Abstract
Sarcomas are cancers of the bone and soft tissue often defined by their gene fusions. However, the timing, context, and processes by which these pathogenic fusions arise are unknown. We explored this in Ewing sarcoma, a cancer driven by EWSR1-ETS fusions, with very few cooperating mutations. Combining whole-genome sequencing with enhanced informatics, we found that the EWSR1-ETS fusion arose from striking rearrangement clusters in 42% of cases (52/124). Notably, these were organized in loops that universally contained the fusion at their center, while also weaving up to 18 genes together with it. We found the same pattern of rearrangements in three additional types of sarcoma. From these data, we define a new signature for sarcoma fusions that precedes other somatic changes, in the earliest replicating DNA of the genome. This dramatic, sudden process impinges on many genes – generating multiple coding changes that profoundly affect the transcriptome, with the disease-defining gene fusion at its core. These rearrangement loops arise in an early ES clone from which both the primary tumor and the lethal relapse emerged, and then evolved in parallel until clinically detected.
Genomic rearrangements (structural variants) are a ubiquitous source of somatic mutation in human cancer. They arise from breaks in chromosomes, which are aberrantly rejoined. Rearrangements may occur in isolation or in the context of complex genomic catastrophes that shatter (chromothripsis (1)) or join chromosomes in chains or loop-structures (chromoplexy (2)). Rearrangements can generate cancer-driving mutations through several mechanisms, including the formation of gene fusions. Typically, fusions are fashioned by translocations that are often reciprocal. An exception is the prostate cancer fusion gene, TMPRSS2–ERG, that can occur in the context of chromoplexy(2).
Oncogenic gene fusions are particularly common in leukemia and bone and soft tissue tumors (3), often acting as the sole driver mutation and delineating clinically relevant tumor entities and subgroups. In leukemia, RAG-mediated recombination has been identified as the leading mutational process that creates canonical gene fusions and drives oncogenesis through translocations and deletions (4). Here, we sought to investigate processes and timing of oncogenic fusions in human bone and soft tissue tumors.
The starting point of our investigation was Ewing sarcoma (ES), a bone and soft tissue cancer predominantly diagnosed in adolescents and young adults. It represents the prototypical fusion-driven sarcoma, defined by fusions between EWSR1 and an ETS transcription factor, including FLI1 and ERG (5). Although the downstream consequences of EWS-ETS are well established (6), the timing and mechanism by which it arises are unknown.
Burden and signatures of small mutations in Ewing sarcoma
We sequenced the gene-containing portions of, or whole genomes of 50 ES tumors and their matched normal DNA (complete sequencing details in table S1). We used a conventional analysis pipeline to call somatic substitutions and rearrangements, with additional custom software to remove recurrent artefacts and sources of false positives (see Methods and fig. S1). Overall, and consistent with previous reports (7–10), the ES genome is genetically quiet, with few somatic substitutions identified (Median: <1 Mut/Mb; Fig. 1A). The number of small coding mutations was also low.
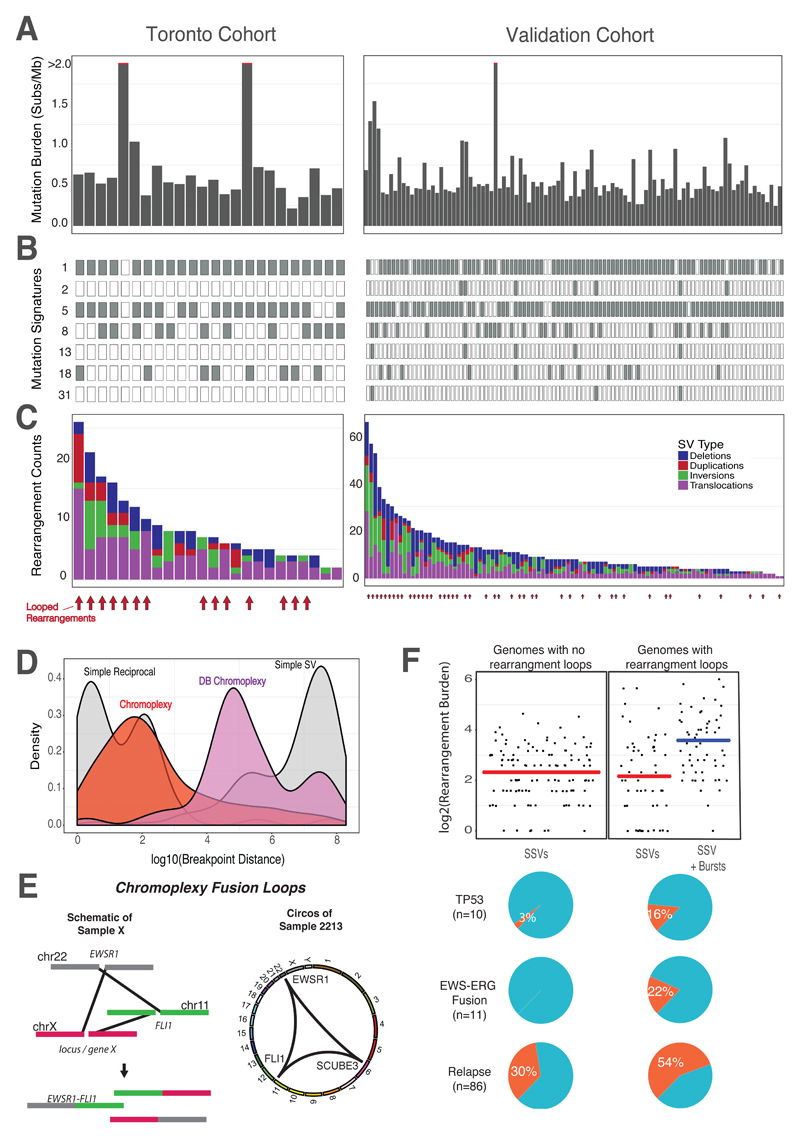
Fig. 1. Mutation Landscape of Ewing Sarcoma.
The initial cohort consisted of 50 primary ES tumors, of which, 23 underwent whole genome sequencing (Toronto cohort, left). One rearrangement screen sample (sample 4462) is included in this figure. The validation cohort consisted of 100 ES whole-genomes from Tirode et al. 2014 (right). (A) Somatic mutation burden for Ewing sarcoma. The mutation burden of all genome samples are shown. Three outlier samples with >2 mutations/MB, are indicated by the red line. (B) Ewing sarcoma mutation signatures. Mutation signature analysis, defined by the proportion of 96 possible trinucleotides, identified common mutation patterns in most samples (Age-associated, “clock-like” signature 1). Other signatures included #2, 5, 8, 13, 18, and 31. Signatures 2 and 13 are associated with the activity of the AID/APOBEC family of cytidine deaminase, while Signature 5 is also clock-like in some cancers, but not ES (11, 13). Signatures 8 and 18 have an unknown molecular aetiology, however it has been suggested that Signature 18 is caused by reactive oxygen species (ROS)(43). Signature 31 is believed to be the result of exposure to platinum-based antineoplastic therapy (24).(C) Rearrangement profiles for Ewing sarcoma. Shown are the burden of deletions (blue), duplications (red), inversions (green) and translocations (purple) in individual ES genomes. Samples with chained complex rearrangements (looped rearrangements) are highlighted by red arrows (14/24 for Toronto, 38/100 for Validation, aggregated prevalence: 52/124). (D) Rearrangement breakpoint clusters. The aggregated density distributions of the genomic distance between consecutive rearrangement breakpoints are shown. Reciprocal breakpoints are close together (~102 bp) because there is an equal exchange of genetic material arising from a single break on each chromosome. Chromoplectic rearrangements (red) overlap this range due to the proximity breakpoints involved in looped rearrangements. Deletion bridge (DB) chromoplexy (purple) are looped rearrangement clusters in which a deletion spans two breakpoints, resulting in breakpoint distances that are farther apart (illustrated in fig. S11). Non-complex breakpoints (simple structural variants) are far apart (~108 bp). (E) Schematic diagram of chromoplexy fusion loops. Illustrative example of chromoplexy in Ewing sarcoma shows three chromosomes undergoing double-strand breakage, shuffling and religation in an aberrant configuration. This phenomenon generates the canonical fusion, EWSR1-FLI1 (ERG or ETV1) and disrupts a third locus, X, in a one-off burst of rearrangements. In reality, up to 8 chromosomes may be disrupted in this looping pattern. A representative genome-wide Circos plots depicting genomic rearrangements in an Ewing sarcoma tumor (from the discovery cohort), which are organized in a loop. (F) Genomic correlates and clinical impact of looped rearrangements. In genomes without rearrangement loops, only simple structural variants (SSV) exist with an average rearrangement burden of 7 rearrangements/sample. This rate is similar to the background SSV rate (determined by removing rearrangements involved in a loop) in genomes with rearrangement bursts (compare the two red lines). The additional complexity of looped rearrangements results in higher genomic instability in these tumors. The most common genomic alterations include somatic TP53 mutations, which are rare, but enriched in patients with complex genomes (top pie chart, p < 0.05). EWS-ERG fusions are also rare, as they represent 10% of all Ewing sarcoma diagnoses, however all EWS-ERG fusion Ewing tumors are either chomothriptic or chromoplectic (middle pie chart). Lastly, patients with complex genomes tend to relapse (bottom pie chart, p < 0.05). All the markers of aggressive disease (high genomic instability, somatic TP53 and relapse) are present in tumors with complex genomes.
We next asked if the collection of all mutations, when considered together, could help highlight consistent mutagenic processes in ES. We extracted mutational signatures using an established method that allows for the discovery of new signatures. Despite their young age and quiet genomes, ES patients’ tumors contained at least seven distinct signatures, all of which matched patterns found in adult cancer (COSMIC # 1, 2, 5, 8, 13, 18 and 31; Fig. 1B, fig. S2A) (11, 12). Two of these (#1 and 5) were nearly universal, and associated with patient age. Signature 1 generated a steady rate of 7 mutations per Gb per year, which is similar to that of adult ovary and breast cancer (fig. S2B) (13). An overview of the somatic architecture and mutational signatures of each tumor in our discovery cohort is shown in Fig 1A-C (left panels, Toronto Cohort).
Chromoplexy rearrangement loops are common in aggressive Ewing sarcoma
Having observed few small mutations, we then focused our attention on structural rearrangements. We applied a bespoke analysis tool to detect clustered rearrangements from whole-genome data, defined as having an inter-rearrangement distance of <10 kbp (see Methods). Using a computational data structure that modeled adjacent breakpoints as vertices and inter-connected rearrangements as edges in a graph, we uncovered several distinct configurations of rearrangement clusters (Fig. 1D). As expected, one configuration of rearrangement clusters was a result of reciprocal rearrangements, where there is an equal exchange of genetic material and overlapping breakpoints. These were isolated rearrangements that occurred without additional breakpoints nearby. A second configuration, seen in 15/24 tumor genomes, was a distinctive pattern of focal clustered events with nearly overlapping junctions, organized as closed loops (distance < 30 bp; Fig. 1D, red distribution). That is, if one follows these complex rearrangements across their multiple constituent chromosomes, one is ultimately brought back to the point of departure. Importantly, the loops were nearly always centered on EWSR1-ETS (Fig. 1E). These abutting rearrangements that occur in a loop resemble a pattern of chromoplexy, akin to the loops of the prostate cancer fusion gene, TMPRSS2–ERG. Of note, the EWSR1-ERG fusion was always generated by a complex mechanism, whereas EWSR1-FLI1 arose with or without this mechanism (fig. S3A). This is likely due to the opposite gene orientation of EWSR1 relative to ERG on their respective chromosome arms. A simple two-chromosome break rearrangement cannot place the genes in the correct transcriptional orientation, necessitating more complex chromosomal rearrangements for fusion formation. Besides this, ERG and FLI1-driven chromoplexy were highly similar (fig. S3B).
In all cases, we resolved the breakpoints and found, primarily, positions consistent with ‘Type I’ or ‘Type II’ ES (14). In the most complex case of chromoplexy, up to 18 genes were brought together with the canonical fusion on the same derivative chromosome (fig. S3C, the full list of genes affecting all samples is shown in fig. S3D). We validated chromoplectic looped rearrangements by deep sequencing or by cytogenetic analysis using standard G-banding and spectral karyotyping (Methods and fig. S4). Using RNA sequencing, we found that chromoplectic loops universally disrupted the reciprocal fusion (FLI1-EWSR1); 52% of the cancers with simple rearrangements expressed the reciprocal fusion, but none of the chromoplectic tumors expressed it (n=27, fig. S5). For further validation of chromoplexy in ES, we re-analyzed a published, independent cohort of 100 ES genomes using our informatics pipeline (10). The somatic architecture and mutational signature of the validation cohort is shown in Fig. 1 (right panels, Validation Cohort). Both cohorts harbored copy number profiles consistent with previous reports (fig. S6)(10). With this series, the aggregated prevalence of chromoplectic EWSR1-ETS gene fusions was 42% (52/124).
The survival for relapsed ES is poor and new prognostic markers are needed. We evaluated the association between chromoplexy, patient outcomes, as well as known markers of worse prognosis. We found that higher overall genomic complexity, a marker of aggressive ES (10, 15), was almost completely explained by chromoplectic rearrangements (Fig. 1F). In contrast, there was no difference in the burden of non-chromoplectic rearrangements. Similarly, TP53 mutations, another established marker of poor prognosis (10, 16), were enriched in chromoplexy ES (16% vs. 3%, p < 0.05). There was no enrichment for CDKN2A or STAG2 mutations (fig. S7). Finally, and consistent with the above, patients with chromoplexy ES were more likely to relapse (54% vs. 30%, p < 0.05), strongly suggesting that it marks a more aggressive variant of ES.
Chromoplexy generates the key fusion in many cancer types
We next widened our search across four different benign and malignant bone and soft tissue tumor types, for which canonical gene fusions have been identified (table S2). We subjected 13 tumors to high or low coverage whole genome sequencing, plus RNA sequencing where feasible. In three tumor types - chondromyxoid fibroma, synovial sarcoma, and phosphaturic mesenchymal tumors - we found that chromoplectic rearrangements (occurring in a similar looped formation) did indeed generate canonical gene fusions (Fig. 2). Furthermore, in one of the chondromyxoid fibroma cases, the fusion emerged from chromothripsis across seven different chromosomes (fig. S8, CMF #2). Chromothripsis was seen in five ES cases, of which four involved the canonical fusion. Taken together, these findings in human bone and soft tissue tumors show that canonical gene fusions are frequently caused by complex rearrangement processes, predominantly chromoplexy, but also chromothripsis.
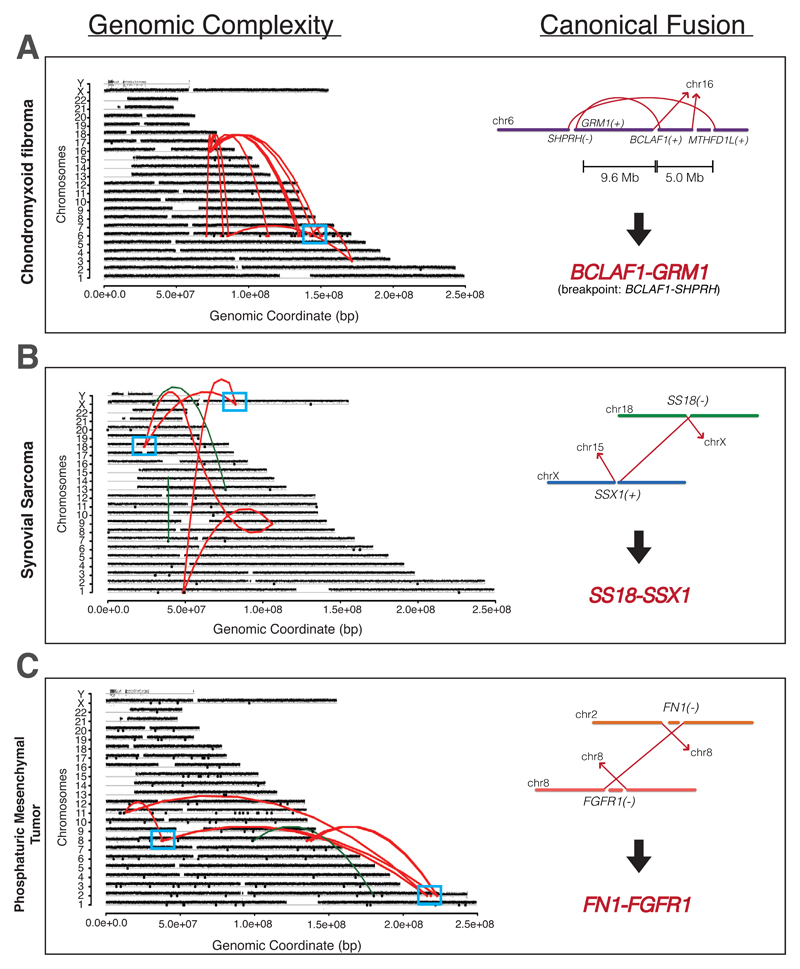
Fig. 2. Genomic Catastrophes are Common in Sarcomas.
Copy number profile for fusion-driven sarcomas with chromoplexy are shown. Rearrangements are colored red, and the loci with the canonical fusion are highlighted (blue box) and enlarged on the right. (A) Chrondromyxoid fibroma (CMF) with chromoplexy. The genomic breakpoint lies in the upstream SHPRH gene, while the BCLAF1-GRM1 fusion was detected by RNA sequencing. Further complex CMFs, which also show a SHRPRH genomic breakpoint but GRM1 fusion, can be found in fig. S8. (B) Synovial sarcoma with chromoplexy. Chromoplexy generating the SS18-SSX1 pathognomonic canonical fusion is shown. (C) Phosphaturic mesenchymal tumor (PMT) with chromoplexy. Genome sequencing of PMTs revealed deletion bridges occurring across the genome at chromoplectic loci, generating the canonical FN1-FGFR1 fusion.
We examined the microanatomy of chromoplexy fusion loops at base pair resolution, comparing ES to a published series of prostate cancers (2). EWSR1-ETS Ewing loops were less complex than TMPRSS2–ERG prostate cancer loops with fewer rearrangements and individual loops involved in their generation (2 to 10 rearrangements in 1- 2 loops compared with up to 130 rearrangements in up to 25 loops in prostate cancer). This may be a consequence of the ES genome having a shorter time frame to mutate compared to prostate cancer patients. Consistent with this proposition, multiple independent chromoplexy loops can exist in older prostate cancers, compared to the one simple loop seen in ES (17). In contrast to ES, where chromoplexy is virtually synonymous with the disease-defining fusion, several chromoplexy fusion loops occur in prostate cancer without necessarily forming the TMPRSS2–ERG fusion. When a loop was present in ES, it almost always generated the EWSR1-ETS fusion (47/52 cases, 90%) (Fig. 3A-B, fig. S9 and S10).
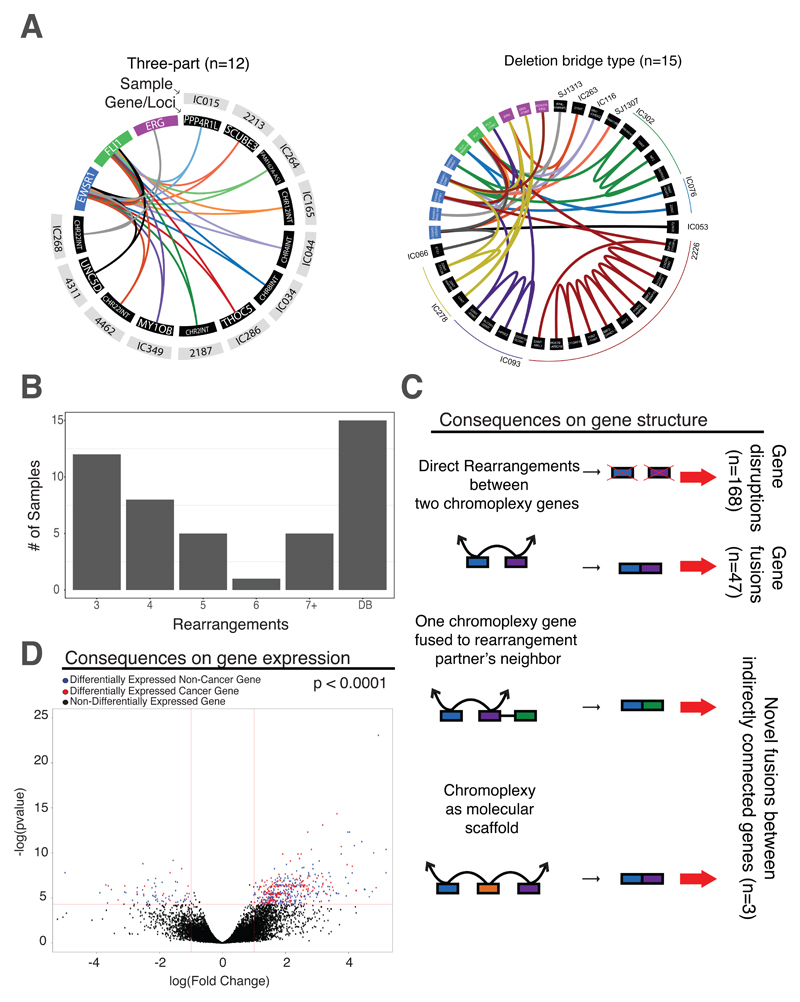
Fig. 3. Characterizing chromoplexy loops that generate EWSR1-ETS in ES.
(A) Patterns of looped rearrangements. Chromoplexy circos webs demonstrate that patterns of looped rearrangements are conserved across samples, while different genes or loci are affected in each cancer (black panels). In each web, individual samples are denoted using a different color (and named in the grey panel). In all cases, central to chromoplexy fusion loops were the key driver genes: EWSR1 (blue), FLI1 (green) and ERG (purple). The most frequent patterns of chromoplexy in Ewing sarcoma are those with a three-way looping structure as well as the presence of deletion bridges. For those with deletion bridges, “adj” refers to adjacent loci affected. An enlarged Circos web can be found in fig. S9 for readability. Three samples have structures only involving EWSR1, FLI1 and adjacent loci. Sample 4004 has deletion bridge chromoplexy and is described in fig. S3C. (B) Summary of chromoplexy types. Bar chart showing the number of rearrangements in a loop (x-axis) and the number of samples with that rearrangement pattern. Other chromoplexy web structures can be found in fig. S10. (C) Transcriptional consequences of chromoplexy: gene expression. Volcano plot illustrating the differential gene expression in chromoplexy vs non-chromoplexy ES, revealing 504 differentially expressed genes. Points greater than 1 or less than -1 and above the 1.3 (as indicated by the red lines) are genes that are significantly differentially expressed (blue dots). Red dots highlight genes that are differentially expressed and involved in a cancer hallmark pathway. (D) Transcriptional consequences of chromoplexy: gene disruptions and fusions. There are three mechanisms of gene dysregulation via RNA fusion when chromoplexy occurs. The first involves two genes (blue and purple boxes) brought together by chromoplectic rearrangements (black arrowed lines) leading to gene disruptions (top scenario) and novel inframe fusions (2nd from top scenario). This was detected in the 3/10 cases where there was genome (+chromoplexy) and transcriptome sequencing available. When RNA sequencing was not available, these are predicted to cause fusions (n=47, excluding the EWSR1-ETS driver) and gene disruptions by fusing genes in opposite transcriptional orientation or fusing a gene to an intergenic sequence (n=168). The second mechanism involves two chromoplexy genes brought together by a rearrangement at the genomic level, but one of the partner’s neighboring genes (green box) is transcriptionally fused to the other chromoplexy partner in its place (3rd from top scenario). This is also the predominant mechanism of GRM1 fusion generation in chrondromyxoid fibromas (fig. S8). Lastly, the final mechanism of gene dysregulation occurs when chromoplexy facilitates the production of a fusion by acting as a molecular scaffold (bottom scenario; illustrated in fig. S12). Two genes are both rearranged to a third locus (orange) and are then, transcriptionally, fused together. No direct genomic link exists between these two genes. These phenomena can only be detected if both whole-genome and RNA-Seq are available.
Significant transcriptional disruptions are associated with chromoplexy
These loops also led to targeted disruptions or fusions between genes brought together directly through chromoplexy (n=168 gene disruptions and n=47 fusions; Fig. 3C). Given that chromoplexy appeared to mark an aggressive form of ES, we wondered if its gene expression program was globally different - above and beyond the immediate, focal, structural consequences listed above. We identified 504 differentially expressed genes in chromoplexy compared to simple ES (p < 0.001, Fig. 3D). Gene set enrichment analysis of well curated pathways (18), uncovered a significant enrichment of dysregulated genes in established cancer hallmark pathways (table S3).
Both prostate cancer and ES loops were characterized by focal intra-chromosomal rearrangements - deletion bridges (2) - that acted as local mediators of large-scale loops (illustrated in fig. S11). We found deletion bridges in ~60% (30/52) of chromoplectic ES. Unlike prostate cancer, more than a third of bridges are utilized in ES in a highly consistent manner. That is, if a deletion bridge was found in one component of the loop, it would occur on all chromosomes. For example, in sample 2226 we observed 13 rearrangements, spanning three chromosomes, all of which involved deletion bridges. These bridged chromoplectic rearrangements fused EWSR1-FLI1, and disrupted the neighboring gene, AP1B1, as well as the known cancer gene, ARID1B. Thus, deletion bridges can create further oncogenic disruptions.
We also observed a remarkable pattern of splicing, whereby the transcriptional machinery further refined the looped rearrangements found in the genome. In chondromyxoid fibromas with chromoplectic GRM1 fusions (3/4 cases), the rearrangement breakpoint did not actually reside within the GRM1 gene body. Rather, the breakpoint was instead found in the upstream gene, SHPRH, within a narrow window (fig. S8). Thus, chromoplexy plus conventional splicing leads to the promoter swap that is characteristic of this cancer (see (19)). Interestingly, we also observed the transcriptional generation of gene fusions in ES. Examination of the transcriptomic consequences of loops showed that genes that were unconnected at the DNA level were brought together, in cis, at the mRNA level. This included examples of the EWSR1-ETS fusion itself (Fig. 3D, fig. S12). In the cases reported here, no direct rearrangement links EWSR1 and FLI1, however they are linked via two rearrangements to a third locus. In this way, chromoplexy generates the canonical fusion driver via a chromoplexy scaffolding event.
Chromoplexy is among the primary, clonal, mutations in Ewing sarcoma, and enriched in early-replicating regions of the genome
Our next line of enquiry examined the timing of chromoplexy rearrangements in tumor evolution. Chromoplexy may arise from a one-off sudden event, generating many breakpoints simultaneously, or through step-wise progressive bursts of mutations in succession (2). To differentiate between these two modes of evolution, we used DNA copy number profiling associated with the breakpoints of chromoplexy rearrangements to assess the copy number of neochromosomes. A low number of copy number state (three or fewer) is associated with a one-off mutational event because breakage and ligation can only involve a small number of chromosomes inside a cell at any given time (20, 21). In contrast, stepwise progression would result in multiple copy number states due to the possibility of copy number alterations arising within older copy number alterations. Chromoplectic breakpoints involve many chromosomes and are not associated with any copy number alterations (fig. S13). That is, these looped rearrangements across the genome are balanced. In addition, using a novel algorithm, we found that the allele frequency of chromoplectic breakpoints was higher than that of simple structural rearrangements, providing further evidence that these breakpoints occurred together and early in tumor development (Methods and fig. S14). Given their extremely tight clustering, low number of copy number state transitions, and consistent clonal variant allele frequency, EWSR1-ETS loops are likely to have arisen from singular bursts of rearrrangements.
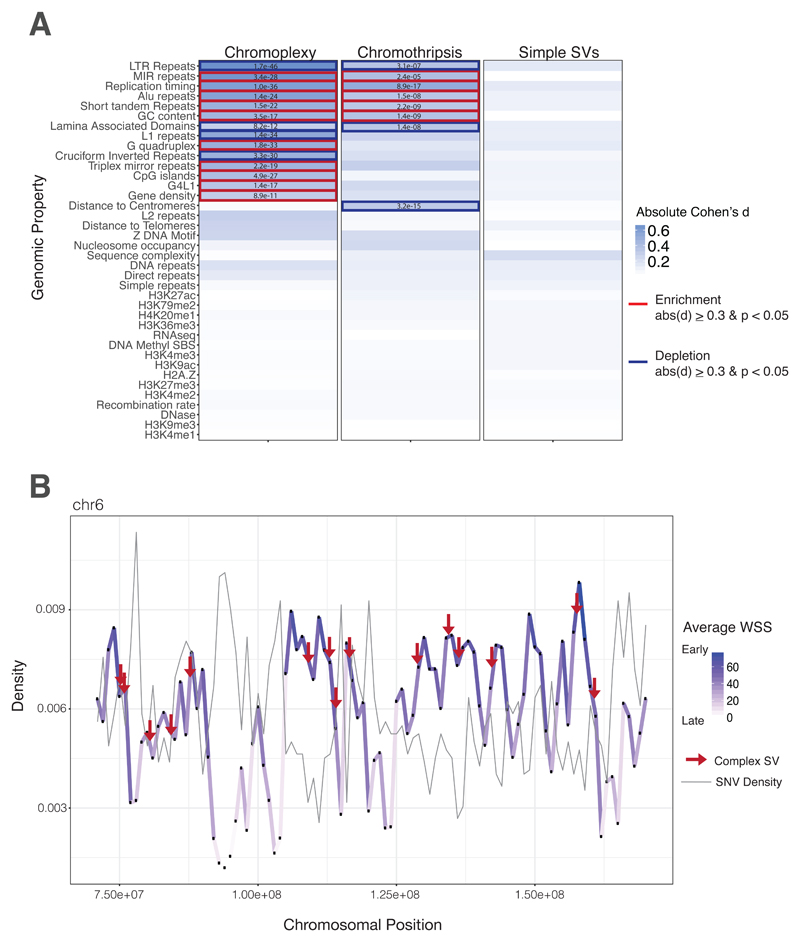
We then examined whether genomic regions of loop breakpoints share genomic properties predisposing these regions to simultaneous rearrangement. We performed a comprehensive analysis of 38 genomic properties, including adjacency to histone marks, association with replication timing, as well as proximity to genes, repetitive or transposable elements (table S4). Of these properties, early replicating DNA, and features consistent with this, were the most strongly associated with chromoplexy loops (p < 1.0 x 10-36, Fig. 4A-B). In stark contrast, neither non-looped simple breakpoints of ES, nor simulated simple breakpoints, were significantly associated with replication timing, or indeed any other feature (see Methods). Replication timing is known to be strongly correlated with gene activity, chromatin accessibility and nuclear position (22). Accordingly, chromoplectic breakpoint positions were also strongly associated with high gene density and high GC content (fig. S15A). Conversely, lamina-associated domains, enriched in late-replication regions and repressive chromatin environments, were found to be negatively associated with chromoplectic rearrangements. These significant associations were upheld when breakpoints directly residing in EWS, FLI1 or ERG were removed from the analyses. Remarkably, the same associations were found for looped rearrangements of ETS+ prostate cancers, but not for simple prostate cancer rearrangements (fig. S15B). Of further interest, we noted that the genes impacted by chromoplexy, were amongst the most highly expressed in ES, across all patients (top 20%; fig. S16). Most expressed genes are found in early replicating DNA (23). These data are consistent with the proposed model of chromoplexy where DNA is co-localized in transcription hubs allowing for multiple genes from many chromosomes to be broken, shuffled and aberrantly ligated, as proposed (2).
Fig. 4. Early Replicating DNA and Chromoplexy.
(A) Heatmap of genomic property associations. The genomic properties listed in supplementary table 4 were calculated for all rearrangements in both cohorts. Complex rearrangements (chromoplexy and chromothripsis), exclusively, are strongly associated with early replication timing, and other genomic features consistent with this feature (gene density, CpG density, Alu density etc.). Table values are indicative of FDR-corrected p-values compared to a million random points in the genome. Blue highlights are indicative of a Cohen’s d equal to or greater than 0.3. Bold boxes indicate a positive (red, enrichment) or negative (blue, depletion) association with the feature. All features were evaluated in 1 kb bins across the genome. For feature density metrics, associations were calculated in 1MB sliding windows centered in 1 kb bins. (B) Density distribution of the average wavelet-smoothed signal and SNVs on representative chromosome. The average wavelet-smoothed signal, of replication timing, is plotted for a subset of chromosome 6 to illustrate changes between early and late replication timing and the co-association with mutations in ES. The positional variation of replication timing across the chromosome is depicted as changes in density and color. Point mutations peak in late-replicating regions (dip in WSS, light purple), whereas complex rearrangements peak in regions of early replication timing (peak in WSS, dark purple).
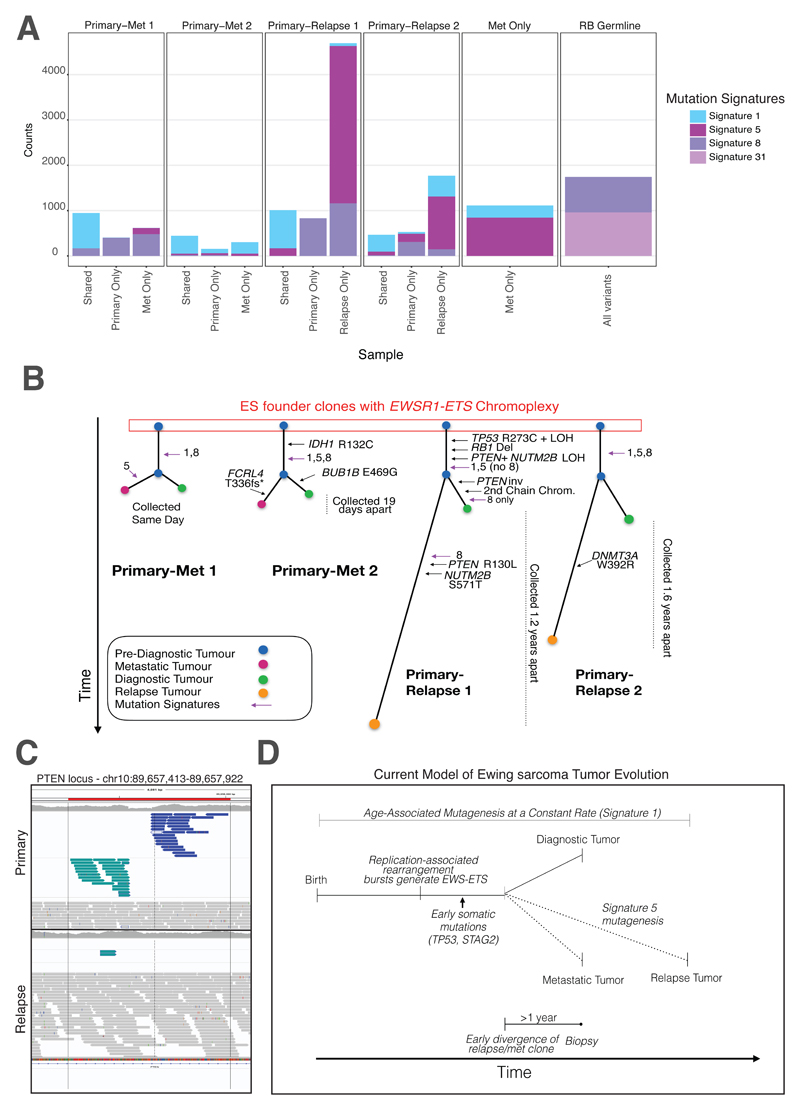
Mutation Patterns of Relapsed and Metastatic Ewing sarcoma
Taken together, we have seen that chromoplexy arises early in the evolutionary history of ES, through a replication-associated mechanism, portending a worse prognosis and possible relapse. However, the genetic makeup of relapsed ES is unknown, since standard of care for ES does not typically involve re-biopsy of the cancer when the disease returns or has metastasized. Therefore, whether further mutations - chromoplectic or otherwise - emerge at relapse is unknown, since very few samples have been available. However, re-biopsies were performed for a small number of our patients, which we profiled by WGS and performed full mutation and signature analysis (Fig. 5A). Strikingly, every relapse or metastatic tumor contained the chromoplexy-associated fusion, whether it was from a metastasis at the time of diagnosis or a relapse arising later (Fig. 5B). The pattern of point mutations was also distinct. There was an enrichment of signatures 8 and/or 18, in addition to the clock-like signature seen at diagnosis, suggesting that new processes drive relapse and metastatic ES (Fig. 5B). For example, in one patient’s tumor we found a striking increase of COSMIC Signature 31, which has been recently associated with exposure to platinum therapy in chronic myelomonocytic leukemia (24). Notably, our patient had been treated with carboplatin for an early retinoblastoma three years prior to their ES. At least three other patients in the validation cohort had a similar signature in their ES, which we believe were also treatment induced (Fig. 1B).
Fig. 5. Mutation Signatures and Relapse and Metastatic ES Tumors.
(A) Prevalence of mutation signatures in relapse and metastatic tumors. Shared and private mutations for four primary-metastatic or relapse pairs are shown (first four columns). Signatures 1 and 5 are common throughout, with signature 5 contributing significantly to the mutations that arise at relapse. Signature 8 was also common throughout the cohort. One metastatic tumor (no paired primary) is also shown to have similar mutation signature patterns as other metastatic/relapse tumors. Lastly, a secondary Ewing sarcoma tumor to a primary retinoblastoma (germline RB1 mutation identified) was also sequenced in this cohort. This patient harbored the rare Signature 31, which likely resulted from the patient’s prior exposure to carboplatin for their primary RB (only patient to receive this treatment in the Toronto cohort). (B) Phylogenetic trees of primary-relapse/metastatic ES. Using the shared and private mutations, we identified the mutational order in ES. Known cancer-driver mutations (IDH1, TP53 etc.) arise early (shared branches). (C) A clonal PTEN inversion. A PTEN inversion was found only in the primary and not in the relapse tissue, suggesting the inversion arose after early divergence of a common clonal ancestor. However, a pathogenic PTEN SNV can be found in the relapse tissue. Together, these point towards parallel, convergent evolution on this gene. (D) Proposed model of Ewing sarcoma tumor evolution. After birth, Signature 1 is operative in all somatic tissues throughout life. ES patients’ cells experience a replication-associated burst of rearrangements that generates the canonical fusion driver. Early somatic cancer gene mutations occur before clonal bifurcation. This occurs 1-2 years before an ES diagnosis, thus the cells that would give rise to the relapse existed years before diagnosis. Signature 5 contributes significantly to the number of mutations seen at relapse.
Early divergence and parallel evolution of Ewing sarcoma tumors
The most commonly held model for progression of cancer is that a metastasis originates directly from the primary tumor - it may have acquired new mutations but, since it derived by linear clonal evolution, most of the properties of the primary will be found in the metastasis (25). A different model was suggested in ES, proposing that the metastasis diverged early, based on mutation data from two primary-metastasis pairs whose exomes were sequenced, although the timing of this divergence was not established (8). We compared coding, non-coding and structural rearrangements across the genome within four ES pairs. As is the cases in most tumor types, relapse and metastatic ES tumors acquired many new mutations (average 50% private). A strikingly high number of clonal mutations from the primary were lost in the relapse (average 20%), confirming that the latter diverged early, evolving in parallel. For example, a disruptive clonal PTEN inversion was found in all tumor cells of one primary ES, but was absent from the relapse (Fig. 5C). We also confirmed the same model of parallel evolution in one additional primary-metastatic pair, profiled using microarrays (fig. S17). The clinical implication for this model is that one should also be searching for therapeutically targetable mutations arising in parallel to the primary ES, using methods like circulating tumor DNA, not necessarily in the primary tumor itself.
To determine when the divergence of the lethal clone occurred, we used the number of COSMIC Signature 1 mutations, which emerge at a steady rate in ES (see Methods and fig. S18). We first confirmed our approach by comparing the number Signature 1 mutations between established time intervals, such as the dates of diagnosis and recurrence. In all cases, the observed number of mutations was extremely close (75-90%) of what would be expected (fig. S18). Using the established rate, we calculated the amount of time between the divergence of the primary and relapse / metastatic tumors. Notably, the common ancestor in ES clonally diverges 1-2 years before diagnosis. Therefore, the cells that give rise to the primary and relapse tumor can exist in the patient years before diagnosis, providing a window for early cancer detection and surveillance. ES is often difficult to diagnose and time-to-diagnosis is notoriously long (26). These findings provide a plausible biological mechanism for this latency.
Discussion
Overall, our analyses have revealed rearrangement bursts (chromoplectic loops) as a source of gene fusion in human bone and soft tissue tumors. It is known that ES with complex karyotypes have worse prognosis, and here we show chromoplexy as the mechanism in 42% of tumors (27). It is possible that it is the chromoplectic tumor’s additional gene disruptions and fusions that contribute to this survival difference. Our whole genome data supports a model in which there is an early clone of ES, containing EWS-ETS and chromoplexy, arising at least 1 year pre-diagnosis, which gives rise to both the primary and metastatic or relapse tumors (Fig. 5D). Whether the bursts described here are chance events or driven by specific mutational processes, akin to RAG-machinery operative in leukemia, remains to be established. As an increasing and diverse number of tumor genome sequences become available, we may be able to define further rearrangement processes that underlie fusion genes and thus unravel the causes of fusion-driven human cancers.
Materials and Methods
Patient and sample collection
Ewing sarcoma tumor and matched blood samples were collected from the Hospital for Sick Children (SickKids) and Mount Sinai Hospital in Toronto, Canada in accordance with each institution’s Research Ethical Board (REB) guidelines. Detailed clinical information (age at presentation, gender, tumor site, stage, etc.) were obtained from the corresponding institutional tumor banks (table S5). Overall, the patients’ clinical features and demographics were typical of Ewing sarcoma: the average age of diagnosis was 14.8 years (2.8 to 36.6 yrs.); the male to female ratio was 1.38:1; and 14 patients had relapsed, with 13 having died from their disease. Additional samples (n=3) were also obtained from Universitätsklinikum Erlangen, Erlangen, Germany. All metastatic or relapse Ewing sarcoma tumors were collected from SickKids tumor bank or the SickKids clinical cancer sequencing program (KiCS). Detailed information on KiCS is available at https://www.kicsprogram.com.
Of the 25 high-coverage genomes sequenced, EWSR1-ETS fusions were detected in all patients except for a 37-year-old who was instead found to have a FUS-ERG translocation. This patient’s gene expression profile (by RNA-Seq) was also discrepant and so they were removed from subsequent analyses (fig. S19). One additional genome was removed due to poor sequencing quality. We also performed low pass (~10X) rearrangement screens on 19 ES samples. However, as we required breakpoint resolution, all but one of the rearrangement screens were excluded from this study due to insufficient coverage (see Table S1, orange row). Taken together, our discovery cohort consisted of 23 standard genomes (30-60X) and one rearrangement screen genome (20X). The validation cohort consisted of 119 tumor-normal samples sequenced by Tirode F. et al (10), which we downloaded from the European Genome-phenome Archive (accessions: EGAS00001000855 and EGAS00001000839). Of these, 19 patient samples were omitted either because the EWSR1-ETS fusion was not detected by our pipeline and manual inspection of the aligned reads, or because they harbored an excess of artefactual small inversions or deletions.
Code availability
Custom code described here is available at github.com/shlienlab
High-throughput sequencing and alignment
Exome, genome and transcriptome (RNA-Seq) sequencing were performed using established protocols on Illumina instruments. For exome and genomes, paired-end FASTQ files were aligned to the human genome (hg19/GRCh37) using BWA-MEM (v.0.7.8), Picard MarkDuplicates (v.1.108) was used to mark PCR duplicates. Indel realignment and base quality scores were recalibrated using the Genome Analysis Toolkit (v.2.8.1).
Detection of high quality somatic substitutions and rearrangements
We detected somatic mutations using established tools (MuTect2 (part of GATK v.3.5) (28) and Delly v.0.7.1 (29)). To evaluate and validate our WGS substitution pipeline, we used a “gold standard” cancer genome tumor/normal dataset, COLO829 (30). Using this somatic reference standard, we determined our precision to be 0.885 and our sensitivity to be 0.971. Copy-number was detected for genomes and rearrangement screens using BIC-seq v.1.2.1 (31). When no matched normal was available (in the case of rearrangement screens), an Ewing sarcoma normal was used. We then developed custom code to increase specificity of putative substitution and rearrangement detection, as follows:
Somatic and Depth Filter. No mutation should exist in the matched-normal sequence. For substitutions, we removed common single-nucleotide polymorphisms (SNPs) as previously described (32) and a required >10X coverage at the mutated locus (10 kb window), in both tumor and normal. For rearrangements, this filter required >= 4 discordant read-pairs in the tumor. We then directly interrogated the normal BAM file, at each putative somatic rearrangement; to ensure no germline variants existed near the breakpoint, on either side of the rearrangement.
Panel of Normals Filtering. To remove common germline variants, we created a panel of normal, non-neoplastic, samples that had been sequenced using the same technology and to a similar depth of coverage (n=133). We removed any putative substitutions or rearrangements if present in ≥ 2 normals. For rearrangements, breakpoints must exist on both sides of the junction within a 1 kb window. We found that as we increased the number of normals in our panel, our specificity increased (fig. S1C).
- Quality Control Filtering. Putative rearrangements were removed if supported by reads with MAPQ < 30. Both putative rearrangements and substitutions were also removed if they met any two of the following criteria:
- Non-unique mapping. <70% of the reads at the locus map uniquely.
- Multi-mapping clusters. At the same locus (200 bp up and downstream), a pattern of multiple overlapping groups of discordant reads whose paired-ends align to different chromosomes (> 3 reads in each group, mapping to > 4 chromosomes). Seen in both the tumor and paired normal.
- High depth. Excessively high depth alignments in difficult to align regions of the genome, as described (33). We apply a maximum depth threshold of d+4*sqrt (d), where d is the average normal mean read depth of the chromosome in the corresponding normal.
- Low-complexity regions. Overlap with a highly repetitive sequence (using DUST (34) with score > 60).
Mutation signature extractions and analysis
First, a de novo extraction was performed on the catalogue of Ewing sarcoma point mutations to produce novel consensus mutational signatures. These signatures were deciphered using a previously described computational framework that optimally explains the proportion of each mutation type found in the catalogue and then estimates the contribution of each signature to the mutation catalogue (11). Overall, we identified 11 consensus mutational signatures. 4 of these signatures were previously found to be attributed to sequencing artefacts. We then compared our true consensus mutational signatures to the previously curated COSMIC list and quantified their similarity using a cosine similarity as previously done (13). We report > 0.9 cosine similarity between the Ewing signatures and the COSMIC list.
Validation by targeted custom-capture sequencing
A custom targeted-capture enrichment system was designed to capture 1 Mb of DNA (Nextera, Illumina) with custom probes for the whole of EWSR1, FLI1, and ERG genes as well as the exons of TP53, STAG2 and ATRX. We also targeted known complex breakpoints from the discovery cohort, achieving between 900 to 1000-fold coverage. We reasoned that paired end sequencing would capture any locus joined to the three core genes, even if the panel did not specifically target it. In this way, we validated rearrangements in samples where chromoplexy was already known from the whole genome, and uncovered new instances in samples that had not been whole genome sequenced (n=7 and 4, respectively). Each tumor had three or four rearrangements validated using the panel. All had the same breakpoint (as found by the whole genome sequence) and were found to harbour looped rearrangements are on the same derivative chromosomes.
Validation by FISH, G-banding or Spectral Karyotyping
We further validated these looped rearrangements by karyotyping Ewing sarcomas using standard G-banding as well as spectral karyotyping (n=17 and 3; fig. S4). By cytogenetics we found additional complexity - beyond the canonical chr22-chr11 translocation - in eight cases. Of these, six tumors had been sequenced and found to be complex. Additionally, there were 5 cases for which chromoplexy was detected by genome sequencing yet not found by cytogenetics techniques, indicating that routine cytogenetics may miss chromosomal complexity present in these genomes due to the nature of these submicroscopic complexities (fig. S20).
Timing of rearrangements using breakpoint allele fraction
To determine the timing of the chromoplectic loops, we developed a tool to accurately measure the breakpoint allele fraction (BAF) of each rearrangement. The BAF is the proportion of reads containing a rearrangement breakpoint divided by the total number of reads, analogous to the variant allele fraction (VAF) for point mutations (illustrated in fig. S14A). This is analogous to the variant allele frequency of substitution mutations and, similarly, can be used to infer the relative order of rearrangement mutations. The tool accurately counts all reads supporting each rearrangement, even if these had not been used to nominate the rearrangement in the first place. From the raw aligned reads, we first collected all split reads near the breakpoint (within 20 bp) from one side of the rearrangement. Next, we extracted the clipped sequence (i.e. the non-aligned portion) from these reads and attempted to map it to the other side of the rearrangement (within 70 bp of the breakpoint) using a Smith-Waterman algorithm (35). Clipped sequences shorter than 5 bp were discarded, as were those that failed to map to the other side of the rearrangement (<= 80% similarity). Since the retained sequences can map at slightly different position, due to microhomology near the breakpoint, we considered all those close to one another as supportive of the same rearrangement. Overall, we found that most rearrangements are supported by re-mapped reads that less than 10 bp apart. Finally, the total number of split and realigned reads were divided by the average coverage between the two breakpoints per side of each rearrangement. This allowed us to arrive at an accurate measure of the breakpoint allele fraction. To validate our tool, we applied it to a curated list of known polymorphic copy number variants (CNVs) (36). As expected, the BAF of germline CNV deletions followed a bimodal distribution with peaks at 0.5 and 1.0, for heterozygous and homozygous rearrangements, respectively (fig. S14B, green line). We then compared the BAF of somatic rearrangements in chains to those without. Chained rearrangements had higher BAFs than simple structural variants (fig. S14B, red vs blue line), confirming that chromoplectic rearrangements are in fact earlier.
Detection of Breakpoint Clusters of Chained Rearrangements
Using their inter-breakpoint distance, we identified rearrangements within 10 kbp of one another. Using these, we created an undirected graph in which two rearrangement breakpoints within 10 kbp of one another (a breakpoint cluster) were represented as a vertex and connected to other breakpoint clusters (rearrangements are edges in the graph). We selected connected components of the graph, and identified components with greater than one vertex as inter-connected rearrangements. In most of our cases, these interconnected rearrangements formed chains or loops, where one could follow the edges around the graph and return to the initial vertex of departure. These were further filtered for reciprocal rearrangements or overlapping intra-chromosomal rearrangements. Chromoplexy rearrangements were validated by manual inspection and using the ChainFinder algorithm (2).
Association of Rearrangements with Genomic Features
We formally evaluated the association of rearrangement position with 38 properties of the human genomes (table S4). We separately evaluated these each of these associations in 1 kb bins across the genome. Feature density properties were calculated as densities in various sliding windows (1kb, 10 kb, 100kb, 1 MB) centered on each 1 kb bin or as the log2 distance, as indicated in table S4. The positions of Ewing sarcoma rearrangements were compared to million random positions that had been uniformly sampled from regions of the genome where confident genotypes could be determined (i.e. the “callable” genome). We limited our analysis to chromosomes 1 to 22 and X. To test for significant associations between our rearrangements and these genomic properties, we performed a Mann Whitney U test and Benjamin and Hochberg FDR correction to raw p-values. We used the Cohen’s d metric to determine the effect size between the two groups to account for differences in sample size. We applied an absolute Cohen’s d cut-off of 0.3, a medium effect size (37, 38). Genomic properties were considered significantly different between rearrangements and random positions if absolute (d) >= 0.3 and the corrected p < 0.05.
Detection of Gene Fusions
We detected gene fusions in regions of genomic complexity using an approach that integrates multiple independent fusion algorithms, and then removes those found in normal tissue (Fuligni et al., Under preparation). Putative fusions were validated by de novo assembly. A total of 1277 normal (non-neoplastic) samples from 43 different tissues were obtained from the NHGRI GTEx consortium (database version 4) and used to remove artefacts. All fusions were visually inspected if one or both genes involved chromoplexy or were adjacent (up to 1 Mbp). Fusions were further filtered by quality of the realigned transcript, breakpoint coverage and gene expression.
Detection of Gene Expression
Gene expression for fusions, differential gene expression analysis, and principal component analysis was performed using HT-Seq (39) to count the reads aligning to every gene. PCR duplicates and reads mapping to ribosomal RNA, miRNA and small nucleolar RNA were removed. We used Trimmed Mean of M-value (TMM) method in the EdgeR package to perform normalization on genes with at least 1 read per million bases in at least 3 samples (40, 41). Differential expression analysis in chromoplexy vs non-chromoplexy samples was performed using a Generalized Linear Model (GLM) likelihood ratio test, taking in consideration different sources of variation like batch, gender and age. P-values for GLM test were adjusted for multiple testing using the Benjamini and Hochberg method for controlling the False Discovery Rate (FDR). Differentially expressed genes in chromoplexy vs non-chromoplexy were considered statistically significant if FDR <= 0.05 and absolute value of log(Fold Change) >=1. Pathway analysis was performed on genes differentially expressed in samples with and without chromoplexy using Gene Set Enrichment Analysis (GSEA) software (javaGSEA v2.2.4). Cancer gene signatures were selected from the hallmark collection from the Molecular Signature DataBase (MsigDB)(18). Enrichment scores for the hallmark pathways were considered statistically significant if FDR < 0.01.
Evaluation of Replication Timing in Prostate Cancer Rearrangements
We obtained prostate cancer rearrangements, including chained and others, from the Baca et al. publication (Supplemental table S3C and S5 from (2)). Samples were annotated as ‘ETS+’ or ‘ETS-’ using Supplementary Table 1. ETS+ fusions include any ETS fusion detected by sequencing (including ERG and ETV1). Using this list we performed the same test for genomic property enrichment as we did in Ewing sarcomas.
Molecular Inversion Probe (MIP) Microarray
Raw MIP data from three additional primary-metastatic ES pairs were obtained from the Huntsman Cancer Institute, Salt Lake City, Utah (42). The original source material was clinically-archived, formalin-fixed paraffin-embedded (FFPE) scrolls that were retrieved from 3 individual patients diagnosed with ES. Primary tumor samples were from diagnostic biopsies prior to chemotherapy. The raw MIP data from the completed assay was loaded into Nexus Copy Number (BioDiscovery, Inc., El Segundo, CA) for copy number detection using default settings.
Supplementary Material
One Sentence Summary.
Disease-defining fusions in sarcoma frequently emerge by rearrangement burst, creating complex genomic loops and disrupting additional genes, in an early clone that may develop multiple years before diagnosis.
Acknowledgments
This manuscript is dedicated to:
Simon Hajjar, a brilliant and fearless friend, colleague and student in our lab, who died of sarcoma.
Ana Novokmet, a friend and colleague with a big heart who held all of us to high standards and was committed to helping children with cancer. Ana died of a brain tumor.
We thank the Centre for Applied Genomics (TCAG) NGS and Biobanking facility for sequencing services. We thank Jodi Lees, Noa Alon, Dr. Grace Collord, and Dr. Roland Arnold for their contributions towards this work.
Funding:
A.S. and D.M. received financial support from the C17 Ewings Cancer Foundation of Canada Grant and the SickKids Garron Family Cancer Centre. N.D.A is personally supported by an NSERC Canada Graduate Scholarship (M) and a SickKids Restracomp Award.
Footnotes
Author Contributions:
A.S., S.B., and D.M. designed the study. N.D.A., R.D., M.D.Y., M.L., A.R., N.D.R., F.F., S.H., P.E.K., B.I., L.C.S., R.M., and J.S. performed experiments. N.D.A. R.D., M.Y., A.R., F.F., M.A., M.S., and L.B.A.collected and analyzed data. L.B., S.D., N.G., A.N., A.F., N.P., A.Y., T.S., M.M., G.R.S., S.W.S., A.M.F., M.S., J.S.W., I.L.A., P.J.C., J.D.S., D.M., S.B and A.S. contributed reagents, tissue and clinical data. N.D.A., A.S., and S.B. wrote the manuscript. M.Z., P.E.K., G.T.G, J.A.T., M.S., I.L.A., D.M., S.B and A.S. provided technical support and conceptual advice. A.S. oversaw the study. All authors have approved the manuscript.
Competing Interests
The authors declare no competing interests.
Data and materials availability
Raw sequencing data has been deposited at the European Genome-phenome Archive (EGA) under accession number EGAS00001003062.
References and Notes
- 1.Stephens PJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baca SC, et al. Punctuated evolution of prostate cancer genomes. Cell. 2013;153:666–677. doi: 10.1016/j.cell.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7:233–245. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- 4.Papaemmanuil E, et al. RAG-mediated recombination is the predominant driver of oncogenic rearrangement in ETV6-RUNX1 acute lymphoblastic leukemia. Nat Genet. 2014;46:116–125. doi: 10.1038/ng.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sankar S, Lessnick SL. Promiscuous partnerships in Ewing's sarcoma. Cancer Genet. 2011;204:351–365. doi: 10.1016/j.cancergen.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kovar H. Blocking the road, stopping the engine or killing the driver? Advances in targeting EWS/FLI-1 fusion in Ewing sarcoma as novel therapy. Expert Opin Ther Targets. 2014;18:1315–1328. doi: 10.1517/14728222.2014.947963. [DOI] [PubMed] [Google Scholar]
- 7.Brohl AS, et al. The genomic landscape of the Ewing Sarcoma family of tumors reveals recurrent STAG2 mutation. PLoS Genet. 2014;10:e1004475. doi: 10.1371/journal.pgen.1004475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crompton BD, et al. The genomic landscape of pediatric Ewing sarcoma. Cancer Discov. 2014;4:1326–1341. doi: 10.1158/2159-8290.CD-13-1037. [DOI] [PubMed] [Google Scholar]
- 9.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tirode F, et al. Genomic landscape of Ewing sarcoma defines an aggressive subtype with co-association of STAG2 and TP53 mutations. Cancer Discov. 2014;4:1342–1353. doi: 10.1158/2159-8290.CD-14-0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alexandrov LB, Nik-Zainal S, Wedge DC, Campbell PJ, Stratton MR. Deciphering signatures of mutational processes operative in human cancer. Cell Rep. 2013;3:246–259. doi: 10.1016/j.celrep.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alexandrov LB, et al. Clock-like mutational processes in human somatic cells. Nat Genet. 2015;47:1402–1407. doi: 10.1038/ng.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zoubek A, et al. Variability of EWS chimaeric transcripts in Ewing tumours: a comparison of clinical and molecular data. Br J Cancer. 1994;70:908–913. doi: 10.1038/bjc.1994.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hattinger CM, et al. Prognostic impact of chromosomal aberrations in Ewing tumours. Br J Cancer. 2002;86:1763–1769. doi: 10.1038/sj.bjc.6600332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neilsen PM, Pishas KI, Callen DF, Thomas DM. Targeting the p53 Pathway in Ewing Sarcoma. Sarcoma. 2011;2011 doi: 10.1155/2011/746939. 746939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cooper CS, et al. Analysis of the genetic phylogeny of multifocal prostate cancer identifies multiple independent clonal expansions in neoplastic and morphologically normal prostate tissue. Nat Genet. 2015;47:367–372. doi: 10.1038/ng.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liberzon A, et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nord KH, et al. GRM1 is upregulated through gene fusion and promoter swapping in chondromyxoid fibroma. Nat Genet. 2014;46:474–477. doi: 10.1038/ng.2927. [DOI] [PubMed] [Google Scholar]
- 20.Malhotra A, et al. Breakpoint profiling of 64 cancer genomes reveals numerous complex rearrangements spawned by homology-independent mechanisms. Genome Res. 2013;23:762–776. doi: 10.1101/gr.143677.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Korbel JO, Campbell PJ. Criteria for inference of chromothripsis in cancer genomes. Cell. 2013;152:1226–1236. doi: 10.1016/j.cell.2013.02.023. [DOI] [PubMed] [Google Scholar]
- 22.Hansen RS, et al. Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proc Natl Acad Sci U S A. 2010;107:139–144. doi: 10.1073/pnas.0912402107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sima J, Gilbert DM. Complex correlations: replication timing and mutational landscapes during cancer and genome evolution. Curr Opin Genet Dev. 2014;25:93–100. doi: 10.1016/j.gde.2013.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Merlevede J, et al. Mutation allele burden remains unchanged in chronic myelomonocytic leukaemia responding to hypomethylating agents. Nat Commun. 2016;7 doi: 10.1038/ncomms10767. 10767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gray JW. Evidence emerges for early metastasis and parallel evolution of primary and metastatic tumors. Cancer Cell. 2003;4:4–6. doi: 10.1016/s1535-6108(03)00167-3. [DOI] [PubMed] [Google Scholar]
- 26.Brasme JF, Chalumeau M, Oberlin O, Valteau-Couanet D, Gaspar N. Time to diagnosis of Ewing tumors in children and adolescents is not associated with metastasis or survival: a prospective multicenter study of 436 patients. J Clin Oncol. 2014;32:1935–1940. doi: 10.1200/JCO.2013.53.8058. [DOI] [PubMed] [Google Scholar]
- 27.Roberts P, et al. Ploidy and karyotype complexity are powerful prognostic indicators in the Ewing's sarcoma family of tumors: a study by the United Kingdom Cancer Cytogenetics and the Children's Cancer and Leukaemia Group. Genes Chromosomes Cancer. 2008;47:207–220. doi: 10.1002/gcc.20523. [DOI] [PubMed] [Google Scholar]
- 28.Cibulskis K, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rausch T, et al. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012;28:i333–i339. doi: 10.1093/bioinformatics/bts378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Craig DW, et al. A somatic reference standard for cancer genome sequencing. Sci Rep. 2016;6:24607. doi: 10.1038/srep24607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xi R, et al. Copy number variation detection in whole-genome sequencing data using the Bayesian information criterion. Proc Natl Acad Sci U S A. 2011;108:E1128–1136. doi: 10.1073/pnas.1110574108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shlien A, et al. Combined hereditary and somatic mutations of replication error repair genes result in rapid onset of ultra-hypermutated cancers. Nat Genet. 2015;47:257–262. doi: 10.1038/ng.3202. [DOI] [PubMed] [Google Scholar]
- 33.Li H. Toward better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics. 2014;30:2843–2851. doi: 10.1093/bioinformatics/btu356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morgulis A, Gertz EM, Schaffer AA, Agarwala R. A fast and symmetric DUST implementation to mask low-complexity DNA sequences. J Comput Biol. 2006;13:1028–1040. doi: 10.1089/cmb.2006.13.1028. [DOI] [PubMed] [Google Scholar]
- 35.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 36.Zarrei M, MacDonald JR, Merico D, Scherer SW. A copy number variation map of the human genome. Nat Rev Genet. 2015;16:172–183. doi: 10.1038/nrg3871. [DOI] [PubMed] [Google Scholar]
- 37.Cohen J. The statistical power of abnormal-social psychological research: a review. J Abnorm Soc Psychol. 1962;65:145–153. doi: 10.1037/h0045186. [DOI] [PubMed] [Google Scholar]
- 38.Cohen J. A power primer. Psychol Bull. 1992;112:155–159. doi: 10.1037//0033-2909.112.1.155. [DOI] [PubMed] [Google Scholar]
- 39.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jahromi MS, et al. Molecular inversion probe analysis detects novel copy number alterations in Ewing sarcoma. Cancer Genet. 2012;205:391–404. doi: 10.1016/j.cancergen.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pilati C, et al. Mutational signature analysis identifies MUTYH deficiency in colorectal cancers and adrenocortical carcinomas. J Pathol. 2017;242:10–15. doi: 10.1002/path.4880. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.