Abstract
Store-operated Ca2+ entry (SOCE) constitutes a major Ca2+ influx pathway in mammals to regulate a myriad of physiological processes, including muscle contraction, synaptic transmission, gene expression, and metabolism. In non-excitable cells, the Ca2+ release-activated Ca2+ (CRAC) channel, composed of ORAI and stromal interaction molecule (STIM), represents a prototypical example of SOCE to mediate Ca2+ entry at specialized membrane contact sites (MCSs) between the endoplasmic reticulum (ER) and the plasma membrane (PM). The key steps of SOCE activation include the oligomerization of the luminal domain of the ER-resident Ca2+ sensor STIM1 upon Ca2+ store depletion, subsequent signal propagation toward the cytoplasmic domain to trigger a conformational switch and overcome the intramolecular autoinhibition, and ultimate exposure of the minimal ORAI-activating domain to directly engage and gate ORAI channels in the plasma membrane. This exquisitely coordinated cellular event is also facilitated by the C-terminal polybasic domain of STIM1, which physically associates with negatively charged phosphoinositides embedded in the inner leaflet of the PM to enable efficient translocation of STIM1 into ER-PM MCSs. Here, we present recent progress in recapitulating STIM1-mediated SOCE activation by engineering CRAC channels with optogenetic approaches. These STIM1-based optogenetic tools make it possible to not only mechanistically recapture the key molecular steps of SOCE activation, but also remotely and reversibly control Ca2+-dependent cellular processes, inter-organellar tethering at MCSs, and transcriptional reprogramming when combined with CRISPR/Cas9-based genome-editing tools.
Keywords: Optogenetics, Ion channel, ORAI, Stromal interaction molecule, CRISPR/Cas9, Membrane contact sites
Graphical Abstract:
1. STIM1 and ORAI1 as two major players in the SOC field
Calcium ions (Ca2+) participate in various aspects of cellular activity and act as versatile chemical signals to control a myriad of biological processes, ranging from short-term responses such as muscle contraction, exocytosis, and synaptic transmission, to long-lasting effects on gene expression, metabolism, cell division, and cell death [1, 2]. The fact that binding Ca2+ to its targets can alter local electrostatic fields and trigger changes in protein conformation makes it a well-suited second messenger for signal transduction [3, 4]. Ca2+ influx is mediated by distinct Ca2+ channels, including voltage-gated, ligand-gated, store-operated and second messenger-operated channels [1, 2]. In excitable cells such as neurons and cardiomyocytes, extracellular Ca2+ flows into the cytosol through the activation of voltage-gated Ca2+ channels [5, 6], whereas in non-excitable cells, store-operated Ca2+ entry (SOCE) constitutes a major route of Ca2+ influx from the extracellular space into the cytoplasm [7-13]. SOCE is regarded as a unique pathway that connects the Ca2+ store depletion in the endoplasmic reticulum (ER) with the subsequent opening of highly selective Ca2+ channels in the plasma membrane (PM) [7]. SOCE is best exemplified by the Ca2+ release-activated Ca2+ channels (CRAC) that are initially characterized in cells of the immune system [14]. Rare gene alleles that compromise CRAC channel expression or activation (e.g., R91W) can lead to severe combined immunodeficiency in human patients [15-18]. Furthermore, gain-of-function mutations in the CRAC channel complex are regarded as the genetic culprits for Stormorken syndrome and tubular aggregate myopathy [19-21]. Aberrant CRAC channel activity is also implicated in tumorigenesis, cancer metastasis, and cardiovascular disorders [13, 22, 23].
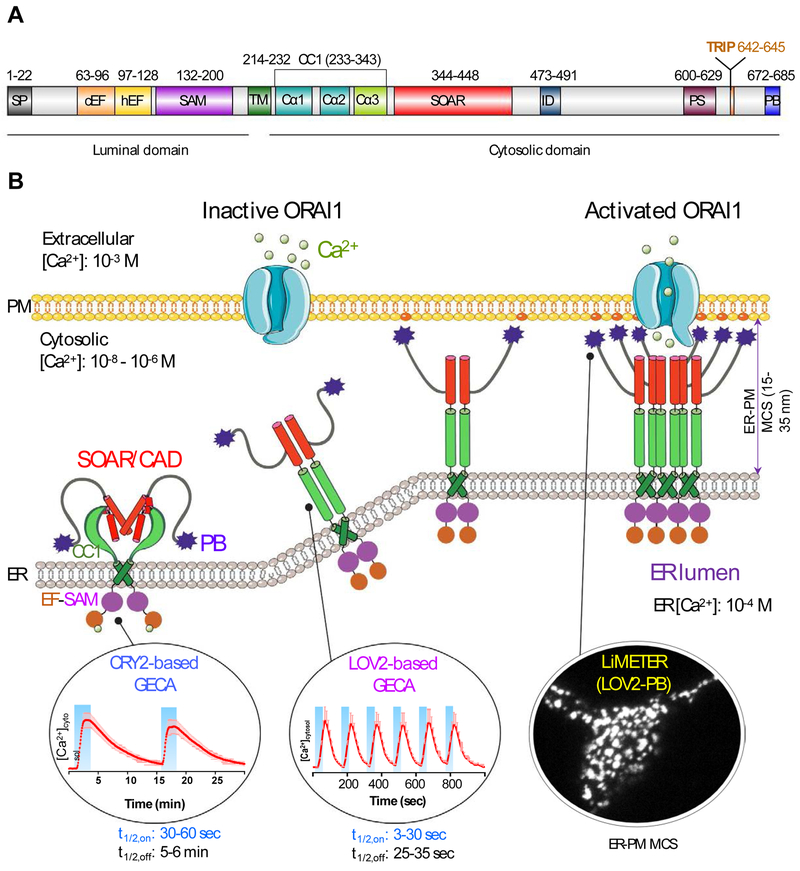
Store-operated CRAC channels are composed of a class of four-pass transmembrane proteins known as ORAI (named after the keepers of heaven's gate in Greek mythology; with three homologs ORAI1, ORAI2, and ORAI3), which function as pore-forming subunits [15, 24, 25]. ORAI proteins are directly gated by type I single-pass transmembrane proteins termed stromal interaction molecules (STIM1 and STIM2) [26-28]. As an ER-resident Ca2+ sensor and activator of ORAI Ca2+ channels, STIM1 contains an ER-luminal domain, a single transmembrane domain (TM), and a cytoplasmic domain (CT). The ER-luminal domain, consisting of EF-hand motifs and a sterile alpha motif (SAM) domain, is responsible for sensing ER Ca2+ fluctuation and initiating Ca2+-depletion induced oligomerization [29, 30]. The TM domain transduces ER luminal signals [31, 32] toward the cytoplasmic domain that comprises a putative coiled-coil region (CC1), a minimal ORAI-activating region (SOAR or CAD or OASF; see abbreviations) [33-35], a Pro/Ser-rich region, an EB1-binding S/TxIP motif, and a C-terminal polybasic (PB) tail (Figure 1A). Following the identification of these two protein families as the molecular identities of the two-component CRAC channel complex, the critical steps and regulatory mechanisms involved in coordinating the exquisite molecular choreography of SOCE have been largely worked out through collective efforts from multiple groups (Figure 1B) [10-13, 36-38].
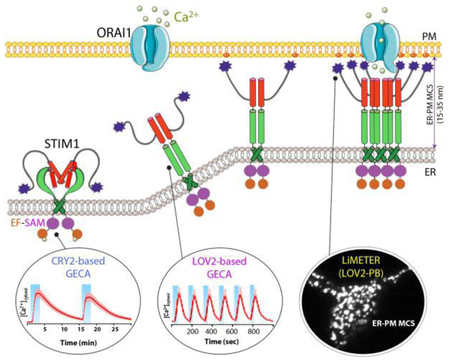
Figure 1 ∣. Engineering STIM1 to convert SOCE at ER-PM membrane contact sites (MCSs) into light-operated Ca2+ entry.
(A) Schematics of the domain architecture of human STIM1. The luminal domain of STIM1 contains a signal peptide (SP), a hidden non-Ca2+ binding EF-hand motif (hEF), a canonical Ca2+ binding EF-hand motif (cEF), and a sterile alpha motif domain (SAM). The cytoplasmic domain comprises a putative coiled-coil region (CC1), a minimal STIM1 ORAI-activating region (SOAR or CAD or OASF), an inactivation domain (ID), a proline/serine-rich region (PS), an EB1-binding sequence (TRIP), and a C-terminal polybasic tail (PB).
(B) Cartoon illustration of the dynamic coupling between STIM1 and ORAI1 during SOCE activation at ER-PM apposition, a specialized membrane contact site that is separated by a distance of approximately 25-35 nm without membrane fusion. The major steps in this tentative model include: (i) Ca2+ depletion induces oligomerization of the luminal EF-SAM domain to initiate STIM1 activation; (ii) the luminal signal is transduced toward the cytoplasmic side to overcome STIM1 autoinhibition mediated by the intramolecular CC1-SOAR interaction, thereby triggering a conformational switch to expose SOAR/CAD/OASF and the C-terminal PB domain; (iii) activated STIM1 undergoes further oligomerization and its subsequent migration toward the PM is facilitated by the association between the positively charged PB domain and PM-embedded, negatively-charged phosphoinositides. SOAR/CAD/OASF is responsible for directly engaging and activating ORAI channels to mediate Ca2+ flux from the extracellular space into the cytosol. When coupled to plant-derived photosensory domains, such as cryptochrome 2 (CRY2) and light-oxygen-voltage domain 2 (LOV2), these critical steps can be individually mimicked with engineered Opto-CRAC constructs (CRY2-STIM1 chimeras or LOV2-STIM1 chimeras) and ER-tethered LOV2-PB proteins (designated LiMETER for light-inducible membrane-tethered peripheral ER). Representative Ca2+ signals generated by Opto-CRAC variants following repeated light-dark cycles are shown with the circles, along with a typical image showing the light-induced assembly of ER-PM MCSs in HeLa cells revealed by total internal reflection fluorescence (TIRF) microscopy. These constructs are available at Addgene (accession ID: #101245, #101246, #113933 and #113934).
SOCE activation involves a dynamic intermembrane coupling between STIM1 and ORAI1 that culminates at ER-plasma membrane contact sites (MCSs), a specialized subcellular structure that is separated by a distance of approximately 15-35 nm [39, 40]. When the ER Ca2+ store remains full at rest, the full-length STIM1 presumably exists as a dimer or a lower-order oligomer maintained by its cytosolic region [41-44]. The Ca2+-bound STIM1 luminal domain, when isolated in vitro by itself, stays as a monomer, with the aggregation-prone SAM domain fitting into the hydrophobic EF-hand cleft to form a compact structure [30, 45]. At the cytosolic side, the intramolecular coiled-coil clamp formed through CC1-SOAR interactions locks the cytoplasmic domain of STIM1 (STIM1-CT) in an inactive conformation [31, 32, 46], thereby preventing SOAR/CAD/OASF from encountering ORAI channels that would consequently lead to a constitutive Ca2+ influx [33-35]. Upon ER Ca2+ store depletion, Ca2+ release from the ER Ca2+ store leads to the dissociation of Ca2+ from EF-hand motifs, which induces EF-SAM oligomerization and stabilizes the conformation as an entropy-favored transition [29, 30]. Subsequently, the ER luminal signals are transmitted toward the STIM1-CT via the rearrangement of the TM domains [32], which prompts a conformational switch in the cytosolic juxta-membrane region and weakens CC1-SOAR association [31, 32, 46]. As a result, the minimal ORAI-activating domain, SOAR or CAD, is fully exposed and further oligomerized STIM1 molecules migrate toward the ER-plasma membrane contact sites to directly recruit and activate ORAI1 channels [33-35, 43]. Given the relatively weak strength of STIM1-ORAI1 interactions under physiological conditions, the redistribution of STIM1 towards the PM is further facilitated by the physical association between its own C-terminal PB domain with PM-embedded phophoinositides (PIs) [39, 44, 47, 48]. Sustained Ca2+ influx is required to further activate downstream effectors such as calcineurin, a Ca2+/calmodulin-dependent phosphatase that dephosphorylates a master transcription factor, the nuclear factor of activated T-cells (NFAT). This process ultimately leads to NFAT nuclear import, which turns on key genes essential for lymphocyte activation [49, 50].
During the course of unraveling of STIM-ORAI coupling at ER-plasma membrane contact sites, a combination of traditional biochemical, pharmacological, structural and genetic approaches has been employed [30, 42, 51-56]. In this review, we aim to reconstruct the molecular choreography of SOCE by taking an unconventional optogenetic approach. Genetically-encoded photoswitches, such as the light-oxygen-voltage domain 2 (LOV2) from Avena sativa [57, 58] and cryptochrome 2 (CRY2) from Arabidopsis thaliana [59, 60], can be installed into STIM1 to mimic critical steps of CRAC channel activation, including STIM1 oligomerization, conformational switch, and ER-PM junction formation (Figures 1-3). We highlight herein the latest progress in this endeavor, with a concentration on the design and use of genetically-encoded Ca2+ actuators (GECAs; OptoSTIM1, Opto-CRAC, and BACCS) derived from STIM1 [61-63] and optical tethers (LiMETER [64] or OptoPBer [48]) that enable real-time photo-inducible ER-PM MCS assembly in living cells. We hope that this review will stimulate further thoughts on incorporating optogenetic approaches towards efforts to aid the mechanistic dissection of cell signaling and spur the development of next-generation optogenetic actuators for both research and therapeutic purposes.
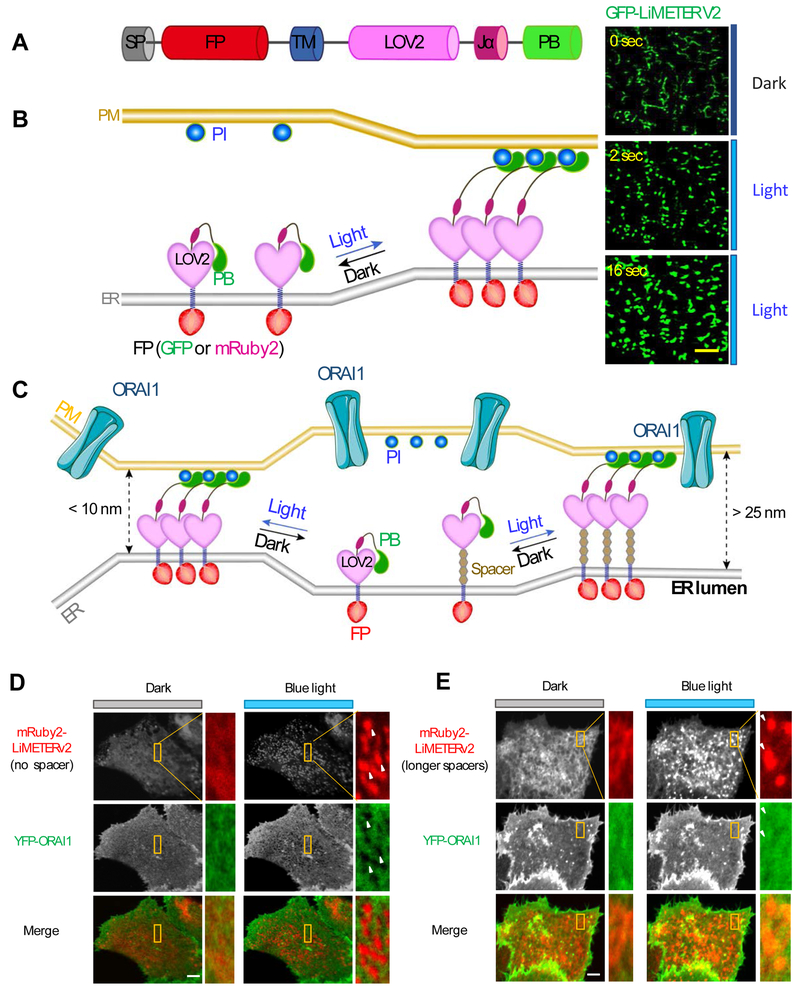
Figure 3 ∣. Mimicking reversible ER-PM MCS formation with LiMETER.
(A) LiMETER is engineered from STIM1 by replacing the luminal EF-SAM with a fluorescent protein (FP) and substituting the majority of its cytosolic domain with the LOV2 photosensitive module. The ER-targeting signal peptide (SP) and transmembrane domain (TM) were used to retain the synthetic protein to ER, whereas the C-terminal PB affords the tethering function through protein-phosphoinositide (PI) interaction. In the prototypical design, PB is derived from the Rit GTPase. In a recently improved version (also termed as OptoPBer or LiMETERv2), the PB is isolated from STIM1.
(B) Schematic representation of the working principle of LiMETER. In the dark, the Jα helix tightly docks to the core body of LOV2, thus imposing steric hindrance to the C-terminally fused PB domain to prevent its association with PM-resident Pis. Upon light illumination, the Jα helix unwinds and releases the constraints on PB, allowing itself to bind to the Pis to bridge the gap between ER and PM apposition. Shown on the right are typical images of GFP-tagged LiMETERv2 under a TIRF microscope following blue light irradiation.
(C) LiMETER can be engineered to manipulate the gap distance between ER-PM MCSs. Spacers with varying lengths can be modularly inserted between the TM and LOV2 domains to bridge a range of gap distances at MCSs. A gap distance of over 25 nm is required to allow efficient diffusion of ORAI1 proteins into MCS to evoke Ca2+ influx.
(D-E) Representative images of HeLa cells co-expressing YFP-ORAI (green) and mRuby-LiMETERv2 (red) with no spacer (D) or a long spacer (E). The yellow boxed regions were enlarged for a better visualization of the relative distribution of the two components in the same cell. YFP-ORAI1 was excluded from ER-PM MCSs (arrows) marked by mRuby-LiMETERv2 in the absence of the spacer (D). By contrast, YFP-ORAI1 can freely diffuse into ER-PM MCSs labeled by mRuby-LiMETERv2 with an appropriate spacer (E).
2. CRY2-STIM1 chimeras to mimic inducible oligomerization of STIM1 luminal domain
Given that the dissociation of Ca2+ from the canonical EF-hand motif can trigger STIM1 oligomerization and cause CRAC channels activation [29], Luik et al. speculated that replacing the EF-SAM domain with a chemical-inducible dimerization system could similarly activate STIM1 to open ORAI channels without store depletion [53]. To test this hypothesis, they substituted the luminal Ca2+-sensing domain of STIM1 with either the FKBP-rapamycin binding (FRB) domain or the FK506 binding protein (FKBP) [65]. Indeed, the addition of rapamycin prompted the rapid heterodimerization of FKBP-STIM1/FRB-STIM1 chimeric proteins, which was sufficient to drive STIM1 puncta formation and CRAC channel activation as seen with wild-type STIM1 upon store depletion [26-28]. This elegant study demonstrated the feasibility of bypassing store depletion to activate STIM1 with a chemical approach. In an independent study, Zhou et al. showed that covalently crosslinking STIM1-CT at residue 233 (where the cytosolic portion of STIM1 emerges from the ER membrane) to bring the N-terminus of CC1 region into close proximity could switch on STIM1-CT to adopt an active conformation [44]. These findings have paved the way for the employment of plant CRY2-based optical multimerizers to reversibly control STIM1 oligomerization and ultimately generate Ca2+ influx upon photostimulation.
Arabidopsis cryptochromes (AtCRY) belong to a family of evolutionarily conserved flavoproteins (with flavin adenine dinucleotide as chromophore; Figure 2A) that are homologous to DNA photolyases [66-68]. AtCRY2 controls photomorphogenesis in response to blue light in plants [68]. The activation of CRY2 is driven by light-induced electron transfer followed by flavin photoreduction [69]. CRY2, which rapidly undergoes monomer-to-oligomer clustering upon blue light illumination, has been successfully used to manipulate protein oligomerization-dependent cell signaling [70, 71], including growth factor receptor activation and Ca2+ signaling. Compared to traditional chemical-based methods, the optogenetic approach offers two obvious advantages. First, the use of chemicals is associated with undesirable effects in living cells such as cytotoxicity and undesired perturbation to host cells due to potential off-target effects. Optogenetics can circumvent these caveats since only a simple pulse of visible light is needed to achieve similar functions. Second, the use of light makes it possible to manipulate signaling at a high spatial and temporal resolution and meet various kinetic requirements.
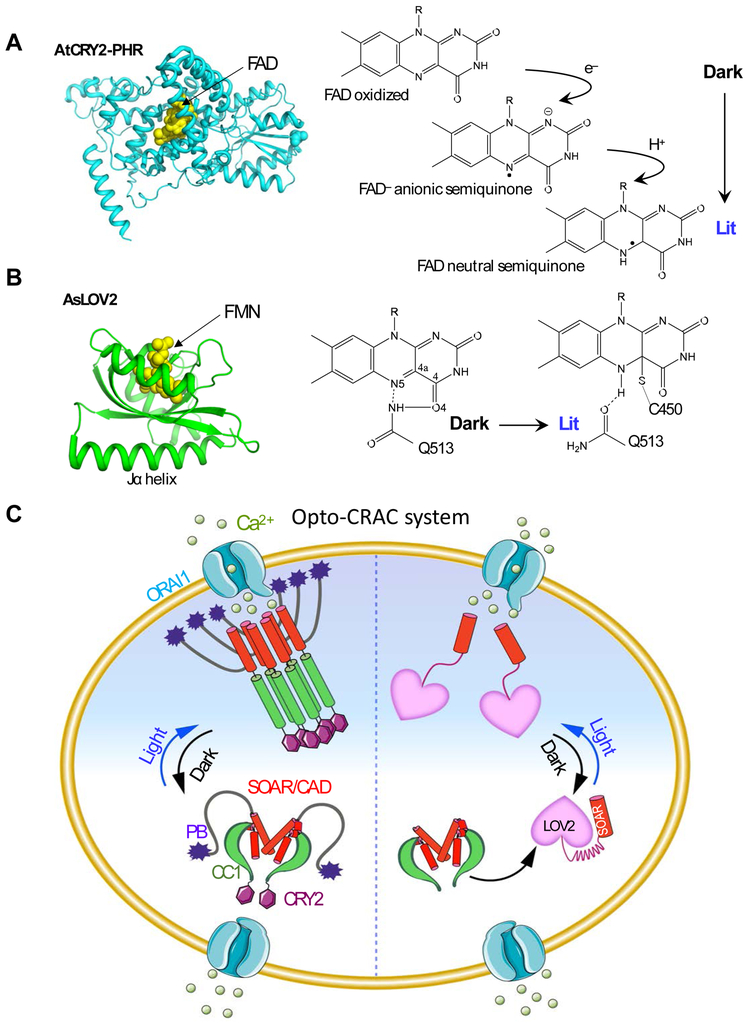
Figure 2 ∣. Opto-CRAC for optical control of Ca2+ influx in mammalian cells.
(A-B) The 3D structures and simplified photocycle reactions of AtCRY2-PHR (modeled from CRY1; PDB entry: 1U3D; A) and AsLOV2 (PDB entry: 2V0W; B). FAD, flavin adenine dinucleotide; FMN: flavin mononucleotide.
(C) The left semi-circle depicts an Opto-CRAC construct made of a CRY2-STIM1 fusion. Upon light illumination, CRY2 undergoes oligomerization, mimicking Ca2+-depletion induced EF-SAM multimerization, to trigger STIM1 activation with subsequent opening of ORAI Ca2+ channels situated in the plasma membrane. The right semi-circle illustrates the engineering strategy of an alternative form of Opto-CRAC, in which the CC1-SOAR interaction-mediated intramolecular autoinhibition is recapitulated by LOV2-SOAR fusion in the dark. Following photostimulation, LOV2 undergoes rapid conformational changes to expose the C-terminal effector domain, thereby restoring the potent ORAI-activating activity of SOAR to elicit Ca2+ influx.
To enable the light-dependent oligomerization of STIM1, STIM1-CT (residues 238-685 [63] or 233-685 [72]) was fused after the N-terminal photolyase homology region of AtCRY2 (termed OptoSTIM1). When expressed in mammalian cells, OptoSTIM1 largely remained inactive in the dark but efficiently translocated toward the plasma membrane to evoke Ca2+ influx through endogenous ORAI channels upon blue light illumination (t1/2, on: ~30-60 sec; t1/2, off: ~5-6 min; Figures 1B). OptoSTIM1-mediated photoactivatable Ca2+ entry has been confirmed in a number of mammalian cells originating from different tissues (e.g., HeLa, HEK293, NIH3T3, COS-7 cells, astrocyte, human umbilical vein endothelial cells, and human embryonic stem cells), as well as in zebrafish embryos and the CA1 hippocampus of mice [63]. These findings, along with results obtained using the aforementioned chemical approaches [44, 53], have reinforced the notion that chemical or optogenetic manipulation is used to force close apposition of the juxtamembrane end of CC1, which likely phenocopies the consequence of Ca2+ depletion-induced EF-SAM luminal domain oligomerization, can activate STIM1 to open ORAI channels.
3. LOV2-STIM chimeras to mimic conformational switch within STIM1 cytoplasmic domain
The SOAR/CAD domain within STIM1-CT, when expressed alone, can fully engage and potently activate ORAI channels [33-35]. By contrast, longer STIM1-CT fragments, particularly those bearing the upstream CC1 region (residues 233-343), seem to act as substantially weaker ORAI activators [33-35]. This early observation led to the speculation that STIM1-CT might be kept in a quiescent state by self-caging the SOAR domain through an intramolecular autoinhibitory mechanism. Korzeniowski et al. claimed that a stretch of acidic amino acids within CC1 (residues 318-322; EEELE) might form electrostatic interactions with a polybasic region (residues 382-386; KIKKKR) in SOAR/CAD, thereby keeping the SOAR/CAD domain buried to prevent ORAI1 activation [73]. Neutralizing the acidic regions within CC1 (the 4EA mutant, EEELE>AAALA) was able to switch on STIM1-CT to cause a constitutive activation of SOCE [73]. Although the proposed physical contacts were not observed in a STIM1-ORAI1 complex structure that included both the acidic and basic regions of STIM1-CT (residues 312-387) [74], the notion of intramolecular conformational switch continued to resonate in the field and inspired follow-up studies to unveil the underlying molecular determinants [31, 32, 41, 44, 46, 75, 76].
To gain a more quantitative view on the conformational rearrangements within STIM1-CT, Muik et al. developed a fluorescence resonance energy transfer (FRET)-based biosensor that detected the folding status of a STIM1-CT fragment (YFP-OASF-CFP; residues 233-474) [41]. Cells expressing the wild-type YFP-OASF-CFP exhibited robust FRET signals, suggesting that OASF adopts a relatively compact structure in close proximity to the donor and acceptor fluorophores in space. In contrast, the introduction of constitutively-activating STIM1 mutations into the initial segment of CC1 (L251S) or the ending helix of SOAR (L416S/L423S) led to a substantial reduction in FRET signals, implying the existence of an intramolecular clamp that locks STIM1 in an inactive conformation at rest [41]. This finding was further corroborated by an independent study using recombinant proteins, in which Zhou et al. used Tb3+-based luminescence resonance energy transfer to demonstrate the existence of distinct conformations in WT and activated (L251S) STIM1-CT molecules [44]. Furthermore, artificial crosslinking of the CC1 domain could trigger STIM1-CT to adopt a more extended configuration [44]. These findings provide compelling evidence to support the conformational switch hypothesis underlying STIM1 activation. To map out key residues accounting for the intramolecular STIM1 autoinhibition, Ma et al. invented a two-component FRET-based assay by splitting STIM1 into two parts: Part I was composed of the EF-SAM-TM-CC1 domains that reside at the ER membrane (residues 1-310 or 1-342; with CFP attached to the C-terminus as donor), and Part II contained SOAR or its C-terminal extension variants (with YFP fused to the N-terminus as an acceptor) [31, 32]. When expressed alone, YFP-SOAR is known to be evenly distributed in the cytosol with partial decoration on the PM due to its interaction with endogenous levels of ORAI channels [33]. However, in the presence of part I, YFP-SOAR tightly docked toward the ER membrane, thereby confirming the interaction between CC1 and SOAR domains as part of the autoinhibitory mechanism proposed in earlier studies [41, 44, 73]. Through a series of truncation and deletion studies, Ma et al. further delineated key minimal regions to support the physical existence of a coiled-coil clamp, which involved the juxtamembrane CC1 region (residues 248-261) and the ending helix of SOAR (residues 408-437). This finding was further supported by FRET results from a complementary study by Fahrner et al., who employed a so-called FIRE (FRET Interaction in a Restricted Environment) assay to dissect key structural elements mediating the CC1-SOAR interaction in cellulo [46]. Overall, these studies have yielded mechanistic insights into the structural determinants that govern STIM1 conformational switch during SOCE activation.
Inspired by the autoinhibitory property of CC1, He et al. attempted to use a genetically-encoded photoswitch derived from Avena sativa phototropin 1, the LOV2 domain [57, 58], to cage the minimal ORAI-activating region (Figure 2B). AsLOV2 has been extensively used to control protein actions because it undergoes remarkable conformational changes upon blue light stimulation. The AsLOV2 module consists of a flavin mononucleotide (FMN)-binding core domain known as a Period-ARNT-Single (PAS core) motif [77] and a C-terminal Jα helix (Figure 2B). A flavin chromophore is an endogenous cofactor that is widely accessible in mammalian cells, thus obviating the need of supplementing exogenous factors. Effector domains can be conveniently appended after the Jα helix of the AsLOV2 domain. In the dark, the Jα helix tightly docks to the PAS scaffold, thereby imposing potential steric hindrance to mask the active site or binding interfaces of the fused effector. Upon blue light illumination, photoexcitation creates a covalent adduct between the LOV2 residue C450 and the cofactor FMN, allowing the undocking of the Jα helix with subsequent exposure of the effector domain [57, 78]. Solution NMR measurements have been conducted to estimate the free energy alterations between the dark and lit states [78]. The available free energy in a LOV2 photoswitch is estimated to be ~3.8 kcal mol−1, which is sufficient enough to induce allosteric changes in polypeptides [79].
In the initial engineering of the LOV2-based GECAs, LOVS1K was constructed by fusing a STIM1-CT fragment (residues 233-450; including both the CC1 and SOAR domains) immediately after the C-terminal Jα helix [80]. LOVS1K was able to generate both global and local Ca2+ signals in response to blue light stimulation. This construct, nonetheless, was found to exhibit non-negligible dark activity. It led to constitutive Ca2+-dependent NFAT nuclear translocation even when shielded from light [63], thereby preventing further applications in physiological processes that demand more precise control of Ca2+ signaling. Given that LOV2 and CC1 might compete for the same binding interface of SOAR and hence reduce the caging efficiency, He et al. decided to use STIM1-CT fragments without the CC1 region. After screening dozens of constructs by optimizing the LOV2-Jα helix, STIM1-CT fragments, and the linker in between, they eventually evolved a photoactivatable LOV2-STIM chimera (LOV2404-546 + STIM1336-486; designated Opto-CRAC) with a higher dynamic range of Ca2+ response while minimizing the dark activity [61]. This construct could rapidly translocate between the cytosol and the PM to reversibly induce Ca2+ influx following repeated dark-light cycle treatments (t1/2, on = ~8 s; t1/2, off = ~ 20 s; Figure 1B) [61]. Substitution of the human STIM1-CT fragment with a homologous region from zebrafish led to further reduction in background activity without sacrificing the high dynamic range of light-induced Ca2+ response [81]. In a third study, Ishii et al. employed a similar strategy to develop a set of tools known as BACCS (Blue light-Activated Ca2+ Channels Switch), composed of LOV2404-538 and STIM1347-448, as well as the corresponding Drosophila version dmBACCS2 that exhibited more potent ORAI-activating capability [62]. The latter two studies presented further diversified variants by generating PM-tethered or dimeric, tandemly-linked versions of LOV2-STIM1 chimeras, as well as fusion with ORAI1 to enable optogenetic control of Ca2+ influx in cells or tissues with low or no expression of endogenous ORAI channels [61, 62]. To confer the tightest caging of SOAR in the dark state, Nguyen et al. developed the second generation Opto-CRAC constructs by further appending Zdark (Zdk) downstream of STIM1-CT fragments. Zdk is a family of engineered affibodies that preferentially recognizes LOV2 in its dark state but dissociates from LOV2 in the presence of blue light [82]. Thus, the introduced Zdk can serve as an additional “lock” to further block the activity of SOAR to reduce the background activation [81].
The successful engineering of LOV2-STIM1 chimeras to photo-control Ca2+ flux furnishes an unambiguous piece of evidence to support the conformational switch hypothesis underlying STIM1 activation. Compared to CRY2-based GECAs, LOV2-STIM1 chimeras exhibit faster deactivation kinetics (~30 sec vs. ~300 sec) with comparable potency in photo-activating Ca2+ entry, thus making them most ideal for mimicking Ca2+ oscillations evoked by physiological stimulations [61]. LOV2-based GECAs have been rigorously tested in a variety of immortalized cell lines and primary cells derived from the nervous and immune systems [61, 62].
4. LiMETER for reversible control of MCS assembly
Membrane contact sites (MCS) are ubiquitous subcellular structures in eukaryotes and serve as the platform for Ca2+ signaling, lipid exchange, organelle fission and inter-organellar communications [39, 40, 83-85]. The initial report of MCS between ER/SR and the plasma membrane can be dated back to the observation of the so-called “triads” or “dyads” in excitable cells by electron microscopy [86]. Triads and dyads are formed by PM-localized voltage-gated Ca2+ channels and the SR-resident ryanodine receptors. They constitute the molecular basis for excitation-contraction coupling, converting changes in PM action potentials into Ca2+ release from the sarcoplasmic reticulum [87]. The gap distance in triads is estimated to be 9 to 12 nm [39, 88]. In comparison, in non-excitable cells, a fraction of ER-PM contact sites is marked by STIM and ORAI proteins during SOCE activation, with the gap distance determined to be 15 to 35 nm.
As described above, an activated STIM1 molecule translocates to ER-PM junctions and recruits ORAI molecules in the plasma membrane to mediate Ca2+ influx (Figure 1B). Notably, STIM1 redistribution toward the PM is facilitated by the interaction between the exposed C-terminal PB domain and the negatively-charged PIs in the inner leaflet of the PM [39, 44, 89-91]. Deletion of the PB domain (residues 672-685) prevents STIM1 translocation to form puncta at ER-PM junctional sites after Ca2+ store depletion despite intact self-oligomerization of STIM1 in the ER [34, 92]. To recapitulate the MCS assembly process in living cells, Jing et al. developed an optogenetic tool (designated LiMETER for light-inducible membrane tethered peripheral ER) by using STIM1 as the engineering scaffold [48, 64]. In the prototypical design, LiMETER was inserted into the ER membrane by keeping the signal peptide and the single transmembrane domain from STIM1, while the Ca2+-sensing and oligomerizable EF-SAM domain was replaced by a fluorescent protein (Figure 3A-B). On the cytosolic side, only the PB domain in the C-tail was retained, and the CC1-SOAR region was substituted with spacers of varying lengths and LOV2. In the dark, the PB domain was shielded because of the steric hindrance imposed by the upstream LOV2 domain. Upon blue light illumination, the PB domain was exposed to restore its PI-interacting capability. Therefore, LiMETER can be expressed in mammalian cells to reversibly label and manipulate the formation of ER-PM MCSs within seconds (t1/2, on = 14 s; t1/2, off = 25 s) [48]. This result clearly attests to the critical role of STIM1-PB in mediating STIM1-PI interaction, which efficiently facilitates the targeting of STIM1 toward PM.
LiMETER can be used as a scaffold to pinpoint residues that are essential for protein-PI interactions in living cells without the purification of recombinant proteins. Polypeptide sequences predicted to interact with PM-embedded PIs, such as PI(4,5)P2 and PI(3,4,5,)P3, can be conveniently grafted into the LiMETER construct, with subsequent introduction of mutations to unveil molecular determinants driving protein-PI associations. He et al. replaced the lysine residues of STIM1-PB with single or double alanine mutations to neutralize the positive charges of the PB domain. They found that single Lys-to-Ala mutations within STIM1-PB modestly decreased puncta formation. By contrast, the replacement of two adjacent lysine residues by alanine (e.g., K672A/K673A, K680A/K681A, or K684A/K685A) in the PB domain abolished photo-inducible puncta formation at ER-PM junctions [48]. This site-directed mutagenesis study has provided an unequivocal line of evidence to support the indispensable role of these lysine residues in mediating protein-lipid association and facilitating the assembly of ER-PM MCSs.
LiMETER could further be repurposed as a molecular gauge to estimate the distance requirements between two organellar membranes. In the prototypical design of LiMETER without insertion of long spacers, the bridge connecting the ER and the PM spanned an estimated distance of 10 nm. ORAI1 molecules were completely excluded from the ER-PM contact sites marked by LiMETER puncta (Figure 3D, left). In contrast, when the inter-membrane gap distance was increased to above 25 nm via the introduction of repeated helical spacers (Figure 3C), ORAI1 molecules were able to freely diffuse into these contact sites (Figure 3E). This finding was in line with the observation that the intracellular regions of ORAI1 might project into the cytoplasm and require at least a gap distance of 10 nm. It also lends support to the tacit belief in the field that the efficient formation of STIM1-ORAI complexes during SOCE probably requires a distance of at least 25 nm (27.1 ± 2.8 nm) in the gap between the ER and the plasma membrane [93]. Collectively, the design of LiMETER to reversibly control the formation of ER-PM contact sites provides a generic engineering strategy to not only probe protein-lipid interactions in living cells, but also tune the intermembrane distance at a nanoscale level in order to manipulate the behavior of cell signaling proteins at high temporal and spatial resolution [48]. Ideally, optogenetic tools like LiMETER can be further exploited as a shuttle to deliver cargo into MCSs, which can aid in the dissection of molecular mechanisms governing the biogenesis, maintenance, and dynamic of membrane contact sites.
5. Applications of STIM1-based optogenetic tools
CRY2- and LOV2-based GECAs enable precise spatial and temporal control of Ca2+ signaling with tailored functions [72]. Compared to conventional microbial opsin-based optical tools [94], STIM1-derived GECAs offer higher specificity for Ca2+ and are more suitable for manipulating biological processes with relatively slower kinetics (seconds to hours) in non-excitable tissues. These tools have been successfully applied to faithfully phenocopy a variety of Ca2+-modulated cellular events and physiological processes (Figure 4). Both CRY2-STIM1ct and LOV2-SOAR can be used to photo-induce Ca2+ influx in various cell types, including T cells, macrophages, dendritic cells, neurons, embryonic stem cells, and cancer cell lines. These cells are derived from different tissues and species as well, including human, mice, zebrafish, fruit flies, and worms [61-63]. Even though the light-triggered magnitude of Ca2+ signals seems to be heterogeneous in different cells due to the varied expression levels of endogenous ORAI channels, STIM1-based GECAs have been used to control hallmark Ca2+-dependent physiological responses (Figure 4B). For instance, both OptoSTIM1 and Opto-CRAC were expressed in model cellular systems to drive the expression of Ca2+/NFAT-dependent genes [61-63]. The induction of NFAT-mediated gene transcription, such as NFAT-dependent luciferase and insulin expression in HEK293T cells, indicates that Opto-CRAC has the capacity to photoactivate transfected cells via the cooperation of NFAT proteins with other transcriptional partners. As expected, transducing primary mouse CD4+ T cells with Opto-CRAC led to a significant increase in the production of signature cytokines, such as IL2 and IFN-γ, in activated T cells [61]. Likewise, Human THP-1-derived macrophages expressing Opto-CRAC secreted large quantities of IL-1β and processed caspase-1 in response to light stimulation, thus demonstrating the possibility of photo-tunable augmentation of inflammasome activation [61].
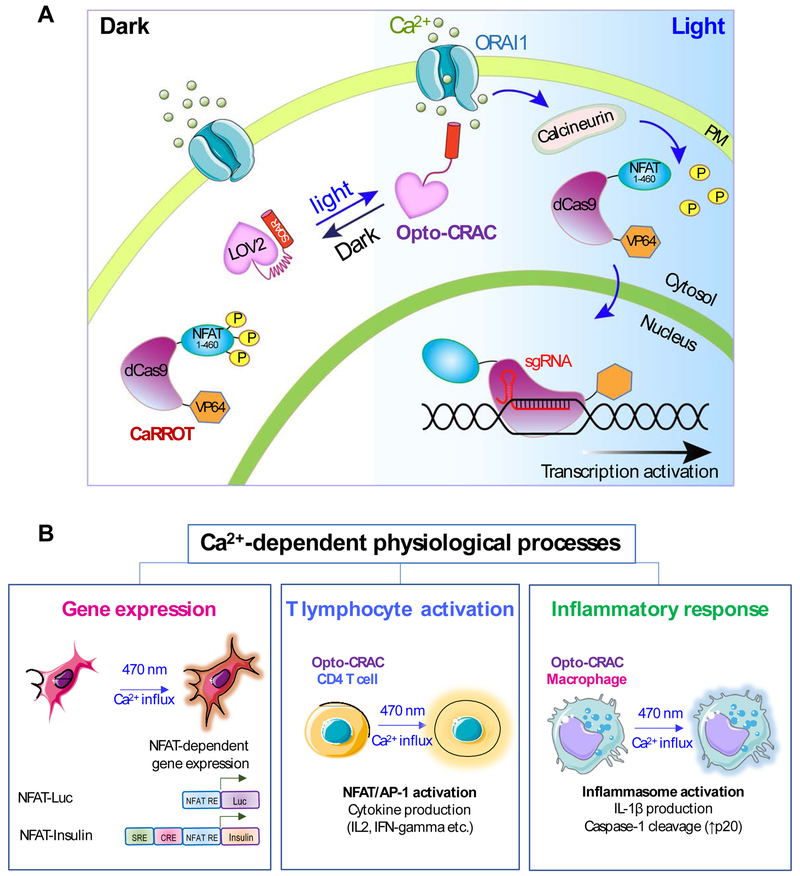
Figure 4 ∣. Examples illustrating potential applications of STIM1-derived optogenetic tools.
(A) Rewiring photoactivatable Ca2+ influx for light-inducible modulation of gene expression. Upon light stimulation, Opto-CRAC induces cytosolic Ca2+ influx to activate the Ca2+/CaM-dependent phosphatase, calcineurin, which in turn dephosphorylates the N-terminal fragment of NFAT (NFAT1-460) to drive the nuclear entry of a synthetic transcription regulatory device made of catalytically-dead Cas9 and the transcriptional coactivator VP64 (NFAT1-460-dCas9-VP64; designated as CaRROT for Ca2+-responsive transcriptional reprogramming tool). In the presence of small guide RNAs (sgRNAs), CaRROT can be precisely targeted to specific genomic loci to photo-tune the expression of endogenous genes to achieve tailored function. Likewise, other effector domains such as transcriptional repressors (e.g., KRAB) and epigenetic regulators (DNA or histone modifying enzymes) [95, 111, 112] can be installed to replace VP64, thereby enabling photoswitchable inhibition of gene expression and epigenetic remodeling.
(B) Opto-CRAC can be applied to photo-control Ca2+-dependent gene expression (left), cytokine production in T lymphocytes (middle), and inflammasome activation in macrophage (right). Optogenetic immunomodulation is likely to be of translational values for remote and personalized control of the immune system to fight invading pathogens and cancer with high precision.
Apart from photo-manipulating hallmark Ca2+/NFAT-dependent gene expression, Opto-CRAC has been further repurposed to control the expression of any endogenous genes (Figure 4). This was made possible by the invention of a light-inducible transcriptional reprogramming device that combines CRISPR/dCas9 with Opto-CRAC [81]. This synthetic device consists of two components. The first component contains a second-generation Opto-CRAC construct made of LOV2 and Danio rerio STIM1-CT (341-442) chimeras, which provide tighter control over Ca2+ signals than the first Opto-CRAC prototype. The second component, termed CaRROT (Figure 4A), consists of an N-terminal fragment of NFAT (residues 1-460) fused to dCas9 and transcriptional coactivators (VP64/VP160). Since the NFAT fragment used in CaRROT lacks a C-terminal DNA binding domain, it is unable to bind to endogenous NFAT targets. In the dark, CaRROT stays exclusively in the cytosol due to the phosphorylation of NFAT. Upon blue light illumination, Opto-CRAC induces Ca2+ influx from the extracellular space into the cytosol to trigger the dephosphorylation of NFAT and the subsequent nuclear entry of CaRROT. After entering the nuclei, dCas9-VP64 is directed by small guide RNAs (sgRNAs) to target sequences in the genome and turn on gene transcription. CaRROT thus allows for inducible transcriptional reprogramming. Theoretically, it can be further used in precision regenerative medicine by controlling the expression of genes that are critical for inducing the differentiation of induced pluripotent cells (iPSC) towards a defined cell fate. Likewise, CaRROT can also be reconfigured for the remote intervention of cancer progression. CaRROT, when combined with transcriptional repressors such as KRAB [95], can be repurposed to shut down the expression of oncogenes in cancer cells. In addition, when coupled with newly developed base editing tools [96], CaRROT can be used to correct mutations in the genome to reverse disease progression.
The afore-described in vitro data strongly supports the feasibility of using these GECAs for in vivo studies. However, the inability of blue light-absorbing photoreceptors to penetrate deep in tissues greatly limits the applications of these tools at organismal levels [97]. Neuroscientists often adopt a rather invasive approach to photoactivate microbial opsins by implanting invasive indwelling fiber optic probes into the brain. This protocol seems to be unfeasible with cells that are constantly circulating within the body. To tackle this challenge, He et al. first explored the use of lanthanide-doped upconversion nanoparticles (UCNPs) as a near-infrared (NIR) light transducer that could convert NIR light (980 nm) into blue light (470 nm) [98-100]. NIR light can penetrate at a depth greater than 2-3 cm [101-103]. When coupled with upconversion nanoparticles, the optogenetic operation window of Opto-CRAC could be shifted from the visible range to NIR wavelengths to enable wireless and remote photoactivation of Ca2+-dependent signaling and optogenetic modulation of immunoinflammatory responses. In a mouse model of melanoma that used ovalbumin as a surrogate tumor antigen, Opto-CRAC in dendritic cells acted as a genetically-encoded “photoactivatable adjuvant” that improved antigen-specific immune responses and specifically destructed tumor cells [61]. A more recent proof-of-concept study has been used a similar approach to achieve deep brain stimulation in the central nervous system [104].
6. Summary and future directions
Overall, the optogenetic engineering of CRAC channels turns out to be an extremely rewarding journey, in that it not only yields insight into the working mechanism underlying STIM1-mediated SOCE activation but also affords unconventional tools to study cell physiology. STIM1-derived GECAs can faithfully recapture the key steps involved in STIM1-ORAI coupling at membrane contact sites to ultimately evoke Ca2+ influx from the extracellular space. As illustrated in the preceding sections, optogenetic tools engineered from STIM1 also open new opportunities for remotely interrogating Ca2+-dependent biological processes, dynamically examining lipid-protein interaction and delivering proteins of interest to this specialized subcellular compartment, and screening protein and chemical modulators that are involved in maintaining and remodeling cell signaling at MCSs.
Aside from CRAC channels, more tools have been recently invented to control the activity of other types of Ca2+ channels, including TRPC channels [105] and voltage-gated Ca2+ channels [106]. Nonetheless, existing efforts are almost exclusively directed to engineer ion channel regulatory units or synthesize photo-sensitive chemical modulators. Direct engineering of a photoswitchable bona fide Ca2+ channel, like those done with channelrhodopsin [107] or potassium channels [108], is still missing in the literature. Capitalizing on naturally-evolved photosensory domains that can be installed into signaling proteins, we hope that optogenetic engineering approaches described above can be adopted to control ion channels per se and be further extended to manipulate other types of MCSs formed between the ER and various intracellular organelles, including mitochondria, lysosomes, endosomes, peroxisomes, and lipid droplets [39, 40, 83-85]. In the near future, we anticipate the continuing expansion of the optogenetic toolkit tailored for the Ca2+ signaling field with on-demand kinetics and high specificity. Finally, we hope that near-infrared light-stimulatable nanomaterials and optogenetic tools, which are expected to have deeper tissue penetration [61, 104, 109, 110], will further accelerate the translational application of optogenetic actuators in vivo.
Highlights.
CRY2-STIM1 chimeras to mimic STIM1 oligomerization for optical activation of ORAI
LOV2-STIM1 chimeras to recapitulate STIM1 conformational switch
LiMETER to manipulate protein-lipid interactions and MCS assembly
STIM1-derived optogenetic tools to phenocopy hallmark Ca2+-dependent responses
Rewiring photo-inducible Ca2+ signaling for precise transcriptional reprogramming
Acknowledgements
We also thank the financial support from the National Institutes of Health (R01GM112003, R21GM126532 and R01HL134780), the Welch Foundation (BE-1913), the American Cancer Society (RSG-16-215-01-TBE and RSG-18-043-01-LIB), the Cancer Prevention and Research Institute of Texas (RR140053 and RP170660), the John S. Dunn Foundation, and by an allocation from the Texas A&M University Start-up Fund. STIM1-derived optogenetic tools mentioned in this article were deposited at Addgene (Opto-CRAC: #101245 and #101246; OptoSTIM1: #70159; LiMETER: #113933 and 113934).
Abbreviations
- ER
endoplasmic reticulum
- PM
plasma membrane
- MCS
membrane contact site
- SOCE
store-operated calcium entry
- CRAC
calcium release-activated calcium channel
- STIM
stromal interaction molecule
- CRY2
cryptochrome 2
- LOV2
light-oxygen-voltage domain 2
- PB
polybasic domain
- SOAR
STIM1 Orai-activating region
- CAD
CRAC activation domain
- OASF
Orai-activation small fragment
- CRISPR
Clustered Regularly Interspaced Short Palindromic Repeats
- LiMETER
light-inducible membrane-tethered peripheral ER
- CaRROT
calcium-responsive transcriptional reprogramming tool
Footnotes
Conflicts of interest
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Berridge MJ, Bootman MD, Roderick HL, Calcium signalling: dynamics, homeostasis and remodelling, Nature reviews. Molecular cell biology, 4 (2003) 517–529. [DOI] [PubMed] [Google Scholar]
- [2].Berridge MJ, Lipp P, Bootman MD, The versatility and universality of calcium signalling, Nature reviews. Molecular cell biology, 1 (2000) 11–21. [DOI] [PubMed] [Google Scholar]
- [3].Clapham DE, Calcium signaling, Cell, 131 (2007) 1047–1058. [DOI] [PubMed] [Google Scholar]
- [4].Zhou Y, Xue S, Yang JJ, Calciomics: integrative studies of Ca2+-binding proteins and their interactomes in biological systems, Metallomics, 5 (2013) 29–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Nowycky MC, Fox AP, Tsien RW, Three types of neuronal calcium channel with different calcium agonist sensitivity, Nature, 316 (1985) 440–443. [DOI] [PubMed] [Google Scholar]
- [6].Zamponi GW, Striessnig J, Koschak A, Dolphin AC, The Physiology, Pathology, and Pharmacology of Voltage-Gated Calcium Channels and Their Future Therapeutic Potential, Pharmacol Rev, 67 (2015) 821–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Putney JW Jr., A model for receptor-regulated calcium entry, Cell Calcium, 7 (1986) 1–12. [DOI] [PubMed] [Google Scholar]
- [8].Parekh AB, Putney JW Jr., Store-operated calcium channels, Physiological reviews, 85 (2005) 757–810. [DOI] [PubMed] [Google Scholar]
- [9].Feske S, Calcium signalling in lymphocyte activation and disease, Nature reviews. Immunology, 7 (2007) 690–702. [DOI] [PubMed] [Google Scholar]
- [10].Hogan PG LR, and Rao A, Molecular basis of calcium signaling in lymphocytes: STIM and ORAI, Annual review of immunology, 28 (2010) 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Soboloff J, Rothberg BS, Madesh M, Gill DL, STIM proteins: dynamic calcium signal transducers, Nature reviews. Molecular cell biology, 13 (2012) 549–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Prakriya M, Lewis RS, Store-Operated Calcium Channels, Physiological reviews, 95 (2015) 1383–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Nguyen NT, Han W, Cao WM, Wang Y, Wen S, Huang Y, Li M, Du L, Zhou Y, Store-Operated Calcium Entry mediated by ORAI and STIM, Compr Physiol, 8 (2018) 981–1002. [DOI] [PubMed] [Google Scholar]
- [14].Zweifach A, Lewis RS, Mitogen-regulated Ca2+ current of T lymphocytes is activated by depletion of intracellular Ca2+ stores, Proceedings of the National Academy of Sciences of the United States of America, 90 (1993) 6295–6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Feske S, Gwack Y, Prakriya M, Srikanth S, Puppel SH, Tanasa B, Hogan PG, Lewis RS, Daly M, Rao A, A mutation in Orai1 causes immune deficiency by abrogating CRAC channel function, Nature, 441 (2006) 179–185. [DOI] [PubMed] [Google Scholar]
- [16].McCarl CA, Picard C, Khalil S, Kawasaki T, Rother J, Papolos A, Kutok J, Hivroz C, Ledeist F, Plogmann K, Ehl S, Notheis G, Albert MH, Belohradsky BH, Kirschner J, Rao A, Fischer A, Feske S, ORAI1 deficiency and lack of store-operated Ca2+ entry cause immunodeficiency, myopathy, and ectodermal dysplasia, The Journal of allergy and clinical immunology, 124 (2009) 1311–1318 e1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Picard C, McCarl CA, Papolos A, Khalil S, Luthy K, Hivroz C, LeDeist F, Rieux-Laucat F, Rechavi G, Rao A, Fischer A, Feske S, STIM1 mutation associated with a syndrome of immunodeficiency and autoimmunity, The New England journal of medicine, 360 (2009) 1971–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Feske S, ORAI1 and STIM1 deficiency in human and mice: roles of store-operated Ca2+ entry in the immune system and beyond, Immunological reviews, 231 (2009) 189–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Misceo D, Holmgren A, Louch WE, Holme PA, Mizobuchi M, Morales RJ, De Paula AM, Stray-Pedersen A, Lyle R, Dalhus B, Christensen G, Stormorken H, Tjonnfjord GE, Frengen E, A dominant STIM1 mutation causes Stormorken syndrome, Hum Mutat, 35 (2014) 556–564. [DOI] [PubMed] [Google Scholar]
- [20].Morin G, Bruechle NO, Singh AR, Knopp C, Jedraszak G, Elbracht M, Bremond-Gignac D, Hartmann K, Sevestre H, Deutz P, Herent D, Nurnberg P, Romeo B, Konrad K, Mathieu-Dramard M, Oldenburg J, Bourges-Petit E, Shen Y, Zerres K, Ouadid-Ahidouch H, Rochette J, Gain-of-Function Mutation in STIM1 (P.R304W) Is Associated with Stormorken Syndrome, Hum Mutat, 35 (2014) 1221–1232. [DOI] [PubMed] [Google Scholar]
- [21].Nesin V, Wiley G, Kousi M, Ong EC, Lehmann T, Nicholl DJ, Suri M, Shahrizaila N, Katsanis N, Gaffney PM, Wierenga KJ, Tsiokas L, Activating mutations in STIM1 and ORAI1 cause overlapping syndromes of tubular myopathy and congenital miosis, Proceedings of the National Academy of Sciences of the United States of America, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Zhang W, Trebak M, STIM1 and Orai1: novel targets for vascular diseases?, Science China. Life sciences, 54 (2011) 780–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Xie J, Pan H, Yao J, Zhou Y, Han W, SOCE and cancer: Recent progress and new perspectives, Int J Cancer, 138 (2016) 2067–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zhang SL, Yeromin AV, Zhang XH, Yu Y, Safrina O, Penna A, Roos J, Stauderman KA, Cahalan MD, Genome-wide RNAi screen of Ca(2+) influx identifies genes that regulate Ca(2+) release-activated Ca(2+) channel activity, Proceedings of the National Academy of Sciences of the United States of America, 103 (2006) 9357–9362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Vig M, Peinelt C, Beck A, Koomoa DL, Rabah D, Koblan-Huberson M, Kraft S, Turner H, Fleig A, Penner R, Kinet JP, CRACM1 is a plasma membrane protein essential for store-operated Ca2+ entry, Science, 312 (2006) 1220–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Roos J, DiGregorio PJ, Yeromin AV, Ohlsen K, Lioudyno M, Zhang S, Safrina O, Kozak JA, Wagner SL, Cahalan MD, Velicelebi G, Stauderman KA, STIM1, an essential and conserved component of store-operated Ca2+ channel function, The Journal of cell biology, 169 (2005) 435–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zhang SL, Yu Y, Roos J, Kozak JA, Deerinck TJ, Ellisman MH, Stauderman KA, Cahalan MD, STIM1 is a Ca2+ sensor that activates CRAC channels and migrates from the Ca2+ store to the plasma membrane, Nature, 437 (2005) 902–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Liou J, Kim ML, Heo WD, Jones JT, Myers JW, Ferrell JE Jr., Meyer T, STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx, Curr Biol, 15 (2005) 1235–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Stathopulos PB, Li GY, Plevin MJ, Ames JB, Ikura M, Stored Ca2+ depletion-induced oligomerization of stromal interaction molecule 1 (STIM1) via the EF-SAM region: An initiation mechanism for capacitive Ca2+ entry, The Journal of biological chemistry, 281 (2006) 35855–35862. [DOI] [PubMed] [Google Scholar]
- [30].Stathopulos PB, Zheng L, Li GY, Plevin MJ, Ikura M, Structural and mechanistic insights into STIM1-mediated initiation of store-operated calcium entry, Cell, 135 (2008) 110–122. [DOI] [PubMed] [Google Scholar]
- [31].Ma G, Zheng S, Ke Y, Zhou L, He L, Huang Y, Wang Y, Zhou Y, Molecular determinants for STIM1 activation during store-operated Ca2+ entry, Curr Mol Med, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ma G, Wei M, He L, Liu C, Wu B, Zhang SL, Jing J, Liang X, Senes A, Tan P, Li S, Sun A, Bi Y, Zhong L, Si H, Shen Y, Li M, Lee MS, Zhou W, Wang J, Wang Y, Zhou Y, Inside-out Ca(2+) signalling prompted by STIM1 conformational switch, Nat Commun, 6 (2015) 7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Yuan JP, Zeng W, Dorwart MR, Choi YJ, Worley PF, Muallem S, SOAR and the polybasic STIM1 domains gate and regulate Orai channels, Nature cell biology, 11 (2009) 337–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Park CY, Hoover PJ, Mullins FM, Bachhawat P, Covington ED, Raunser S, Walz T, Garcia KC, Dolmetsch RE, Lewis RS, STIM1 clusters and activates CRAC channels via direct binding of a cytosolic domain to Orai1, Cell, 136 (2009) 876–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Frischauf I, Muik M, Derler I, Bergsmann J, Fahrner M, Schindl R, Groschner K, Romanin C, Molecular determinants of the coupling between STIM1 and Orai channels: differential activation of Orai1-3 channels by a STIM1 coiled-coil mutant, The Journal of biological chemistry, 284 (2009) 21696–21706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Frischauf I, Schindl R, Derler I, Bergsmann J, Fahrner M, Romanin C, The STIM/Orai coupling machinery, Channels (Austin), 2 (2008) 261–268. [DOI] [PubMed] [Google Scholar]
- [37].Muik M, Schindl R, Fahrner M, Romanin C, Ca(2+) release-activated Ca(2+) (CRAC) current, structure, and function, Cell Mol Life Sci, 69 (2012) 4163–4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Gudlur A, Zhou Y, Hogan PG, STIM-ORAI Interactions That Control the CRAC Channel, Current topics in membranes, 71 (2013) 33–58. [DOI] [PubMed] [Google Scholar]
- [39].Carrasco S, Meyer T, STIM proteins and the endoplasmic reticulum-plasma membrane junctions, Annual review of biochemistry, 80 (2011) 973–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Saheki Y, De Camilli P, Endoplasmic Reticulum-Plasma Membrane Contact Sites, Annual review of biochemistry, 86 (2017) 659–684. [DOI] [PubMed] [Google Scholar]
- [41].Muik M, Fahrner M, Schindl R, Stathopulos P, Frischauf I, Derler I, Plenk P, Lackner B, Groschner K, Ikura M, Romanin C, STIM1 couples to ORAI1 via an intramolecular transition into an extended conformation, The EMBO journal, 30 (2011) 1678–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Stathopulos PB, Schindl R, Fahrner M, Zheng L, Gasmi-Seabrook GM, Muik M, Romanin C, Ikura M, STIM1/Orai1 coiled-coil interplay in the regulation of store-operated calcium entry, Nat Commun, 4 (2013) 2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Zhou Y, Meraner P, Kwon HT, Machnes D, Oh-hora M, Zimmer J, Huang Y, Stura A, Rao A, Hogan PG, STIM1 gates the store-operated calcium channel ORAI1 in vitro, Nature structural & molecular biology, 17 (2010) 112–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Zhou Y, Srinivasan P, Razavi S, Seymour S, Meraner P, Gudlur A, Stathopulos PB, Ikura M, Rao A, Hogan PG, Initial activation of STIM1, the regulator of store-operated calcium entry, Nature structural & molecular biology, 20 (2013) 973–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Stathopulos PB, Ikura M, Structurally delineating stromal interaction molecules as the endoplasmic reticulum calcium sensors and regulators of calcium release-activated calcium entry, Immunological reviews, 231 (2009) 113–131. [DOI] [PubMed] [Google Scholar]
- [46].Fahrner M, Muik M, Schindl R, Butorac C, Stathopulos P, Zheng L, Jardin I, Ikura M, Romanin C, A coiled-coil clamp controls both conformation and clustering of stromal interaction molecule 1 (STIM1), The Journal of biological chemistry, 289 (2014) 33231–33244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Korzeniowski MK, Popovic MA, Szentpetery Z, Varnai P, Stojilkovic SS, Balla T, Dependence of STIM1/Orai1-mediated calcium entry on plasma membrane phosphoinositides, The Journal of biological chemistry, 284 (2009) 21027–21035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].He L, Jing J, Zhu L, Tan P, Ma G, Zhang Q, Nguyen NT, Wang J, Zhou Y, Huang Y, Optical control of membrane tethering and interorganellar communication at nanoscales, Chem Sci, 8 (2017) 5275–5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Hogan PG, Chen L, Nardone J, Rao A, Transcriptional regulation by calcium, calcineurin, and NFAT, Genes Dev, 17 (2003) 2205–2232. [DOI] [PubMed] [Google Scholar]
- [50].Muller MR, Rao A, NFAT, immunity and cancer: a transcription factor comes of age, Nature reviews. Immunology, 10 (2010) 645–656. [DOI] [PubMed] [Google Scholar]
- [51].Navarro-Borelly L, Somasundaram A, Yamashita M, Ren D, Miller RJ, Prakriya M, STIM1-Orai1 interactions and Orai1 conformational changes revealed by live-cell FRET microscopy, J Physiol, 586 (2008) 5383–5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Muik M, Frischauf I, Derler I, Fahrner M, Bergsmann J, Eder P, Schindl R, Hesch C, Polzinger B, Fritsch R, Kahr H, Madl J, Gruber H, Groschner K, Romanin C, Dynamic coupling of the putative coiled-coil domain of ORAI1 with STIM1 mediates ORAI1 channel activation, The Journal of biological chemistry, 283 (2008) 8014–8022. [DOI] [PubMed] [Google Scholar]
- [53].Luik RM, Wang B, Prakriya M, Wu MM, Lewis RS, Oligomerization of STIM1 couples ER calcium depletion to CRAC channel activation, Nature, 454 (2008) 538–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Hou X, Pedi L, Diver MM, Long SB, Crystal structure of the calcium release-activated calcium channel Orai, Science, 338 (2012) 1308–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Yang X, Jin H, Cai X, Li S, Shen Y, Structural and mechanistic insights into the activation of Stromal interaction molecule 1 (STIM1), Proceedings of the National Academy of Sciences of the United States of America, 109 (2012) 5657–5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Zheng L, Stathopulos PB, Schindl R, Li GY, Romanin C, Ikura M, Auto-inhibitory role of the EF-SAM domain of STIM proteins in store-operated calcium entry, Proceedings of the National Academy of Sciences of the United States of America, 108 (2011) 1337–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Harper SM, Neil LC, Gardner KH, Structural basis of a phototropin light switch, Science, 301 (2003) 1541–1544. [DOI] [PubMed] [Google Scholar]
- [58].Wu YI, Frey D, Lungu OI, Jaehrig A, Schlichting I, Kuhlman B, Hahn KM, A genetically encoded photoactivatable Rac controls the motility of living cells, Nature, 461 (2009) 104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Liu H, Yu X, Li K, Klejnot J, Yang H, Lisiero D, Lin C, Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis, Science, 322 (2008) 1535–1539. [DOI] [PubMed] [Google Scholar]
- [60].Kennedy MJ, Hughes RM, Peteya LA, Schwartz JW, Ehlers MD, Tucker CL, Rapid blue-light-mediated induction of protein interactions in living cells, Nat Methods, 7 (2010) 973–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].He L, Zhang Y, Ma G, Tan P, Li Z, Zang S, Wu X, Jing J, Fang S, Zhou L, Wang Y, Huang Y, Hogan PG, Han G, Zhou Y, Near-infrared photoactivatable control of Ca(2+) signaling and optogenetic immunomodulation, eLife, 4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Ishii T, Sato K, Kakumoto T, Miura S, Touhara K, Takeuchi S, Nakata T, Light generation of intracellular Ca(2+) signals by a genetically encoded protein BACCS, Nat Commun, 6 (2015) 8021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Kyung T, Lee S, Kim JE, Cho T, Park H, Jeong YM, Kim D, Shin A, Kim S, Baek J, Kim J, Kim NY, Woo D, Chae S, Kim CH, Shin HS, Han YM, Kim D, Heo WD, Optogenetic control of endogenous Ca(2+) channels in vivo, Nat Biotechnol, 33 (2015) 1092–1096. [DOI] [PubMed] [Google Scholar]
- [64].Jing J, He L, Sun A, Quintana A, Ding Y, Ma G, Tan P, Liang X, Zheng X, Chen L, Shi X, Zhang SL, Zhong L, Huang Y, Dong MQ, Walker CL, Hogan PG, Wang Y, Zhou Y, Proteomic mapping of ER-PM junctions identifies STIMATE as a regulator of Ca(2)(+) influx, Nature cell biology, 17 (2015) 1339–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Fegan A, White B, Carlson JC, Wagner CR, Chemically controlled protein assembly: techniques and applications, Chem Rev, 110 (2010) 3315–3336. [DOI] [PubMed] [Google Scholar]
- [66].Sancar A, Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors, Chem Rev, 103 (2003) 2203–2237. [DOI] [PubMed] [Google Scholar]
- [67].Cashmore AR, Jarillo JA, Wu YJ, Liu D, Cryptochromes: blue light receptors for plants and animals, Science, 284 (1999) 760–765. [DOI] [PubMed] [Google Scholar]
- [68].Lin C, Shalitin D, Cryptochrome structure and signal transduction, Annu Rev Plant Biol, 54 (2003) 469–496. [DOI] [PubMed] [Google Scholar]
- [69].Liu H, Liu B, Zhao C, Pepper M, Lin C, The action mechanisms of plant cryptochromes, Trends Plant Sci, 16 (2011) 684–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Bugaj LJ, Choksi AT, Mesuda CK, Kane RS, Schaffer DV, Optogenetic protein clustering and signaling activation in mammalian cells, Nat Methods, 10 (2013) 249–252. [DOI] [PubMed] [Google Scholar]
- [71].Taslimi A, Vrana JD, Chen D, Borinskaya S, Mayer BJ, Kennedy MJ, Tucker CL, An optimized optogenetic clustering tool for probing protein interaction and function, Nat Commun, 5 (2014) 4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Ma G, Wen S, He L, Huang Y, Wang Y, Zhou Y, Optogenetic toolkit for precise control of calcium signaling, Cell Calcium, 64 (2017) 36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Korzeniowski MK, Manjarres IM, Varnai P, Balla T, Activation of STIM1-Orai1 involves an intramolecular switching mechanism, Science signaling, 3 (2010) ra82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Stathopulos PB, Schindl R, Fahrner M, Zheng L, Gasmi-Seabrook GM, Muik M, Romanin C, Ikura M, STIM1/Orai1 coiled-coil interplay in the regulation of store-operated calcium entry, Nat Commun, 4 (2013) 2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Feske S, Prakriya M, Conformational dynamics of STIM1 activation, Nature structural & molecular biology, 20 (2013) 918–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Korzeniowski MK, Wisniewski E, Baird B, Holowka DA, Balla T, Molecular anatomy of the early events in STIM1 activation - oligomerization or conformational change?, J Cell Sci, 130 (2017) 2821–2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Nash AI, Ko WH, Harper SM, Gardner KH, A conserved glutamine plays a central role in LOV domain signal transmission and its duration, Biochemistry, 47 (2008) 13842–13849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Yao X, Rosen MK, Gardner KH, Estimation of the available free energy in a LOV2-J alpha photoswitch, Nat Chem Biol, 4 (2008) 491–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Dagliyan O, Tarnawski M, Chu PH, Shirvanyants D, Schlichting I, Dokholyan NV, Hahn KM, Engineering extrinsic disorder to control protein activity in living cells, Science, 354 (2016) 1441–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Pham E, Mills E, Truong K, A synthetic photoactivated protein to generate local or global Ca(2+) signals, Chem Biol, 18 (2011) 880–890. [DOI] [PubMed] [Google Scholar]
- [81].Nguyen NT, He L, Martinez-Moczygemba M, Huang Y, Zhou Y, Rewiring Calcium Signaling for Precise Transcriptional Reprogramming, ACS Synth Biol, 7 (2018) 814–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Wang H, Hahn KM, LOVTRAP: A Versatile Method to Control Protein Function with Light, Curr Protoc Cell Biol, 73 (2016) 21 10 21–21 10 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Prinz WA, Bridging the gap: membrane contact sites in signaling, metabolism, and organelle dynamics, The Journal of cell biology, 205 (2014) 759–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Gallo A, Vannier C, Galli T, Endoplasmic Reticulum-Plasma Membrane Associations:Structures and Functions, Annu Rev Cell Dev Biol, 32 (2016) 279–301. [DOI] [PubMed] [Google Scholar]
- [85].Phillips MJ, Voeltz GK, Structure and function of ER membrane contact sites with other organelles, Nature reviews. Molecular cell biology, 17 (2016) 69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Porter KR, Palade GE, Studies on the endoplasmic reticulum. III. Its form and distribution in striated muscle cells, J Biophys Biochem Cytol, 3 (1957) 269–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Dirksen RT, Bi-directional coupling between dihydropyridine receptors and ryanodine receptors, Front Biosci, 7 (2002) d659–670. [DOI] [PubMed] [Google Scholar]
- [88].Flucher BE, Structural analysis of muscle development: transverse tubules, sarcoplasmic reticulum, and the triad, Dev Biol, 154 (1992) 245–260. [DOI] [PubMed] [Google Scholar]
- [89].Heo WD, Inoue T, Park WS, Kim ML, Park BO, Wandless TJ, Meyer T, PI(3,4,5)P3 and PI(4,5)P2 lipids target proteins with polybasic clusters to the plasma membrane, Science, 314 (2006) 1458–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Ercan E, Momburg F, Engel U, Temmerman K, Nickel W, Seedorf M, A conserved, lipid-mediated sorting mechanism of yeast Ist2 and mammalian STIM proteins to the peripheral ER, Traffic, 10 (2009) 1802–1818. [DOI] [PubMed] [Google Scholar]
- [91].Li L, Shi X, Guo X, Li H, Xu C, Ionic protein-lipid interaction at the plasma membrane: what can the charge do?, Trends Biochem Sci, 39 (2014) 130–140. [DOI] [PubMed] [Google Scholar]
- [92].Liou J, Fivaz M, Inoue T, Meyer T, Live-cell imaging reveals sequential oligomerization and local plasma membrane targeting of stromal interaction molecule 1 after Ca2+ store depletion, Proceedings of the National Academy of Sciences of the United States of America, 104 (2007) 9301–9306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Fernandez-Busnadiego R, Saheki Y, De Camilli P, Three-dimensional architecture of extended synaptotagmin-mediated endoplasmic reticulum-plasma membrane contact sites, Proceedings of the National Academy of Sciences of the United States of America, 112 (2015) E2004–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Fenno L, Yizhar O, Deisseroth K, The development and application of optogenetics, Annu Rev Neurosci, 34 (2011) 389–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, Lim WA, Weissman JS, Qi LS, CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes, Cell, 154 (2013) 442–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, Liu DR, Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage, Nature, 551 (2017) 464–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Tan P, He L, Han G, Zhou Y, Optogenetic Immunomodulation: Shedding Light on Antitumor Immunity, Trends Biotechnol, 35 (2017) 215–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Sun Y, Feng W, Yang P, Huang C, Li F, The biosafety of lanthanide upconversion nanomaterials, Chem Soc Rev, 44 (2015) 1509–1525. [DOI] [PubMed] [Google Scholar]
- [99].Gnach A, Lipinski T, Bednarkiewicz A, Rybka J, Capobianco JA, Upconverting nanoparticles: assessing the toxicity, Chem Soc Rev, 44 (2015) 1561–1584. [DOI] [PubMed] [Google Scholar]
- [100].Wu X, Chen G, Shen J, Li Z, Zhang Y, Han G, Upconversion nanoparticles: a versatile solution to multiscale biological imaging, Bioconjug Chem, 26 (2015) 166–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Henderson TA, Morries LD, Near-infrared photonic energy penetration: can infrared phototherapy effectively reach the human brain?, Neuropsychiatr Dis Treat, 11 (2015) 2191–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Chen G, Shen J, Ohulchanskyy TY, Patel NJ, Kutikov A, Li Z, Song J, Pandey RK, Agren H, Prasad PN, Han G, (alpha-NaYbF4:Tm(3+))/CaF2 core/shell nanoparticles with efficient near-infrared to near-infrared upconversion for high-contrast deep tissue bioimaging, ACS Nano, 6 (2012) 8280–8287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Jayakumar MK, Idris NM, Zhang Y, Remote activation of biomolecules in deep tissues using near-infrared-to-UV upconversion nanotransducers, Proc Natl Acad Sci U S A, 109 (2012) 8483–8488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Chen S, Weitemier AZ, Zeng X, He L, Wang X, Tao Y, Huang AJY, Hashimotodani Y, Kano M, Iwasaki H, Parajuli LK, Okabe S, Teh DBL, All AH, Tsutsui-Kimura I, Tanaka KF, Liu X, McHugh TJ, Near-infrared deep brain stimulation via upconversion nanoparticle-mediated optogenetics, Science, 359 (2018) 679–684. [DOI] [PubMed] [Google Scholar]
- [105].Lichtenegger M, Tiapko O, Svobodova B, Stockner T, Glasnov TN, Schreibmayer W, Platzer D, de la Cruz GG, Krenn S, Schober R, Shrestha N, Schindl R, Romanin C, Groschner K, An optically controlled probe identifies lipid-gating fenestrations within the TRPC3 channel, Nat Chem Biol, 14 (2018) 396–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Ma G, Liu J, Ke Y, Liu X, Li M, Wang F, Han G, Huang Y, Wang Y, Zhou Y, Optogenetic Control of Voltage-Gated Calcium Channels, Angew Chem Int Ed Engl, 57 (2018) 7019–7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Hegemann P, Moglich A, Channelrhodopsin engineering and exploration of new optogenetic tools, Nat Methods, 8 (2011) 39–42. [DOI] [PubMed] [Google Scholar]
- [108].Cosentino C, Alberio L, Gazzarrini S, Aquila M, Romano E, Cermenati S, Zuccolini P, Petersen J, Beltrame M, Van Etten JL, Christie JM, Thiel G, Moroni A, Optogenetics. Engineering of a light-gated potassium channel, Science, 348 (2015) 707–710. [DOI] [PubMed] [Google Scholar]
- [109].Zhang Y, Huang L, Li Z, Ma G, Zhou Y, Han G, Illuminating Cell Signaling with Near-Infrared Light-Responsive Nanomaterials, ACS Nano, 10 (2016) 3881–3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Chernov KG, Redchuk TA, Omelina ES, Verkhusha VV, Near-Infrared Fluorescent Proteins, Biosensors, and Optogenetic Tools Engineered from Phytochromes, Chem Rev, 117 (2017) 6423–6446. [DOI] [PubMed] [Google Scholar]
- [111].Lee M, Li J, Liang Y, Ma G, Zhang J, He L, Liu Y, Li Q, Li M, Sun D, Zhou Y, Huang Y, Engineered Split-TET2 Enzyme for Inducible Epigenetic Remodeling, J Am Chem Soc, 139 (2017) 4659–4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Rost BR, Schneider-Warme F, Schmitz D, Hegemann P, Optogenetic Tools for Subcellular Applications in Neuroscience, Neuron, 96 (2017) 572–603. [DOI] [PubMed] [Google Scholar]