Abstract
Background
This study aims to evaluate the impact of tumor location on key molecular alterations on a single voxel level in patients with newly diagnosed glioma.
Methods
A consecutive series of n = 237 patients with newly diagnosed glioblastoma and n = 131 patients with lower-grade glioma was analyzed. Volumetric tumor segmentation was performed on preoperative MRI with a semi-automated approach and images were registered to the standard Montreal Neurological Institute 152 space. Using a voxel-based lesion symptom mapping (VLSM) analysis, we identified specific brain regions that were associated with tumor-specific molecular alterations. We assessed a predefined set of n = 17 molecular characteristics in the glioblastoma cohort and n = 2 molecular characteristics in the lower-grade glioma cohort. Permutation adjustment (n = 1000 iterations) was used to correct for multiple testing, and voxel t-values that were greater than the t-value in >95% of the permutations were retained in the VLSM results (α = 0.05, power > 0.8).
Results
Tumor location predilection for isocitrate dehydrogenase (IDH) mutant tumors was found in both glioblastoma and lower-grade glioma cohorts, each showing a concordant predominance in the frontal lobe adjacent to the rostral extension of the lateral ventricles (permutation-adjusted P = 0.021 for the glioblastoma and 0.013 for the lower-grade glioma cohort). Apart from that, the VLSM analysis did not reveal a significant association of the tumor location with any other key molecular alteration in both cohorts (permutation-adjusted P > 0.05 each).
Conclusion
Our study highlights the unique properties of IDH mutations and underpins the hypothesis that the rostral extension of the lateral ventricles is a potential location for the cell of origin in IDH-mutant gliomas.
Keywords: glioblastoma, glioma, IDH, radiogenomics, VLSM
Key Points
IDH-mutant gliomas show a predilection around the rostral extension of the lateral ventricles.
The identified niche may be a potential location for the cell of origin in IDH-mutant gliomas.
Importance of the Study
Increasing evidence suggests that tumor location is linked to the genetic profile of gliomas. In this study, we investigated the association of key molecular alterations and tumor locations in patients with newly diagnosed glioma on a single voxel level. We demonstrate concordant tumor location predilection for IDH-mutant tumors in 2 independent cohorts of patients with lower-grade glioma and glioblastoma, specifically adjacent to the rostral extensions of the lateral ventricles. Since mutations of the IDH genes are considered an important event at early stages during gliomagenesis, our study substantiates the possibility of neural stem cells in this region as the origin of IDH-mutant gliomas, potentially advancing personalized treatment and clinical management of patients with this tumor entity.
Gliomas are the most common type of primary brain tumor and represent a histologically similar yet molecularly and clinically heterogeneous entity.1,2 The 2016 World Health Organization classification of CNS tumors therefore integrated molecular parameters in addition to the histology to better define this disease.3 As a collection of molecularly distinct diseases, each subtype of glioma could potentially derive from a distinct cell of origin. Therefore, understanding of the early events that may lead to gliomagenesis is pivotal and carries great promise for the development of effective therapies.4 Numerous epidemiological studies, large-scale sequencing efforts, as well as genetically engineered mouse models have contributed to better understanding the molecular events that most likely lead to gliomagenesis.5 In this context it was reported that tumor location correlates with distinct molecular subtypes of gliomas, clinical outcomes, and growth patterns and therefore may reflect the genetic profile of tumor precursor cells.5–8 Applying non-invasive mapping techniques to better define the association of tumor location with the presence of key molecular characteristics is therefore of great interest, and several neuroimaging studies indeed reported tumor location predilection for specific molecular alterations in glioma patients, with the most compelling evidence currently existing for frontal predilection of isocitrate dehydrogenase (IDH)-mutated tumors.5,7,9–11
The aim of this study was to assess tumor location predilection for key molecular parameters in both glioblastoma and lower-grade glioma on a single voxel level with MRI using voxel lesion symptom mapping (VLSM). This approach allows much better spatial localization and discrimination inference compared with subdividing patients according to gross lesion location or using a classical anatomical atlas.12 Specifically we aimed to independently validate the existing body of evidence on tumor location predilection, but also apply this technique to a broader range of key molecular characteristics.
Patients and Methods
The study was approved by the local ethics committee, and informed consent was obtained from all patients. All patients with newly diagnosed histologically confirmed glioblastoma (in the period from August 2009 to May 2016) or newly diagnosed histologically confirmed lower-grade glioma (ie, diffuse astrocytoma and oligodendroglioma in the period from February 2009 to September 2017) were screened. Inclusion criteria were:
(i) Availability of key molecular parameters obtained from tissue specimens of the initial surgery (from patients undergoing surgical resection or biopsy at the Department of Neurosurgery, University of Heidelberg Medical Center and gathered according to the research procedures approved by the institutional review board at the Medical Faculty Heidelberg). For the glioblastoma cohort, key molecular parameters were derived from the Illumina Infinium HumanMethylation450k or HumanMethylation850k (EPIC) array. For the lower-grade glioma cohort, key molecular parameters were derived either from Illumina Infinium HumanMethylation450k or EPIC array or from targeted molecular analysis as described below.
(ii) Availability of corresponding pretreatment MRI study (acquired at the Department of Neuroradiology, University of Heidelberg Medical Center) prior to surgery. Patients were excluded if the MRI data were of insufficient quality resulting from motion artifacts or poor contrast injection.
A total of 237 patients in the glioblastoma cohort and 131 patients in the lower-grade glioma cohort met the outlined inclusion and exclusion criteria and served as the final cohorts for the present study.
MR Imaging and Postprocessing
Images were acquired in the routine clinical workup using a 3 tesla MR system (Magnetom Trio TIM/Prisma fit, Verio or Skyra, Siemens Healthcare) with a 12-channel head-matrix coil. Briefly, the protocol included T1-weighted 3D magnetization-prepared rapid acquisition with gradient echo (MPRAGE) images both before (T1) and after (cT1) administration of a 0.1 mmol/kg dose of gadoterate meglumine (Dotarem, Guerbet) as well as axial fluid attenuated inversion recovery (FLAIR) and axial T2-weighted images, as described previously.13 Sequence parameters for T1 and cT1 MPRAGE (3D sagittal or axial) were as follows: inversion time (TI) = 900–1100 ms, echo time (TE) = 3–4 ms, repetition time (TR) = 1710–2250 ms, and flip angle = 15°; for T2 (2D, axial): TE = 85–88 ms; TR = 2740–5950 ms; section thickness, 5 mm; spacing, 5.5 mm; for FLAIR (2D, axial): TI = 2400–2500 ms; TE = 85–135 ms; TR = 8500–10000 ms; section thickness, 5 mm; spacing, 5.5 mm.
Postprocessing of MRI data was performed as described previously.13,14 Briefly, the FMRIB software library (FSL, http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/FSL; accessed August 23, 2018) was used for image registration. First, brain voxels were isolated by generating a binary brain mask from the T1 volume using the brain extraction tool15 and transferred to all other imaging volumes (cT1, FLAIR, T2) for each patient. These image volumes were then registered to the brain extracted T1 volume using the linear image registration tool16,17 with a mutual information algorithm and a 6-degree of freedom transformation. T1 subtraction volumes (subT1) were generated by voxel-wise subtraction of the T1 from the cT1 volume. Tumor segmentation was performed by U.N., a radiology resident with 3 years of experience, and subsequently checked by P.K., a radiology resident with 5 years of experience, and D.B., a board-certified radiologist and neuro-radiologist with 15 years of experience in image processing (discrepancies were resolved through a consensus discussion). Specifically, we selected the contrast-enhancing (CE) portion of the whole tumor (on the subT1 images), the non-enhancing (NE) FLAIR hyperintense portion (defined as FLAIR hyperintense abnormality excluding the CE and necrotic [NEC] tumor portion, that is, including both FLAIR hyperintense tumor and potentially vasogenic edema), and the NEC portion of the tumor (on the cT1 images) using a region-growing segmentation algorithm implemented in ITK-SNAP (www.itksnap.org; accessed August 23, 201818) as described previously.19–21 The final tumor segmentation masks from each patient were either based on the (i) CE segmentation mask if CE but not NEC tumor was present, (ii) from a combined CE+NEC segmentation mask if both CE and NEC tumor were present, or (iii) from the NE segmentation mask if only NE but not CE and NEC tumor were present. A holefilling algorithm (using the imdilate function in Matlab) was applied to slightly dilate the segmentation mask and thus include single-voxel holes within the segmentation mask. Next, the precontrast T1 volumes from all patients were registered to a 2.0-mm isotropic brain atlas (Montreal Neurological Institute 152 [MNI-152] template) by using the linear image registration tool of FSL with a mutual information algorithm and a 12-degree of freedom transformation. The obtained coregistration matrices were applied to the corresponding final tumor segmentation masks to allow transformation of the tumor segmentation masks into the common stereotaxic MNI-152 space.
Molecular Analysis
For the glioblastoma cohort, key molecular parameters from the Illumina Infinium HumanMethylation450k or EPIC array (Illumina) were processed according to the manufacturer’s instructions at the Genomics and Proteomics Core Facility of the German Cancer Research Center as described previously.22 The following parameters were assessed:
O6-methylguanine DNA methyltransferase (MGMT) promoter methylation status (methylated vs unmethylated) was determined as described previously, using the “mgmtstp27” library in R version 3.4.0 (R Foundation for Statistical Computing, Vienna, Austria).23,24 For a subset of samples (n = 28) the “mgmtstp27” package failed to predict the MGMT promoter methylation status, and additional MGMT pyrosequencing was performed (n = 13/28).25
Copy-number variation analysis was performed (as described previously with the “conumee” package) of epidermal growth factor receptor (EGFR, amplified vs non-amplified), platelet-derived growth factor receptor alpha (PDGFRA, amplified vs non-amplified), MDM4 (amplified vs non-amplified), MDM2 (amplified vs non-amplified), MET (amplified vs non-amplified), N-myc proto-oncogene protein (MYCN, amplified vs non-amplified), cyclin-dependent kinase 4 (CDK4, amplified vs non-amplified), CDK6 (amplified vs non-amplified), phosphatase and tensin homolog (PTEN, loss vs balanced), cyclin-dependent kinase inhibitor 2A (CDKN2A, loss vs balanced), neurofibromin 1 (NF1, loss vs balanced), retinoblastoma 1 (RB1, loss vs balanced), tumor protein p53 (TP53, loss vs balanced), telomerase reverse transcriptase (TERT, gain vs balanced), and G1/S-specific cyclin-D2 (CCND2, gain vs balanced).22
A random forest algorithm compared each case with a brain tumor DNA methylation profile reference bank consisting of more than 2800 brain tumor cases to assign each patient to a molecular subgroup based on the individual global DNA methylation pattern (see www.molecularneuropathology.org/mnp/classifier/1; accessed August 23, 2018).22
For the lower-grade glioma cohort, key molecular parameters (IDH mutation and 1p/19q codeletion status) from the Infinium HumanMethylation450k or EPIC array (available in n = 46 patients) were obtained as described previously.22 For the remaining patients (n = 85), IDH mutation status was assessed with immunohistochemistry (IHC) for IDH1-R132H and DNA sequencing for IHC-negative cases, as described previously.26 Furthermore, detection of chromosome arms 1p and 19q deletions was performed in n = 76 cases with fluorescence in situ hybridization, as described previously.27
Voxel-Based Lesion Symptom Mapping
The relationship between tumor location and the assessed key molecular parameters (see Tables 1 and 2) was evaluated in both the glioblastoma and lower-grade glioma cohorts on a voxel-by-voxel basis by employing a VLSM approach implemented in Matlab (VLSM toolbox, version 2.55—http://www.aphasialab.org/vlsm; accessed August 23, 2018).28 We performed a separate VLSM analysis for each molecular parameter to identify tumor voxels that were strongly correlated with a specific molecular alteration. The results were corrected using a multiple comparison permutation test, with n = 1000 iterations. Finally, voxel t-values that were greater than the t-value in >95% of the permutations were retained in the VLSM results (with α = 0.05, power > 0.8).12
Table 1.
Analyzed key molecular parameters in the glioblastoma cohort
| Assessed Molecular Alteration | Assessed Number of Patients | Patients | VLSM | ||
|---|---|---|---|---|---|
| Presenting Alteration (1) | Not Presenting Alteration (0) | Proportion (%) Presenting Alteration | Permutation-Adjusted P-value | ||
| MGMT promoter methylation | 221 | 103 | 118 | 46.6 | 0.401 |
| Methylation class1 | |||||
| IDH glioma, subclass high-grade astrocytoma | 237 | 9 | 228 | 3.8 | 0.021 |
| Glioblastoma, IDH wildtype, subclass mesenchymal “mesenchymal” | 237 | 72 | 165 | 30.4 | 0.739 |
| Glioblastoma, IDH wildtype, subclass RTK I “PGFRA” | 237 | 36 | 201 | 15.2 | 0.353 |
| Glioblastoma, IDH wildtype, subclass RTK II “classic” | 237 | 108 | 129 | 45.6 | 0.276 |
| Glioblastoma, IDH wildtype, subclass midline “Midline” | 237 | 5 | 232 | 2.1 | 0.197 |
| Diffuse midline glioma H3 K27M mutant | 237 | 5 | 232 | 2.1 | 0.994 |
| Copy number variation | |||||
| EGFR amplification | 237 | 108 | 129 | 45.6 | 0.325 |
| PDGFRA amplification | 237 | 12 | 225 | 5.1 | 0.762 |
| MDM4 amplification | 236 | 22 | 214 | 9.3 | 0.515 |
| CDK4 amplification | 237 | 174 | 63 | 73.4 | 0.681 |
| PTEN loss | 233 | 186 | 47 | 79.8 | 0.570 |
| TP53 loss | 234 | 19 | 215 | 8.1 | 0.419 |
| CDKN2A loss | 237 | 108 | 129 | 45.6 | 0.747 |
| NF1 loss | 233 | 38 | 195 | 16.3 | 0.722 |
| RB1 loss | 233 | 67 | 166 | 28.8 | 0.148 |
| MYCN amplification | 237 | 5 | 232 | 2.1 | 0.586 |
| TERT gain | 232 | 10 | 222 | 4.3 | 0.289 |
| CDK6 amplification | 237 | 5 | 232 | 2.1 | 0.834 |
| MET amplification | 237 | 7 | 230 | 3.0 | 0.137 |
| CCND2 gain | 230 | 12 | 218 | 5.2 | 0.089 |
| MDM2 amplification | 237 | 14 | 223 | 5.9 | 0.622 |
Abbreviation: RTK, receptor tyrosine kinase.
Annotations: 1 = detailed explanation of methylation classes available via https://www.molecularneuropathology.org/mnp/classifier/1; accessed August 23, 2018.
Table 2.
Analyzed key molecular parameters in the lower-grade glioma cohort
| Assessed Molecular Alteration | Assessed Number of Patients | Patients: | VLSM: | ||
|---|---|---|---|---|---|
| Presenting Alteration (1) | Not Presenting Alteration (0) | Proportion (%) Presenting Alteration | Permutation-Adjusted P-value | ||
| IDH mutation | 131 | 114 | 17 | 87.0% | 0.013 |
| 1p/19q codeletion | 122 | 50 | 72 | 41.0% | 0.097 |
Statistical Analysis
Subsequent analysis was performed using R version 3.4.0. For those molecular parameters where the VLSM analysis revealed a significant association with a specific tumor location, we performed additional analysis. Specifically, we quantified the ratio of tumor voxels from the identified VLSM cluster located inside the patient’s individual final tumor segmentation mask. Normal distribution of the ratios was tested by the Shapiro–Wilk test. A paired t-test (for normally distributed measurements) or a Wilcoxon signed-rank test (for nonnormally distributed data) was used for evaluation. P-values <0.05 were considered significant.
Results
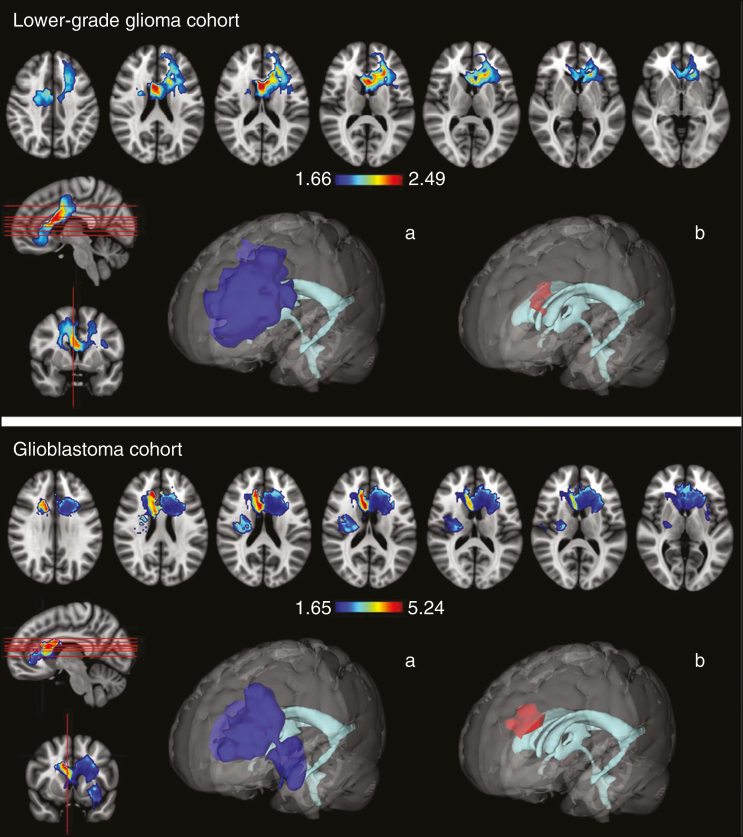
In the glioblastoma cohort, the VLSM analysis identified a significant tumor location predilection for IDH-mutant tumors (P = 0.021) with 2 significant voxel clusters, one located in the frontal lobe adjacent to the anterior horn of the lateral ventricles and the anterior portion of the corpus callosum (with t-values ranging from 1.65 up to 5.24), while the second one was located at the left temporal lobe near the insula (with t-values ranging from 1.65 to 2.50) (Fig. 1, upper panel). None of the remaining assessed key molecular parameters in the glioblastoma cohort (see Table 1) showed a significant tumor location predilection (permutation-adjusted P > 0.05 for all comparisons).
Fig. 1.
Heatmap of tumor voxel clusters that were significantly associated with the presence of IDH mutations in the glioblastoma (upper panel) and lower-grade glioma cohort (lower panel). Voxel-wise threshold of P < 0.05 based on a permutation test (n = 1000 iterations). 3D models of the identified tumor voxel cluster, including (a) all identified voxels and (b) those showing the highest t-values.
In the lower-grade glioma cohort, the VLSM analysis identified, concordant with the results obtained in the glioblastoma cohort, again a significant tumor location predilection for IDH-mutant tumors (P = 0.013) with a similarly located voxel cluster in the frontal lobe, adjacent to the anterior horn of the lateral ventricles and the anterior portion of the corpus callosum (with t-values ranging from 1.66 to 2.45) (Fig. 1, lower panel). No significant tumor location predilection was found for 1p/19q codeleted tumors (permutation-adjusted P = 0.097).
The median ratio of tumor voxels within the identified IDH-related VLSM clusters in the glioblastoma cohort was 9.5% (interquartile range [IQR], 0.9–24.9%) for patients with IDH-mutant and 0.0% (IQR, 0.0–3.1%) for IDH-wildtype tumors (Wilcoxon rank-sum test P = 0.008). Similarly, in the lower-grade glioma cohort, the median ratio of tumor voxels within the identified IDH-related VLSM cluster was 5.7% (IQR, 0.1–26.0%) for patients with IDH-mutant and 0.0% (IQR, 0.0–0.2%) for IDH-wildtype tumors (Wilcoxon rank-sum test P < 0.001).
Beyond the outlined results based on the final tumor segmentation masks, we found no significant tumor location predilection for any of the remaining tumor segmentation combinations (ie, CE tumor segmentation mask only, NE tumor segmentation mask only, or total tumor segmentation mask consisting of CE + NE + NEC) in both cohorts (permutation-adjusted P > 0.05 for all comparisons).
Discussion
We applied a statistically robust voxel-by-voxel analysis to identify whether key molecular parameters in 2 large cohorts of patients with glioblastoma and lower-grade glioma are associated with a predilection for specific brain regions. The principal finding of our study is that IDH-mutated tumors in both glioblastoma and lower-grade glioma cohorts show a significant and concordant predilection that included the rostral extension of the lateral ventricles (Fig. 1). Our results support findings from previous studies7,8,29 and consequently underpin the possibility of the presence of neural stem cells (NSCs) in the subventricular zone (SVZ) as the origin of IDH-mutant gliomas. Specifically, NSCs have been isolated in the tissues of the SVZ, which lines the lateral ventricles, the dentate gyrus of the hippocampus, and the subcortical white matter tracts.30–34 Since mutations of the IDH genes are considered an important event that occurs at an early stage during gliomagenesis, the observed predilection of IDH-mutant gliomas around the rostral extension of the lateral ventricles may resemble the consequence of early gliomagenesis. This location may serve as a nidus for tumor cell repopulation and recurrence and thus could potentially impact treatment decisions (eg, for the delivery of dose-escalated radiotherapy). Chen et al recently provided an intriguing explanation for why the NSCs in the prefrontal cortex around the rostral lateral ventricles, which was also identified as the single most significant tumor location predilection in IDH-mutant tumors within our study, are especially vulnerable to the formation and growth of IDH-mutant gliomas. They suggested that the evolutionary specialization of the prefrontal cortex for high glutamate neurotransmitter flux, which contributes to human cognitive abilities, creates a metabolic niche that inadvertently supports the growth of IDH-mutant gliomas in this specific location.35 Indirectly, the findings from our study also support previous evidence showing a higher incidence of proneural tumors around the rostral extension of the lateral ventricles,8 since IDH mutation is known to be one of the constitutive mutations of this molecular subgroup.36
The presence of 1p/19q codeleted tumors is limited to a subset of tumors with IDH mutations and thus may show a similar anatomical predilection as shown for IDH-mutant tumors in general. The lack of significance for this comparison (1p/19q codeleted vs non-codeleted tumors) in our study can be attributed to the confounding effect introduced by IDH-mutant, 1p/19q non-codeleted tumors.
Contrasting previous neuroimaging studies, we did not find tumor location predilection for the remaining assessed key molecular parameters, most notably not for MGMT promoter methylation, PTEN loss or EGFR amplification, where a tumor location predilection has been postulated.8 Nevertheless, the findings from our study are in line with previous molecular studies indicating that EGFR amplification is not a driver of initial oncogenic events, but rather contributes to tumor progression or maintenance through amplification arising at later stages of tumor formation. Moreover, loss of PTEN was also reported to play only a supportive role rather than serving as a tumor initiator during gliomagenesis.37,38 Overall, founder events in IDH-wildtype gliomas remain to be more clearly identified39 and the role of a biologically driven tumor location predilection for specific key molecular characteristics in IDH-wildtype gliomas therefore remains to be established.
Despite integrating molecular and imaging data from 2 large cohorts of lower-grade glioma and glioblastoma, we acknowledge the following limitations: due to the presence of class imbalance for low-frequency molecular alterations, further tumor location predilections may have been detected in larger study cohorts. Moreover, molecular parameters were primarily obtained from DNA methylation arrays, thereby limiting the assessment of tumor-location predilection for somatic point mutations (which was confined to the assessment of IDH mutation status in the present study). Future multiplatform analysis beyond DNA methylation arrays are required to assess the relevance of additional hallmark genomic abnormalities (such as mutation status of alpha thalassemia/mental retardation syndrome X-linked40). Finally, the presence of intratumoral heterogeneity may have affected the generalizability of the extracted molecular features from a single tumor specimen. Nevertheless, several studies found that, for instance, IDH mutation status41 and MGMT methylation level42–45 are consistent throughout individual gliomas. Whether this homogeneity is also applicable to the assessed copy-number variations and/or global DNA methylation subgroups remains to be investigated in future studies.
In conclusion, our study highlights the unique properties of IDH mutations and underpins the hypothesis that the rostral extension of the lateral ventricles is a potential location for the cell of origin in IDH-mutant gliomas.
Funding
P.K. is funded through the Medical Faculty Heidelberg Postdoc Program and the Else-Kröner Memorial Scholarship of the Else Kröner-Fresenius Foundation.
Acknowledgments
DNA methylation analysis was performed as part of the National Center for Tumor Diseases (NCT) Precision Oncology Program of the Heidelberg Center for Personalized Oncology (German Cancer Research Center–HIPO). We would further like to thank the Genomics and Proteomics Core Facility of the German Cancer Research Center (DKFZ) for excellent technical assistance.
Conflict of interest statement. None declared.
Authorship statement. Conceived and designed the research: Miguel Angel Tejada Neyra, Ulf Neuberger, Philipp Kickingereder. Acquired the data: Ulf Neuberger, Annekathrin Reinhardt, David Bonekamp, Martin Sill, Antje Wick, David T. W. Jones, Alexander Radbruch, Andreas Unterberg, Jürgen Debus, Sabine Heiland, Christel Herold-Mende, Stefan Pfister, Andreas von Deimling, Wolfgang Wick, David Capper, Martin Bendszus, Philipp Kickingereder. Analyzed and interpreted the data: Miguel Angel Tejada Neyra, Ulf Neuberger, Annekathrin Reinhardt, Gianluca Brugnara, David Bonekamp, Martin Sill, David T. W. Jones, Christel Herold-Mende, Stefan Pfister, Andreas von Deimling, Wolfgang Wick, David Capper, Martin Bendszus, Philipp Kickingereder. Performed statistical analysis: Miguel Angel Tejada Neyra, Ulf Neuberger, Philipp Kickingereder. Handled funding and supervision: Philipp Kickingereder. Drafted the manuscript: Miguel Angel Tejada Neyra, Ulf Neuberger, Gianluca Brugnara, Philipp Kickingereder. Made critical revision of the manuscript for important intellectual content: all authors
References
- 1. Brat DJ, Verhaak RG, Aldape KD, et al. ; Cancer Genome Atlas Research Network Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372(26):2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ceccarelli M, Barthel FP, Malta TM, et al. ; TCGA Research Network Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164(3):550–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 4. Visvader JE. Cells of origin in cancer. Nature. 2011;469(7330):314–322. [DOI] [PubMed] [Google Scholar]
- 5. Cahill D, Turcan S. Origin of gliomas. Semin Neurol. 2018;38(1):5–10. [DOI] [PubMed] [Google Scholar]
- 6. Alcantara Llaguno SR, Wang Z, Sun D, et al. Adult lineage-restricted CNS progenitors specify distinct glioblastoma subtypes. Cancer Cell. 2015;28(4):429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lai A, Kharbanda S, Pope WB, et al. Evidence for sequenced molecular evolution of IDH1 mutant glioblastoma from a distinct cell of origin. J Clin Oncol. 2011;29(34):4482–4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ellingson BM, Lai A, Harris RJ, et al. Probabilistic radiographic atlas of glioblastoma phenotypes. AJNR Am J Neuroradiol. 2013;34(3):533–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ellingson BM, Zaw T, Cloughesy TF, et al. Comparison between intensity normalization techniques for dynamic susceptibility contrast (DSC)-MRI estimates of cerebral blood volume (CBV) in human gliomas. J Magn Reson Imaging. 2012;35(6):1472–1477. [DOI] [PubMed] [Google Scholar]
- 10. Sun ZL, Chan AK, Chen LC, et al. TERT promoter mutated WHO grades II and III gliomas are located preferentially in the frontal lobe and avoid the midline. Int J Clin Exp Pathol. 2015;8(9):11485–11494. [PMC free article] [PubMed] [Google Scholar]
- 11. Gorovets D, Kannan K, Shen R, et al. IDH mutation and neuroglial developmental features define clinically distinct subclasses of lower grade diffuse astrocytic glioma. Clin Cancer Res. 2012;18(9):2490–2501. [DOI] [PubMed] [Google Scholar]
- 12. Kimberg DY, Coslett HB, Schwartz MF. Power in Voxel-based lesion-symptom mapping. J Cogn Neurosci. 2007;19(7):1067–1080. [DOI] [PubMed] [Google Scholar]
- 13. Kickingereder P, Neuberger U, Bonekamp D, et al. Radiomic subtyping improves disease stratification beyond key molecular, clinical, and standard imaging characteristics in patients with glioblastoma. Neuro Oncol. 2018;20(6):848–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kickingereder P, Götz M, Muschelli J, et al. Large-scale radiomic profiling of recurrent glioblastoma identifies an imaging predictor for stratifying anti-angiogenic treatment response. Clin Cancer Res. 2016;22(23):5765–5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Smith SM. Fast robust automated brain extraction. Hum Brain Mapp. 2002;17(3):143–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage. 2002;17(2):825–841. [DOI] [PubMed] [Google Scholar]
- 17. Jenkinson M, Smith S. A global optimisation method for robust affine registration of brain images. Med Image Anal. 2001;5(2):143–156. [DOI] [PubMed] [Google Scholar]
- 18. Yushkevich PA, Piven J, Hazlett HC, et al. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage. 2006;31(3):1116–1128. [DOI] [PubMed] [Google Scholar]
- 19. Kickingereder P, Radbruch A, Burth S, et al. MR perfusion-derived hemodynamic parametric response mapping of bevacizumab efficacy in recurrent glioblastoma. Radiology. 2015;151172. [DOI] [PubMed] [Google Scholar]
- 20. Bonekamp D, Mouridsen K, Radbruch A, et al. Assessment of tumor oxygenation and its impact on treatment response in bevacizumab-treated recurrent glioblastoma. J Cereb Blood Flow Metab. 2017;37(2):485–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kickingereder P, Neuberger U, Bonekamp D, et al. Radiomic subtyping improves disease stratification beyond key molecular, clinical, and standard imaging characteristics in patients with glioblastoma. Neuro Oncol. 2018;20(6):848–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Capper D, Jones DTW, Sill M, et al. DNA methylation-based classification of central nervous system tumours. Nature. 2018;555(7697):469–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bady P, Sciuscio D, Diserens AC, et al. MGMT methylation analysis of glioblastoma on the Infinium methylation BeadChip identifies two distinct CpG regions associated with gene silencing and outcome, yielding a prediction model for comparisons across datasets, tumor grades, and CIMP-status. Acta Neuropathol. 2012;124(4):547–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bady P, Delorenzi M, Hegi ME. Sensitivity analysis of the MGMT-STP27 model and impact of genetic and epigenetic context to predict the MGMT methylation status in gliomas and other tumors. J Mol Diagn. 2016;18(3):350–361. [DOI] [PubMed] [Google Scholar]
- 25. Wiestler B, Capper D, Hovestadt V, et al. Assessing CpG island methylator phenotype, 1p/19q codeletion, and MGMT promoter methylation from epigenome-wide data in the biomarker cohort of the NOA-04 trial. Neuro Oncol. 2014;16(12):1630–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kickingereder P, Sahm F, Radbruch A, et al. IDH mutation status is associated with a distinct hypoxia/angiogenesis transcriptome signature which is non-invasively predictable with rCBV imaging in human glioma. Sci Rep. 2015;5:16238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lass U, Hartmann C, Capper D, et al. Chromogenic in situ hybridization is a reliable alternative to fluorescence in situ hybridization for diagnostic testing of 1p and 19q loss in paraffin-embedded gliomas. Brain Pathol. 2013;23(3):311–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bates E, Wilson SM, Saygin AP, et al. Voxel-based lesion-symptom mapping. Nat Neurosci. 2003;6(5):448–450. [DOI] [PubMed] [Google Scholar]
- 29. Wang Y, Zhang T, Li S, et al. Anatomical localization of isocitrate dehydrogenase 1 mutation: a voxel-based radiographic study of 146 low-grade gliomas. Eur J Neurol. 2015;22(2):348–354. [DOI] [PubMed] [Google Scholar]
- 30. Sanai N, Alvarez-Buylla A, Berger MS. Neural stem cells and the origin of gliomas. N Engl J Med. 2005;353(8):811–822. [DOI] [PubMed] [Google Scholar]
- 31. Li G, Fang L, Fernández G, Pleasure SJ. The ventral hippocampus is the embryonic origin for adult neural stem cells in the dentate gyrus. Neuron. 2013;78(4):658–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Muñoz DM, Guha A. Mouse models to interrogate the implications of the differentiation status in the ontogeny of gliomas. Oncotarget. 2011;2(8):590–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nunes MC, Roy NS, Keyoung HM, et al. Identification and isolation of multipotential neural progenitor cells from the subcortical white matter of the adult human brain. Nat Med. 2003;9(4):439–447. [DOI] [PubMed] [Google Scholar]
- 34. Jacques TS, Swales A, Brzozowski MJ, et al. Combinations of genetic mutations in the adult neural stem cell compartment determine brain tumour phenotypes. EMBO J. 2010;29(1):222–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen R, Nishimura MC, Kharbanda S, et al. Hominoid-specific enzyme GLUD2 promotes growth of IDH1R132H glioma. Proc Natl Acad Sci U S A. 2014;111(39):14217–14222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Verhaak RG, Hoadley KA, Purdom E, et al. ; Cancer Genome Atlas Research Network Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ozawa T, Riester M, Cheng YK, et al. Most human non-GCIMP glioblastoma subtypes evolve from a common proneural-like precursor glioma. Cancer Cell. 2014;26(2):288–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hu X, Pandolfi PP, Li Y, Koutcher JA, Rosenblum M, Holland EC. mTOR promotes survival and astrocytic characteristics induced by Pten/AKT signaling in glioblastoma. Neoplasia. 2005;7(4):356–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Reitman ZJ, Winkler F, Elia AEH. New directions in the treatment of glioblastoma. Semin Neurol. 2018;38(1):50–61. [DOI] [PubMed] [Google Scholar]
- 40. Wiestler B, Capper D, Holland-Letz T, et al. ATRX loss refines the classification of anaplastic gliomas and identifies a subgroup of IDH mutant astrocytic tumors with better prognosis. Acta Neuropathol. 2013;126(3):443–451. [DOI] [PubMed] [Google Scholar]
- 41. Lass U, Nümann A, von Eckardstein K, et al. Clonal analysis in recurrent astrocytic, oligoastrocytic and oligodendroglial tumors implicates IDH1-mutation as common tumor initiating event. PLoS One. 2012;7(7):e41298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hamilton MG, Roldán G, Magliocco A, McIntyre JB, Parney I, Easaw JC. Determination of the methylation status of MGMT in different regions within glioblastoma multiforme. J Neurooncol. 2011;102(2):255–260. [DOI] [PubMed] [Google Scholar]
- 43. Grasbon-Frodl EM, Kreth FW, Ruiter M, et al. Intratumoral homogeneity of MGMT promoter hypermethylation as demonstrated in serial stereotactic specimens from anaplastic astrocytomas and glioblastomas. Int J Cancer. 2007;121(11):2458–2464. [DOI] [PubMed] [Google Scholar]
- 44. Dunn J, Baborie A, Alam F, et al. Extent of MGMT promoter methylation correlates with outcome in glioblastomas given temozolomide and radiotherapy. Br J Cancer. 2009;101(1):124–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. van Thuijl HF, Mazor T, Johnson BE, et al. Evolution of DNA repair defects during malignant progression of low-grade gliomas after temozolomide treatment. Acta Neuropathol. 2015;129(4):597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]