEGF)-C and VEGF-D in the progression of cutaneous melanoma. Int J Surg Pathol. 2015;23(8):629–637. [DOI] [PubMed] [Google Scholar]
- 45. Raimondi C, Ruhrberg C. Neuropilin signalling in vessels, neurons and tumours. Semin Cell Dev Biol. 2013;24(3):172–178. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.