Abstract
Metastasis remains a leading cause of cancer mortality due to the lack of specific inhibitors against this complex process. To identify compounds selectively targeting the metastatic state, we used the perinuclear compartment (PNC), a complex nuclear structure associated with metastatic behaviors of cancer cells, as a phenotypic marker for a high-content screen of over 140,000 structurally diverse compounds. Metarrestin, obtained through optimization of a screening hit, disassembles PNCs in multiple cancer cell lines, inhibits invasion in vitro, blocks metastatic development in three mouse models of human cancer, and extends survival of mice in a metastatic pancreatic cancer xenograft model with no organ toxicity or discernable adverse effects. Metarrestin disrupts the nucleolar structure and inhibits RNA polymerase (Pol) I transcription, at least in part by interacting with the translation elongation factor eEF1A2. Altogether, metarrestin represents a potential therapeutic approach for the treatment of metastatic cancer.
One-sentence summary:
A compound that reduces the prevalence of perinucleolar compartment in cancer cells inhibits metastasis in vivo.
Introduction:
Cancer is a leading cause of death in the US and worldwide, with metastasis to other organs the primary cause of mortality for many cancer types (1). Cancer survival in general has steadily increased due to enhanced screening, early detection, and treatment improvements for primary tumors. However, cancers prone to metastatic progression remain stubbornly lethal, in part due to the lack of therapeutic approaches that block metastatic processes. For example, pancreatic cancer with a tendency for early dissemination has a 5-year survival rate of 5–7%, which has not changed over the last three decades (2). Current chemo- and molecular therapy options for this disease, or other metastatic solid tissue cancers in general, provide only modest gains in improving clinical outcome (1, 2), making the metastatic stage an incurable condition with little promise for prolonged remission or survival.
The lack of effective therapeutics for the treatment of metastasis is primarily due to the complexity of the disease process and an incomplete understanding, making treatments targeting a single gene or gene product less likely to be successful. In this regard, the complex metastatic disease state could be better represented by comprehensive phenotypic markers that reflect the metastatic capability. One such marker is the perinucleolar compartment (PNC) (3), a subnuclear body that is: 1) selectively detected in cancer cells but not in normal cells, including embryonic stem cells, 2) a multi-component nuclear structure highly enriched with non-coding RNAs and RNA-binding proteins (4-6), 3) a structure associated with chromatin (7), and 4) involved in RNA metabolism and Pol III function (7–10). Analyses of cancer cells from different tissues and metastatic lesions demonstrated that PNC prevalence (percentage of non-mitotic cells containing at least 1 PNC) positively correlates with metastatic potential and disease progression (11, 12). High PNC prevalence in primary tumors is associated with poor patient outcomes, including overall survival of patients with breast, colorectal (11, 12), and ovarian cancer. Thus, being a complex, multi-component structure, PNC prevalence may better reflect the complex nature of malignant transformation than a single gene or gene product. Using PNC reduction as a surrogate metastatic phenotypic marker, we developed a lead compound upon optimization of hits from a high-content screen (13). Here we report the evidence for metarrestin as a promising anti-cancer compound.
Results:
Metarrestin disrupts the perinucleolar compartment in multiple cancer cells.
To screen for small molecules that disassemble PNCs, we engineered a metastatic prostate cancer cell line, PC3M (14), with a PNC prevalence of 75-85% to stably express GFP-PTB (polypyrimidine track binding protein) (Fig. 1A, top panels), an essential component of PNCs (15). Compounds able to reduce PNC prevalence by 50% (13) were filtered in secondary assays to eliminate those that induced apoptosis, DNA damage, generic cytotoxicity, or cell-cycle blockage (13, 16). Remaining hits were evaluated for invasion inhibition through Matrigel and anchorage-independent growth inhibition using soft agar assays. Compound MLS000556915 was selected for optimization based on its potency as a PNC inhibitor, soft agar growth inhibitor, ability to block invasion, and lack of cytotoxicity (13, 16).
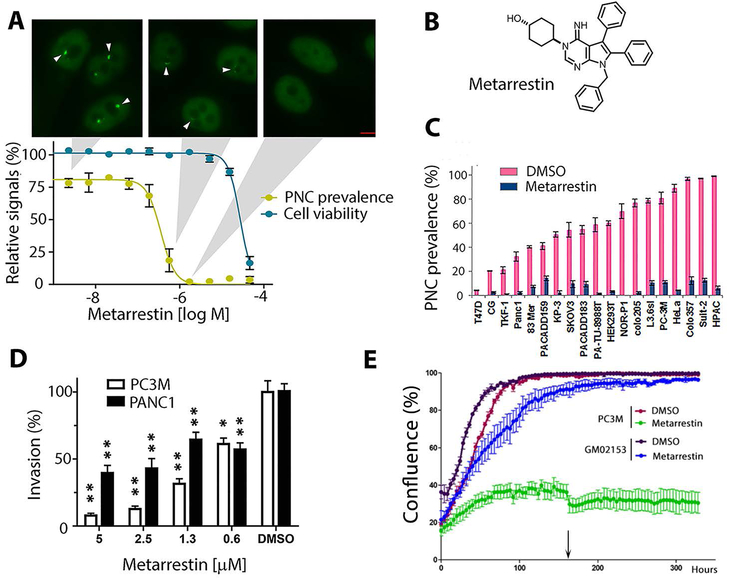
Figure 1: Metarrestin reduces PNCs at a sub-micromolar concentration and inhibits invasion of cancer cells.
(A) Metarrestin concentration-response curve against PNC prevalence in PC3M-GFP-PTB cells (yellow, PC3M, has a PNC prevalence of 75-85% in the absence of treatment) and cytotoxicity as measured by cellular ATP (CellTiter Glo) (blue). Shown above are representative PTB-GFP images for cells at the indicated concentrations (arrowheads indicate PNCs) (scale bar = 5 μm). (B) The structure of metarrestin. (C) 1 μM metarrestin was effective at reducing PNCs in a range of cancer cell lines (p<0.05 for PNC reduction in all cell lines; the list of cell lines is in table S1). (D) Metarrestin inhibits Matrigel invasion at below micromolar concentrations (0.6 μM) within 24 hours of treatment (* p<0.05 and ** p<0.01 in comparison to DMSO). (E) Metarrestin at 1 μM impacts cell growth in cancer cell line PC3M but not in normal fibroblasts (GM02153) (arrow indicates the time of medium change). * P < 0.05, ** P < 0.01.
Medicinal chemistry studies explored the structure-activity relationships in the series and optimized the pharmacokinetic profile for in vivo use, ultimately producing metarrestin (Fig. 1B, fig. S1 for synthetic scheme). Metarrestin disrupted PNCs in PC3M-GFP-PTB cells with an IC50 of 0.39 μM (Fig. 1A, yellow line) without acute cytotoxicity (Fig. 1A, blue line). Treatment at 1 μM (IC100 for PC3M cells) for 24 hours reduced PNC prevalence in different human cancer cell lines (Fig. 1C, cell lines described in table S1). Matrigel invasion assay analyses showed that metarrestin effectively blocked the invasion of PC3M and PANC1 cells (Fig. 1D) within 24 hours at a concentration (0.6 μM) which did not affect PC3M or PANC1 cell growth after 24 hours of treatment (Fig. 1E for PC3M cells and fig. S2 for PANC1 cells). When PC3M cells and normal human fibroblasts (GM02153) were treated at 1 μM concentration for an extended time using the Incucyte system, metarrestin preferentially inhibited cell growth in the PC3M cancer cells (Fig. 1E).
Metarrestin suppresses metastasis and extends survival in mouse models of human pancreatic cancer xenografts.
Because pancreatic cancer patients suffer particular high mortality from metastasis, we initially evaluated the in vivo efficacy of metarrestin against metastasis in an orthotopic pancreatic cancer metastasis model. The 3D PANC1 cell spheres model, deployed in NOD/interleukin 2 receptor common gamma chain (Il2ry)null (NSG) PANC-1 mice (17), recapitulates human cancer progression and metastatic phenotype of the disease without the limitations of early death due to complications of local invasion, such as gastric outlet obstruction or impingement of the common bile duct. Sixty thousand 3D luciferase-expressing PANC1-luc cell spheres were injected orthotopically into the pancreas of NSG mice (18, 19). Histopathological examination revealed that measurable metastasis, in the form of occasional peri-portal infiltrates and micrometastatic deposits, had developed in livers at ~4 weeks after implantation, then in the form of parenchymal infiltration and visible macrometastasis on the liver surface at 8 weeks, and mice had a lifespan of 10–14 weeks (fig. S3).
To determine whether the preclinical model used retained the features of PNCs observed previously in clinical specimens (11, 12), we first measured PNC prevalence in a panel of pancreatic cancer cell lines derived from primary tumors and metastatic lesions from various organs in NSG PANC1 mice. The results showed that PNCs were more numerous in human cancer lineages from a metastatic origin and in metastatic lesions compared to those from primary tumors (Fig. 2A, cell line explanation, table S2). When cryopreserved tissue sections were examined, PNCs were detected by immunolabeling with a monoclonal anti-human specific PTB antibody (6), SH54, which labels PNCs and allows for specific identification of human xenograft tissues over mouse tissues. PNC prevalence was higher in metastatic lesions than in primary tumors harvested 8 weeks after implantation (Fig. 2B).
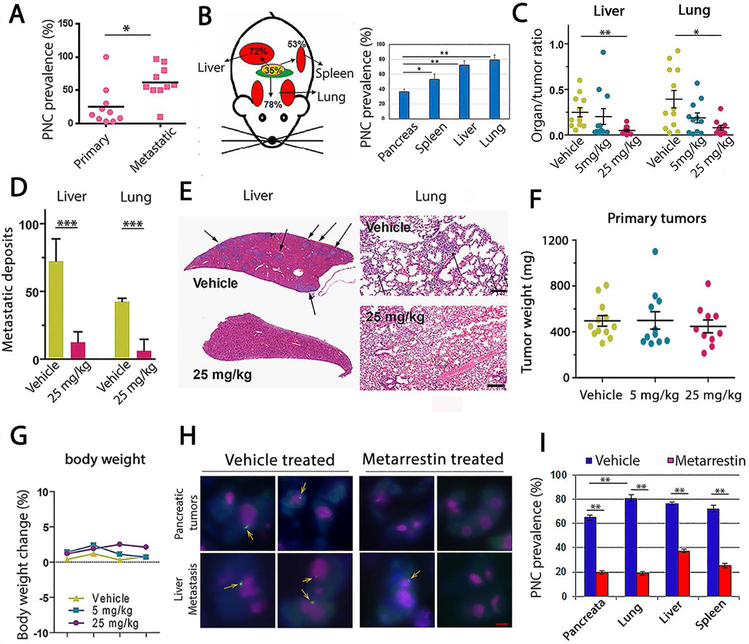
Figure 2: Metarrestin treatment reduces metastasis to the lungs and liver in NOD/IL2 gamma (null) PANC1 mice.
(A) A panel of pancreatic cancer cell lines derived from either primary pancreatic tumors or metastatic lesions showed a higher PNC prevalence in cells derived from metastasis than from primary tumors (cell line explanations in table S2). (B) PNC prevalence increased in metastatic tissues (red) from NOD/IL2 gamma (null) PANC1 mice over primary tumor tissues (yellow), harvested 8 weeks after implantation. PNC prevalence was determined on frozen tissue sections stained with SH54 antibodies. (C) After six weeks of treatment, metastatic deposits measured by organ/tumor ratio in liver and lungs decreased in mice treated once daily with 25 mg/kg of metarrestin compared to vehicle-treated animals (n=10 mice were randomized to each cohort). (D) Pathology and (E) histological examinations demonstrated that livers and lungs from metarrestin-treated animals have a reduced metastatic burden compared to those treated with vehicle (bar=250 μm; n=4 animals per group analyzed). (F) The primary tumors in treated animals were not changed. (G) Treatment was well tolerated, and there were no significant weight differences between treatment groups across the duration of the experiment. (H) Metarrestin disassembles PNCs in primary pancreatic tumors and metastases of NSG PANC1 mice. PNCs in tumors were visualized via immunofluorescence (PNCs labeled green and marked with arrows, nucleoli labeled pink, DAPI, blue) 12 weeks after inoculation. Images from the primary tumors and liver metastases are shown (scale bar = 5 μm). Vehicle-treated animals showed typical, easily detectable PNCs. PNC prevalence was reduced and remaining PNCs appeared smaller in metarrestin-treated animals (25 mg/kg IP daily for 6 weeks; n=4 animals per group analyzed). (I) Metarrestin effect on PNC prevalence in primary pancreatic tumors and sites of metastasis. PNC prevalence was reduced with metarrestin treatment (25 mg/kg IP daily for 6 weeks) in the primary tumor (pancreas) and in metastatic tumors in the lung, liver, and spleen. * P < 0.05, ** P < 0.01, *** P < 0.001.
Pharmacokinetic studies using single and multiple daily dosing via intra-peritoneal (IP) administration of metarrestin in mice (fig. S4A) at 5 and 25 mg/kg indicated good exposure, distribution, and tolerability in vivo, with a half-life of 4.6 to 5.5 hours and a predicted moderate risk of accumulation (fig. S4A). Metarrestin showed high bioavailability, with concentrations in plasma above its PNC disassembling IC50 of 0.39 μM in P3CM cells for an extended period of time (Fig. 1A and fig. S4A), and concentrations more than 10-fold above the IC90 of 0.75 μM (Fig. 1A) in primary tumors and even higher in metastatic deposits of tumor-bearing NSG PANC1 animals 1 hour after discontinuation of 10 mg/kg metarrestin given for 7 days via PO gavage (fig. S4B).
Four weeks after inoculation, mice were treated once daily with metarrestin (5 mg/kg or 25 mg/kg) or vehicle via IP injections, continuing for six weeks. At the end of the tenth week after initial inoculation, the cohort exposed to daily administration of 25 mg/kg metarrestin displayed a decrease in metastatic burden in both liver (p <0.01) and lung (p <0.05) compared to vehicle-treated control as measured by Xenogen photon organ/tumor ratios (Fig. 2C) and by standard histopathological examination (Fig. 2D and E, p <0.001). Primary tumor weight was not significantly changed across cohorts (Fig. 2F). The treatment was well-tolerated, and animals maintained their body weight (Fig. 2G). Dedicated veterinary histopathology review of 12 organ systems as well as standard clinical chemistry after three months of chronic, uninterrupted dosing of autochthonous KPC mice with 10 mg/kg metarrestin-infused chow (70 ppm) (table S3 and S4) or two weeks of 25 mg/kg metarrestin via oral gavage failed to discern any histopathological or laboratory abnormalities in treated animals compared to control mice.
To determine whether PNCs in treated animals were impacted by metarrestin, we examined PNC prevalence in primary tumors and metastatic lesions from control and metarrestin-treated animals (25 mg/kg) at 12 weeks after tumor inoculation. The results demonstrated a significant reduction of PNC prevalence (Fig. 2, H and I, p < 0.01) in metastatic and primary tumor tissues, suggesting that PNC suppression is associated with the anti-metastatic activity of the compound. PNC prevalence in non-treated animals was higher in these groups of tumors and metastatic lesions than in those harvested at an earlier time (Fig. 2B) or in cultured PANC1 cells (Fig. 1C), consistent with previous findings that late-stage cancers have higher PNC prevalence (11, 12).
To evaluate whether metarrestin treatment can provide a survival advantage by preventing metastatic progression, the above experiment was repeated, but mice were followed until death or when animals reached study endpoint. NSG mice were injected with 60,000 3D PANC1 cells, and daily treatment with metarrestin-infused chow (10 mg/kg; 70 ppm, micronized particles added to NIH 31 Haslan rodent diet) began 6 weeks after tumor cell implantation, when metastasis was limited to micrometastasis by histopathological examination (fig. S3) (15). Animals in the control group receiving regular chow started to die 25 days after the start of treatment, whereas the metarrestin treatment group had no mortality more than 90 days after the start of treatment (Fig. 3A).
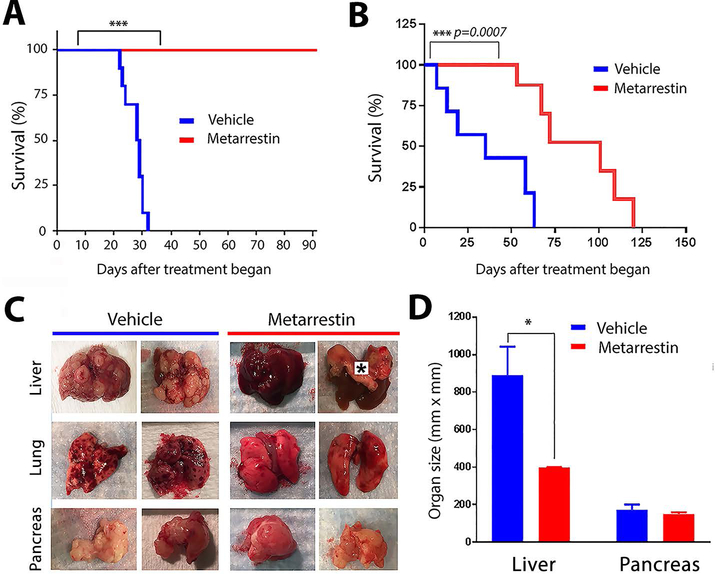
Figure 3: Metarrestin treatment extends survival in the NSG PANC1 pancreatic cancer metastasis model.
(A) Metarrestin treatment through drug-infused chow (70 ppm designed to administer 10 mg/kg daily) starting 6 weeks after 3D PANC1 tumor cell inoculation, when animals generally do not show macrometastasis on organ surfaces, prevented mortality beyond 90 days of treatment. (B) Metarrestin treatment (10 mg/kg daily) in NSG mice starting after mice developed macrometastasis, including visible liver surface deposits, extends survival compared to vehicle-treated (NIH 31 Haslan diet) mice. (C) Full necropsy of mice on the survival study at time of death demonstrates decreased metastatic disease burden in the liver of metarrestin-treated animals without detectable impact on primary tumor size. Animals in the control group showed near complete or complete organ replacement with tumors, particularly in the liver (*indicates thick right hemidiaphragm) and to a lesser degree in the lung. Pancreatic tumors, in comparison, were similar in both groups. (D) Average sizes of livers (containing metastatic tumors) and of primary pancreatic tumors in vehicle and metarrestin-treated mice from the study in Fig. 3B. * P < 0.05, ** P < 0.01, *** P < 0.001.
To evaluate whether metarrestin treatment impacts survival of mice with further evolved metastasis, we injected NSG mice with 60,000 3D PANC1 cells. After macrometastasis was visible on the liver surface, animals were randomized to receive metarrestin-infused chow (10 mg/kg; 70 ppm) or vehicle diet. Mice on the vehicle diet began to die within the first week of the treatment course (Fig. 3B). Metarrestin treatment extended median overall survival by more than two-fold over the control group. Full necropsy at the study endpoint revealed significantly greater metastatic disease burden (p < 0.05) in the vehicle-treated animals (Fig. 3, C and D). Most animals in the control group showed near-complete or complete organ replacement with tumors, particularly in the liver and to a lesser degree in the lungs, whereas metarrestin-fed mice had preserved organs (Fig. 3C). There was no significant difference in primary tumor growth in the pancreas between treated and untreated animals (Fig. 3D). These findings suggest a survival gain due to suppression of metastasis-related death in metarrestin-treated mice.
To test whether metarrestin’s anti-metastasis activity observed in the NSG PANC1 mice and in vitro pan-cancer PNC suppression translates into anti-metastasis activity in additional cancer models, we evaluated the compound in PC3M xenograft mice. Two weeks after subcutaneous implantation of PC3M cells, mice were started on 5 mg/kg or 25 mg/kg metarrestin or vehicle by daily IP injection for 4 additional weeks. Tumor progression was tracked by both in vivo imaging systems (IVIS) spectroscopy and tumor volume. Metastasis to the lungs was evaluated ex vivo through IVIS spectroscopy and by histopathology at the experimental endpoint. Metarrestin treatment at 25 mg/kg (P < 0.05) decreased development of lung metastasis compared to vehicle-treated mice (Fig. 4A) and modestly reduced growth of primary tumor xenografts (Fig. 4B). Body weight and behavior of the treated animals were not significantly different from control animals (Fig. 4C). To evaluate the effect of metarrestin on a malignant xenograft considered closest to in vivo conditions, we used a patient-derived breast cancer xenotransplantation model (PDX). Metastatic breast cancer cells harvested from the pleural fluid of a stage IV ductal breast cancer patient were inoculated into mice, and third generation mouse-passaged tumors were transplanted into the mammary fat pad of NOD.Cg-Prkdcscid Il2rgtmlSug/JicTac mice. Treatment with metarrestin started after the tumors reached 150–200 mm3. Metarrestin (25 mg/kg) or vehicle was administered daily by IP injection for 4 weeks with a 5-day on and 2-day off schedule. Tumor size and total body weight were measured twice a week, and tumor weight was measured at experimental end-point. The results show that metarrestin effectively inhibited the PDX growth, which formed entirely from metastatic cells from a patient without passing through culture in vitro (Fig. 4D). The treatment was also well-tolerated, without apparent impact on animal body weight and behavior (Fig. 4E).
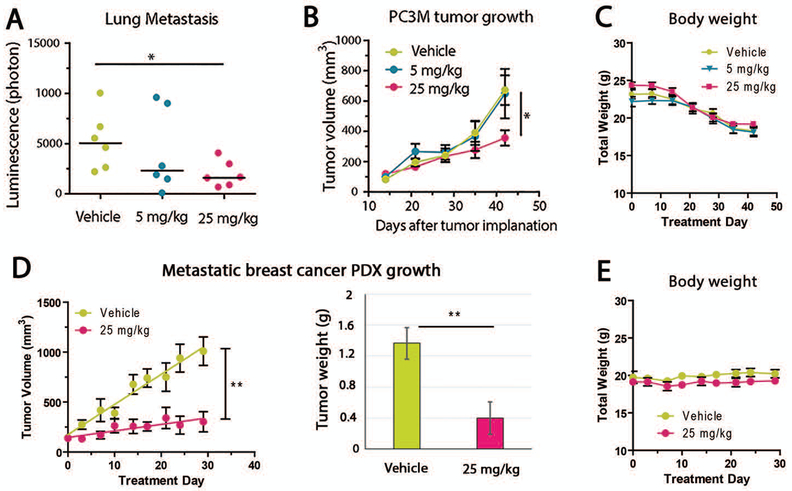
Figure 4: Metarrestin treatment reduces metastasis of prostate cancer (PC3M) and growth of metastatic breast cancer PDX models.
(A) Daily treatment with metarrestin significantly reduced lung metastasis as measured by quantitative IVIS (n=6 for each group), (B) but only a small effect on the growth of PC3M primary tumors inoculated SC, as measured by tumor volume. (C) Weekly body weight evaluation did not show significant differences between treated and control groups. (D) Metarrestin treatment effectively inhibited the growth of a PDX consisting of metastatic cells from a patient’s pleural fluid (n=5 for each group), as measured by tumor volume and tumor weight at the end of the experiment. (E) The body weight of treated animals was not changed. Animals treated with metarrestin remained agile and well-groomed, in contrast to vehicle-treated animals. * P < 0.05, ** P < 0.01.
Metarrestin treatment disrupts nucleolar structure and inhibits Pol I transcription
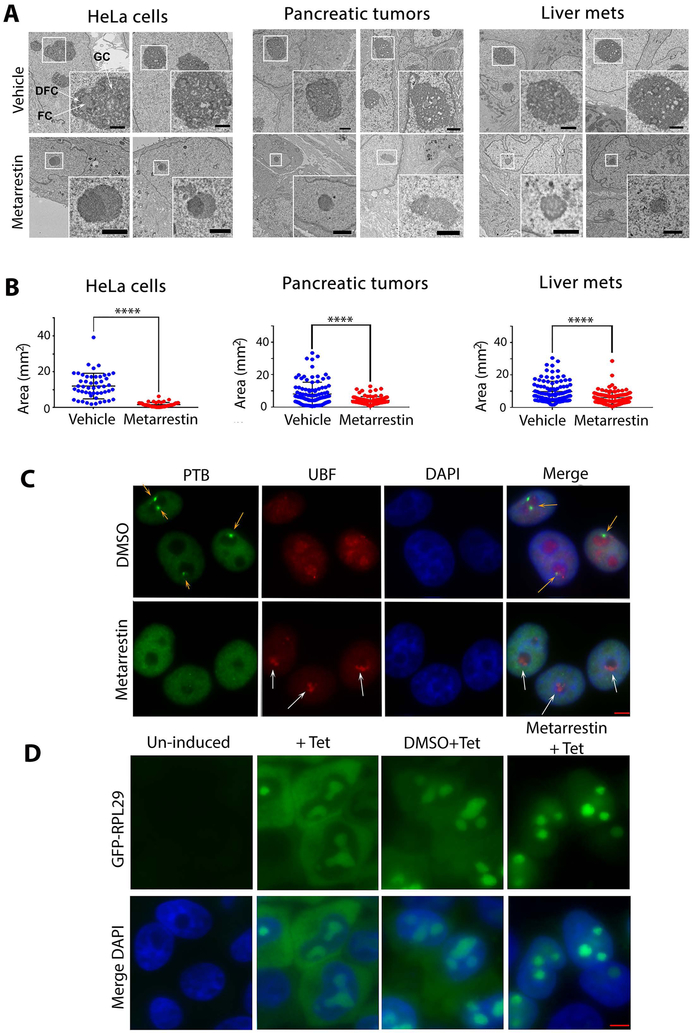
To address the mechanisms by which metarrestin inhibits metastasis, we examined cellular changes upon metarrestin treatment. At 1 μM concentration, metarrestin induced collapse of the nucleolus from the typically integrated three substructures (20-23) (arrows in fig. S5A): fibrillar centers (FC), dense fibrillar components (DFC), and granular components (GC), resulting in a segregation of the fibrillar from the granular components in the nucleus as observed by electron microscopy (EM) (Fig. 5A, lower panels). The nucleolar disruption was reversible within 48 hours upon withdrawal of metarrestin (fig. S5A). Similar disruptions were observed among other treated cell lines, including PANC1 and PC3M cells (fig. S5B), primary tumor, and liver metastatic tissues in the PANC1 xenograft model (Fig. 5A, right panels). Quantitative evaluation of the EM images demonstrated that nucleolar volume was significantly reduced (p < 0.0001) in the treated cell lines and cancer tissues (Fig. 5B, fig. S5C), but was not changed in treated normal mouse hepatocytes (fig. S5C, right panel). The segregation of the nucleolar compartments observed by EM is believed to correspond to the nucleolar capping structures detected by immunofluorescence of nucleolar proteins (24-26) (Fig. 5A and C, fig. S6). The coupling of nucleolar segregation with PNC loss was confirmed across several cell lines by immunolabeling with antibodies against the nucleolar Pol I transcription initiation factors, UBF (upstream binding transcription factor) (Fig. 5C, fig. S6), or Pol I polymerase, RPA194 (fig. S7A). Factors involved in ribosome maturation, such as fibrillarin or NOPP140 (fig. S7A and B) were similarly reorganized. In contrast, UBF distribution was not changed when the normal human fibroblast GM02153 cell line was treated with metarrestin at the same concentration (fig. S7C).
Figure 5. Metarrestin treatment induces nucleolar structure changes.
(A) Nucleoli lose their typical three substructures, as seen in untreated or DMSO-treated cells (arrows indicate DFC, dense fibrillary components, FC, fibrillar components, and GC, granular components), and develop nucleolar capping (enlarged inserts) upon treatment with metarrestin in HeLa cells and tumor tissues. Representative electron micrographs are shown for HeLa cells (treated at 1 μM for 24 hours), primary pancreatic tumors, and liver metastases from NSG PANC1 mice treated with vehicle (top) or 10 mg/kg metarrestin (bottom) for 7 days Mice were harvested 1 hour after last metarrestin dose (inset shows nucleoli). Scale bar = 1 μm. (B) Quantitative evaluation of the EM images demonstrated that average nucleolar area was reduced in metarrestin-treated cell lines and tissues compared to vehicle control (P<0.0001). 100 nucleoli were randomly selected and analyzed to calculate nucleolar area as ((largest+shortest diameter)/2)2×π). The comparison of mean nucleolar areas was performed using Mann Whitney U test, 2-tailed, n=4 animals per group. (C) The changes in nucleolar architecture induced by metarrestin treatment were reflected in the redistribution of Pol I transcription factor, UBF, into cap-like structures (capping) (white arrows), corresponding to the loss of PNCs (green panel, orange arrows). As shown in the merge panel, the capping of UBF reflects the segregation of the fibrillar components from the granular components, as seen in EM images in A. (D) Metarrestin interferes with ribosomal biogenesis. An inducible RPL29-GFP-expressing cell line synthesizes RPL29-GFP when treated with tetracyclin (2nd panels). When cells were treated with 1 μM metarrestin before tetracycline induction, the newly synthesized proteins accumulated in the nucleoli and nuclei (4th panels) compared to the DMSO control treated cells (3rd panels). Scale bars = 5 μm (C and D).
To examine whether the disruption of the nucleolar structure impacts ribosome synthesis, we used a cell line expressing inducible GFP-RPL29 (a ribosomal subunit) (27). GFP-RPL29 induced by the addition of tetracycline (Fig. 5D) showed similar cellular distribution to the endogenous ribosomal subunit (27), localized to the nucleolus and the cytoplasm as ribosomes assemble and traffic into the cytoplasm (Fig. 5D). When GFP-RPL29 expression was induced after exposure to metarrestin for 5 hours (PNCs were disassembled and nucleolar structure altered), the production of GFP-RPL29 (fig. S8A) and its nucleolar localization appeared unchanged (Fig 5D, right panel) in spite of the alterations in nucleolar structure (fig. S8B), indicating that metarrestin did not broadly impact Pol II transcription, translational, and nuclear import functions in treated cells within this time frame. However, the newly synthesized GFP-RPL29 was largely undetected in the cytoplasm of treated cells (Fig. 5D, right panels, fig. S8B, lower panels), indicating a failure of the newly synthesized ribosomal protein to be incorporated into ribosomal particles and to be exported to the cytoplasm, which is a phenotype of ribosome synthesis defect (27). These findings show that metarrestin induces nucleolar ultrastructural disruption in vitro and in vivo with alterations of cellular distribution of ribosomal subunits, suggesting the involvement of ribosome synthesis modulation in the mechanism of action of the compound.
Metarrestin treatment reduces pre-RNA synthesis and Pol I occupancy at rDNA promoters.
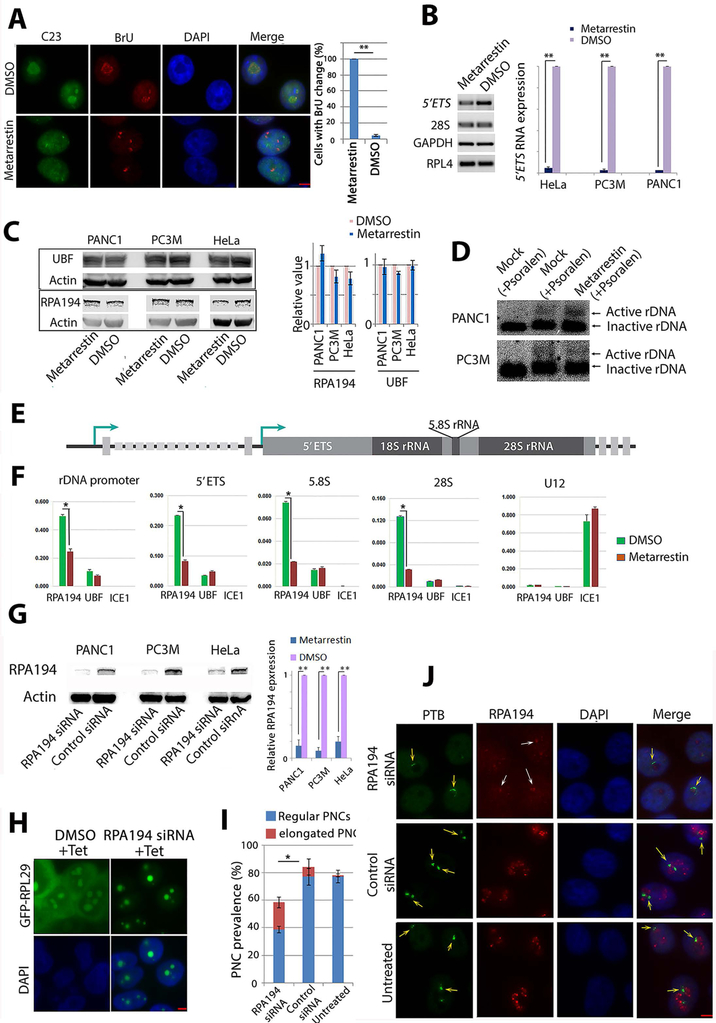
To investigate the step(s) in ribosomal biogenesis that are disrupted by metarrestin, rDNA transcription was evaluated using a BrU incorporation assay, an in situ run-on assay that detects the localization pattern of newly synthesized RNA (8). A five-minute incubation of semi-permeabilized cells with a transcription cocktail containing BrU (Fig. 6A, red) showed that the organization of newly synthesized rRNA (reflecting transcription sites) was altered into tight small clusters and nucleolar structure was disrupted, as represented by the dispersion of C23/nucleolin, a protein involved in many aspects of nucleolar function (28), from nucleoli (Fig. 6A, green). To evaluate the impact on rDNA transcription in metarrestin-treated cells, we quantified the amounts of 5’ETS RNA. Because 5’ETS in nascent pre-rRNA transcripts is rapidly removed (29, 30), the amount of 5’ETS correlates with the amount of rDNA transcription. Quantitative RT-PCR of 5’ETS in metarrestin-treated and DMSO-treated control cells showed a substantial reduction of 5’ETS RNA in metarrestin-treated cells (Fig. 6B).
Figure 6: Metarrestin treatment reduces pre-RNA synthesis and Pol I occupancy at rDNA without changing rDNA chromatin states.
(A) Pol I transcription patterns are altered, as demonstrated by a redistribution of BrU incorporation signals from typical nucleolar labeling to pin-points at 5 min (red panel), corresponding to the changes in nucleolar structure shown as a loss of nucleolar labeling of C23/nucleolin (green panel). All cells treated with metarrestin show alterations of BrU labeling in the nucleoli compared to a small fraction of cells in DMSO treated cells (right panel). (B) RT-PCR (left panel) and qRT-PCR (right panel) show the reduction in 5’ETS of the pre-rRNA in metarrestin-treated cells. (C) Western blot analyses show no changes in protein expression of RPA194, the large subunit of Pol I, and UBF in metarrestin-treated cells. (D) Psoralen-crosslinking experiments show that the ratio of active to inactive rDNA chromatin appears unchanged upon exposure to metarrestin. (E) A diagram of rDNA structure. (F) Quantitative ChIP evaluations demonstrate that metarrestin treatment reduces the occupancy of RPA194, but not UBF, on rDNA through the promoter and the coding region. (G) Knockdown of pol I by siRNA showed reduction of RPA194 by Western blots, and the amount was quantified in relation to control siRNA treated cells (set as 1). (H) Correspondingly, ribosome synthesis was reduced in RPA194 knockdown cells, and induced GFP-RPL29 expression 72 hours after transfection of the siRNA showed the absence of cytoplasmic localization of the protein. (I) Knockdown of RPA194 by siRNA reduced PNC prevalence and increased the number of PNCs with a crescent shape (red portion). (J) RPA194 knockdown also disrupts the nucleolus (top red panel, white arrows) compared to untreated and control oligo treated cells (lower two panels). PNC structures were altered into crescent shapes (top green panel, orange arrows) compared to untreated or control oligo treated cells (lower two panels). Scale bars for all images = 5 μm. * p<0.05, ** p<0.01.
To examine the mechanisms by which metarrestin disrupts Pol I transcription, we evaluated the overall state of the transcription machinery and rDNA chromatin structure in treated cells. Western blot demonstrated that the amounts of Pol I large subunit RPA194 and UBF did not change significantly after 24 hours of treatment with metarrestin at 1 μM concentration in the three cell lines, PANC1, PC3M, and HeLa (Fig. 6C). To examine whether an altered chromatin state of the rDNA clusters could be involved in the reduction of rRNA, we carried out psoralen-crosslinking experiments to determine the active/inactive rDNA chromatin ratios in metarrestin- and DMSO-treated cells. Psoralen crosslinks active accessible DNA under UV light without displacing nucleosomes or transcription factors, or changing chromatin states (31, 32). Psoralen-crosslinked DNA moves slower on agarose gel and can be distinguished from the non-crosslinked DNA by Southern blots (32). The results showed that the ratio of active/inactive rDNA chromatin is similar in treated and untreated cells (Fig. 6D), suggesting that rDNA chromatin is not grossly altered. To examine the interactions between Pol I transcription factors and rDNA, we carried out quantitative ChIP experiments using primer sets across promoter and coding regions of rDNA (Fig. 6, E and F). Metarrestin treatment significantly reduced RPA194 occupancy on rDNA promoter and coding regions without significant impact on UBF binding (Fig. 6F, p <0.01). As a control, a component of the small elongation complex, ICE1, did not show significant associations with rDNA sequences or altered association with its target gene U12 transcribed by Pol II. These observations show that metarrestin impairs Pol I-rDNA interaction.
Genotoxic agents (33) that intercalate into or alkylate DNA, such as actinomycin D (34), induce similar nucleolar segregation to that observed with metarrestin treatment. To determine whether metarrestin induces nucleolar changes through genotoxic effects or general cytotoxicity, we evaluated the impact of metarrestin on DNA damage repair, cell cycle, and general Pol II transcription status in three cell lines, PANC1, PC3M, and HeLa. Cells were treated with 1 μM metarrestin or DMSO for 24 hours before fixation for flow cytometry, immunolabeling, or Western blot analyses. Evaluation of DNA damage response signature factors in treated cells (fig. S9A) demonstrated that neither phosphorylated γH2AX or p53BPl, nor phosphorylated p53 (HeLa and PC3M have little to no p53) were altered in these cell lines upon metarrestin treatment (fig. S9B). Cell cycle analyses of DNA content through flow cytometry did not show a significant alteration of cell cycle pattern upon metarrestin treatment at two different concentrations within 24 hours (fig. S9C). Evaluation of apoptotic index showed less than 1% of cells undergoing apoptosis in response to metarrestin treatment.
To further determine whether metarrestin interferes with Pol II transcription in general, we performed immunolabeling of CUGBP. The steady state nuclear distribution of CUGBP, a multifunctional hnRNP protein (35, 36) highly enriched in the PNC (15), is dependent upon Pol II transcription (15). Selective inhibition of Pol II by α-amanitin induces a localization shift to the cytoplasm except for PNC-localized CUGBP (fig. S9D top right panel). If metarrestin treatment significantly impacted Pol II transcription, one would expect a predominant cytoplasmic redistribution of CUGBP, as shown in α-amanitin-treated cells, however, that was not the case (fig. S8D, top middle panel). In addition, SC35, an essential pre-mRNA splicing factor normally has an intranuclear, interconnected speckle-distribution pattern (37-39) (fig. S9D, lower left panel), which changed to isolated large dots in α-amanitin-treated cells (fig. S9D, lower right panel). In comparison, the SC35 pattern was not significantly changed in metarrestin-treated cells (fig. S9D, lower middle panel). Together with the observation that metarrestin treatment (Fig. 5D) does not impact the induction of GFP-RPL29 expression (fig. S8) and its nucleolar localization (Fig. 5D and S8), these findings suggest that metarrestin does not globally inhibit Pol II transcription, general protein synthesis, or nucleocytoplasmic trafficking within the effective concentration range, but has a selective effect on Pol I transcription and ribosome synthesis.
Reduction of RPA194 partially phenocopies the disruption of nucleoli and PNCs by metarrestin.
We next asked whether metarrestin disassembles PNCs by inhibiting Pol I function and disrupting nucleoli. The large subunit of Pol I, RPA194, was knocked down by a specific siRNA, as evident by the reduction of the protein 72 hours after transfection with the siRNA oligos in cell lines PANC1, HeLa, and PC3M (Fig. 6G). The reduction of RPA194 was sufficient to inhibit ribosome synthesis at a single cell level, as shown in HeLa cells expressing inducible GFP-RPL29, where RPA194 knockdown rendered cytoplasmic GFP-RPL29 near undetectable (Fig. 6H right panels). The reduction of RPA194 decreased PNC prevalence (Fig. 6I), although not to the same extent as in metarrestin-treated cells (Fig. 1C). Some PNCs also showed structural changes (Fig. 6I), becoming crescent shaped around distorted nucleoli in RPA194-reduced cells (Fig. 6J top panels, yellow arrows), similar to cells treated with metarrestin at a concentration that did not completely eliminate PNCs (Fig. 1A, middle panel). The siRNA against RPA194 also disrupted the nucleolar structure into capping structure, as shown by labeling with anti-RPA194 for the residual RPA194 (Fig. 6J, white arrows at cap-like red signals in the top 2nd panel). The changes in PNC and nucleoli in RPA194-silenced cells were similar to those observed with metarrestin treatment (Fig. 5C, fig. S7). These findings demonstrate that downregulating Pol I activity partially recapitulates the disruption of nucleolar and PNC structures induced by metarrestin treatment, and support the idea that metarrestin disrupts PNCs, at least in part, through inhibition of Pol I function.
Metarrestin binds the translation elongation factor eEF1A
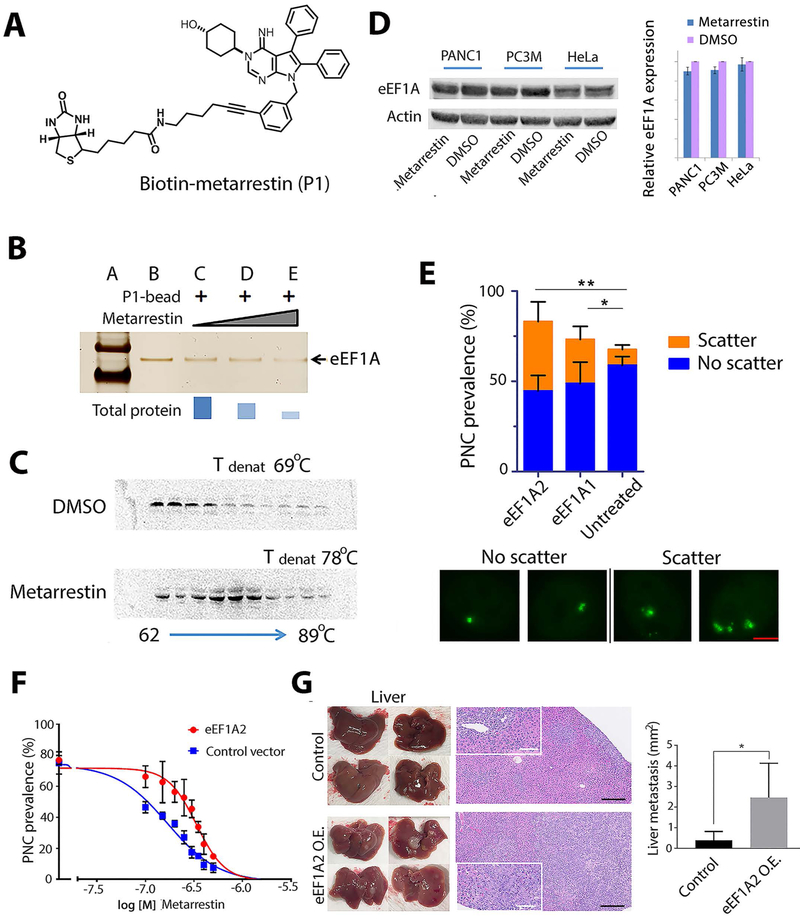
To identify the molecular target(s) of metarrestin and upstream factors involved in ribosomal and Pol I function, we identified proteins that bind metarrestin through affinity purification using biotin-conjugated metarrestin (Fig. 7A), which maintains its efficacy for PNC disassembly in cultured cells (fig. S10). Competition studies using untagged metarrestin and proteomic analysis identified eukaryotic translation elongation factor (eEF1A) as a metarrestin-binding protein (Fig. 7B). eEF1A has two isoforms, eEF1A1 and eEF1A2 (40), and whereas eEF1A1 is ubiquitously expressed in all tissues, eEF1A2 is only expressed in brain, heart, and skeletal muscle, although its over- or re-expression, including in a stage-specific manner, has been described in a number of cancers, including pancreatic cancer (40, 41). Binding of metarrestin to eEF1A was further confirmed in cells using a cellular thermal shift assay (CETSA), which showed an increase in the aggregation temperature of eEF1A upon metarrestin treatment (Fig. 7C). However, the binding did not significantly change the amounts of the proteins, and western blot analyses showed that total eEF1A protein remained similar after 1 μM metarrestin treatment for 24 hours (Fig. 7D).
Figure 7: Metarrestin specifically binds eEF1A2, and increased eEF1A2 enhances PNCs and metastasis formation.
(A) The structure of metarrestin tagged with biotin. (B) Metarrestin effectively outcompeted recombinant eEFl A from binding to anchored biotinylated metarrestin-eEF1A complex. (C) Metarrestin treatment stabilized eEF1A in a thermal stability assay using PC3M cell lysate. (D) Western blot analyses do not show changes in the amount of eEF1A proteins upon metarrestin treatment at 1 μM for 24 hours. (E) Overexpression of HA-eEF1A2 enhanced PNC structures in cells (image panels, each containing a single nucleus). Although it did not significantly increase overall PNC prevalence (top), overexpression of HA-eEF1A2 increased the number of PNCs per nucleus (scattered PNC prevalence: the number of cells containing 2 or more PNCs); n = 300 cells (bar= 2 μm). (F) Overexpression of eEF1A2 in PC3M cells increased the IC50 of metarrestin for PNC disassembly. (G) 6×104 PANC1 3D spheres transduced with empty vector (control) or eEF1A2 (eEF1A2 O.E.) were injected into the tail of the pancreas of NSG mice. Mice from both groups were harvested 6 weeks after implantation and subjected to necropsy. Macroscopic images of anterior (top) and posterior (bottom) liver surfaces showed higher metastatic burden in PANC eEF1A2 animals than in empty vector control (left, harvested livers). Histopathological images (H&E) of livers (black scale bar=250 μm, white scale bar=100 μm) are shown on the right. Insets depict representative metastatic lesions. Quantification of liver metastasis showed a higher metastatic burden in PANC1 eEF1A2 O.E. animals (n=4 animals analyzed per group). * P < 0.05, ** P < 0.01.
eEF1As are multi-functional proteins, implicated in translation elongation, actin bundling, nuclear transport, and tRNA export (41-44). To evaluate whether eEF1A mediates the effect of metarrestin on cancer cells, eEF1A was either overexpressed or reduced through siRNA silencing. Whereas overexpression of HA-eEF1A1 or HA-EFF1A2 did not significantly increase total PNC prevalence (Fig. 7E), overexpression of HA-eEF1A2, more so than HA-eEF1A1, increased the scattering patterns (number of PNCs per cell) of existing PNCs in PNC-containing cells (Fig. 7E). Thus, eEF1A2 enhances PNC structures, but is alone not sufficient to induce significant formation of PNCs in PNC-negative cells. To evaluate the functional link between the phenotype of metarrestin-induced PNC disassembly and eEF1A2, we studied PNC disassembly in wild type and eEF1A2-overexpressing cancer cells. Cells with and without over expression of eEF1A2-HA (48 hours after transfection with an efficiency at around 70%, fig. S11) were treated with metarrestin at 10 concentrations, and PNC prevalence was evaluated at 63× magnification. The results showed that increased expression of eEF1A2 increased the inhibitory concentration 50 (IC50) of metarrestin for PNC disassembly (Fig. 7F). To evaluate whether eEF1A2 indeed regulates the metastatic phenotype, PANC1 cells with (PANC1 eEF1A2 O.E.) or without (PANC1 control) overexpression of eEF1A2 were orthotopically injected into NSG mice. Six weeks after implantation, organs were subjected to histopathological evaluation. NSG PANC1 eEF1A2 O.E. mice showed a significantly increased metastatic disease burden compared to PANC1 control animals (Fig. 7G, P <0.05), suggesting that metarrestin engages a target governing the metastatic phenotype.
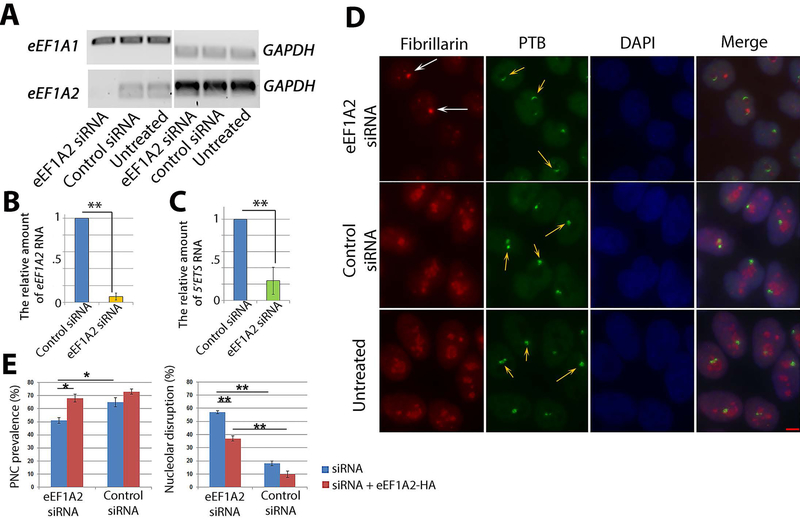
To determine whether the reduction of eEF1A2 phenocopies metarrestin’s impact on PNCs and nucleolar structure, eEF1A2 expression was reduced using siRNA oligos. Seventy-two hours after transfection, qRT-PCR showed a selective reduction of eEF1A2 (Fig. 8, A and B). To evaluate whether reduction of eEF1A2 impacts Pol I transcription, 5’ETS RNA expression was compared in cells with and without eEF1A2 knockdown, and the results showed a significant reduction (Fig. 8C, P <0.01) of rDNA transcription. Correspondingly, immunofluorescence detection of nucleoli by anti-fibrillarin antibodies and of PNCs by SH54 showed that reduction of eEF1A2 disrupted nucleolar (nucleolar segregation) and PNC structure (loss or becoming elongated and crescent shaped, arrows in Fig. 8, D and E) similarly to metarrestin-treated (Fig. 1A) or Pol I-knockdown cells (Fig. 6J), although with lower efficacy compared to metarrestin (near 100% PNC disassembly and nucleolar distortion at 1 μM, Fig. 1A, Fig. 5C). The disruption could be partially rescued by overexpression of HA-eEF1A2 (Fig. 8E, red bars). Overall, these findings support the notion that eEF1A2 is involved in metastatic progression in pancreatic cancer and that interfering with its expression largely phenocopies the nucleolar and PNC structural alterations and Pol I inhibition induced by metarrestin, supporting the idea that eEF1A2, at least in part, mediates the nucleolar and PNC alterations induced by metarrestin treatment.
Figure 8: eEF1A2 reduction induces similar nucleolar and PNC disruption as metarrestin.
(A) Seventy-two hours after siRNA transfection into HeLa cells, eEF1A2 RNA, but not eEF1A1 RNA, was reduced, as measured by RT-PCR. (B) qRT-PCR showed a reduction of eEF1A2 RNA in siRNA-transfected cells. (C) Seventy-two hours after transfection with eEF1A2 siRNA, the amount of 5’ETS RNA was reduced, as measured by qRT-PCR. (D) Nucleolar and PNC disruption was detected using immunofluorescence in siRNA transfected cells. The PNCs in eEF1A2 knock-down cells generally showed crescent shapes (orange arrows), as well as segregated nucleoli (capping, white arrows) immunolabeled with an antibody recognizing a pre-RNA processing factor fibrillarin; n = 500 cells. Scale bar = 5 μm. (E) PNC prevalence was modestly reduced (left graph, blue bars), and the rate of nucleolar disruption was increased (right graph, blue bars). Transfection of HA-eEF1A2 (red bars) after siRNA for additional 24 hours partially rescued PNC prevalence (left graph, red bars) and nucleolar disruption (right graph, red bars). * p < 0.05, ** p < 0.01.
Discussion
Despite decades of intensive research efforts, the key mechanisms of cancer progression and metastasis are still not fully understood. Metastasis involves complex, multi-step processes that allow for the selection of cancer cells with the ability to escape immune system surveillance, propagate, migrate, infiltrate, and colonize different organs (45, 46). Thus, complex phenotypic markers that reflect the malignant capacity of cancer cells could potentially be useful to identify effective treatments. PNCs are formed almost exclusively in cancer cells, and their prevalence is closely associated with metastasis in vitro and in vivo, and therefore they are believed to be such a marker (3, 11, 12, 15). As a multi-component complex subnuclear body, PNCs may reflect the complex characteristics specific to metastasis better than a single gene or gene product.
Through a strategy of screening followed by chemical optimization, metarrestin emerged as a compound with potent PNC-disassembly efficacy and attractive pharmacokinetic characteristics. In vitro, metarrestin effectively disassembles PNCs across multiple human cancer cell lines, reducing their invasion capabilities. In vivo, metarrestin treatment inhibited metastasis in mouse models of human cancer without the adverse effects typically associated with chemotherapeutic agents. Metarrestin had a modest impact on primary tumor growth in both pancreatic and prostate cancer models, but effectively inhibited the growth of inoculated PDX tumors derived from metastatic breast cancer cells from pleural fluid of a stage IV breast cancer patient. Plasma PK measures correlated with the anti-metastasis effect of the drug; mice dosed with 5 mg metarrestin did not have 24-hour coverage above the cell-based IC50 value of metarrestin in the plasma, a finding in line with the reduced anti-metastatic activity in NSG PANC1 mice compared to the 25 mg/kg cohort. Metarrestin’s selectivity for cancer vs. normal cells in general, selectivity of metastasis vs. primary lesions, pan-cancer applicability, lack of overt toxicity, and favorable PK profile support metarrestin as an attractive candidate for clinical translation.
The disruption of PNCs by metarrestin is closely coupled with the disruption of nucleolar structure, reduction of Pol I occupancy on rDNA, and rDNA transcription. siRNA knockdown of the Pol I large subunit, RPA194, partially recapitulates the impact of metarrestin on PNCs, suggesting that nucleolar functional changes could in part mediate the disruption of PNCs by metarrestin. The nucleolar segregation in metarrestin-treated cells is in many respects similar to that observed in cells treated by actinomycin D (25, 34) and other agents that impact DNA or inhibit topoisomerases (33), all of which are genotoxic and induce nucleolar stress, causing cell-cycle arrest and apoptosis (47-49). In comparison, treatment by metarrestin for 24 hours did not induce detectable DNA damage-repair responses, appreciable apoptosis in either p53 wild type or mutant metarrestin-treated cells, global Pol II transcription, translation, or nucleocytoplasmic trafficking. These findings distinguish metarrestin from the many genotoxic Pol I transcription inhibitors that are currently used as cancer therapeutics with substantial adverse effects.
Although it has long been used as a parameter in pathology for cancer diagnosis and prognosis (50), the role of nucleoli and their possible value as a therapeutic target in malignancy have only come into focus over the past few years (21, 22, 51-56). Increasing evidence indicates that nucleoli play key roles in tumorigenesis beyond ribosome biogenesis. These include cell-cycle regulation, apoptosis, gene expression regulation, response to genomic stress, partition of cellular factors, and RNA transport (22, 57-59). Correspondingly, many of the key factors in ribosome synthesis, including ribosomal proteins, are multifunctional and act as secondary regulators of epigenetic modulation and cancer progression (22, 57-59). The disruption of rDNA transcription by metarrestin reported here is in line with nucleoli as a potential therapeutic target for cancer.
Nucleolar stress-induced apoptosis has been shown as the mechanism for antitumor effects of recently developed anti-cancer compounds, such as CX5461, which inhibits Pol I transcription and induces p53-mediated apoptosis (53, 54). BMH-21, another compound that binds to GC-rich sequences and degrades RNA Pol I, also induces cell death with yet to be defined pathways (55, 60, 61). In comparison, metarrestin modulates Pol I transcriptional function through different mechanisms as evidenced by the following: 1) metarrestin does not induce p53-mediated or - independent apoptosis or p53-activated cell-cycle arrest in cells (with or without wild type p53), 2) metarrestin does not inhibit pol I transcription through downregulation of Pol I machinery components and has no apparent impact on the rDNA chromatin state, 3) metarrestin interferes with Pol I occupancy on rDNA without affecting rDNA interaction with another Pol I factor, UBF, and 4) Pol I knockdown partially recapitulates the cellular phenotype on nucleoli and PNC structures observed in metarrestin-treated cells, supporting the idea that metarrestin inhibits Pol I function without direct interference with rDNA or reduction of the transcription machinery at the protein level.
Repeated affinity purification and proteomic analyses did not show RPA194 binding to biotin-tagged metarrestin, suggesting an indirect modulation of Pol I activity by metarrestin. A search for protein targets of metarrestin identified eEF1A as a binding partner, as supported by the results of competition binding assays and changes in eEF1A-protein thermal stability in cells. Manipulation of eEF1A2 expression through either overexpression or reduction by siRNA showed that eEF1A2 plays a role in PNC structures. Even though expression of eEF1A2 alone was insufficient to increase PNC prevalence in PNC-null cells, it increased the number of PNCs per cell and increased the IC50 of metarrestin against PNC. Furthermore, overexpression of eEF1A2 increased metastatic burden in PANC1 xenograft mice. Together with the observation that eEF1A2 knockdown partially recapitulated the nucleolar and PNC disruption induced by metarrestin treatment, our findings support the concept that eEF1A2 in part mediates the function of metarrestin in cancer cells.
Many biological functions are described for eEF1As, including translation elongation, actin bundling and polymerization, tRNA nuclear export, viral cycle, protein degradation, and stress response (40, 53, 54, 62-66). Overexpression of eEF1A2 and functional association have been documented in a number of cancers such as breast, prostate, pancreatic, and ovarian (66-70). Several clinical studies have associated increases in eEF1A2 expression with poor prognosis (69, 70), and previous preclinical studies have described anti-eEF1A disrupting translation and protein synthesis (71-73). In contrast, metarrestin treatment at effective concentrations, as observed with the inducible expression of GFP-RPL29, does not interfere with general translation of existing ribosomes even through it reduces new ribosome synthesis. Although our findings suggest that metarrestin selectively interferes with non-translational functions of eEF1A2, its role in PDX and other types of tumors in vivo, the precise mechanisms of its involvement in pol I transcription, and potential other functional targets of metarrestin require further investigations.
In summary, metarrestin is a small molecule which is selective against metastasis across different preclinical cancer histologies, with high intratumoral exposure and without appreciable toxicity. In contrast to the classical, single target-driven anti-cancer drug discovery paradigm, metarrestin was obtained from an alternative approach, screening against a complex phenotypic marker for malignancy. Metarrestin inhibits Pol I transcription, induces nucleolar segregation, reduces nucleolar volume, and disrupts PNCs, in part by interfering with eEF1A2 function. Because metastasis is a key cause of lethality in cancer patients, the selective anti-metastasis activity with limited toxicity makes metarrestin a promising chemotherapeutic candidate.
Materials and Methods
Study design
The goal of this study was to develop and validate an alternative approach to identify effective anti-metastasis lead compounds. PNC, a complex nuclear body that forms only in cancer cells, was used as a phenotypic marker and surrogate of metastatic behavior in a screen to identify small molecules that disassemble the nuclear body without killing the cells at the effective concentrations. Secondary and tertiary screens were used to identify compounds, which were further optimized through chemical modification and synthesis.
In vitro assays and in vivo xenograft modeling were used to determine the efficacy of a lead compound, metarrestin, for inhibiting PNC, cancer cell growth, cell invasion, tumor growth, and metastasis in pancreatic, prostate, and breast cancer xenograft models. Cultured cells, 3D spheres, or PDX tissues were used for these animal models. A biotin-tagged metarrestin derivative was synthesized and confirmed to have a similar PNC disassembly IC50 as the untagged compound. Biochemical and mass-spec approaches were used to identify its cellular binding proteins. Cellular and molecular approaches were used to validate the potential targets of metarrestin. Standard practices involved in pharmacokinetic analyses were used to determine metarrestin exposure in mice. A dedicated veterinary pathology review of 12 organ systems from male and female genetically engineered tumor-bearing and wild type mice exposed for 3 months to metarrestin was conducted to evaluate the toxicity of the compound. The majority of the findings included in the article were evaluated by more than 1 method, and experiments were repeated multiple times (the specifics are included in the supplemental materials and methods section). In vivo experiments were randomized, unblinded, and conducted in accordance with the NIH Guide for the Care and Use of Laboratory Animals. The animals were sacrificed at the predetermined time point or upon signs of animal distress (immobility or weight loss greater than 15 %). Animal data were excluded as outliers in cases of unspecified death or sacrifice prior to the predetermined endpoint, which affected less than 10 percent of initially enrolled mice. Individual trial details (sample size, mouse type, trial duration, and specimen handling) are provided in the Supplementary Materials and Methods.
Statistical analyses
Data are presented as means +/− SD. Student’s t tests were performed for all the experiments, except where indicated differently in the figure legends. Mean nucleolar areas were analyzed using Mann Whitney U test, 2-tailed.
Supplementary Material
Synthetic scheme to metarrestin
Growth inhibition of PANC1 by metarrestin
Metastatic cancer progression in NOD/IL2Rgnull (NSG) PANC1 mice
Single and multiple dose metarrestin plasma and tumor pharmacokinetics
Metarrestin treatment induced nucleolar structure changes as observed by electron microscopy.
Nucleolar structure changes (UBF labeling in red, blue arrows) were closely associated with the loss of PNC (PTB labeling in green, orange arrows) in metarrestin-treated cells.
Pol I transcription factors, UBF, RPA194, and pre-ribosomal RNA processing factor, Nopp140 and fibrillarin showed similar capping structure in metarrestin-treated cells.
Metarrestin treatment did not significantly impact Pol II transcription, translation, or nucleocytoplasmic trafficking.
Metarrestin treatment did not induce DNA damage response, cell cycle blockage, or global inhibition of pol II transcription.
Biotin-metarrestin was similarly effective in disassembling PNCs in treated PC3M cells at 1 μM for 18 hours.
Transfection of eEF1A2-HA showed the expression of the protein in red in transfected cells.
Synthesis of 2-amino-1-benzyl-4,5-diphenyl-1H-pyrrole-3-carbonitrile.
Synthesis of (E)-ethyl N-(1-benzyl-3-cyano-4,5-diphenyl-1H-pyrrol-2-yl)formimidate.
Synthesis of trans-4-(7-benzyl-4-imino-5,6-diphenyl-4,7-di hydro-3 H-pyrrolo[2,3-d]pyrimidin-3-yl)cyclohexanol (Metarrestin).
Synthesis of 2-amino-1-(3-bromobenzyl)-4,5-diphenyl-1H-pynole-3-carbonitrile.
Synthesis of (E)-ethyl N-(1-(3-bromobenzyl)-3-cyano-4,5-diphenyl-1H-pyrrol-2-yl)formimidate.
Synthesis of trans-4-(7-(3-bromobenzyl)-4-imino-5,6-diphenyl-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-3-yl)cyclohexanol.
Synthesis of N-(6-(3-((3-(trans-4-hydroxycyclohexyl)-4-imino-5,6-diphenyl-3,4-dihydro-7H-pyrrolo[2,3-d]pyrimidin-7-yl)methyl)phenyl)hex-5-yn-1-yl)-5-((3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl)pentanamide (biotin-metarrestin, P1).
Cancer cell lines examined for PNC prevalence with or without treatment of metarrestin (from Fig. 1C).
Pancreatic cancer cell lines evaluated for PNC prevalence in Fig. 2A.
Veterinary pathology read of organs derived from tumor-bearing KPC and wild type mice treated by metarrestin for 3 months.
Blood biochemistry and hematology toxicity of metarrestin in tumor bearing KPC and wild type mice.
Acknowledgements:
We would like to thank the Dr. Ulrike Kutay for the GFP-RPL29 cell line, Dr. Meredith S. Irwin for the HA-eEF1A1 and HA-eEF1A2 constructs, and Dr. Shi-Yuan Cheng for cell lines. We thank the Center for Therapeutic Development, Genomic Core Facility and Cell Imaging Facility of NU for the support of some of the work. The opinions expressed in this article are the authors’ own and do not reflect the view of the National Institutes of Health, the Department of Health and Human Services, or the United States government.
Funding: This study was supported by the intramural research program of NIH (JM, MF, WZ, UR), including ZIA BC 011267 for UR. Other support included U54HG005031 for JA, NIHR01GM115710 for SB, NIH F31DE026946 for KF, NIH R01GM078555 for SH, IDP, H, and V foundation funding through Robert H. Lurie Comprehensive Cancer Center at NU for SH, Donations from the Baskes family to the Robert H. Lurie Comprehensive Cancer Center, NU, from ‘Running for Rachel’ to the National Cancer Institute (UR), Cancer Center Support grant 2 P30 CA060553-19 to APM, Robert H. Lurie Comprehensive Cancer Center – Translational Bridge Program Fellowship in Lymphoma Research for DH, and Molecular Libraries Initiative funding to the University of Kansas Specialized Chemistry Center (U54HG005031) for KF.
Footnotes
Supplementary materials:
Materials and methods
Competing interests: Patent entitled “Compounds and methods for the prevention and treatment of tumor metastasis and tumorigenesis”, U.S. Patent Application # 61/576,780, is pending for metarrestin. The inventors include Samarjit Patnaik, Kevin Frankowski, Sui Huang, Chen Wang, Juan Jose Marugan, John Norton, Steven Titus, and Wei Zheng. A disclosure application entitled “Metarrestin for the Treatment of Pancreatic Cancer” is being prepared. Inventors include Udo Rudloff, Serguei Kozlov, Juan J Marugan, Samarjit Patnaik, Noel Southall, Marc Ferrer, Michael Baltezor, John Haslam, Sui Huang.
Materials and data availability: Currently metarrestin is under preclinical development toward IND filling. Metarrestin is available from AOBIOUS under the name of ML246.
References and notes:
- 1.Jiang WG, Sanders AJ, Katoh M, Ungefroren H, Gieseler F, Prince M, Thompson SK, Zollo M, Spano D, Dhawan P, Sliva D, Subbarayan PR, Sarkar M, Honoki K, Fujii H, Georgakilas AG, Amedei A, Niccolai E, Amin A, Ashraf SS, Ye L, Helferich WG, Yang X, Boosani CS, Guha G, Ciriolo MR, Aquilano K, Chen S, Azmi AS, Keith WN, Bilsland A, Bhakta D, Halicka D, Nowsheen S, Pantano F, Santini D, Tissue invasion and metastasis: Molecular, biological and clinical perspectives. Semin Cancer Biol 35 Suppl, S244–75 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Weber GF, Why does cancer therapy lack effective anti-metastasis drugs? Cancer Lett 328, 207–11 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Norton JT, Pollock CB, Wang C, Schink JC, Kim JJ, Huang S, Perinucleolar compartment prevalence is a phenotypic pancancer marker of malignancy. Cancer 113, 861–9 (2008a). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matera AG, Frey MR, Margelot K, Wolin SL, A perinucleolar compartment contains several RNA polymerase III transcripts as well as the polypyrimidine tractbinding protein, hnRNP I. J Cell Biol 129, 1181–93 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee B, Matera AG, Ward DC, Craft J, Association of RNase mitochondrial RNA processing enzyme with ribonuclease P in higher ordered structures in the nucleolus: a possible coordinate role in ribosome biogenesis. Proc Natl Acad Sci U S A 93, 11471–6 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang S, Deerinck TJ, Ellisman MH, Spector DL, The dynamic organization of the perinucleolar compartment in the cell nucleus. J Cell Biol 137, 965–74 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Norton JT, Wang C, Gjidoda A, Henry RW, Huang S, The perinucleolar compartment is directly associated with DNA. Journal of Biological Chemistry 284, 4090–101 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang C, Politz JC, Pederson T, Huang S, RNA polymerase III transcripts and the PTB protein are essential for the integrity of the perinucleolar compartment. Mol Biol Cell 14, 2425–35 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pollock C, Daily K, Nguyen VT, Wang C, Lewandowska MA, Bensaude O, Huang S, Characterization of MRP RNA-protein interactions within the perinucleolar compartment. Molecular Biology of the Cell 22, 858–66 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pollock C, Huang S, The perinucleolar compartment. J Cell Biochem 107, 189–93 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Slusarczyk A, Kamath R, Wang C, Anchel D, Pollock C, Lewandowska MA, Fitzpatrick T, Bazett-Jones DP, Huang S, Structure and Function of the Perinucleolar Compartment in Cancer Cells. Cold Spring Harb Symp Quant Biol (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamath RV, Thor AD, Wang C, Edgerton SM, Slusarczyk A, Leary DJ, Wang J, Wiley EL, Jovanovic B, Wu Q, Nayar R, Kovarik P, Shi F, Huang S, Perinucleolar compartment prevalence has an independent prognostic value for breast cancer. Cancer Research 65, 246–53 (2005). [PubMed] [Google Scholar]
- 13.Norton JT, Titus SA, Dexter D, Austin CP, Zheng W, Huang S, Automated high-content screening for compounds that disassemble the perinucleolar compartment. J Biomol Screen 14, 1045–53 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pettaway CA, Pathak S, Greene G, Ramirez E, Wilson MR, Killion JJ, Fidler IJ, Selection of highly metastatic variants of different human prostatic carcinomas using orthotopic implantation in nude mice. Clin Cancer Res 2, 1627–36 (1996). [PubMed] [Google Scholar]
- 15.Huang S, Deerinck TJ, Ellisman MH, Spector DL, The perinucleolar compartment and transcription. Journal of Cell Biology 143, 35–47 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frankowski K, Patnaik S, Schoenen F, Huang S, Norton J, Wang C, Titus S, Ferrer M, Zheng W, Southall N, Day VW, Aube J, Marugan JJ, Discovery and Development of Small Molecules That Reduce PNC Prevalence, in Probe Reports from the NIH Molecular Libraries Program. Bethesda (MD)(2010). [PubMed] [Google Scholar]
- 17.Suemizu H, Monnai M, Ohnishi Y, Ito M, Tamaoki N, Nakamura M, Identification of a key molecular regulator of liver metastasis in human pancreatic carcinoma using a novel quantitative model of metastasis in NOD/SCID/gammacnull (NOG) mice. Int J Oncol 31, 741–51 (2007). [PubMed] [Google Scholar]
- 18.Yin T, Wei H, Gou S, Shi P, Yang Z, Zhao G, Wang C, Cancer stem-like cells enriched in Panc-1 spheres possess increased migration ability and resistance to gemcitabine. Int J Mol Sci 12, 1595–604 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mathews LA, Cabarcas SM, Hurt EM, Zhang X, Jaffee EM, Farrar WL, Increased expression of DNA repair genes in invasive human pancreatic cancer cells. Pancreas 40, 730–9 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raska I, Shaw PJ, Cmarko D, Structure and function of the nucleolus in the spotlight. Curr Opin Cell Biol 18, 325–34 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Pederson T, The nucleolus. Cold Spring Harb Perspect Biol 3, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grummt I, Life on a planet of its own: regulation of RNA polymerase I transcription in the nucleolus. Genes Dev 17, 1691–702 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Busch H, Smetana K, The nucleolus. New York: Academic Press; (1970). [Google Scholar]
- 24.Sirri V, Urcuqui-Inchima S, Roussel P, Hernandez-Verdun D, Nucleolus: the fascinating nuclear body. Histochemistry and Cell Biology 129, 13–31 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shav-Tal Y, Blechman J, Darzacq X, Montagna C, Dye BT, Patton JG, Singer RH, Zipori D, Dynamic sorting of nuclear components into distinct nucleolar caps during transcriptional inhibition. Mol Biol Cell 16, 2395–413 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Daskal Y, Smetana K, Busch H, Evidence from studies on segregated nucleoli that nucleolar silver staining proteins C23 and B23 are in the fibrillar component. Exp Cell Res 127, 285–91 (1980). [DOI] [PubMed] [Google Scholar]
- 27.Wild T, Horvath P, Wyler E, Widmann B, Badertscher L, Zemp I, Kozak K, Csucs G, Lund E, Kutay U, A protein inventory of human ribosome biogenesis reveals an essential function of exportin 5 in 60S subunit export. PLoS Biol 8, e1000522 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ugrinova I, Monier K, Ivaldi C, Thiry M, Storck S, Mongelard F, Bouvet P, Inactivation of nucleolin leads to nucleolar disruption, cell cycle arrest and defects in centrosome duplication. BMC Mol Biol 8, 66 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Popov A, Smirnov E, Kováčik L, Raška O, Hagen G, Stixová L, Raška I, Duration of the first steps of the human rRNA processing. Nucleus 4, 134–141 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Houseley J, Tollervey D, The many pathways of RNA degradation. Cell 136, 763–76 (2009). [DOI] [PubMed] [Google Scholar]
- 31.Stefanovsky V, Moss T, Regulation of rRNA synthesis in human and mouse cells is not determined by changes in active gene count. Cell Cycle 5, 735–9 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Sogo JM, Ness PJ, Widmer RM, Parish RW, Roller T, Psoralen-crosslinking of DNA as a probe for the structure of active nucleolar chromatin. J Mol Biol 178, 897–919 (1984). [DOI] [PubMed] [Google Scholar]
- 33.Burger K, Muhl B, Harasim T, Rohrmoser M, Malamoussi A, Orban M, Kellner M, Gruber-Eber A, Kremmer E, Holzel M, Eick D, Chemotherapeutic drugs inhibit ribosome biogenesis at various levels. J Biol Chem 285, 12416–25 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schoefl GI, The Effect of Actinomycin D on the Fine Structure of the Nucleolus. J Ultrastruct Res 10, 224–43 (1964). [DOI] [PubMed] [Google Scholar]
- 35.Perrotti D, Neviani P, From mRNA metabolism to cancer therapy: chronic myelogenous leukemia shows the way. Clin Cancer Res 13, 1638–42 (2007). [DOI] [PubMed] [Google Scholar]
- 36.House RP, Talwar S, Hazard ES, Hill EG, Palanisamy V, RNA-binding protein CELF1 promotes tumor growth and alters gene expression in oral squamous cell carcinoma. Oncotarget 6, 43620–34 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spector DL, Fu XD, Maniatis T, Associations between distinct pre-mRNA splicing components and the cell nucleus. EMBO J 10, 3467–81 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spector DL, Higher order nuclear organization: three-dimensional distribution of small nuclear ribonucleoprotein particles. Proc Natl Acad Sci U S A 87, 147–51 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fu XD, Maniatis T, Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature 343, 437–41. (1990). [DOI] [PubMed] [Google Scholar]
- 40.Sasikumar AN, Perez WB, Kinzy TG, The many roles of the eukaryotic elongation factor 1 complex. Wiley Interdiscip Rev RNA 3, 543–55 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abbas W, Kumar A, Herbein G, The eEF1A Proteins: At the Crossroads of Oncogenesis, Apoptosis, and Viral Infections. Front Oncol 5, 75 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu G, Grant WM, Persky D, Latham VM Jr., Singer RH, Condeelis J, Interactions of elongation factor 1alpha with F-actin and beta-actin mRNA: implications for anchoring mRNA in cell protrusions. Mol Biol Cell 13, 579–92 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khacho M, Mekhail K, Pilon-Larose K, Pause A, Cote J, Lee S, eEF1A is a novel component of the mammalian nuclear protein export machinery. Mol Biol Cell 19, 5296–308 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Doyle A, Crosby SR, Burton DR, Lilley F, Murphy MF, Actin bundling and polymerisation properties of eukaryotic elongation factor 1 alpha (eEF1A), histone H2A-H2B and lysozyme in vitro. J Struct Biol 176, 370–8 (2011). [DOI] [PubMed] [Google Scholar]
- 45.Yachida S, White CM, Naito Y, Zhong Y, Brosnan JA, Macgregor-Das AM, Morgan RA, Saunders T, Laheru DA, Herman JM, Hruban RH, Klein AP, Jones S, Velculescu V, Wolfgang CL, Iacobuzio-Donahue CA, Clinical significance of the genetic landscape of pancreatic cancer and implications for identification of potential long-term survivors. Clin Cancer Res 18, 6339–47 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Waclaw B, Bozic I, Pittman ME, Hruban RH, Vogelstein B, Nowak MA, A spatial model predicts that dispersal and cell turnover limit intratumour heterogeneity. Nature 525, 261–4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.James A, Wang Y, Raje H, Rosby R, Dimario P, Nucleolar stress with and without p53. Nucleus 5, 402–26 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Holmberg Olausson K, Nister M, Lindstrom MS, p53 -Dependent and -Independent Nucleolar Stress Responses. Cells 1, 774–98 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boulon S, Westman BJ, Hutten S, Boisvert FM, Lamond AI, The nucleolus under stress. Mol Cell 40, 216–27 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pianese G, Beitrag zur histologie und aetiologie der carcinoma. Histologische und experimentelle untersuchungen. Beitr Pathol Anat Allgem Pathol 142, 1:193 (1896). [Google Scholar]
- 51.Hannan RD, Drygin D, Pearson RB, Targeting RNA polymerase I transcription and the nucleolus for cancer therapy. Expert Opin Ther Targets 17, 873–8 (2013). [DOI] [PubMed] [Google Scholar]
- 52.Drygin D, O'brien SE, Hannan RD, Mcarthur GA, Von Hoff DD, Targeting the nucleolus for cancer-specific activation of p53. Drug Discov Today 19, 259–65 (2014). [DOI] [PubMed] [Google Scholar]
- 53.Drygin D, Lin A, Bliesath J, Ho CB, O'brien SE, Proffitt C, Omori M, Haddach M, Schwaebe MK, Siddiqui-Jain A, Streiner N, Quin JE, Sanij E, Bywater MJ, Hannan RD, Ryckman D, Anderes K, Rice WG, Targeting RNA polymerase I with an oral small molecule CX-5461 inhibits ribosomal RNA synthesis and solid tumor growth. Cancer Res 71, 1418–30 (2011). [DOI] [PubMed] [Google Scholar]
- 54.Drygin D, Rice WG, Grummt I, The RNA polymerase I transcription machinery: an emerging target for the treatment of cancer. Annu Rev Pharmacol Toxicol 50, 131–56 (2010). [DOI] [PubMed] [Google Scholar]
- 55.Colis L, Peltonen K, Sirajuddin P, Liu H, Sanders S, Ernst G, Barrow JC, Laiho M, DNA intercalator BMH-21 inhibits RNA polymerase I independent of DNA damage response. Oncotarget 5, 4361–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bywater MJ, Poortinga G, Sanij E, Hein N, Peck A, Cullinane C, Wall M, Cluse L, Drygin D, Anderes K, Huser N, Proffitt C, Bliesath J, Haddach M, Schwaebe MK, Ryckman DM, Rice WG, Schmitt C, Lowe SW, Johnstone RW, Pearson RB, Mcarthur GA, Hannan RD, Inhibition of RNA polymerase I as a therapeutic strategy to promote cancer-specific activation of p53. Cancer Cell 22, 51–65 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pederson T, The plurifunctional nucleolus. Nucleic Acids Research 26, 3871–6 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boisvert FM, Van Koningsbruggen S, Navascues J, Lamond AI, The multifunctional nucleolus. Nat Rev Mol Cell Biol 8, 574–85 (2007). [DOI] [PubMed] [Google Scholar]
- 59.Zhou X, Liao WJ, Liao JM, Liao P, Lu H, Ribosomal proteins: functions beyond the ribosome. J Mol Cell Biol 7, 92–104 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Peltonen K, Colis L, Liu H, Trivedi R, Moubarek MS, Moore HM, Bai B, Rudek MA, Bieberich CJ, Laiho M, A targeting modality for destruction of RNA polymerase I that possesses anticancer activity. Cancer Cell 25, 77–90 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peltonen K, Colis L, Liu H, Jaamaa S, Zhang Z, Af Hallstrom T, Moore HM, Sirajuddin P, Laiho M, Small molecule BMH-compounds that inhibit RNA polymerase I and cause nucleolar stress. Mol Cancer Ther 13, 2537–46 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shamovsky I, Ivannikov M, Kandel ES, Gershon D, Nudler E, RNA-mediated response to heat shock in mammalian cells. Nature 440, 556–60 (2006). [DOI] [PubMed] [Google Scholar]
- 63.Huang HY, Hopper AK, In vivo biochemical analyses reveal distinct roles of beta-importins and eEF1A in tRNA subcellular traffic. Genes Dev 29, 772–83 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gross SR, Kinzy TG, Translation elongation factor 1A is essential for regulation of the actin cytoskeleton and cell morphology. Nat Struct Mol Biol 12, 772–8 (2005). [DOI] [PubMed] [Google Scholar]
- 65.Chuang SM, Chen L, Lambertson D, Anand M, Kinzy TG, Madura K, Proteasome-mediated degradation of cotranslationally damaged proteins involves translation elongation factor 1A. Mol Cell Biol 25, 403–13 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mateyak MK, Kinzy TG, eEF1A: thinking outside the ribosome. J Biol Chem 285, 21209–13 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tomlinson VA, Newbery HJ, Bergmann JH, Boyd J, Scott D, Wray NR, Sellar GC, Gabra H, Graham A, Williams AR, Abbott CM, Expression of eEF1A2 is associated with clear cell histology in ovarian carcinomas: overexpression of the gene is not dependent on modifications at the EEF1A2 locus. Br J Cancer 96, 1613–20 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tomlinson VA, Newbery HJ, Wray NR, Jackson J, Larionov A, Miller WR, Dixon JM, Abbott CM, Translation elongation factor eEF1A2 is a potential oncoprotein that is overexpressed in two-thirds of breast tumours. BMC Cancer 5, 113 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Scaggiante B, Dapas B, Bonin S, Grassi M, Zennaro C, Farra R, Cristiano L, Siracusano S, Zanconati F, Giansante C, Grassi G, Dissecting the expression of EEF1A1/2 genes in human prostate cancer cells: the potential of EEF1A2 as a hallmark for prostate transformation and progression. Br J Cancer 106, 166–73 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kawamura M, Endo C, Sakurada A, Hoshi F, Notsuda H, Kondo T, The prognostic significance of eukaryotic elongation factor 1 alpha-2 in non-small cell lung cancer. Anticancer Res 34, 651–8 (2014). [PubMed] [Google Scholar]
- 71.Van Goietsenoven G, Hutton J, Becker JP, Lallemand B, Robert F, Lefranc F, Pirker C, Vandenbussche G, Van Antwerpen P, Evidente A, Berger W, Prevost M, Pelletier J, Kiss R, Kinzy TG, Kornienko A, Mathieu V, Targeting of eEF1A with Amaryllidaceae isocarbostyrils as a strategy to combat melanomas. FASEB J 24, 4575–84 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dang Y, Schneider-Poetsch T, Eyler DE, Jewett JC, Bhat S, Rawal VH, Green R, Liu JO, Inhibition of eukaryotic translation elongation by the antitumor natural product Mycalamide B. RNA 17, 1578–88 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ahuja D, Vera MD, Sirdeshpande BV, Morimoto H, Williams PG, Joullie MM, Toogood PL, Inhibition of protein synthesis by didemnin B: how EF-1alpha mediates inhibition of translocation. Biochemistry 39, 4339–46 (2000). [DOI] [PubMed] [Google Scholar]
- 74.Roth HJ, Eger K, [Synthesis of 2-amino-3-cyano-pyrroles (author's transl)]. Arch Pharm (Weinheim) 308, 179–85 (1975). [DOI] [PubMed] [Google Scholar]
- 75.Mohamed MS, Rashad AE, Zaki ME, Fatahala SS, Synthesis and antimicrobial screening of some fused heterocyclic pyrroles. Acta Pharm 55, 237–49 (2005). [PubMed] [Google Scholar]
- 76.Kittur N, Zapantis G, Aubuchon M, Santoro N, Bazett-Jones DP, Meier UT, The nucleolar channel system of human endometrium is related to endoplasmic reticulum and R-rings. Mol Biol Cell 18, 2296–304 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lee TI, Johnstone SE, Young RA, Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc 1, 729–48 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Blanch A, Robinson F, Watson IR, Cheng LS, Irwin MS, Eukaryotic translation elongation factor 1-alpha 1 inhibits p53 and p73 dependent apoptosis and chemotherapy sensitivity. PLoS One 8, e66436 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Arastu-Kapur S, Ponder EL, Fonovic UP, Yeoh S, Yuan F, Fonovic M, Grainger M, Phillips CI, Powers JC, Bogyo M, Identification of proteases that regulate erythrocyte rupture by the malaria parasite Plasmodium falciparum. Nature Chemical Biology 4, 203–213 (2008). [DOI] [PubMed] [Google Scholar]
- 80.Shevchenko A, Wilm M, Vorm O, Mann M, Mass spectrometric sequencing of proteins from silver stained polyacrylamide gels. Analytical Chemistry 68, 850–858 (1996). [DOI] [PubMed] [Google Scholar]
- 81.Ye XY, Johann DJ, Hakami RM, Xiao Z, Meng ZJ, Ulrich RG, Issaq HJ, Veenstra TD, Blonder J, Optimization of protein solubilization for the analysis of the CD14 human monocyte membrane proteome using LC-MS/MS. Journal of Proteomics 73, 112–122 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Molina DM, Jafari R, Ignatushchenko M, Seki T, Larsson EA, Dan C, Sreekumar L, Cao Y, Nordlund P, Monitoring Drug Target Engagement in Cells and Tissues Using the Cellular Thermal Shift Assay. Science 341, 84 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Synthetic scheme to metarrestin
Growth inhibition of PANC1 by metarrestin
Metastatic cancer progression in NOD/IL2Rgnull (NSG) PANC1 mice
Single and multiple dose metarrestin plasma and tumor pharmacokinetics
Metarrestin treatment induced nucleolar structure changes as observed by electron microscopy.
Nucleolar structure changes (UBF labeling in red, blue arrows) were closely associated with the loss of PNC (PTB labeling in green, orange arrows) in metarrestin-treated cells.
Pol I transcription factors, UBF, RPA194, and pre-ribosomal RNA processing factor, Nopp140 and fibrillarin showed similar capping structure in metarrestin-treated cells.
Metarrestin treatment did not significantly impact Pol II transcription, translation, or nucleocytoplasmic trafficking.
Metarrestin treatment did not induce DNA damage response, cell cycle blockage, or global inhibition of pol II transcription.
Biotin-metarrestin was similarly effective in disassembling PNCs in treated PC3M cells at 1 μM for 18 hours.
Transfection of eEF1A2-HA showed the expression of the protein in red in transfected cells.
Synthesis of 2-amino-1-benzyl-4,5-diphenyl-1H-pyrrole-3-carbonitrile.
Synthesis of (E)-ethyl N-(1-benzyl-3-cyano-4,5-diphenyl-1H-pyrrol-2-yl)formimidate.
Synthesis of trans-4-(7-benzyl-4-imino-5,6-diphenyl-4,7-di hydro-3 H-pyrrolo[2,3-d]pyrimidin-3-yl)cyclohexanol (Metarrestin).
Synthesis of 2-amino-1-(3-bromobenzyl)-4,5-diphenyl-1H-pynole-3-carbonitrile.
Synthesis of (E)-ethyl N-(1-(3-bromobenzyl)-3-cyano-4,5-diphenyl-1H-pyrrol-2-yl)formimidate.
Synthesis of trans-4-(7-(3-bromobenzyl)-4-imino-5,6-diphenyl-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-3-yl)cyclohexanol.
Synthesis of N-(6-(3-((3-(trans-4-hydroxycyclohexyl)-4-imino-5,6-diphenyl-3,4-dihydro-7H-pyrrolo[2,3-d]pyrimidin-7-yl)methyl)phenyl)hex-5-yn-1-yl)-5-((3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl)pentanamide (biotin-metarrestin, P1).
Cancer cell lines examined for PNC prevalence with or without treatment of metarrestin (from Fig. 1C).
Pancreatic cancer cell lines evaluated for PNC prevalence in Fig. 2A.
Veterinary pathology read of organs derived from tumor-bearing KPC and wild type mice treated by metarrestin for 3 months.
Blood biochemistry and hematology toxicity of metarrestin in tumor bearing KPC and wild type mice.