Abstract
Enzymes in the respiratory chain are increasingly seen as potential targets against multi-drug resistance of human pathogens and cancerous cells. However, a detailed understanding of the mechanism and specificity determinants of known inhibitors is still lacking. Oligomycin, for example, has been known to be an inhibitor of the membrane motor of the mitochondrial ATP synthase for over five decades, and yet little is known about its mode of action at the molecular level. In a recent breakthrough, a crystal structure of the S. cerevisiae c-subunit ring with bound oligomycin revealed the inhibitor docked on the outer face of the proton-binding sites, deep into the transmembrane region. However, the structure of the complex was obtained in an organic solvent rather than detergent or a lipid bilayer, and therefore it has been unclear whether this mode of recognition is physiologically relevant. Here, we use molecular dynamics simulations to address this question and gain insights into the mechanism of oligomycin inhibition. Our findings lead us to propose that oligomycin naturally partitions into the lipid/water interface, and that in this environment the inhibitor can indeed bind to any of the c-ring proton-carrying sites that are exposed to the membrane, thereby becoming an integral component of the proton-coordinating network. As the c-ring rotates within the membrane, driven either by downhill proton permeation or ATP hydrolysis, one of the protonated, oligomycin-bound sites eventually reaches the subunit-a interface and halts the rotary mechanism of the enzyme.
Keywords: Multi-drug resistance, respiratory chain, membrane bioenergetics, membrane plasticity, molecular simulation
1. INTRODUCTION
The ATP synthase is a paradigmatic membrane-bound enzyme whose function and basic architecture are conserved from prokaryotes to humans [1, 2]. The enzyme produces the majority of cellular ATP, synthesized from ADP and Pi, through a mechanism that is energized by the transmembrane flux of protons down an electrochemical gradient (or, in some bacterial species, sodium ions [3–5]). This mechanism is reversible, and thus the enzyme can also pump protons uphill, driven by ATP hydrolysis. In either mode, catalysis and proton transport occur in two distinct but mechanically coupled domains, one exposed to the mitochondrial matrix (or the cytosol/stroma), and another embedded in the membrane. Proton transport in particular involves two membrane-integral proteins [6–8]: the c-ring, which is a circular oligomer of subunits-c, the number of which varies across species [9]; and directly adjacent to the ring but barely in contact, subunit-a [10]. Owing to its structural symmetry, the c-ring has the inherent ability to rotate against subunit-a, but despite its pore-like architecture, none of the ATP synthase c-rings of known structure are known to form transmembrane channels; their interior, which is highly hydrophobic, is blocked by lipids [11–13]. On the outer face of the ring, however, approximately halfway across the transmembrane span, each of the c-subunits harbors a proton-binding site featuring a conserved acidic residue [14–18]. At any given time, the majority of these sites are exposed to the core of the lipid bilayer, and proton-loaded [19]. However, as the c-ring rotates these sites become sequentially exposed to the interface with subunit-a, where they first unload the bound proton, to one side of the membrane, and subsequently reload another, from the other side [10, 19]. Thus, the trajectory of each proton as it permeates the membrane follows the c-ring as it carries out a near complete revolution, through a series of discrete rotational steps, in each of which a new proton is captured while another is released.
Owing to its essential role in cellular bioenergetics, the ATP synthase is considered a potential last-resort target against drug-resistant forms of aerobic pathogenic bacteria and cancer cells. The membrane domain is particularly appealing, as any viable drug must be able to partition into the lipid bilayer. Indeed, a number of inhibitors of this domain are known, some of which display significant specificity. Oligomycin and apoptolidin, for example, inhibit the mitochondrial ATP synthase but not the bacterial enzymes, and have been reported to have anti-cancer activity [20–22]. By contrast, the compound known as bedaquiline, FDA-approved as Sirturo, has been reported to selectively target mycobacterial enzymes, including that of M. tuberculosis [23–26], although evidence of mitochondrial function inhibition [27] suggest this drug might be less selective than originally thought.
Arguably, efforts to develop novel drugs targeting the membrane domain of the ATP synthase would be fostered by a better understanding of the mechanism of action and specificity determinants of known inhibitors. It remains to be clarified, for example, where these inhibitors are first recognized, and where the site of inhibition is; these two sites do not necessarily coincide in an inherently dynamic structure. A case in point is that of oligomycin, which has been known to be an inhibitor of the ATP synthase for over five decades [28, 29]. A high-resolution crystal structure of the S. cerevisiae c10−ring was recently determined in complex with oligomycin, showing four inhibitor molecules bound to four of the proton-binding sites, on the outer surface of the ring [30]. This structure is unlike that of other c-rings in that it was obtained in an organic solvent, consisting mainly of 2-methyl-2,4-pentanediol and water, rather than in detergent or a lipidic phase. By providing a more aqueous medium, this solvent might resemble the environment of the a-c interface more than the interior of the lipid membrane [17]. Taken together with earlier genetic evidence that mutations in both subunits-a and -c confer S. cerevisiae with resistance to inhibition [31, 32], this structural work has led to the proposal that the oligomycin-binding site is at the face of the c-ring that is positioned at the proton-access channel formed by subunit-a, and that the inhibitor does not bind to the c-subunits facing the lipid bilayer [30]. Seemingly at odds with this interpretation, however, is a subsequent crystal structure of the c9−ring from a mycobacterial species in complex with bedaquiline [33]. Like oligomycin, this inhibitor is seen docked onto the outer face of the proton-binding sites of the c-ring, but unlike the S. cerevisiae study, the mycobacterial structure was obtained in a detergent that more clearly resembles a membrane. Thus, it was suggested that bedaquiline is first recognized by c-subunits exposed to the lipid bilayer, and that it subsequently inhibits the enzyme by stalling the rotation of the c-ring, as the drug-bound sites would be unable to enter the a-c interface [33]. The take-away message from these studies is therefore that the modes of recognition of bedaquiline and oligomycin are very different. This notion seems plausible, given the fact that these inhibitors are chemically and structurally also very different, and indeed this different mode of recognition might contribute to their reported specificity.
The above reasoning notwithstanding, we set out to examine in more detail the mode of oligomycin recognition by the mitochondrial c10−ring, using atomically-detailed molecular dynamics simulations. First, we will gain insights into the physicochemical properties of the inhibitor and in particular its inherent solvation preferences. Then, we seek to reproduce the experimental finding that oligomycin binds to the c-ring in an aqueous organic solvent, and to rationalize it at the molecular level. We then evaluate whether this same mode of interaction is viable and stable in the lipid membrane, both for the wild-type protein as well as for a variant with mutations that confer S. cerevisiae with resistance to oligomycin, which serves as a negative control. Finally, we discuss the mechanistic implications of our findings.
2. RESULTS
2.1. Oligomycin is strongly amphipathic -
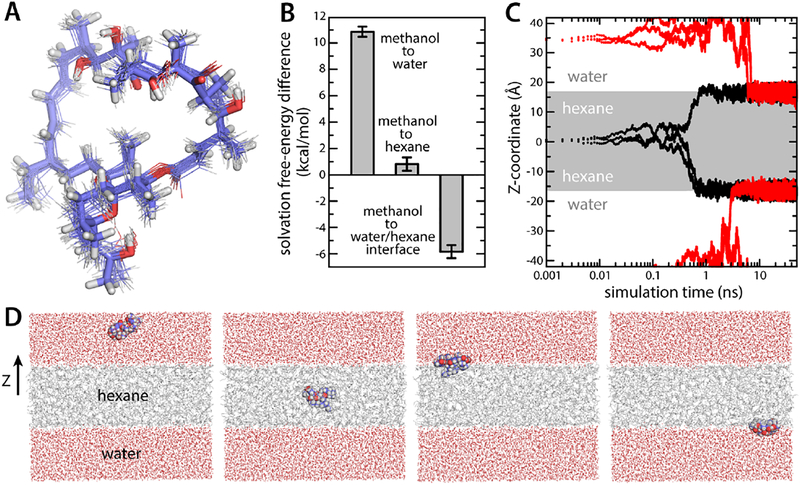
To begin to understand how the ATP synthase recognizes oligomycin, we first sought to characterize the intrinsic solvation preferences of the inhibitor, using molecular dynamics (MD) simulations. To our knowledge, no experimental partition coefficients are known any of the isoforms of this compound. At the onset of this study, no force-field parameters were available either. Thus, a set of parameters was first developed, using the GAAMP portal [34]; this parameterization followed a fully automated procedure, except for a few dihedral-angle parameters, which were individually adjusted so that the energy-minimized molecule would better reproduce the geometry observed in the crystal structure of isolated oligomycin. Accordingly, in room-temperature MD simulations, either in methanol or water, the inhibitor geometry remained close to the experimental structure, on average (Fig. 1A). The solvation free energies of the inhibitor in water, methanol and hexane were then computed and compared. This comparison reveals a strong preference for methanol over water (~11 kcal/mol, Fig. 1B), consistent with the fact that oligomycin is insoluble in water. By contrast, purely hydrophobic hexane is slightly disfavored over methanol, but only by about ~1 kcal/mol (Fig. 1B), consistent with the empirical observation that oligomycin permeates the plasma and mitochondrial membranes before reaching the ATP synthase. The preferred state of the inhibitor, however, is at the interface between a hydrophobic and a hydrophilic phase. As shown in Fig. 1CD, MD simulations using an artificial hexane layer flanked by water consistently show oligomycin migrating to the interface, irrespective of its initial location; indeed, this interfacial state is more favorable than pure methanol by about ~6 kcal/mol (Fig. 1B). At low dilution, therefore, partitioning of oligomycin into the water-lipid interface is effectively irreversible, but the inhibitor can permeate the hydrophobic core of the membrane, only hampered by an energy barrier of ~7 kcal/mol. Escape from the membrane, however, is energetically much more costly, and likely requires prior accumulation of the inhibitor beyond a certain threshold. This process might also entail the formation of some kind of molecular aggregate, to counter the pronounced water insolubility of the inhibitor.
Figure 1. Structure and amphipathic character of oligomycin in simulation.
(A) Twenty randomly selected configurations of oligomycin from a simulation are overlaid onto its crystallized structure (thick sticks). The average RMS deviation in water and methanol is 0.7 and 0.8 Å, respectively. (B) Solvation free energies in pure water, pure hexane and at a water/hexane interface, relative to that in methanol. (C) Migration of oligomycin to the water-hexane interface in MD simulations. Oligomycin was initially placed either in the center of a hydrated hexane layer (indicated in grey) (black traces) or in bulk water (red traces). (D) Selected snapshots of the simulations analyzed in panel (C). The two figures on the left show two initial configurations, and the two figures on the right are snapshots at 50 ns.
2.2. Oligomycin binds to the protonated form of the c-ring binding sites -
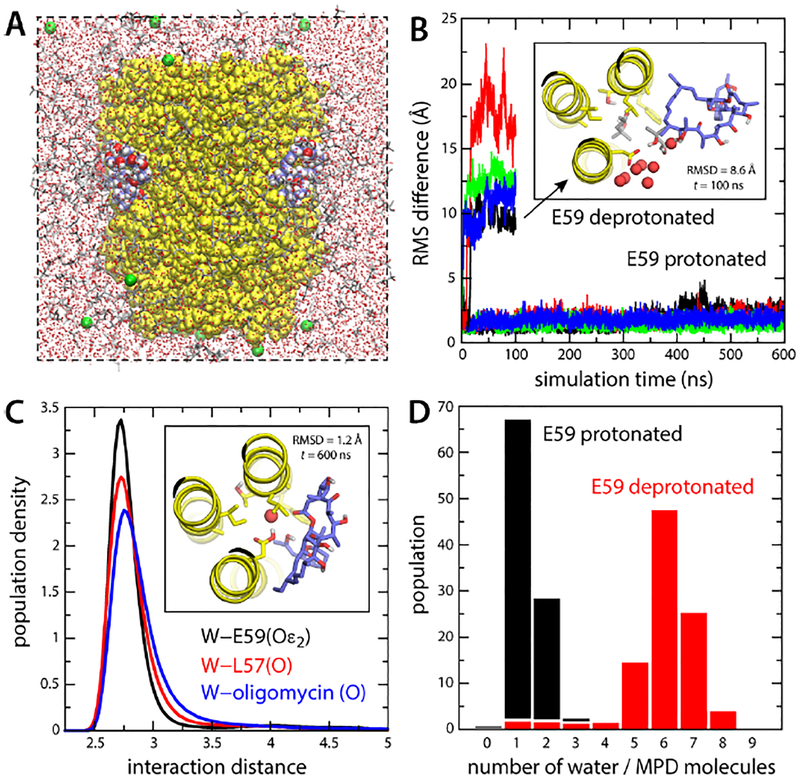
As mentioned, a high-resolution structure of the S. cerevisiae c10−ring in complex with oligomycin has been determined in an aqueous solvent whose primary component is 2-methyl-2,4-pentanediol (MPD) [30]. This compound is much more amphipathic than, for example, N-dodecyl-beta-D-maltoside (DDM), which is often used in membrane-protein structural studies; PubChem predicts the logP value for MPD is 0.3, while that of DDM is 1.4, i.e. MPD is 10 times more hydrophilic that DDM. Despite the presence of this precipitant in the crystallization buffer, the c10−ring assembly was found to be stable, and its structure could be determined at 1.9 Å resolution [30]. In this structure, four inhibitor molecules can be clearly seen bound on the outer face of four proton-binding sites. However, it cannot be discerned from the crystallographic data whether oligomycin selectively recognizes the protonated or deprotonated form of these sites, both of which are thought to co-exist in this aqueous, amphipathic solvent [17]. If, hypothetically, oligomycin required the sites to be deprotonated, this finding would imply the site of recognition is at the interface between the c-ring and subunit-a. To determine which form is recognized, we carried out two independent MD simulations of protein-inhibitor complex (Fig. 2A). In the sites with bound oligomycin, the key residue mediating proton transport, Glu59, was constitutively protonated in one simulation and deprotonated in the other (the other six sites were protonated in both cases). In both simulations, the protein-inhibitor complex was immersed in a mixture of 30% MPD and 70% water (volume percentages), to mimics the buffer used in the structural study.
Figure 2. Simulations of the S. cerevisiae c10 ring with bound oligomycin, in an organic solvent.
(A) Simulation system comprising the c-ring (yellow) and four bound oligomycin molecules (blue/red/white spheres). The solvent is a mixture of MPD (red/grey sticks) and water (small red spheres). K+ ions (green spheres) neutralize the total charge. Hydrogen atoms in the solvent are omitted for clarity. (B) RMS deviation of each of the oligomycin molecules relative to the experimental binding pose (no hydrogens), as a function of simulation time, when Glu59 is protonated or deprotonated. The inset shows a snapshot of an oligomycin molecule partially dissociated when Glu59 is deprotonated, viewed from the matrix. Only 3 helices of the c-ring are shown for clarity (yellow). Residues lining the binding site, including Glu59, are highlighted (sticks). Neighboring water and MPD molecules are highlighted. (C) Persistence of the water-mediated network stabilizing oligomycin for protonated Glu59. The plot shows probability distributions, from a 600-ns trajectory, for the distance between this water molecule (oxygen atom) and the H-bond donors/acceptors in Glu59 (black), Leu57 (red) and oligomycin (blue). The inset shows one of the bound inhibitors at the end of the simulation. (D) Histogram of the number of water and MPD oxygen atoms coordinating Glu59, averaged over the four oligomycin sites, when Glu59 is protonated or deprotonated.
As shown in Fig. 2B, the simulation in which Glu59 is protonated revealed no significant changes in the arrangement of the protein-inhibitor complex over 600 ns of unrestricted dynamics. All four oligomycin molecules remained stably bound to the c-ring, in the same binding pose as that observed in the X-ray structure. A seemingly important, water-mediated interaction between the inhibitor, the side-chain of Glu59 and the backbone carbonyl of Leu57, already present in the experimental structure, was fully preserved in this time-scale for all four inhibitor molecules (Fig. 2C). In contrast, when Glu59 was deprotonated all four oligomycin molecules dissociated from the c-ring within 100 ns of simulation time (Fig. 2B). Inspection of individual snapshots during this simulation revealed that the inhibitor molecules were displaced by water and MPD entering the binding site, seemingly to better solvate the ionized carboxyl group of Glu59 (Fig. 2B). Indeed, the number of water and/or MPD molecules in proximity to this group increased from 1–2 in the protonated form to 6–7 in the deprotonated state, on average (Fig. 2D). We therefore conclude that in the crystal structure of the S. cerevisiae c 10-ring in complex with oligomycin, the proton-binding sites bound to the inhibitor are protonated, and that oligomycin would not bind to a deprotonated c-subunit site. This finding begins to define the context in which the inhibitor can be recognized in the operating enzyme. The protonated state is constitutive for the c-subunits that are exposed to the lipid membrane, while those at the a-c interface may exchange between the protonated and deprotonated forms reversibly.
2.3. Oligomycin binding requires a partially aqueous environment -
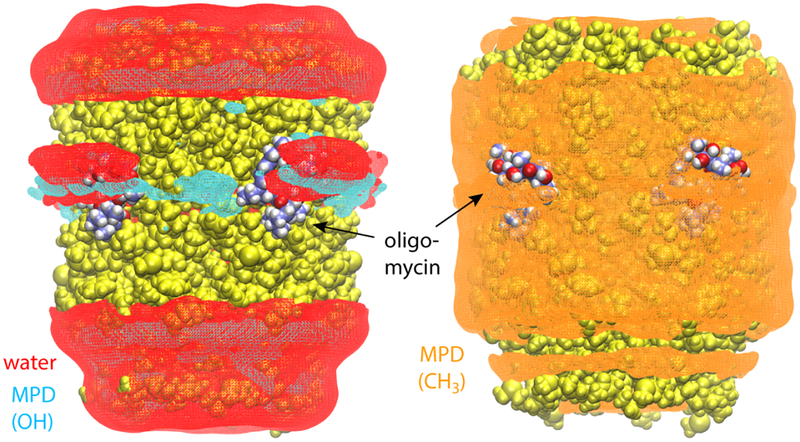
That oligomycin is strongly amphipathic seems to be at odds with the observation that it binds to sites on the c-ring that are deep within the hydrophobic span of the protein (Fig. 2A). This observation, however, stems from a crystal structure obtained in a medium that is much more hydrophilic than a lipid membrane [30]. Indeed, using MD simulations analogous to those carried out here, but in the absence of oligomycin, we have previously shown that the MPD/water buffer used during crystallization of the c10−ring is a viable protein solvent because it becomes non-homogenous on the protein surface [17]. Specifically, the solvent molecules redistribute so that the MPD methyl groups coat the hydrophobic transmembrane region of the protein, both on the outer face and within the central pore, while simultaneously water molecules and MPD hydroxyl groups provide hydrogen-bonding interactions for the polar regions, on both sides of the transmembrane span (as well as the proton-binding sites). Here, we observe that a similar partitioning of the MPD/water solvent is what appears to stabilize oligomycin on the surface of the c-ring (Fig. 3), by accommodating its strong amphipathic character. Specifically, water molecules and MPD hydroxyl groups preferentially localize around the exposed polar regions of the inhibitor, while methyl groups solubilize the exposed hydrophobic groups. (Note that MPD and water molecules were uniformly distributed and randomly oriented at the beginning of the simulation.) By contrast, a control simulation in which the protein-inhibitor complex was embedded in an artificial hexane layer, which by design does not allow for water penetration, showed all four oligomycin molecules dissociate within 100 ns, even though Glu59 was protonated (Fig. 4).
Figure 3. Partitioning of the MPD/water solvent on the surface of the ring-oligomycin complex.
The figure shows iso-surfaces of calculated 3D density maps for water and MPD within interaction distance from the protein-inhibitor complex (either via hydrogen-bonding or hydrophobic contacts), derived from the simulated trajectory (same as that in Fig. 2). The water-density map is shown in red. Density maps for the MPD hydroxyl and methyl groups, calculated separately, are shown in cyan and orange, respectively. The protein and inhibitor are shown as spheres, colored as in Fig. 2.
Figure 4. Oligomycin binding requires a partially aqueous environment.
(A) Simulation of the S. ceravisiae c10 ring with protonated Glu59, embedded in an artificial water-impermeable hexane layer. The protein and solvent are represented as in Fig. 2A. Initially four oligomycin molecules are bound to the cring. (B) RMS deviation of each of the four oligomycin molecules relative to their binding poses in the crystal structure, as a function of simulation time. Fast, spontaneous dissociation of all four inhibitor molecules indicates the bound state is not stable in the context of a purely hydrophobic environment.
2.4. Membrane plasticity foster oligomycin binding to the c-ring in lipid -
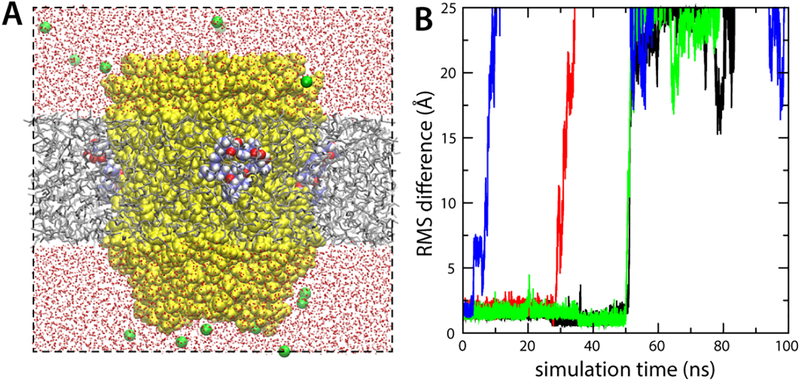
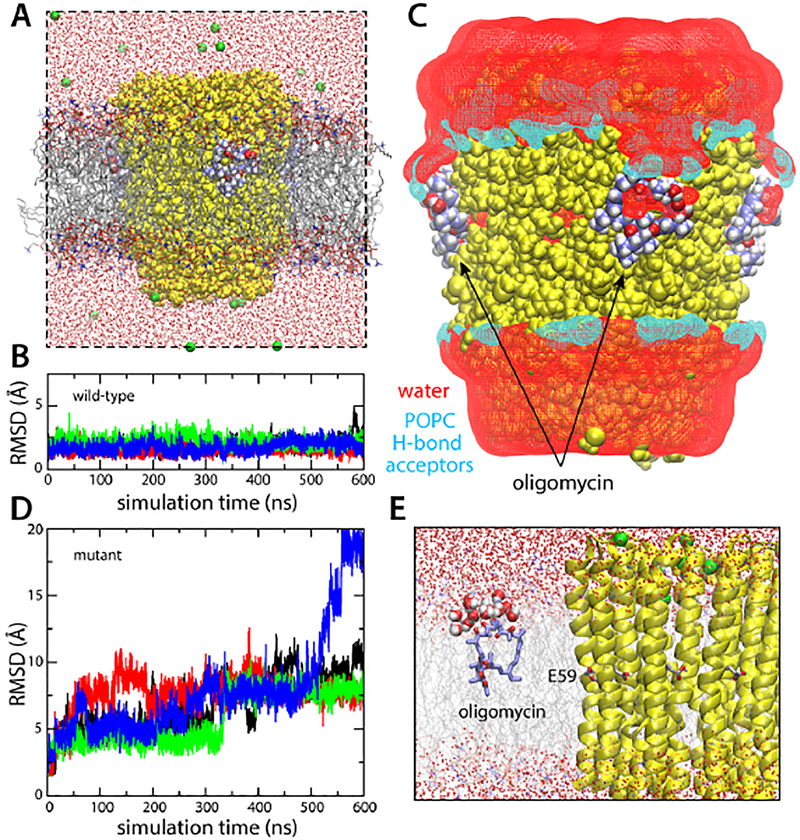
In the operating ATP synthase, most of the proton-binding sites in the c-ring are exposed to the interior of the lipid membrane at any given time. The finding that hydration is required to stabilize the protein-inhibitor interaction appears to rule out that these lipid-exposed sites mediate the recognition of oligomycin, as initially suggested [30]. To examine whether or not oligomycin would bind to these sites, we carried out a simulation of the protein-inhibitor complex embedded in a hydrated phospholipid membrane (Fig. 5A). This simulation was analogous in every other way to that carried out in MPD/water, described above. Unexpectedly, we found that the oligomycin binding pose observed in the available crystal structure was unambiguously stable, with no indication whatsoever of dissociation over 600 ns of simulation time, for any of the four inhibitor molecules bound to the c-ring (Fig. 5B). To rationalize this result, which is in stark contrast to that obtained for the purely hydrophobic hexane layer (Fig. 4), we analyzed the density distribution of hydrophobic and polar groups belonging to phospholipid and water around the protein-inhibitor complex. This analysis revealed that the lipid/water interface in the vicinity of the protein-oligomycin complex is perturbed, so that water and hydrogen-bonding groups from the surrounding lipid molecules solubilize the polar groups exposed in the inhibitor (Fig. 5C).
Figure 5. Simulations of the S. cerevisiae c10 ring with bound oligomycin, in a hydrated phospholipid bilayer.
(A) Simulation system. Lipid (POPC) molecules are shown as gray/red/orange/blue sticks. The rest of the system is represented as in Fig. 2. (B) RMS deviation of each of the four oligomycin molecules bound to the c-ring, relative to the observed binding pose. (C) Lipid/water structure at the protein-inhibitor interface. The figure shows iso-surfaces of 3D density maps for water and lipid, calculated analogously to those shown in Fig. 3. The water density map is shown in red. Separate density maps for POPC were calculated; the map for the potential hydrogen-bond acceptors (POPC has no potential donors) within interaction distance from the protein-inhibitor surface is shown in cyan. (D) Analysis analogous to that in (B), for a control simulation in which a series of mutations known to prevent oligomycin inhibition were introduced in the c-subunit. (E) Close up of the snapshot at the end of the control simulation, showing one of the disassociated oligomycin molecules, at the lipid-water interface.
It should be noted that this oligomycin-induced membrane defect occurs only in the context of a viable protein-inhibitor complex; a control simulation in which a series of c-subunit mutations known to confer the yeast ATP synthase with resistance to oligomycin [31] were introduced in the c-subunits (G23A, L53F, A56T, L57V, F64L and C65S) had an entirely different outcome. Despite careful preparation, this simulation showed all four oligomycin molecules dissociate from the mutant c-ring within 600 ns of simulation time (Fig. 5D), subsequently migrating to the membrane-water interface (Fig. 5E, Movies S1–S2). The structure of the ring itself, however, was not impacted in any discernible way. Thus, the stabilizing effect of the reconfigured lipid-water interface therefore adds to the complementary between protein and inhibitor (in the presence of the proton at Glu59), but by itself it is not sufficient to sustain binding.
3. DISCUSSION
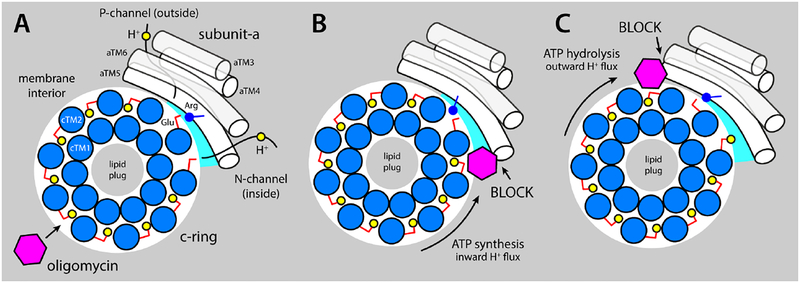
In an operating ATP synthase, the c-ring rotates around its axis against subunit-a, which remains stationary. When the enzyme is driven by downhill proton permeation, resulting in ATP synthesis, the ring rotates counterclockwise, if viewed from the mitochondrial matrix (Fig. 6A). Conversely, ATP hydrolysis drives the c-ring to rotate clockwise, resulting in uphill proton transport. The strict correlation between the direction of rotation and that of proton transport is a fundamental property of these molecular motors, encoded in the structure and chemical make-up of subunit-a and its interface with the c-ring [6–8, 10]. Specifically, upon first entering this interface, each of the binding sites in the c-ring encounters a hydrated crevice formed between the two proteins, decorated with multiple polar and ionizable side-chains, which facilitates the release of the bound proton, towards the mitochondrial interior. Concurrently, a deprotonated site deeper into the interface loads a different proton, reaching from the exterior via a second pathway formed largely within subunit-a. Crucially, a conserved arginine on subunit-a serves as a barrier to preclude direct proton hopping between the releasing and loading sites, by alternatingly locking one of them in the deprotonated state, and by facilitating the step whereby the releasing site takes the position of the loading site [35]. In addition, because the proton pathway to the interior is clockwise relative to that exposed to exterior, proton influx necessarily implies counter-clockwise rotation of the c-ring. This is the direction of rotation associated with ATP production in the catalytic domain, which is why the inward proton electrochemical gradient set by the respiratory chain results in ATP synthesis. Conversely, ATP hydrolysis drives the cring to rotate in a clockwise direction, which necessarily results in efflux.
Figure 6. Proposed mechanism of inhibition of the mitochondrial ATP synthase by oligomycin.
The c-ring (marine circles) and subunit (white cylinders) are viewed from the matrix. (A) Oligomycin (magenta hexagon) binds to proton-binding sites in the c-ring that are exposed to the interior of the lipid membrane (gray area), by integrating into the coordination structure of the proton (yellow circles), which also includes the conserved Glu59 (red sticks). The c-ring rotates against subunit-a (white cylinders represent TM3-TM6, equivalent to aTM2-TM5 in bacteria), exposing two proton-binding sites to two aqueous noncollinear access permeation pathways, referred to as the P- and N-half-channels (from the positive and negative side of the membrane, respectively). A conserved arginine in aTM5 (blue stick/circle) is crucial to preclude direct proton hoping from one site to the other. Stochastic fluctuations permit one of these two sites to interact at close range with the arginine, while the other is able to load/release a proton through one of the half-channels. (B) When the c-ring rotates under a proton-motive-force, i.e. counter-clockwise, a bound oligomycin molecule eventually reaches the subunit-a/c-ring interface, blocking the release of protons into the mitochondrial matrix via the N-channel and preventing further rotation. (C) The clockwise rotation of the c-ring is halted similarly when the enzyme operates as an ATPase.
Intriguingly, oligomycin inhibits both modes of operation of the membrane motor [36]. The results presented in this study suggest a recognition mechanism whereby oligomycin diffuses laterally at the membrane-water interface, and upon encountering the c-ring, docks onto the proton-binding sites exposed to the interior of the lipid bilayer. In the bound state, the inhibitor preserves an amphipathic microenvironment that matches its inherent solvation preferences, and thus the formation of specific interactions with the protein is unhindered. One of these specific interactions is mediated by a water molecule [30] that most likely originates from the hydration shell of the unbound inhibitor, and which ultimately provides a hydrogen-bond bridge between protonated Glu59 and the inhibitor (Fig. 2C). Oligomycin thus becomes an integral component of the proton-coordinating structure within the c-ring binding sites.
Clearly, binding of oligomycin to most of the lipid-exposed sites in the c-ring does not result in inhibition. However, as the c-ring rotates a site carrying oligomycin will ultimately encounter subunit-a (Fig. 6), and at this point further rotation will be halted, for one of two reasons. If the site with bound oligomycin cannot be accommodated at the a/c interface, the inhibitor would simply impair the rotation of the ring sterically. Alternatively, if the inhibitor enters the a/c interface, it will block proton release, as oligomycin binding requires protonation of the site. Specifically, if the c-ring is rotating counter-clockwise (driven by the proton gradient), the inhibitor will prevent proton release into the matrix. In the opposite direction of rotation (driven by ATP hydrolysis), it would be proton release toward the exterior that is precluded.
In either case, our results suggest that the site of oligomycin inhibition, i.e. at or near the a-c interface, does not coincide with the site of initial recognition, i.e. lipid-exposed c-subunits; our data does not rule out that a higher-affinity binding site also exists at the a-c interface, to which oligomycin may bind directly, but it should also be noted that no supporting evidence exists of this possibility, to our knowledge. It is thus worth underscoring that this study and the observations made elsewhere for bedaquiline [33] suggest that hydrophobic inhibitors of the membrane motor of the ATP synthase have a common mechanism of action, and that that efforts to enhance their potency and specificity might be well-served by systematic quantitative analyses of the initial recognition step, by the c-ring, isolated from the rest of the complex.
Finally, our simulation data also illustrates how the plasticity of the lipid bilayer in proximity to the protein-inhibitor complex provides the kind of microenvironment that oligomycin requires. To advance the development of more potent and efficacious inhibitors against this and other membrane proteins, it would be important to keep in mind that the lipid membrane is not a static, passive entity with rigidly defined boundaries; instead, it is adjustable and plastic, and as such, will contribute significantly to the energetics of bi-molecular association processes on a wide range of scales, from that of small-molecule inhibitors to larger supramolecular assemblies [39].
4. METHODS
4.1. Molecular Dynamics -
All simulations were performed with NAMD2 [40], using the CHARMM36 force field for proteins and lipids [41, 42], periodic boundary conditions and constant temperature (298 K) and pressure (1 atm). Long-range electrostatic interactions were calculated using PME, with a real-space cut-off of 12 Å. Van der Waals interactions were computed with a Lennard-Jones potential, cut-off at 12 Å with a smooth switching function taking effect at 10 Å.
4.2. Force-field parameters for oligomycin -
Force field parameters for oligomycin A were derived using the GAAMP server [34]. Briefly, an initial set of parameters was determined based on the CHARMM General Force Field, which includes entries for a broad set of chemical compounds. Parameters deemed to be suboptimal for this specific inhibitor were then optimized relative to ab initio calculations (Hartree–Fock 6–31G*). MD simulations of 100 ns each were then carried out with the inhibitor immersed either in pure methanol or water (in cubic boxes of side 40 and 45 Å, respectively), to evaluate the resulting parameters and make adjustments, where necessary, to the dihedral-angle parameters (to better match the geometries observed in experimental crystal structures). The final parameter set is available upon request to the authors.
4.3. Solvation free-energy calculations -
Solvation free energies for oligomycin A were calculated using the Free Energy Perturbation (FEP) method, as implemented in NAMD [40]. Four different solvents were considered, namely water, hexane, methanol and a water/hexane interface. Oligomycin was gradually decoupled from the solvent using a scaling parameter of all intermolecular interactions, λ, in 38 intermediate steps from 1 to 0. The sampling time for each λ-step was 440 ps, with the first 40 ps discarded as equilibration time. The inhibitor was then re-coupled to the solvent analogously; the reported free-energy values are the mean of the decoupling/coupling values. The geometry of oligomycin during these simulations was maintained using an RMSD restraint (defined relative to the coupled state) with a force constant of 9.6 kcal/mol/Å2. The final solvation free energies were therefore calculated by adding up the solvent coupling/decoupling free energy, and the free-energy cost/gain of imposing/releasing the conformational RMSD restraint. The latter free-energy values were also computed using the FEP method, with the λ parameter scaling up/down the force constant of the RMSD restraining potential, in 50 steps of 1-ns sampling each. To our knowledge, no experimental data with which to directly compare the computed values is available.
4.4. Simulations of oligomycin in hexane/water -
The amphipathic character of oligomycin was further examined using a simulation system consisting of an artificial hexane layer, 30 Å wide, flanked by water on both sides. Oligomycin was initially placed either in the water phase or inside the hexane layer. Three independent trajectories of 100 ns each were calculated. The initial 50-ns fragment consisted of a series of restrained equilibrations, after which the inhibitor was allowed to move freely. The slab-like geometry of the hexane layer was maintained using a confining potential (flat-bottom harmonic function with force constant of 1 kcal/mol/Å2) acting on the Z-coordinate of each hexane molecule (represented by the center-of-mass of the two central carbon atoms).
4.5. Simulations of the mitochondrial c10−ring with oligomycin bound -
The simulations of the protein-inhibitor complex are based on PDB entry 4F4S [30]. The backbone structure of the c10−ring in this entry, at 1.9 Å resolution, is highly similar (RMSD = 0.92 Å) to that of the c10−ring in detergent, in complex with the catalytic domain, at 3.4 Å resolution [43]. Four different simulation systems were prepared. For the wild-type protein, three systems had the protein-inhibitor complex embedded in either a preequilibrated amphipathic mixture of 2-methyl-2,4-pentanediol (MPD) and water (volume ratio 30:70), prepared as detailed elsewhere [18] (Fig. S1); or in a preequilibrated hydrated bilayer of palmitoyl-oleoyl-phosphatidyl-choline (POPC) lipids, prepared using GRIFFFIN [44] (Fig. S1); or in an artificial hexane layer analogous to that described above. A fourth system was prepared in a POPC membrane and where the c-subunits carry the mutations G23A, L53F, A56T, L57V, F64L and C65S. The rationale for using a single-component lipid bilayer is that it leads to results that are more clearly statistically significant. Although a mixed membrane might seem more realistic in terms of its composition, it is in fact more artefactual than a simple homogenous bilayer, in that the spatial distribution of its various components must be postulated a priori, as it is in fact unknown; this distribution, however, will hardly change in the course of an all-atom simulation of hundreds of nanoseconds, owing to the slow lateral diffusion of the lipid molecules. The hypothetical influence that this mixed composition might have on the problem of interest is therefore not adequately characterized; to the contrary, spurious effects might be observed if the initial distribution is unrealistic or unrepresentative. In all simulation systems, four oligomycin A molecules were initially bound to the c-ring, as in the existing crystal structure [30]. In these sites, Glu59 was set either protonated or deprotonated (all other Glu59 were considered to be protonated). Counter-ions were added to neutralize the total charge of the molecular systems. All systems were equilibrated following a staged protocol comprising a series of restrained simulations. The protocol consists of both positional and conformational restraints, gradually weakened over 200 ns, and individually applied to protein side-chains and backbone, the inhibitor and crystallographic water molecules. Subsequently, a trajectory was calculated in the absence of any conformational restraints on the protein-inhibitor complex. For the wild-type systems with protonated Glu59, the calculated trajectories were 600-ns long for the MPD/water solvent and the POPC membrane, and 100 ns for the hexane layer; for the MPD/water system with Glu59 ionized, and the c-ring mutant in a POPC membrane with Glu59 protonated, the length of the unrestrained trajectories was 100 ns.
Supplementary Material
HIGHLIGHTS.
Oligomycin inhibits the ATP synthase by stalling the rotary mechanism of the c-ring
The inhibitor first binds to the surface of the c-ring exposed to the lipid bilayer
The complex is stabilized by local perturbations of the lipid/water interface
Potency, specificity of hydrophobic inhibitors likely determined by c-ring recognition
ACKNOWLEDGEMENTS
This research was funded by the Division of Intramural Research of the National Heart, Lung and Blood Institute, National Institutes of Health, USA. We are grateful to David Mueller (Rosalind Franklin University) for useful discussions, and to Lucy R. Forrest and Vanessa Leone (NINDS/NIH) for their comments on this manuscript.
REFERENCES
- [1].Meier T, Faraldo-Gómez JD, Börsch M, ATP Synthase: a paradigmatic molecular machine, in: Frank J (Ed.) Molecular Machines in Biology, Cambridge University Press, Cambridge, UK, 2011, pp. 208–238. [Google Scholar]
- [2].Walker JE, The ATP synthase: the understood, the uncertain and the unknown, Biochem Soc T, 41 (2013) 1–16. [DOI] [PubMed] [Google Scholar]
- [3].Dimroth P, Primary sodium ion translocating enzymes, Biochim. Biophys. Acta, 1318 (1997) 11–51. [DOI] [PubMed] [Google Scholar]
- [4].Schlegel K, Leone V, Faraldo-Gómez JD, Muller V, Promiscuous archaeal ATP synthase concurrently coupled to Na+ and H+ translocation, Proc. Natl. Acad. Sci. USA, 109 (2012) 947–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Leone V, Pogoryelov D, Meier T, Faraldo-Gómez JD, On the principle of ion selectivity in Na+/H+−coupled membrane proteins: experimental and theoretical studies of an ATP synthase rotor, Proc. Natl. Acad. Sci. USA, 112 (2015) E1057–E1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Vik SB, Antonio BJ, A mechanism of proton translocation by F1Fo ATP synthases suggested by double mutants of the alpha-subunit, J Biol Chem, 269 (1994) 30364–30369. [PubMed] [Google Scholar]
- [7].Junge W, Lill H, Engelbrecht S, ATP synthase: an electrochemical transducer with rotatory mechanics, Trends in biochemical sciences, 22 (1997) 420–423. [DOI] [PubMed] [Google Scholar]
- [8].Fillingame RH, Steed PR, Half channels mediating H+ transport and the mechanism of gating in the Fo sector of Escherichia coli F1Fo ATP synthase, Biochim. Biophys. Acta Bioenerg, 1837 (2014) 1063–1068. [DOI] [PubMed] [Google Scholar]
- [9].Pogoryelov D, Klyszejko AL, Krasnoselska GO, Heller EM, Leone V, Langer JD, Vonck J, Muller DJ, Faraldo-Gómez JD, Meier T, Engineering rotor ring stoichiometries in the ATP synthase, Proc. Natl. Acad. Sci. USA, 109 (2012) E1599–E1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Leone V, Faraldo-Gómez JD, Structure and mechanism of the ATP synthase membrane motor inferred from quantitative integrative modeling, J. Gen. Physiol, 148 (2016) 5–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Meier T, Matthey U, Henzen F, Dimroth P, Muller DJ, The central plug in the reconstituted undecameric c cylinder of a bacterial ATP synthase consists of phospholipids, FEBS Lett, 505 (2001) 353–356. [DOI] [PubMed] [Google Scholar]
- [12].Oberfeld B, Brunner J, Dimroth P, Phospholipids occupy the internal lumen of the c ring of the ATP synthase of Escherichia coli, Biochemistry, 45 (2006) 1841–1851. [DOI] [PubMed] [Google Scholar]
- [13].Zhou W, Marinelli F, Nief C, Faraldo-Gómez JD, Atomistic simulations indicate the c-subunit ring of the F1Fo ATP synthase is not the mitochondrial permeability transition pore, eLife, 6 (2017) 23781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Pogoryelov D, Yildiz O, Faraldo-Gómez JD, Meier T, High-resolution structure of the rotor ring of a proton-dependent ATP synthase, Nat. Struct. Mol. Biol, 16 (2009) 1068–1073. [DOI] [PubMed] [Google Scholar]
- [15].Krah A, Pogoryelov D, Meier T, Faraldo-Gómez JD, On the structure of the proton-binding site in the Fo rotor of chloroplast ATP synthases, J Mol Biol, 395 (2010) 20–27. [DOI] [PubMed] [Google Scholar]
- [16].Leone V, Krah A, Faraldo-Gómez JD, On the question of hydronium binding to ATP-synthase membrane rotors, Biophys J, 99 (2010) L53–L55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Symersky J, Pagadala V, Osowski D, Krah A, Meier T, Faraldo-Gómez JD, Mueller DM, Structure of the c10 ring of the yeast mitochondrial ATP synthase in the open conformation, Nat. Struct. Mol. Biol, 19 (2012) 485–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zhou W, Leone V, Krah A, Faraldo-Gómez JD, Predicted structures of the proton-bound membrane-embedded rotor rings of the Saccharomyces cerevisiae and Escherichia coli ATP synthases, J. Phys. Chem. B, 121 (2017) 3297–3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Pogoryelov D, Krah A, Langer JD, Yildiz O, Faraldo-Gómez JD, Meier T, Microscopic rotary mechanism of ion translocation in the Fo complex of ATP synthases, Nat. Chem. Biol, 6 (2010) 891–899. [DOI] [PubMed] [Google Scholar]
- [20].Salomon AR, Voehringer DW, Herzenberg LA, Khosla C, Understanding and exploiting the mechanistic basis for selectivity of polyketide inhibitors of FOF1−ATPase, Proc. Natl. Acad. Sci. USA, 97 (2000) 14766–14771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Salomon AR, Voehringer DW, Herzenberg LA, Khosla C, Apoptolidin, a selective cytotoxic agent, is an inhibitor of FoF1−ATPase, Chem. Biol, 8 (2001) 71–80. [DOI] [PubMed] [Google Scholar]
- [22].Daniel PT, Koert U, Schuppan J, Apoptolidin: induction of apoptosis by a natural product, Angew Chem. Int. Ed. Engl, 45 (2006) 872–893. [DOI] [PubMed] [Google Scholar]
- [23].Andries K, Verhasselt P, Guillemont J, Gohlmann HW, Neefs JM, Winkler H, Van Gestel J, Timmerman P, Zhu M, Lee E, Williams P, de Chaffoy D, Huitric E, Hoffner S, Cambau E, Truffot-Pernot C, Lounis N, Jarlier V, A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis, Science, 307 (2005) 223–227. [DOI] [PubMed] [Google Scholar]
- [24].Koul A, Dendouga N, Vergauwen K, Molenberghs B, Vranckx L, Willebrords R, Ristic Z, Lill H, Dorange I, Guillemont J, Bald D, Andries K, Diarylquinolines target subunit c of mycobacterial ATP synthase, Nat. Chem. Biol, 3 (2007) 323–324. [DOI] [PubMed] [Google Scholar]
- [25].Koul A, Vranckx L, Dendouga N, Balemans W, Van den Wyngaert I, Vergauwen K, Gohlmann HW, Willebrords R, Poncelet A, Guillemont J, Bald D, Andries K, Diarylquinolines are bactericidal for dormant mycobacteria as a result of disturbed ATP homeostasis, J Biol Chem, 283 (2008) 25273–25280. [DOI] [PubMed] [Google Scholar]
- [26].Haagsma AC, Abdillahi-Ibrahim R, Wagner MJ, Krab K, Vergauwen K, Guillemont J, Andries K, Lill H, Koul A, Bald D, Selectivity of TMC207 towards mycobacterial ATP synthase compared with that towards the eukaryotic homologue, Antimicrob. Agents Chemother, 53 (2009) 1290–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Fiorillo M, Lamb R, Tanowitz HB, Cappello AR, Martinez-Outschoorn UE, Sotgia F, Lisanti MP, Bedaquiline, an FDA-approved antibiotic, inhibits mitochondrial function and potently blocks the proliferative expansion of stem-like cancer cells (CSCs), Aging (Albany NY), 8 (2016) 1593–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Racker E, A mitochondrial factor conferring oligomycin sensitivity on soluble mitochondrial ATPase, Biochem Biophys Res Commun, 10 (1963) 435–439. [DOI] [PubMed] [Google Scholar]
- [29].Kagawa Y, Racker E, Partial resolution of the enzymes catalyzing oxidative phosphorylation. IX. Reconstruction of oligomycin-sensitive adenosine triphosphatase, J Biol Chem, 241 (1966) 2467–2474. [PubMed] [Google Scholar]
- [30].Symersky J, Osowski D, Walters DE, Mueller DM, Oligomycin frames a common drug-binding site in the ATP synthase, Proc. Natl. Acad. Sci. USA, 109 (2012) 13961–13965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Nagley P, Hall RM, Ooi BG, Amino acid substitutions in mitochondrial ATPase subunit 9 of Saccharomyces cerevisiae leading to oligomycin or venturicidin resistance, FEBS Lett, 195 (1986) 159–163. [DOI] [PubMed] [Google Scholar]
- [32].John UP, Nagley P, Amino acid substitutions in mitochondrial ATPase subunit 6 of Saccharomyces cerevisiae leading to oligomycin resistance, FEBS Lett, 207 (1986) 79–83. [DOI] [PubMed] [Google Scholar]
- [33].Preiss L, Langer JD, Yildiz O, Eckhardt-Strelau L, Guillemont JEG, Koul A, Meier T, Structure of the mycobacterial ATP synthase Fo rotor ring in complex with the anti-TB drug bedaquiline, Sci. Adv, 1 (2015) e1500106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Huang L, Roux B, Automated force field parameterization for nonpolarizable and polarizable atomic models based on ab initio target data, J. Chem. Theory Comput, 9 (2013) 3543–3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Matthies D, Zhou W, Klyszejko AL, Anselmi C, Yildiz O, Brandt K, Muller V, Faraldo-Gómez JD, Meier T, High-resolution structure and mechanism of an F/V-hybrid rotor ring in a Na+−coupled ATP synthase, Nat. Commun, 5 (2014) 5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Pagadala V, Vistain L, Symersky J, Mueller DM, Characterization of the mitochondrial ATP synthase from yeast Saccharomyces cerevisae, J. Bioenerg. Biomembr, 43 (2011) 333–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Klusch N, Murphy BJ, Mills DJ, Yildiz O, Kuhlbrandt W, Structural basis of proton translocation and force generation in mitochondrial ATP synthase, eLife, 6 (2017) 33274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Guo H, Bueler SA, Rubinstein JL, Atomic model for the dimeric Fo region of mitochondrial ATP synthase, Science, 358 (2017) 936–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Davies KM, Anselmi C, Wittig I, Faraldo-Gomez JD, Kuhlbrandt W, Structure of the yeast F1Fo ATP synthase dimer and its role in shaping the mitochondrial cristae, Proc. Natl. Acad. Sci. USA, 109 (2012) 13602–13607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K, Scalable molecular dynamics with NAMD, J Comput Chem, 26 (2005) 1781–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Best RB, Zhu X, Shim J, Lopes PE, Mittal J, Feig M, Mackerell AD Jr., Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone phi, psi and side-chain chi1 and chi2 dihedral angles, J. Chem. Theory Comput, 8 (2012) 3257–3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Klauda JB, Venable RM, Freites JA, O’Connor JW, Tobias DJ, Mondragon-Ramirez C, Vorobyov I, MacKerell AD Jr., Pastor RW, Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types, J. Phys. Chem. B, 114 (2010) 7830–7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Dautant A, Velours J, Giraud MF, Crystal structure of the Mg-ADP-inhibited state of the yeast F1c10−ATP synthase, J Biol Chem, 285 (2010) 29502–29510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Staritzbichler R, Anselmi C, Forrest LR, Faraldo-Gómez JD, GRIFFIN: a versatile methodology for optimization of protein-lipid interfaces for membrane protein simulations, J Chem Theory Comput, 7 (2011) 1167–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.