Abstract
Purpose
To assess the impact of varying magnetic resonance imaging (MRI) scanner parameters on the extraction of algorithmic features in breast MRI radiomics studies.
Methods
In this retrospective study, breast imaging data for 272 patients were analyzed with magnetic resonance (MR) images. From the MR images, we assembled and implemented 529 algorithmic features of breast tumors and fibrograndular tissue (FGT). We divided the features into 10 groups based on the type of data used for the feature extraction and the nature of the extracted information. Three scanner parameters were considered: scanner manufacturer, scanner magnetic field strength, and slice thickness. We assessed the impact of each of the scanner parameters on each of the feature by testing whether the feature values are systematically diverse for different values of these scanner parameters. A two-sample t-test has been used to establish whether the impact of a scanner parameter on values of a feature is significant and receiver operating characteristics have been used for to establish the extent of that effect.
Results
On average, higher proportion (69% FGT versus 20% tumor) of FGT related features were affected by the three scanner parameters. Of all feature groups and scanner parameters, the feature group related to the variation in FGT enhancement was found to be the most sensitive to the scanner manufacturer (AUC = 0.81 ± 0.14).
Conclusions
Features involving calculations from FGT are particularly sensitive to the scanner parameters.
Keywords: breast cancer MRI radiomics, computer-extracted features MRI, scanner manufacturer, magnetic field strength, slice thickness, ROC analysis
Introduction
Breast MRI is an imaging modality used in complement to mammography and ultrasound for cancer diagnosis, staging, and gaining additional knowledge about the tumor biology (Brasic, Wisner, & Joe, 2013; Knuttel, Menezes, van den Bosch, Gilhuijs, & Peters, 2014; Sardanelli et al., 2004; Saslow et al., 2007). MR imaging of breasts also finds usage in screening in high risk patients (C. K. Kuhl et al., 2005; Warner et al., 2001). More recently, two research directions, radiomics (Lambin et al., 2012) and radiogenomics (Mazurowski, 2015), focus on extracting a variety of algorithmic imaging features to assess the abnormalities in breast for improved diagnosis (Yang, Li, Zhang, Shao, & Zheng, 2015; Zheng et al., 2009), prediction of outcomes (Cho et al., 2014; Mahrooghy et al., 2015; Mazurowski et al., 2015), and correlation with tumor genomics (Ashraf et al., 2014; Grimm, Zhang, & Mazurowski, 2015; Mazurowski, Zhang, Grimm, Yoon, & Silber, 2014; Wan et al., 2016; Wang et al., 2015).
Breast MRI radiomics and radiogenomics features use dynamic contrast enhanced MR imaging sequences as the primary input for feature extraction. Acquisition of an MRI is a complex procedure and has several factors that affect the image’s appearance, quality and interpretation (Hendrick, 2007; C. K. Kuhl & Schild, 2000; Mann, Kuhl, Kinkel, & Boetes, 2008; Rausch & Hendrick, 2006; Sawyer-Glover & Shellock, 2000). Moreover, breast MR image acquisition protocols vary significantly across different studies, and across and within institutions. As these factors affect the image, they are likely to also affect the features extracted from the images for radiomics and radiogenomics studies. Currently, the imaging features prevalent in the literature are extracted by different investigators from different datasets. This could affect generalizability of the presented results. Therefore, we believe that it is of high importance to systematically evaluate the effect of parameter scanners on extraction of features for radiomics and radiogenomics studies.
Assessing variability of radiomics features for imaging equipment or its different settings is an important topic and has been studied for non small cell lung cancers in different studies (Lu, Ehmke, Schwartz, & Zhao, 2016; Mackin et al., 2015; Zhao et al., 2016) and in breast MRI based lesion classification (Chen et al., 2010). Repeated CT scans of the same patient in the same day were used to assess reproducibility of a set of feature in (Lu et al., 2016; Zhao et al., 2016). The study in (Mackin et al., 2015) focused on the comparison of imaging features from patients and phantoms computed using 17 different scans of the phantom (used as ground truth). While in the first two studies, the datasets are dependent, the third study enables study of independent datasets by using the phantom. While repeated scans subject patients to discomfort, employing a phantom involves increase in cost developing the phantom. The study in (Chen et al., 2010) used on two independent datasets (generated using images from two different scanner manufacturers) to analyze the effect of training a model on with images from a single manufacturer and testing it on the dataset consisting images from a different manufacturer.
In this retrospective study, in the absence of repeated scans and phantoms, we assume there is no significant difference in the independent patient subgroups corresponding to different values of a specific scanner parameter, and these subgroups belong to the same patient population. Hereafter, to conduct this study, we extracted a set of radiomic and radiogenomic imaging features that have been commonly used in literature, and that have exhibited significance in various aspects of breast cancer. We extended this set by more features defined in our laboratory to arrive at a comprehensive description of the tumor and its surrounding. These features were grouped according to their origin (for example, tumor or tissue) and the type of information they conveyed. Then, we investigated how our comprehensive set of breast MRI features and feature groups are affected by different scanner parameters.
Methods
Patient Population
In this institutional review board approved study, we used data for 272 patients at our institution. To arrive at this set, we collected a set of pre-operative MR images from 400 consecutive female breast cancer patients at our institution from September 2007 to June 2009. We excluded 109 patients for the following clinical reasons: (a) for having breast cancer or elective breast surgery (n=38) prior to MRI, (b) for undergoing breast cancer treatment at the time of the MRI (n=29), and (c) for missing pathology data (n=42). The remaining 291 patients were annotated by radiologists. After the annotation, 19 more cases were excluded for the following imaging related reasons: (a) missing MRI sequences required for feature computation (n = 3), (b) discordant numbers of slices in pre and post-contrast sequences (n=1), (c) the features of the tumor could not be confidently described by a reader (n=11), (d) biopsied-lesion was not marked by reader (n=1), and (e) unsuccessful algorithmic extraction of the breast mask (n =3). Thus, 272 patients were used in the analysis. The clinicopathological characteristics for our patient cohort are provided in Table 1.
Table 1.
Clinicopathological characteristics of patients in our cohort
| Characteristics | Pertinent Details | Values for 272 patients |
|---|---|---|
| Age (years) | Median | 53 |
| Range | 22–80 | |
| Race | White | 191 |
| Black | 63 | |
| Asian | 5 | |
| Native | 3 | |
| Hispanic | 2 | |
| Multi-racial | 1 | |
| Hawaiian | 1 | |
| Not Reported | 6 | |
| Menopausal Status | Pre | 111 |
| Post | 159 | |
| Not Reported | 2 | |
| Tumor Grade | Low | 50 |
| Intermediate | 140 | |
| High | 81 | |
| Not Reported | 1 |
Selection of Scanner Parameters
Acquisition of MRI uses several parameters for the optimal acquisition of the image (Huang et al., 2004; C. Kuhl, 2007). Though there are various scanner parameters that can influence the final image, the study in (Kumar et al., 2012) reports dose of contrast agent, method of dose administration, used pulse sequence, magnetic field strength and method of analysis as the principal factors that affect dynamic contrast enhanced MRI. As discussed in the next section, contrast agent dose, method of dose administration and the pulse sequence have not varied for patients included in the study.
In our study, we chose the following three scanner parameters: scanner manufacturer, magnetic field strength, and slice thickness. We chose scanner manufacturer as the first parameter as it captures the intrinsic differences in the acquisition devices and used protocols (Hendrick, 2007; Ikeda et al., 2001). The second parameter is magnetic field strength as it is shown to affect signal-to-noise ratio and spatial resolution (Bernstein, Huston, & Ward, 2006; Haacke, Brown, Thompson, & Venkatesan, 1999; Hendrick, 2007; C. K. Kuhl et al., 2006) in any MRI acquisition. The third selected parameter is the slice thickness, as it is typically the largest dimension of the voxel and captures the poorest spatial resolution (Heywang-Köbrunner, Viehweg, Heinig, & Küchler, 1997). In terms of the scanner manufacturer, our cohort has two categories for magnetic field strength. The slice thickness has four different values and therefore, we bifurcated the set into two categories (high and low), based on the median value of the slice thickness for our cohort. We found that we have sufficient variability for these three parameters in our cohort. The distributions of patients for different scanner parameters are shown in Table 2. The MRI scanning protocol is presented in Table 3.
Table 2.
Distribution of patients for different values of three scanner parameters
| Scanner Parameter |
Technical Details | Other Pertinent Details | Patient Count |
|---|---|---|---|
| Manufacturer | GE Healthcare, Little Chalfont, UK | Model: Signa HDx | 210 |
| Model: Signa HDxt | 5 | ||
| Siemens, Munich, Germany | Model: MAGNETOM Avanto | 33 | |
| Model: MAGNETOM TrioTim | 24 | ||
| Magnetic Field Strength | 1.5T | Signa HDxt, GE Healthcare, Little Chalfont, UK | 5 |
| Signa HDx, GE Healthcare, Little Chalfont, UK | 166 | ||
| Avanto, Siemens, Munich, Germany | 33 | ||
| 3.0T | Signa HDx, GE Healthcare, Little Chalfont, UK | 44 | |
| MAGNETOM Trio Tim, Siemens, Munich, Germany | 24 | ||
| Slice Thickness (mm) | 1.1 | - | 70 |
| 1.2 | - | 21 | |
| 1.3 | - | 1 | |
| 2 | - | 180 |
Table 3.
Typical scanning protocols across different scanner models
| Scanner Model |
Magnetic Field Strength (T) |
Field-of-View FOV(cm) |
Acquisition Matrix |
Echo Time TE (ms) |
Repetition Time TR(ms) |
|---|---|---|---|---|---|
| Signa HDxt/ Signa HDx | 1.5 | 34 | 340 × 340 | 2.3 | 5 |
| Avanto | 1.5 | 36 | 448 × 448 | 1.5 | 4.7 |
| Signa HDx | 3 | 35 | 350 × 350 | 2.4 | 5.5 |
| MAGNETOM Trio Tim | 3 | 34 | 448 × 448 | 1.4 | 4.1 |
Imaging Data and Image Annotation
All of our patients had axial MRI acquired by the scanner in prone position. The following MRI sequences were available: a non-fat saturated T1-weighted sequence, a fat-saturated gradient echo T1-weighted pre-contrast sequence, typically four post-contrast T1-weighted sequences acquired after the intravenous administration of contrast agent using a weight based protocol of 0.2 mL/kg. The contrast agents used were: gadopentetate dimeglumine (Magnevist, Bayer Health Care, Berlin, Germany in 93.75% of our cohort), gadobenate dimeglumine (MultiHance, Bracco, Milan, Italy in 2.94 % of our cohort). This data was not available for 3.31% of the patients. The pulse sequence type of ‘gradient recalled (GR)’ was used for all patients.
The images were annotated by six fellowship-trained breast imagers (6–22 years of post-fellowship experience) using an internally developed graphical user interface. In each imaging study, up to 5 mass or/and non-mass-enhancements were outlined by drawing a rectangle in a slice and then indicating the first and the last slices that had the lesion. For each patient, the annotated mass/ non-mass corresponding to the largest biopsied tumor was used to extract the features.
Image Segmentation
The tumor segmentation was accomplished using a fuzzy C-means based automatic segmentation algorithm (Bezdek, 1981) and the tumor mask was obtained. Two fibrograndular tissue masks were extracted based on two different MRI sequences. For the first mask, the N4-corrected (Tustison et al., 2010) T1-non-fat saturated (T1-NFS) image was used to : (i) segment the breast to obtain the breast mask (Mazurowski et al., 2015), (ii) segment the fibrograndular tissue using the breast mask, (iii) register the FGT mask to the DCE-MRI. For the second mask, first T1- fat saturated post contrast (PostCon) sequence was used to: (i) register the breast mask extracted from T1-NFS to PostCon, and (ii) segment the FGT. From both of these masks, any overlap with the tumor mask was removed. A set of four masks was extracted for each study: (1) breast mask (registered) (2) tumor mask, (3) FGT from T1-NFS, and (4) FGT from PostCon.
Selection and Organization of Imaging Features
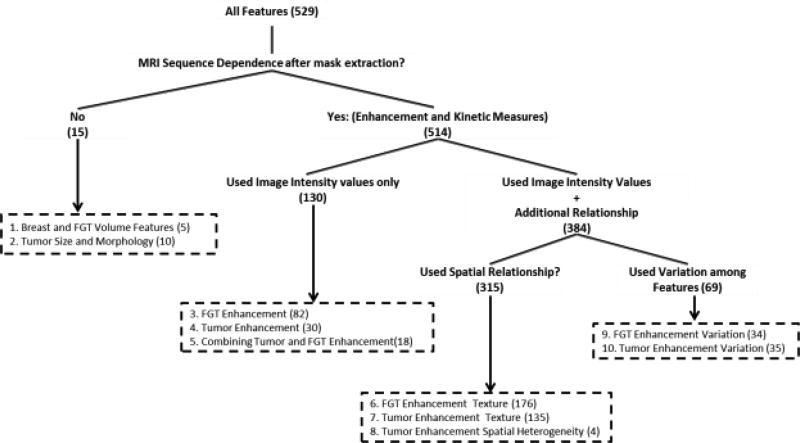
To define a set of imaging features in DCE-MRI, we first reviewed the literature on the topic. We selected features that previously described the breast, tumor and fibrograndular tissue properties and were shown to have significance in terms of their prognostic abilities of outcomes or predictive ability for different genomic factors. Our categorization of these features can be found in Fig.1.
Figure 1.
The grouping of the features used in the analysis
We have 10 categories of features that we believe to be depicting breast cancer MRI substantially with imaging features. At first, we divided the features into two sections that (a) use tumor/FGT masks only and (b) image intensity values along with the tumor/FGT masks. Using the breast and FGT masks only, breast and FGT volumetric features comprising of breast volume, FGT volume and FGT density features (Faermann, Sperber, Schneebaum, & Barsuk, 2014; Klifa et al., 2010) can be computed. Using the tumor mask only, general features of tumor shape and morphology (Czarnek, Clark, Peters, & Mazurowski, 2017; Georgiou, Mavroforakis, Dimitropoulos, Cavouras, & Theodoridis, 2007; Giger, Vyborny, & Schmidt, 1994; Sutton et al., 2015) can be calculated.
Features which use both masks and intensity values are called enhancement and kinetic features as they use intensity values from a subset of post-contrast sequences and pre-contrast sequence (if necessary). These features were further divided into 8 groups: tumor enhancement, FGT enhancement and combining tumor enhancement, tumor enhancement texture, FGT enhancement texture, tumor enhancement spatial heterogeneity, FGT enhancement variation and tumor enhancement variation.
Features of tumor enhancement include mean and volumetric features computed after grouping tumor voxels (Ashraf et al., 2014), and forming signal enhancement ratio (SER) and peak enhancement (PE) maps (Arasu et al., 2011). Similar features from FGT masks were also computed along with features related to background parenchymal enhancement (Wu et al., 2015) as FGT enhancement features. Features of combined tumor and FGT enhancements (Grimm et al., 2015; Mazurowski et al., 2015; Mazurowski et al., 2014) were also included. Different measures of dispersion were computed from grouped tumor/FGT voxels, and SER and PE maps of tumor/FGT to obtain features of tumor/FGT enhancement variation. In addition to these, different properties of the tumor uptake curves (Chen, Giger, Bick, & Newstead, 2006; Chen, Giger, Lan, & Bick, 2004; Gilhuijs, Giger, & Bick, 1998) were also included as tumor enhancement variation features. The 5 enhancement related feature groups, mentioned in the paragraph, do not use the spatial relationships of the voxels included in the tumor/FGT masks.
The feature groups that use spatial relationships of voxels are texture features from tumor and FGT, and features from spatial heterogeneity of tumor. Features of enhancement texture include commonly used texture features (Haralick, Shanmugam, & Dinstein, 1973) from the first post contrast (Bhooshan et al., 2010), PE, SER and washin rate maps. Texture features computed from Discrete Fourier Transform (DFT) (Zheng et al., 2009), Dynamic Local Binary Patterns (DLBP) and Dynamic Histogram of Oriented Gradients (DHOG) (Wan et al., 2016), and spatial variations of tumor enhancement were also included in our feature set. Heterogeneity of spatial enhancement was computed using global Morans’I (Song, Smith, Huang, Jeraj, & Fain, 2009) from different enhancement maps. We are including two new features of spatial heterogeneity: EnhancementClusterNeighborhoodSimilarity_Tumor and EnhancementClusterDiscontinuity_Tumor. To see the list of all features and the definitions of the new features, please refer to the “Supplemental Material A”.
Kinetic curves of breast lesions were affected by MRI scanner protocols as shown in (Jansen et al., 2009). In a previous study using MR scans of a phantom (Collewet, Strzelecki, & Mariette, 2004), different MRI scanner protocols were shown to affect texture features. Therefore, some investigators have used intensity normalization prior to the extraction of texture features by excluding tumor voxels having intensity values beyond outside a predefined range (Karahaliou et al., 2010; Waugh et al., 2016). As our study aims to investigate the effect of scanner parameters on the feature groups discussed, we have not conducted any intensity normalization prior to the extraction of features.
Analysis of the Effect of Scanner Parameter Variation
After the feature extraction, we conducted our analyses that aim at determining whether different scanner parameters affect values of the features. In the absence of repeated scans (at the same time or with a short time gap) of a patient, we analyzed whether the feature values differed for patients given one value of a parameter (e.g. a GE scanner) than for another (e.g. a Siemens scanner). This is based on the assumption that the patients scanned by different scanners come from the same distributions of patients (e.g., that there is nothing inherently different about the population of patients scanned by GE scanners and Siemens scanners). The natural variation between patients is accounted for in our analysis. Please note that this methodology is similar to that used in drug trials where two different conditions (e.g. a drug and a placebo) are applied to two samples of patients to determine whether their eventual outcomes (e.g. survival of cancer patients) will differ under the two conditions. Please see our supplementary materials on analysis of the inherent characteristics of the samples dichotomized by the scanner parameters. The rationale and details of our analyses methods are provided below.
Two sample t-test
For each of the scanner parameters and each feature, we conducted a two-sample t-test to assess, if the feature’s values are significantly diverse for different scanner parameters. For a particular scanner parameter, we bifurcate the values of the feature under consideration in two different sets. Next, we evaluate with t-test, if these sets are from populations with equal means in terms of the feature value. It is desired that the two sample sets for different scanner parameters should belong to populations that have same means.
Receiver operating characteristics (ROC)
ROC analysis is popularly conducted for the evaluation of medical diagnostic systems (Akobeng, 2007). While that is a typical use of ROC and the area under the ROC (AUC), it can also be used to assess a correlation of a binary variable with a continuous one or similarly to assess an impact of a binary variable (such as a scanner manufacturer) on a continuous variable (such as an MRI feature). The latter is the way that we use the ROC analysis. In this interpretation, an AUC value of 0.5 signifies that there is no relationship between a particular scanner parameter and a particular feature and AUC values notably above 0.5 indicate that the distribution of the feature values is different for one value of a scanner parameter (e.g. a scanner manufacturer) than for another value of that scanner parameter implying an effect of the scanner parameter on feature values. Please note that in our study we are not evaluating how predictive the MRI features are of diagnosis or prognosis of a patient.
Specifically, we calculated the area under the receiver operating characteristic (Bradley, 1997) curves for every feature and scanner parameter to assess the ability of the feature to predict the value of the scanner parameter. A high value of this association for a feature is undesirable in our analysis as we need the feature values to be independent of the scanner parameter value.
Pearson’s linear correlation coefficient
In an additional analysis, to assess how the correlation of feature groups are affected by different values of the scanner parameters, we computed the Pearson’s linear correlation coefficient between each pair of features keeping the value of one of the scanner parameters constant at a time. Since we have six values from all of the scanner parameters, we have six sets of correlations. From each set, the within-group correlation was computed as the average correlation of all features belonging to the same group and the between-group correlation was the average correlation from all of the features belonging to the corresponding pair of groups. It is desirable that these correlations remain similar when computed for different values of scanner parameters.
Results
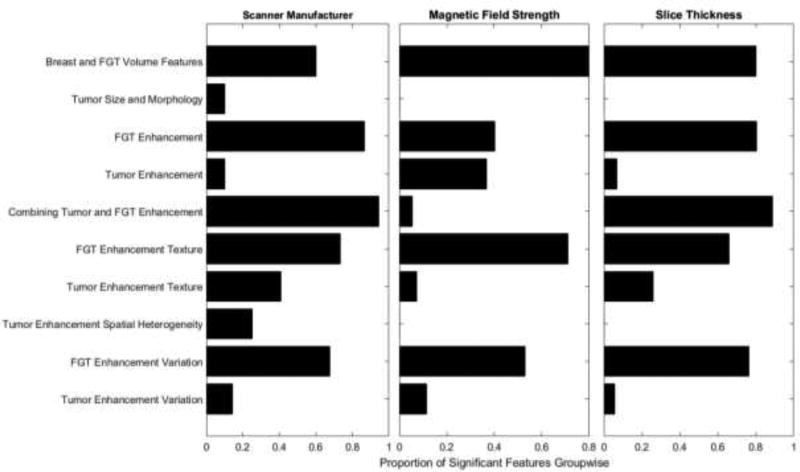
The effect of different scanner parameters on each of the feature groups is shown in Fig. 2. Specifically, this figure shows the proportion of the features in each group that proved to be significantly associated (at the level of significance of p<0.05, please see Supplementary Material B for the p-values obtained from the t-tests for individual features) with scanner parameters. We found that high proportions of features are affected by the scanner manufacturer (58.22%) and slice thickness (50.47%). A lower proportion (38.94%) of the features are affected by the magnetic field strength. In our study, 30.62% of the features are affected by all of the three scanner parameters. Only 3.7% of these features are tumor-related. The proportion of FGT related features is 56.14% in this study and they are more affected by the scanner parameters.
Figure 2.
Effect of different scanner parameters in feature groups in terms of proportion of significant features
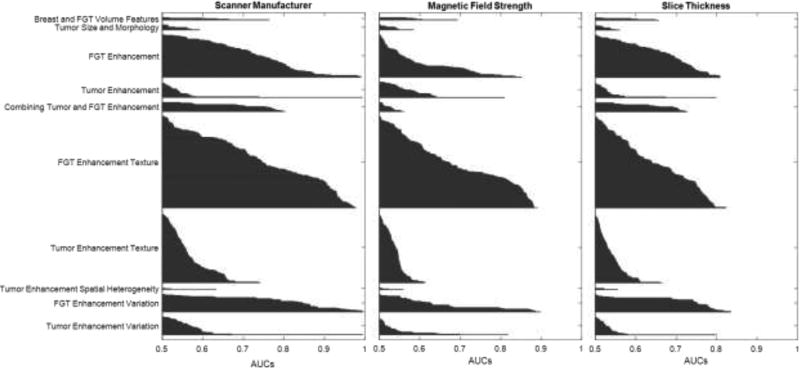
In Fig. 3, we presented the Area Under Curve (AUC) of every feature and arranged them group-wise. The AUC values are sorted in ascending order from top to bottom. This figure helps us to observe the group-wise traits of features. For every group, the highest predictive value is obtained for scanner manufacturer. For magnetic field strengths and slice thickness, the predictive ability of the features decreases. Thus, the effect of scanner manufacturer is more prevalent in the features compared to the effect of magnetic field strength and slice thickness.
Figure 3.
Area under Curves for each group in terms of three scanner parameters
For each of the 529 features and 3 scanner parameters, the AUC for each of the features alongside with the value of the test for association between a scanner parameter and the feature values are included in the “Supplemental Material B”.
Figure 4 shows the correlation of features within and between-groups. To discuss the correlation values shown in this figure, we are considering correlation values greater than 0.6 to be high and those below 0.6 to be low. For the scanner manufacturer, the change in correlation for features belonging to the same group (within group correlation) can be observed by comparing the anti-diagonal elements of figures 4(a) and 4(b). The maximum change of 45% in within group correlation can be observed for the group FGT enhancement variation. However, the correlation value remains low. The correlation changes from low to high for the feature groups Combining Tumor and FGT Features and Tumor Enhancement Spatial Heterogeneity. The correlation values between groups doesn’t change from low to high or vice versa. For change in magnetic field strength values (figures 4(c) and 4(d)), none of the within or between group correlation values change from low to high or vice-versa. For change in slice thickness (figures 4(e) and 4(f)), within group correlation values for features related to groups Tumor Size and Morphology and Combining Tumor and FGT Features change from low to high. For all the scanner parameters, the only a few within group correlations are high (>0.6). Also in 92% of the comparisons, features belonging to one group are more correlated with each other than with features from other groups.
Figure 4.
Average Correlation within and between Feature Groups for (a) GE scanner (b) Siemens Scanner (c) 1.5 T magnetic field strength (d) 3T magnetic field strength (e) high slice thickness (f) low slice thickness
Discussion
We presented a study that assesses the effect of selected scanner parameters on an extensive set of features computed from breast cancer MRI. We demonstrated that there is a significant effect of scanner parameters on the values of the extracted features. Specifically, for different scanner manufacturers, magnetic field strength and slice thicknesses, a high proportion of FGT related features show high sensitivity to scanner parameters (i.e. high variability when the scanner parameter changes). The tumor related features show lower sensitivity to the scanner manufacturer. The features related to both tumor and FGT are sensitive mostly to scanner manufacturer and slice thickness. However, their sensitivity to magnetic field is lower.
This finding is not surprising since FGT features cover more imaging volume than tumor and hence they are more likely to capture changes brought about by the scanning parameters. The features related to the combined usage of tumor masks and tissue masks are also affected by scanner manufacturers and varying slice thicknesses. However, features specifically related to tumor find more usage in clinical context. Hence, their lesser dependence on scanner parameters is a positive trait that we observe. This is consistent with the study (Chen et al., 2010) where the robustness of tumor morphological and kinetic features across scanner manufacturers was reported. This conveys that the tumor related features are representative more of the abnormality than that of the scanner used to acquire the image of the abnormality. This type of effect has been observed earlier in a scanner parameter related study for other disease (Stonnington et al., 2008).
Our study had some limitations. The cases were annotated by six breast imagers and this generates reader variability. However, the cases were randomly assigned to the readers. Also, with one reader, we would have had individual bias which is not present in this study. We have focused on three important scanner parameters for this study. It is likely that other scanner related parameters also introduce variability in the features and they are worthy of studying. However, that study will demand a patient cohort with significant variation in the selected scanner parameters. The scanner parameters were divided into two categories as per the available data. In future, this study can be extended to multiple categories of values for different scanner parameters across multiple institutions for stronger conclusions. As breast cancer radiomics and radiogenomics is expanding with the addition of new features, this type of study needs updating with the progression of time.
This study is of high significance. It demonstrates that the scanner parameters, which vary significantly between institutions and even within an institution, have a significant effect on the extracted radiomics/radiogenomic features from breast. This effect should be considered when interpreting radiomics and radiogenomics studies and careful measures should be taken to address the issue. Our results further underline the importance of validating radiomics and radiogenomics studies in large heterogeneous cohorts, as these validation studies can assess the generalizability of different features. Finally, since we included the measured impact of different scanner parameters on extraction of each of the 529 features and included all of them in the supporting information, this study could serve as a guide for selection of robust features in radiomics and radiogenomics studies in future.
Conclusions
We experimentally assessed the susceptibility of breast cancer MRI features to three types of scanner-related parameters. We found that features related to the fibrograndular tissue of the breast are more sensitive to scanner parameters than the features related to tumor only. We reported the sensitivity of the each of the examined features to each of the scanner parameters as a guide for researchers developing radiomics-based tools in breast cancer.
Supplementary Material
Highlights.
We studied the effect of different MRI scanner parameters on breast radiomics.
We extracted 529 radiomic features from 272 breast cancer patients in our cohort.
The radiomic features were categorized into relevant groups.
Scanner parameters affected tissue radiomics more compared to tumor radiomics.
Acknowledgments
The authors would like to acknowledge the funding from NIH (1R01EB021360) and North Carolina Biotechnology Center (2016-BIG-6520)
Grant Support:
National Institutes of Health, 1R01EB021360 (MAM)
North Carolina Biotechnology Center, 2016-BIG-6520 (MAM).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure:
The authors have no conflict of interest to disclose.
References
- Akobeng AK. Understanding diagnostic tests 3: receiver operating characteristic curves. Acta Pædiatrica. 2007;96(5):644. doi: 10.1111/j.1651-2227.2006.00178.x. [DOI] [PubMed] [Google Scholar]
- Arasu VA, Chen RCY, Newitt DN, Chang CB, Tso H, Hylton NM, Joe BN. Can signal enhancement ratio (SER) reduce the number of recommended biopsies without affecting cancer yield in occult MRI-detected lesions? Academic Radiology. 2011;18(6):716–721. doi: 10.1016/j.acra.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashraf AB, Daye D, Gavenonis S, Mies C, Feldman M, Rosen M, Kontos D. Identification of intrinsic imaging phenotypes for breast cancer tumors: Preliminary associations with gene expression profiles. Radiology. 2014;272(2):374–384. doi: 10.1148/radiol.14131375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein MA, Huston J, Ward HA. Imaging artifacts at 3.0T. Journal of Magnetic Resonance Imaging. 2006;24(4):735–746. doi: 10.1002/jmri.20698. [DOI] [PubMed] [Google Scholar]
- Bezdek JC. Pattern Recognition with Fuzzy Objective Function Algorithms. Kluwer Academic Publishers; 1981. [Google Scholar]
- Bhooshan N, Giger ML, Jansen SA, Li H, Lan L, Newstead GM. Cancerous Breast Lesions on Dynamic Contrast-enhanced MR Images: Computerized Characterization for Image-based Prognostic Markers. Radiology. 2010;254(3):680–690. doi: 10.1148/radiol.09090838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley AP. The use of the area under the ROC curve in the evaluation of machine learning algorithms. Pattern recognition. 1997;30(7):1145–1159. [Google Scholar]
- Brasic N, Wisner DJ, Joe BN. Breast MR Imaging for Extent of Disease Assessment in Patients with Newly Diagnosed Breast Cancer. Magnetic Resonance Imaging Clinics of North America. 2013;21(3):519–532. doi: 10.1016/j.mric.2013.04.012. [DOI] [PubMed] [Google Scholar]
- Chen W, Giger ML, Bick U, Newstead GM. Automatic identification and classification of characteristic kinetic curves of breast lesions on DCE-MRI. Medical Physics. 2006;33(8):2878–2887. doi: 10.1118/1.2210568. [DOI] [PubMed] [Google Scholar]
- Chen W, Giger ML, Lan L, Bick U. Computerized interpretation of breast MRI: investigation of enhancement-variance dynamics. Medical Physics. 2004;31(5):1076–1082. doi: 10.1118/1.1695652. [DOI] [PubMed] [Google Scholar]
- Chen W, Giger ML, Newstead GM, Bick U, Jansen SA, Li H, Lan L. Computerized Assessment of Breast Lesion Malignancy using DCE-MRI: Robustness Study on Two Independent Clinical Datasets from Two Manufacturers. Academic Radiology. 2010;17(7):822–829. doi: 10.1016/j.acra.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho N, Im S-A, Park I-A, Lee K-H, Li M, Han W, Moon WK. Breast Cancer: Early Prediction of Response to Neoadjuvant Chemotherapy Using Parametric Response Maps for MR Imaging. Radiology. 2014;272(2):385–396. doi: 10.1148/radiol.14131332. [DOI] [PubMed] [Google Scholar]
- Collewet G, Strzelecki M, Mariette F. Influence of MRI acquisition protocols and image intensity normalization methods on texture classification. Magnetic Resonance Imaging. 2004;22(1):81–91. doi: 10.1016/j.mri.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Czarnek N, Clark K, Peters KB, Mazurowski MA. Algorithmic three-dimensional analysis of tumor shape in MRI improves prognosis of survival in glioblastoma: a multi-institutional study. Journal of Neuro-Oncology. 2017:1–8. doi: 10.1007/s11060-016-2359-7. [DOI] [PubMed] [Google Scholar]
- Faermann R, Sperber F, Schneebaum S, Barsuk D. Tumor-to-breast volume ratio as measured on MRI: a possible predictor of breast-conserving surgery versus mastectomy. Israel Medical Association Journal. 2014;16(2):101–105. [PubMed] [Google Scholar]
- Georgiou H, Mavroforakis M, Dimitropoulos N, Cavouras D, Theodoridis S. Multi-scaled morphological features for the characterization of mammographic masses using statistical classification schemes. Artificial Intelligence in Medicine. 2007;41(1):39–55. doi: 10.1016/j.artmed.2007.06.004. [DOI] [PubMed] [Google Scholar]
- Giger ML, Vyborny CJ, Schmidt RA. Computerized characterization of mammographic masses: analysis of spiculation. Cancer Letters. 1994;77(2):201–211. doi: 10.1016/0304-3835(94)90103-1. [DOI] [PubMed] [Google Scholar]
- Gilhuijs KGA, Giger ML, Bick U. Computerized analysis of breast lesions in three dimensions using dynamic magnetic-resonance imaging. Medical Physics. 1998;25(9):1647–1654. doi: 10.1118/1.598345. [DOI] [PubMed] [Google Scholar]
- Grimm LJ, Zhang J, Mazurowski MA. Computational Approach to Radiogenomics of Breast Cancer: Luminal A and Luminal B Molecular Subtypes Are Associated With Imaging Features on Routine Breast MRI Extracted Using Computer Vision Algorithms. J Magn Reson Imaging. 2015 doi: 10.1002/jmri.24879. [DOI] [PubMed]
- Haacke EM, Brown RW, Thompson MR, Venkatesan R. Magnetic resonance imaging: physical principles and sequence design. Vol. 82. Wiley-Liss; New York: 1999. [Google Scholar]
- Haralick RM, Shanmugam K, Dinstein IH. Textural Features for Image Classification. IEEE Transactions on Systems, Man, and Cybernetics. 1973;3(6):610–621. doi: 10.1109/TSMC.1973.4309314. [DOI] [Google Scholar]
- Hendrick RE. Breast MRI: fundamentals and technical aspects. Springer Science & Business Media; 2007. [Google Scholar]
- Heywang-Köbrunner SH, Viehweg P, Heinig A, Küchler CH. Contrast-enhanced MRI of the breast: accuracy, value, controversies, solutions. European Journal of Radiology. 1997;24(2):94–108. doi: 10.1016/s0720-048x(96)01142-4. [DOI] [PubMed] [Google Scholar]
- Huang W, Fisher PR, Dulaimy K, Tudorica LA, O’Hea B, Button TM. Detection of Breast Malignancy: Diagnostic MR Protocol for Improved Specificity. Radiology. 2004;232(2):585–591. doi: 10.1148/radiol.2322030547. [DOI] [PubMed] [Google Scholar]
- Ikeda DM, Hylton NM, Kinkel K, Hochman MG, Kuhl CK, Kaiser WA, Viehweg P. Development, standardization, and testing of a lexicon for reporting contrast-enhanced breast magnetic resonance imaging studies. Journal of Magnetic Resonance Imaging. 2001;13(6):889–895. doi: 10.1002/jmri.1127. [DOI] [PubMed] [Google Scholar]
- Jansen SA, Shimauchi A, Zak L, Fan X, Wood AM, Karczmar GS, Newstead GM. Kinetic Curves of Malignant Lesions Are Not Consistent Across MRI Systems: Need for Improved Standardization of Breast Dynamic Contrast-Enhanced MRI Acquisition. American Journal of Roentgenology. 2009;193(3):832–839. doi: 10.2214/AJR.08.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karahaliou A, Vassiou K, Arikidis NS, Skiadopoulos S, Kanavou T, Costaridou L. Assessing heterogeneity of lesion enhancement kinetics in dynamic contrast-enhanced MRI for breast cancer diagnosis. The British Journal of Radiology. 2010;83(988):296–309. doi: 10.1259/bjr/50743919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klifa C, Carballido-Gamio J, Wilmes L, Laprie A, Shepherd J, Gibbs J, Hylton N. Magnetic resonance imaging for secondary assessment of breast density in a high-risk cohort. Magnetic resonance imaging. 2010;28(1):8–15. doi: 10.1016/j.mri.2009.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuttel FM, Menezes GLG, van den Bosch MAAJ, Gilhuijs KGA, Peters NHGM. Current clinical indications for magnetic resonance imaging of the breast. Journal of Surgical Oncology. 2014;110(1):26–31. doi: 10.1002/jso.23655. [DOI] [PubMed] [Google Scholar]
- Kuhl C. The Current Status of Breast MR Imaging Part I. Choice of Technique, Image Interpretation, Diagnostic Accuracy, and Transfer to Clinical Practice. Radiology. 2007;244(2):356–378. doi: 10.1148/radiol.2442051620. [DOI] [PubMed] [Google Scholar]
- Kuhl CK, Jost P, Morakkabati N, Zivanovic O, Schild HH, Gieseke Jr. Contrast-enhanced MR Imaging of the Breast at 3.0 and 1.5 T in the Same Patients: Initial Experience 1. Radiology. 2006;239(3):666–676. doi: 10.1148/radiol.2392050509. [DOI] [PubMed] [Google Scholar]
- Kuhl CK, Schild HH. Dynamic image interpretation of MRI of the breast. Journal of Magnetic Resonance Imaging. 2000;12(6):965–974. doi: 10.1002/1522-2586(200012)12:6<965::AID-JMRI23>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Kuhl CK, Schrading S, Leutner CC, Morakkabati-Spitz N, Wardelmann E, Fimmers R, Schild HH. Mammography, breast ultrasound, and magnetic resonance imaging for surveillance of women at high familial risk for breast cancer. Journal of Clinical Oncology. 2005;23(33):8469–8476. doi: 10.1200/JCO.2004.00.4960. [DOI] [PubMed] [Google Scholar]
- Kumar V, Gu Y, Basu S, Berglund A, Eschrich SA, Schabath MB, Gillies RJ. QIN “Radiomics: The Process and the Challenges”. Magnetic resonance imaging. 2012;30(9):1234–1248. doi: 10.1016/j.mri.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RGPM, Granton P, Aerts HJWL. Radiomics: Extracting more information from medical images using advanced feature analysis. European Journal of Cancer. 2012;48(4):441–446. doi: 10.1016/j.ejca.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Ehmke RC, Schwartz LH, Zhao B. Assessing Agreement between Radiomic Features Computed for Multiple CT Imaging Settings. PLOS ONE. 2016;11(12):e0166550. doi: 10.1371/journal.pone.0166550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackin D, Fave X, Zhang L, Fried D, Yang J, Taylor B, Court L. Measuring Computed Tomography Scanner Variability of Radiomics Features. Investigative radiology. 2015;50(11):757–765. doi: 10.1097/rli.0000000000000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahrooghy M, Ashraf AB, Daye D, McDonald ES, Rosen M, Mies C, Kontos D. Pharmacokinetic tumor heterogeneity as a prognostic biomarker for classifying breast cancer recurrence risk. IEEE Transactions on Biomedical Engineering. 2015;62(6):1585–1594. doi: 10.1109/TBME.2015.2395812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann RM, Kuhl CK, Kinkel K, Boetes C. Breast MRI: guidelines from the European Society of Breast Imaging. European Radiology. 2008;18(7):1307–1318. doi: 10.1007/s00330-008-0863-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazurowski MA. Radiogenomics: What It Is and Why It Is Important. Radiogenomics: What It Is and Why It Is Important. 2015;12(8):862–866s. doi: 10.1016/j.jacr.2015.04.019. [DOI] [PubMed] [Google Scholar]
- Mazurowski MA, Grimm LJ, Zhang J, Macrom PK, Yoon S, Kim C, Johnson K. Recurrence-free survival in breast cancer is associated with MRI tumor enhancement dynamics quantified using computer algorithms. European Journal of Radiology. 2015;84(11):2117–2122. doi: 10.1016/j.ejrad.2015.07.012. [DOI] [PubMed] [Google Scholar]
- Mazurowski MA, Zhang J, Grimm LJ, Yoon SC, Silber JI. Radiogenomic analysis of breast cancer: Luminal B molecular subtype is associated with enhancement dynamics at MR imaging. Radiology. 2014;273(2):365–372. doi: 10.1148/radiol.14132641. [DOI] [PubMed] [Google Scholar]
- Rausch DR, Hendrick RE. How to Optimize Clinical Breast MR Imaging Practices and Techniques on Your 1.5-T System. RadioGraphics. 2006;26(5):1469–1484. doi: 10.1148/rg.265055176. [DOI] [PubMed] [Google Scholar]
- Sardanelli F, Giuseppetti GM, Panizza P, Bazzocchi M, Fausto A, Simonetti G, Del Maschio A. Sensitivity of MRI versus mammography for detecting foci of multifocal, multicentric breast cancer in fatty and dense breasts using the whole-breast pathologic examination as a gold standard. American Journal of Roentgenology. 2004;183(4):1149–1157. doi: 10.2214/ajr.183.4.1831149. [DOI] [PubMed] [Google Scholar]
- Saslow D, Boetes C, Burke W, Harms S, Leach MO, Lehman CD, for the American Cancer Society Breast Cancer Advisory G American Cancer Society Guidelines for Breast Screening with MRI as an Adjunct to Mammography. CA: A Cancer Journal for Clinicians. 2007;57(2):75–89. doi: 10.3322/canjclin.57.2.75. [DOI] [PubMed] [Google Scholar]
- Sawyer-Glover AM, Shellock FG. Pre-MRI Procedure Screening: Recommendations and Safety Considerations for Biomedical Implants and Devices. Journal of Magnetic Resonance Imaging. 2000;12(1):92–106. doi: 10.1002/1522-2586(200007)12:1<92::AID-JMRI11>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Song C, Smith M, Huang Y, Jeraj R, Fain S. Heterogeneity of vascular permeability in breast lesions with dynamic contrast enhanced MRI; Paper presented at the 17th International Symposium for Magnetic Resonance in Medicine.2009. [Google Scholar]
- Stonnington CM, Tan G, Klöppel S, Chu C, Draganski B, Jack CR, Jr, Frackowiak RSJ. Interpreting scan data acquired from multiple scanners: A study with Alzheimer’s disease. NeuroImage. 2008;39(3):1180–1185. doi: 10.1016/j.neuroimage.2007.09.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton EJ, Oh JH, Dashevsky BZ, Veeraraghavan H, Apte AP, Thakur SB, Morris EA. Breast cancer subtype intertumor heterogeneity: MRI-based features predict results of a genomic assay. Journal of Magnetic Resonance Imaging. 2015;42(5):1398–1406. doi: 10.1002/jmri.24890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, Gee JC. N4ITK: Improved N3 Bias Correction. IEEE transactions on medical imaging. 2010;29(6):1310–1320. doi: 10.1109/TMI.2010.2046908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan T, Bloch BN, Plecha D, Thompson CL, Gilmore H, Jaffe C, Madabhushi A. A radio-genomics approach for identifying high risk estrogen receptor-positive breast cancers on DCE-MRI: preliminary results in predicting OncotypeDX risk scores. Scientific Reports. 2016:6. doi: 10.1038/srep21394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Kato F, Oyama-Manabe N, Li R, Cui Y, Tha KK, Shirato H. Identifying triple-negative breast cancer using background parenchymal enhancement heterogeneity on dynamic contrast-enhanced MRI: a pilot radiomics study. PloS one. 2015;10(11):e0143308. doi: 10.1371/journal.pone.0143308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner E, Plewes DB, Shumak RS, Catzavelos GC, Di Prospero LS, Yaffe MJ, Cole DEC. Comparison of breast magnetic resonance imaging, mammography, and ultrasound for surveillance of women at high risk for hereditary breast cancer. Journal of Clinical Oncology. 2001;19(15):3524–3531. doi: 10.1200/JCO.2001.19.15.3524. [DOI] [PubMed] [Google Scholar]
- Waugh SA, Purdie CA, Jordan LB, Vinnicombe S, Lerski RA, Martin P, Thompson AM. Magnetic resonance imaging texture analysis classification of primary breast cancer. European Radiology. 2016;26(2):322–330. doi: 10.1007/s00330-015-3845-6. [DOI] [PubMed] [Google Scholar]
- Wu S, Weinstein SP, DeLeo MJ, Conant EF, Chen J, Domchek SM, Kontos D. Quantitative assessment of background parenchymal enhancement in breast MRI predicts response to risk-reducing salpingo-oophorectomy: preliminary evaluation in a cohort of BRCA1/2 mutation carriers. Breast Cancer Research. 2015;17(1):1. doi: 10.1186/s13058-015-0577-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Li L, Zhang J, Shao G, Zheng B. A new quantitative image analysis method for improving breast cancer diagnosis using DCE-MRI examinations. Medical Physics. 2015;42(1):103–109. doi: 10.1118/1.4903280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Tan Y, Tsai W-Y, Qi J, Xie C, Lu L, Schwartz LH. Reproducibility of radiomics for deciphering tumor phenotype with imaging. Scientific Reports. 2016;6:23428. doi: 10.1038/srep23428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Englander S, Baloch S, Zacharaki EI, Fan Y, Schnall MD, Shen D. STEP: spatiotemporal enhancement pattern for MR-based breast tumor diagnosis. Medical Physics. 2009;36(7):3192–3204. doi: 10.1118/1.3151811. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.