ABSTRACT
Mammalian members of the ErbB family, including the epidermal growth factor receptor (EGFR), can regulate transcription, DNA replication and repair through nuclear entry of either the full-length proteins or their cleaved cytoplasmic domains. In cancer cells, these nuclear functions contribute to tumor progression and drug resistance. Here, we examined whether the single Drosophila EGFR can also localize to the nucleus. A chimeric EGFR protein fused at its cytoplasmic C-terminus to DNA-binding and transcriptional activation domains strongly activated transcriptional reporters when overexpressed in cultured cells or in vivo. However, this activity was independent of cleavage and endocytosis. Without an exogenous activation domain, EGFR fused to a DNA-binding domain did not activate or repress transcription. Addition of the same DNA-binding and transcriptional activation domains to the endogenous Egfr locus through genome editing led to no detectable reporter expression in wild-type or oncogenic contexts. These results show that, when expressed at physiological levels, the cytoplasmic domain of the Drosophila EGFR does not have access to the nucleus. Therefore, nuclear EGFR functions are likely to have evolved after vertebrates and invertebrates diverged.
KEY WORDS: EGFR, Transcription, Nucleus, Cancer
Summary: Nuclear functions of mammalian EGF receptors contribute to cancer. Here, we show that such functions are not conserved in the invertebrate Drosophila.
INTRODUCTION
Direct transcriptional regulation by the cleaved intracellular domain is the primary mode of signal transduction for some cell surface receptors, such as Notch (Ehebauer et al., 2006). Receptor tyrosine kinases of the ErbB family primarily signal through Ras and a kinase cascade that culminates in phosphorylation of transcription factors by mitogen-activated protein kinases (MAPKs) (Fey et al., 2016). However, ErbB receptors can also have direct transcriptional functions (Carpenter and Liao, 2013; Chen and Hung, 2015), and their nuclear localization correlates with poor prognosis in several types of cancer (Brand et al., 2013; Traynor et al., 2013).
Two different mechanisms for nuclear entry by ErbB family members have been described. Binding to neuregulin triggers metalloprotease- and γ-secretase-mediated proteolytic cleavages of ErbB4 that release the cytoplasmic domain, which then enters the nucleus (Lee et al., 2002; Ni et al., 2001). Ligand-bound EGFR and ErbB2 are also endocytosed from the plasma membrane and can undergo retrograde transport to the endoplasmic reticulum and outer nuclear membrane. The full-length proteins are translocated to the inner nuclear membrane through interactions of their nuclear localization signals (NLSs) with importin-β, and extracted into the nucleus through the action of Sec61β (Giri et al., 2005; Lo et al., 2006; Wang et al., 2010).
These nuclear receptors affect transcription by interacting with factors that target them to specific chromatin regions. EGFR positively regulates the expression of cyclin D1, Myc and other genes together with cofactors that include STATs and E2F1 (Bitler et al., 2010; Hanada et al., 2006; Huo et al., 2010; Lo et al., 2005), while ErbB4 can either activate or repress transcription with different cofactors (Beguelin et al., 2010; Clark et al., 2005; Wang et al., 2004). In addition to regulating transcription, nuclear EGFR phosphorylates proteins involved in DNA replication and repair (Chou et al., 2014; Wang et al., 2006).
As the trafficking mechanisms that transport ErbB proteins to the nucleus and their functions there are not fully understood, despite their importance in regeneration and cancer (Chen and Hung, 2015), we hoped to interrogate this process in a genetic model system. We therefore tested whether the single Drosophila ErbB homolog, EGFR, could enter the nucleus. A chimeric EGFR protein fused to exogenous DNA-binding and transcriptional activation domains robustly activated a reporter when overexpressed. However, expression of the same protein at physiological levels did not detectably activate reporters even in oncogenic conditions. These results argue against a significant role for nuclear entry of Drosophila EGFR.
RESULTS AND DISCUSSION
An EGFR–LexAVP16 fusion protein activates transcription when overexpressed
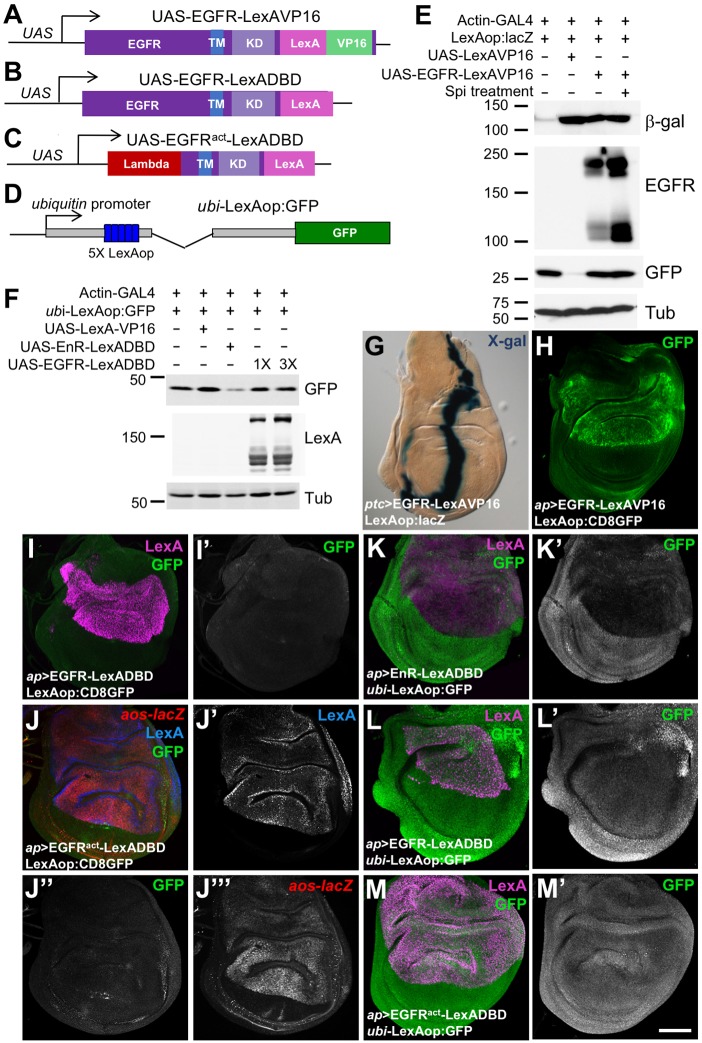
Nuclear entry by mammalian ErbB proteins has been reported primarily in the context of cancer (Brand et al., 2013), although some studies describe transcriptional functions during normal development (Adilakshmi et al., 2011; Sardi et al., 2006; Zscheppang et al., 2011). We hoped to exploit a genetic model system to systematically identify sites and functions of nuclear EGFR. To detect nuclear entry of Drosophila EGFR, we used a sensitive transcriptional reporter system (Kidd et al., 2015; Masuyama et al., 2012; Struhl and Adachi, 1998). A chimeric EGFR protein with a LexA DNA-binding domain and VP16 transcriptional activation domain fused to its cytoplasmic C-terminus (EGFR–LexAVP16; Fig. 1A) activated a LexAop:lacZ reporter when expressed in cultured S2R+ cells (Fig. 1E). Treating the cells with a constitutively secreted form of the ligand Spitz (sSpi) (Schweitzer et al., 1995) enhanced cleavage of full-length EGFR–LexAVP16 to release its cytoplasmic domain, but did not increase reporter activation (Fig. 1E). When expressed in vivo using the GAL4 system, EGFR–LexAVP16 strongly activated the LexAop:lacZ and LexAop:CD8GFP reporters (Lai and Lee, 2006) (Fig. 1G,H). Thus, all or part of the fusion protein can enter the nucleus and activate transcription when it is overexpressed. However, most of the protein localized to cytoplasmic aggregates, and nuclear entry was difficult to detect by immunostaining (Fig. S1A,D).
Fig. 1.
The EGFR activates transcription when fused to DNA-binding and activation domains. (A–D) Diagrams of UAS-EGFRLexAVP16 (A), UAS-EGFRLexADBD (B), UAS-EGFRactLexADBD (C) and ubi-LexAop:GFP (D). TM, transmembrane domain; KD, kinase domain; Lambda, dimerization domain. (E,F) Western blots of lysates from S2R+ cells transfected as indicated, and either treated for 4.5 h with purified sSpiCS or untreated. In E, the β-galactosidase (β-gal) blot reflects expression from LexAop:lacZ. The EGFR blot shows full-length EGFR-LexAVP16 (upper bands) and its cleaved cytoplasmic domain (lower bands). Co-transfected UAS-GFP was used to normalize for transfection efficiency and β-Tubulin (Tub) for total protein. Toxicity of UAS-LexAVP16 resulted in fewer surviving GFP-expressing cells, each of which strongly expressed LexAop:lacZ. In F, the GFP blot reflects ubi-LexAop:GFP reporter expression, which is reduced by transfection of UAS-EnR-LexADBD but not by equal or 3-fold higher amounts of UAS-EGFR-LexADBD. (G,H) Wing discs expressing UAS-EGFRLexAVP16 at the anterior-posterior compartment boundary with patched (ptc)-GAL4 (G) or in the dorsal compartment with ap-GAL4 (H), stained with X-gal, reflecting LexAop:lacZ expression (G) or anti-GFP antibody reflecting LexAop:CD8GFP (H). EGFR–LexAVP16 strongly activates both reporters. (I–M) Wing discs from flies in which ap-GAL4 drove UAS-EGFRLexADBD (I,L), UAS-EGFRactLexADBD (J,M) or UAS-EnRLexADBD (K). Anti-LexA antibody staining (J′, blue in J, magenta in I,K–M) shows the expression of these constructs. Anti-GFP antibody staining (I′,J″,K′–M′, green in K–M) reflects expression from LexAop:GFP (I,J) or ubi-LexAop:GFP (K–M). Anti-β-gal antibody staining shows aos-lacZ expression (J‴, red in J). Scale bar (M′): 100 μm. Fusion of the LexA DNA-binding domain to wild-type or activated EGFR (EGFRact) does not enable it to activate or repress these reporters, although EnRLexADBD represses ubi-LexAop:GFP.
The EGFR does not contain transcriptional activation or repression domains
To determine whether the EGFR contains domains that can activate or repress transcription when targeted to DNA, we fused it to only the LexA DNA-binding domain (EGFR–LexADBD; Fig. 1B). Although its localization appeared identical to that of EGFR–LexAVP16 (Fig. S1B,E), this chimeric protein failed to activate transcription from LexAop:CD8GFP (Fig. 1I). To assay transcriptional repression, we inserted five LexA-binding sites into a ubiquitin promoter driving GFP (Davis et al., 1995) (Fig. 1D). In S2R+ cells, GFP expression from this construct was reduced upon co-transfection of an Engrailed repressor domain and LexA DNA-binding domain fusion (EnR–LexADBD), but not by EGFR–LexADBD (Fig. 1F). EnR–LexADBD also repressed this reporter when expressed in the dorsal wing disc via apterous (ap)-GAL4 (Fig. 1K). In contrast, UAS-EGFR–LexADBD driven by ap-GAL4 did not repress transcription from ubi-LexAop:GFP (Fig. 1L).
To test whether kinase activity would enable the EGFR to regulate transcription, we fused a constitutively active form of the receptor, made by replacing its extracellular domain with the lambda repressor dimerization domain (Queenan et al., 1997), to LexADBD (Fig. 1C). This transgene strongly induced expression of argos (aos), a transcriptional target of the Ras-MAPK pathway (Golembo et al., 1996), when expressed in the dorsal wing disc (Fig. 1J). However, it did not activate or repress transcription through LexA operator sequences (Fig. 1J,M). The EGFR cytoplasmic domain is thus not sufficient to activate or repress transcription when targeted to DNA, regardless of its activation state.
Overexpressed EGFR–LexAVP16 bypasses the normal endocytic pathway
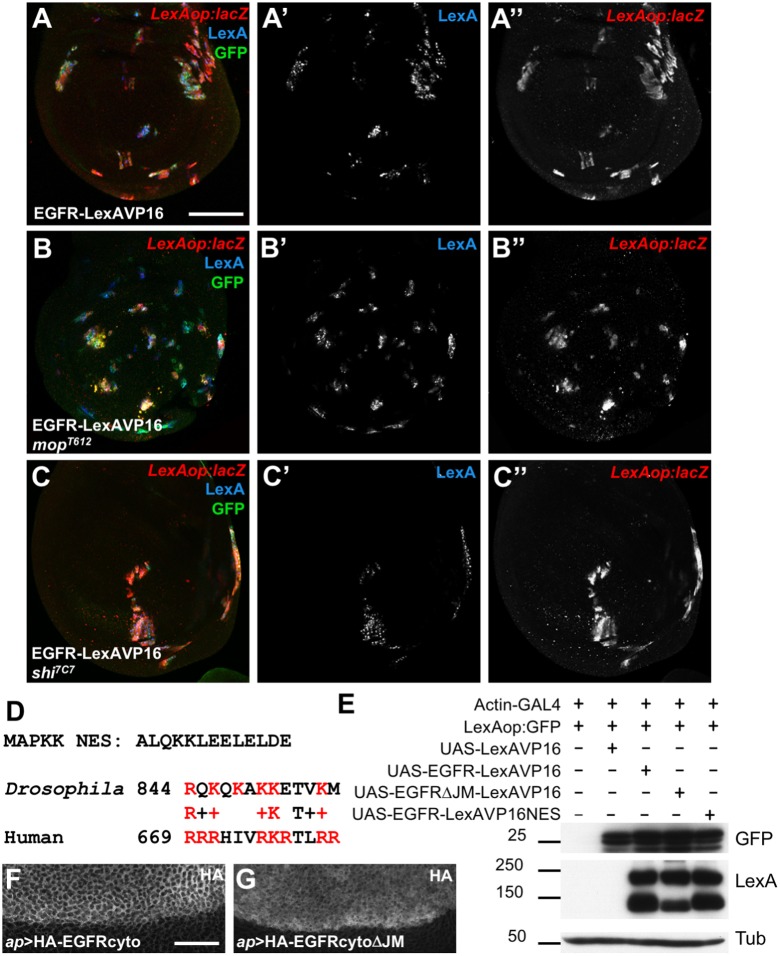
We previously found that EGFR cleavage was dependent on Myopic (Mop), which promotes EGFR progression through endocytosis (Miura et al., 2008). If cleavage is necessary for the cytoplasmic domain to enter the nucleus, then transcriptional activation by EGFR–LexAVP16 should require Mop. However, EGFR–LexAVP16 still activated transcription from LexAop:lacZ when expressed in clones of mop mutant cells in the wing disc (Fig. 2A,B). Reporter activation was unaffected even in clones mutant for shibire (shi), which encodes a Dynamin required for endocytosis (Chen et al., 1991) (Fig. 2C). These data suggest that overexpressed EGFR–LexAVP16 bypasses normal plasma membrane insertion and endocytosis, and enters the nucleus without passing through the secretory pathway.
Fig. 2.
Overexpressed EGFR–LexAVP16 can bypass normal trafficking. (A–C) Wing discs in which UAS-EGFR-LexAVP16 is expressed in clones of cells that are wild type (A), mop mutant (B) or shi mutant (C). Clones are marked by anti-GFP antibody staining (green) and anti-LexA antibody staining (A′–C′, blue). Anti-β-gal antibody staining reflects LexAop:lacZ expression (A″–C″, red), which is not reduced by the blocking of EGFR cleavage that occurs in mop cells or of endocytosis that occurs in shi cells. (D) Sequence of the NES from MAPKK and an alignment of the amino acids immediately following the transmembrane domains of Drosophila and human EGFR. Basic residues are in red. (E) Western blots of S2R+ cell lysates transfected with the indicated constructs. Deleting the juxtamembrane region reduces EGFR–LexAVP16 cleavage, but neither this deletion nor adding an NES reduces reporter expression. (F,G) Portions of wing discs spanning the dorsal-ventral boundary in which UAS-HA-EGFRcyto (F) or UAS-HA-EGFRcytoΔJM (G) is expressed in the dorsal (upper) compartment with ap-GAL4. Anti-HA antibody staining shows cytoplasmic localization. Scale bars: 50 μm (A), 20 μm (F).
The human EGFR contains a tripartite NLS immediately following its transmembrane domain (Hsu and Hung, 2007). The juxtamembrane region of Drosophila EGFR has limited similarity to this sequence, but shares several of its basic residues (Fig. 2D). The cytoplasmic domain contains no other predicted NLS. Deleting this region from EGFR–LexAVP16 did not prevent it from activating LexAop:GFP in S2R+ cells, although production of the smaller EGFR fragment was reduced (Fig. 2E), suggesting that this sequence contributes to proteolytic cleavage. Adding a nuclear export signal (NES) from MAPK kinase (Fukuda et al., 1996) (Fig. 2D) to the C-terminus of EGFR–LexAVP16 likewise did not alter its ability to activate reporter transcription (Fig. 2E).
As another test for potential nuclear functions of the cleaved cytoplasmic domain, we generated transgenic flies expressing the cytoplasmic domain of the EGFR, with or without the juxtamembrane region, driven by UAS sequences. Neither transgene caused lethality or visible phenotypes when expressed ubiquitously with tubulin-GAL4, in the nervous system with elav-GAL4, or in imaginal discs with ap-GAL4 or eyeless-GAL4, and both proteins appeared to be cytoplasmic (Fig. 2F,G). These experiments do not support the model that release of the intracellular domain into the cytoplasm allows it to enter the nucleus and regulate transcription.
Nuclear entry by the endogenous EGFR cannot be detected
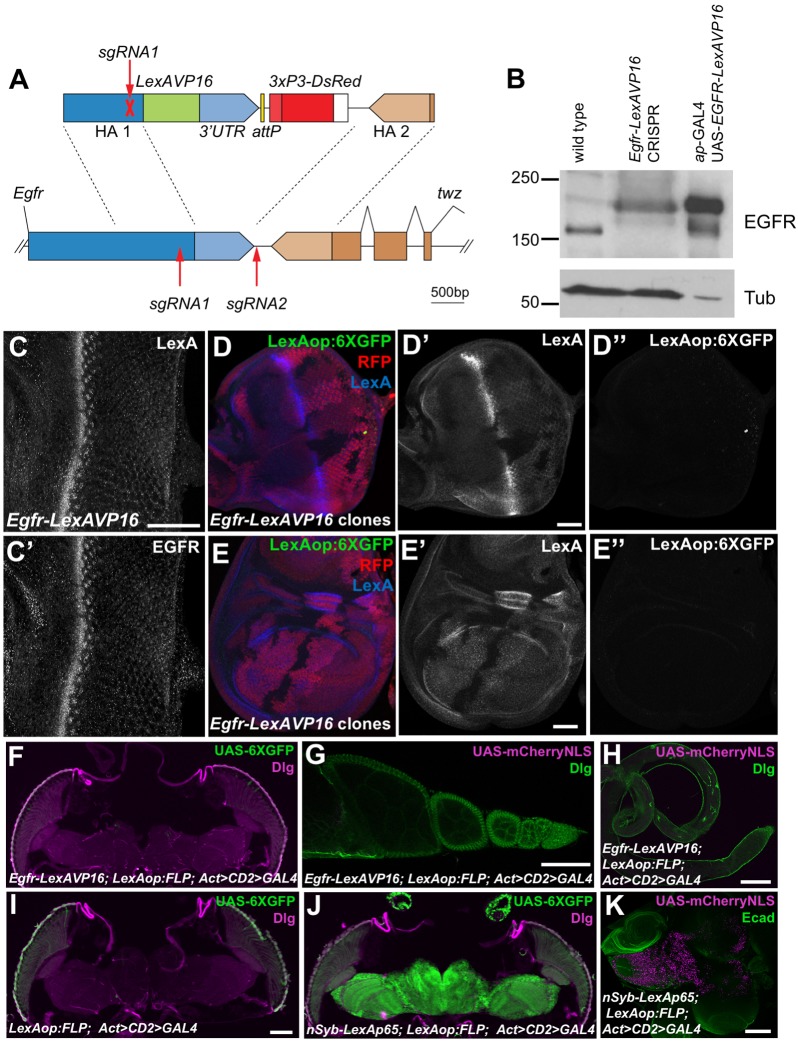
The ability of EGFR–LexAVP16 to activate transcription in cells deficient for endocytosis suggested that its nuclear import might be an artifact of overexpression. To test whether nuclear entry occurs at physiological expression levels, we used CRISPR-Cas9 editing to insert the LexA DNA-binding domain and VP16 activation domain at the C-terminus of the endogenous Egfr gene (Fig. 3A). These domains were followed by the native Egfr 3′UTR and a 3XP3-DsRed element used to screen for editing events (Gratz et al., 2014). Flies homozygous for this insertion were viable, although the females were sterile, indicating that addition of LexAVP16 slightly reduced receptor signaling through the canonical pathway (Price et al., 1989). The EGFR in these homozygous flies migrated at the size expected for EGFR–LexAVP16, and was expressed at the same level as wild-type EGFR, which was much lower than that for the GAL4-driven EGFR–LexAVP16 (Fig. 3B). LexA staining was observed in the same pattern as EGFR in imaginal discs (Fig. 3C) and was not detectable in the nucleus (Fig. S1C). To look for transcriptional activation by the modified Egfr-LexAVP16 gene, we used a very sensitive reporter with six tandem copies of GFP (Shearin et al., 2014) (Fig. 4A) and generated clones lacking the locus to compare background and induced reporter levels within the same disc; however, no activation of the reporter was observed (Fig. 3D,E).
Fig. 3.
EGFR–LexAVP16 does not activate transcription when expressed at physiological levels. (A) The sgRNAs used to edit the endogenous Egfr sequence (lower) and the construct used for homology-directed repair (upper). (B) Western blot of lysates from wild-type and homozygous Egfr-LexAVP16 CRISPR adult flies and from ap-GAL4; UAS-EGFRLexAVP16 larval wing discs. In the CRISPR flies, EGFR is expressed at wild-type levels and migrates at the size for EGFR–LexAVP16. The wing disc extracts had much stronger EGFR–LexAVP16 expression and less protein was loaded. (C) Egfr-LexAVP16 eye disc stained with anti-LexA (C) and anti-EGFR (C′) antibody. Scale bar: 50 μm. (D,E) Eye disc (D) and wing disc (E) with clones lacking the Egfr-LexAVP16-modified locus marked by the absence of RFP (red) and LexA (D′,E′, blue). Anti-GFP antibody staining shows LexAop:6XGFP expression (D″,E″, green), which is not detectable in cells with Egfr-LexAVP16 above the background staining present in wild-type cells. Scale bars: 50 μm. (F–H) Egfr-LexAVP16; LexAop:FLP; Act>CD2>GAL4 adult head (F), ovary (G) and testis (H) showing no expression of UAS-6×GFP stained with anti-GFP antibody (green, F) or of UAS-mCherry-NLS (magenta, G,H). (I) A negative control adult head without Egfr-LexAVP16. (J,K) nSyb-LexAp65; LexAop:FLP; Act>CD2>GAL4 induces strong expression of UAS-6×GFP in adult brain (J) and of UAS-mCherry-NLS in larval brain (K). F–J are stained for Discs large (Dlg, magenta in F,I,J; green in G,H) and K is stained for E-cadherin (Ecad, green). Scale bars: 100 μm.
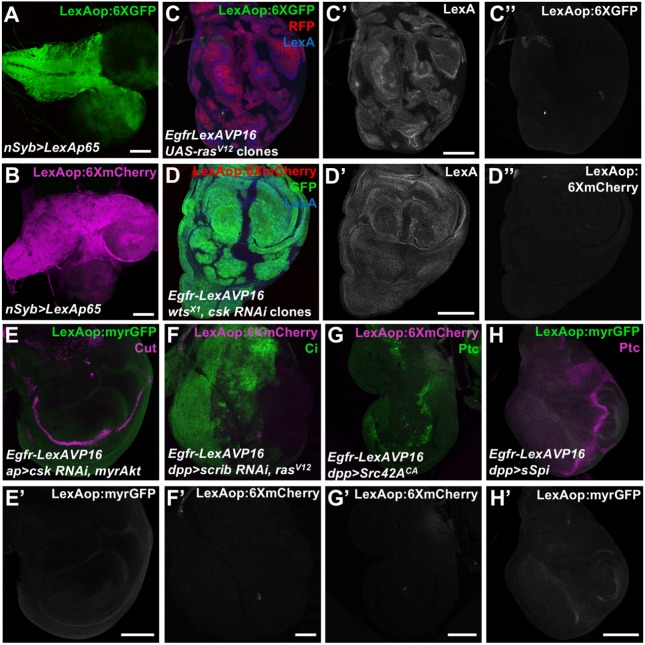
Fig. 4.
Oncogenic transformation does not induce EGFR nuclear localization. (A,B) Larval brains showing LexAop:6XGFP (anti-GFP antibody, A) and LexAop:6XmCherry (B) expression driven by nSyb>LexAp65. (C) Wing disc with UAS-rasV12 expressed in clones homozygous for Egfr-LexAVP16, labeled with RFP (red), showing LexA (C′, blue) and LexAop:6XGFP (C″, green). (D) Egfr-LexAVP16 wing disc with wts mutant clones expressing UAS-csk RNAi and GFP (green), showing LexA (D′, blue) and LexAop:6XmCherry (D″, red) expression. (E–H) Egfr-LexAVP16 wing discs expressing UAS-csk RNAi and UAS-myrAkt (E), UAS-scrib RNAi and UAS-rasV12 (F), UAS-Src42ACA (G) or UAS-sSpi (H) in the dorsal compartment with ap-GAL4 (E) or at the anterior-posterior compartment boundary with decapentaplegic (dpp)-GAL4 (F-H), showing LexAop:myrGFP (anti-GFP, E′,H′, green in E,H) or LexAop:6XmCherry (F′,G′, magenta in F,G) expression. Cut (magenta in E) marks the dorsal-ventral boundary, Cubitus interruptus (Ci, green in F) marks the anterior compartment, and Ptc (green in G, magenta in H) marks the anterior-posterior boundary. None of these oncogenic conditions induce reporter activation by Egfr-LexAVP16. Scale bars: 100 μm.
It remained possible that the chimeric receptor could transiently enter the nucleus at some stage of development. We combined our Egfr-LexAVP16 CRISPR modification with LexAop:FLP, Actin-GAL4 interrupted by an FRT-flanked stop cassette (Act>CD2>GAL4), and UAS-6×GFP or UAS-mCherry-NLS reporters, to permanently label any cells that transiently expressed FLP recombinase. We detected no reporter expression above background in the adult head or reproductive organs (Fig. 3F–I) or in any other tissues examined, although the reporters were activated in the nervous system as expected when LexA:p65 was driven by the neural Synaptobrevin (nSyb) promoter (Fig. 3J,K). As the overexpression experiments show that EGFR–LexAVP16 activates transcription if it can enter the nucleus, we conclude that the cytoplasmic domain of endogenous EGFR does not have access to the nucleus.
Oncogenic transformation does not promote EGFR nuclear entry
Nuclear entry by mammalian EGFR homologs has been observed in a variety of tumors, and is considered a biomarker for poor prognosis (Brand et al., 2013). Phosphorylation by Akt or Src family kinases targets EGFR to the nucleus in breast cancer and lung cancer, respectively (Huang et al., 2011; Iida et al., 2013; Li et al., 2009). We examined whether oncogenic transformations would allow endogenous Egfr-LexAVP16 to activate transcription. We tested overexpression of activated forms of Ras85D, Akt1 or Src42A, warts (wts) mutant clones, and RNAi targeting C-terminal Src kinase (Csk) or scribbled (scrib), as well as combinations of these factors; for example, Ras activation in cells lacking scrib promotes metastasis (Pagliarini and Xu, 2003). None of these manipulations resulted in detectable activation of reporters driven by LexA operator sequences (Fig. 4C–G). Activating the EGFR by overexpressing sSpi also did not produce detectable reporter activity (Fig. 4H).
Nuclear EGFR functions may be specific to chordates
The endocytosis-independent transcriptional activation initiated by overexpressed EGFR-–LexAVP16, coupled with the lack of activation when the same chimeric protein was expressed at physiological levels following genome editing, argue against a nuclear function for Drosophila EGFR. Although it is difficult to exclude the possibility that activation by Egfr-LexAVP16 can occur in some contexts, we have examined epithelial tissues, which are the major site of EGFR expression at embryonic, larval and adult stages, using sensitive reporters and a recombinase-based system to capture transient expression. Oncogenic transformations also failed to induce endogenous EGFR to enter the nucleus. We conclude that nuclear entry by the overexpressed protein did not reflect a normal physiological process, and that studies of nuclear functions of cell surface receptors that are solely based on protein overexpression should be interpreted with caution. The cleavage that releases the intracellular domain of the EGFR may occur in a different subcellular compartment in Drosophila to that in mammals. The failure of endogenous Egfr-LexAVP16 to activate transcription suggests that the receptor is not cleaved until its intracellular domain is sequestered from the cytoplasm in the internal vesicles of multivesicular bodies, consistent with the requirement of mop for both endocytic progression and cleavage (Miura et al., 2008).
Previous studies have found that Drosophila Egfr mutations strongly resemble mutations in downstream components of the Ras-MAPK pathway (Galindo et al., 2005; Jiang et al., 2011; Roch et al., 2002; Yang and Baker, 2001; Yang and Baker, 2003), and thus did not identify candidate functions for nuclear EGFR. Nuclear localization of ErbB proteins may have evolved after the separation of vertebrate and invertebrate lineages. The NLS identified in human EGFR is conserved in other ErbB family members (Hsu and Hung, 2007), but does not appear to be homologous at the sequence or functional level in Drosophila EGFR. Significant conservation of this sequence is present in ErbB homologs in all classes of chordates, including the basal chordate Ciona intestinalis, but not in other phyla. The strong conservation of the EGFR NLS in chordates implies that it has evolved essential physiological functions. Unfortunately, nuclear roles of EGFR have been exploited by cancer cells, enabling them to grow and metastasize more aggressively. Our findings show that signaling through the Ras-MAPK pathway is independent of the ability of EGFR to localize to the nucleus, supporting cancer treatment strategies that specifically target nuclear EGFR (Brand et al., 2013).
MATERIALS AND METHODS
Genetics
Fly lines used were: (1) ptc-GAL4; LexAop:lacZ (Szuts and Bienz, 2000), (2) ap-GAL4; LexAop:CD8GFP (Lai and Lee, 2006), (3) ap-GAL4; ubi-LexAop:GFP, (4) UAS-EGFRLexAVP16, LexAop:lacZ; FRT80, (5) UAS-EGFRLexAVP16, LexAop:lacZ; FRT80, mopT612 (Miura et al., 2008), (6) FRT19, shi7C7 (Legent et al., 2012); UAS-EGFRLexAVP16, LexAop:lacZ, (7) hsFLP122, UAS-CD8GFP; tub-GAL4/CyO; FRT80, tub-GAL80, (8) UbxFLP, UAS-CD8GFP; tub-GAL4/CyO; FRT80, tub-GAL80, (9) FRT42, ubi-RFP, EgfrLexAVP16 CRISPR, (10) hsFLP122; FRT42, (11) LexAop:6XGFP (Shearin et al., 2014), (12) Act>CD2>GAL4; LexAop:FLP, (13) UAS-6XGFP (Shearin et al., 2014), (14) Egfr-LexAVP16; UAS-6XGFP, (15) nSyb-LexAp65 (Shearin et al., 2013); UAS-6XGFP, (16) Egfr-LexAVP16; UAS-mCherryNLS, (17) nSyb-LexAp65; UAS-mCherryNLS, (18) LexAop:6XmCherry (Shearin et al., 2014), (19) FRT42, ubi-RFP, EgfrLexAVP16; UAS-rasV12 (Karim and Rubin, 1998), LexAop:6XGFP, (20) Ubx-FLP; FRT42, tub-GAL80; tub-GAL4/TM6B, (21) UAS-csk RNAi (TRiP.HMS02277), EgfrLexAVP16; LexAop:6XmCherry, FRT82, wtsX1, (22) UbxFLP, UAS-GFP; tub-GAL4, FRT82, tub-GAL80/TM6B, (23) dpp-GAL4 (Hazelett et al., 1998); LexAop:myrGFP, (24) Egfr-LexAVP16; UAS-sSpi (Schweitzer et al., 1995), (25) ap-GAL4; LexAop:myrGFP, (26) UAS-csk RNAi, EgfrLexAVP16; UAS-myrAkt (Stocker et al., 2002), (27) dpp-GAL4; LexAop:6XmCherry, (28) UAS-rasV12, EgfrLexAVP16; UAS-scrib RNAi (TRiP.JF03229), (29) Egfr-LexAVP16; UAS-Srcc42ACA (Tateno et al., 2000), (30) EgfrLexAVP16; Act>CD2>wg (Struhl and Basler, 1993), (31) LexAop:FLP, (32) tub-GAL4, (33) elav-GAL4 and (34) eyeless-GAL4 (Hazelett et al., 1998). Components of these stocks that are not specifically referenced are described in FlyBase. When hs-FLP was used to generate clones, larvae were heat shocked at 38°C for 1 h in both first and second instar. Larval tissues from both males and females were examined and no differences were noted.
Immunohistochemistry
Eye and wing discs, larval brains and adult tissues from both males and females were stained with antibodies and X-gal as previously described (Hazelett et al., 1998; Maurel-Zaffran et al., 2001; Miura et al., 2006; Steinhauer et al., 2009). Antibodies used were chicken anti-GFP (Life Technologies, Carlsbad, CA A10262; 1:400), mouse anti-LexA DNA-binding domain (Santa Cruz Biotechnology, Dallas, TX sc-7544; 1:50), rabbit anti-β-gal (MPBio, Santa Ana, CA 08559761; 1:5000), rat anti-HA (Roche, Basel, Switzerland 3F10; 1:100), rabbit anti-EGFR (Rodrigues et al., 2005) (1:500), mouse anti-Dlg [Developmental Studies Hybridoma Bank (DSHB), Iowa City, IA 4F3; 1:10], rat anti-Ecad (DSHB DCAD2; 1:10), mouse anti-Ptc (DSHB Apa1; 1:10), mouse anti-Cut (DSHB 2B10; 1:10), mouse anti-Lamin (DSHB ADL67.10; 1:10) and rat anti-Ci (DSHB 2A1; 1:10). RFP and mCherry were detected using the intrinsic fluorescence of the proteins. Fluorescent secondary antibodies (1:200) were from Jackson Immunoresearch, West Grove, PA, and images were obtained with a Leica (Wetzlar, Germany) SP5 confocal microscope. For each image shown, the same phenotype was observed in at least five individuals from at least two independent crosses. We expected this to be sufficient to distinguish between the presence and absence of reporter expression.
Molecular biology
UAS-EGFR–LexAVP16 was made by combining the LexA DNA-binding domain amplified from pEG202 (Golemis et al., 2001) with the VP16 activation domain amplified from pUAS-osaAD (Collins et al., 1999) in pUAST (Drosophila Genomics Resource Center) using EcoRI, XhoI and XbaI restriction sites to make UAS-LexAVP16. The LexAVP16 fragment was then amplified and cloned into an XbaI site at the 3′ end of pMT-EGFR-V5-His (a gift from Diego Alvarado, Perelman School of Medicine, University of Pennsylvania, PA). The EGFR–LexAVP16 sequence was transferred to the KpnI and XbaI sites of pUAST following partial digestion. UAS-EGFR–LexADBD was made by replacing the LexA-VP16 fragment of this plasmid with a PCR product encoding only the LexA DNA-binding domain, using XbaI sites. To make UAS-EGFRact–LexADBD, an EcoRI fragment of this construct extending into the cytoplasmic domain of the EGFR was replaced with a fragment from UAS-λtop (Queenan et al., 1997) that ended at the same EcoRI site. UAS-EnR–LexADBD was made by replacing the osa segment of UAS-osaRD (Collins et al., 1999) with an EcoRI/XhoI LexADBD fragment from UAS-LexAVP16. The LexAop:lacZ reporter used in S2R+ cells was made by synthesizing one LexA operator as an oligonucleotide and cloning it into the EcoRI and BamHI sites of pCasper-hs-β-gal (Ronchi et al., 1993). The LexAop:GFP S2R+ cell reporter was pJFRC18-8XLexAop:mCD8GFP (Pfeiffer et al., 2010). To make ubi-LexAop:GFP, five LexAop sites were amplified from this plasmid and cloned into the HindIII site of an XbaI/KpnI fragment of pCasper4-ubi-NLSGFP (Davis et al., 1995) transferred to Bluescript (Stratagene). The same fragment containing the LexAop sites was then cloned back into pCasper4-ubi-NLSGFP. UAS-HA–EGFRcyto and UAS-HA–EGFRcytoΔJM were made by amplifying the cytoplasmic domain of the EGFR (beginning at R844 or M856 respectively) and cloning it into the NdeI and BglII sites of pUAST-HA (a gift from Michael Weir, Wesleyan University, CT). UAS-EGFRΔJMLexAVP16 was made using PCR to delete the RQKQKAKK sequence from UAS-EGFR–LexAVP16. UAS-EGFR–LexAVP16NES was made using PCR to add the sequence ALQKKLEELELDE to the C-terminus of UAS-EGFR–LexAVP16 in the AgeI and SpeI sites. All constructs were verified by sequencing. Transgenic flies were made for UAS-EGFR–LexAVP16, UAS-EGFR–LexADBD, UAS-EGFRact–LexADBD, UAS-EnR–LexADBD, ubi-LexAop:GFP, UAS-HA–EGFRcyto and UAS-HA–EGFRcytoΔJM by Genetic Services, Inc., Cambridge, MA. These constructs were integrated in random locations and at least three lines were tested for each.
To make the CRISPR modification of endogenous Egfr, synthetic-guide (sg)RNA1 (5′-GCCCGTTAGTGTGGACAATC-3′) and sgRNA2 (5′-GTTTACGAGTTAGTAAGGTG-3′) were cloned into pCFD4 (Port et al., 2014) by PCR and Gibson assembly using the BbsI site. A 1.2 kb right homology arm beginning adjacent to the cleavage site of sgRNA2 (and lacking most of the sgRNA sequence) was cloned into the AscI site of pDsRed-attP (Addgene, Cambridge, MA) to make pDsRed-RA. The left arm was synthesized by Genscript and consisted of the last 1.2 kb of the Egfr coding sequence (introducing a silent mutation in the PAM of sgRNA1, from CGG to CAG) in frame with the LexA DNA-binding domain and VP16 activation domain as in UAS-EGFR–LexAVP16, followed by the endogenous Egfr 3′UTR (895 bp), cloned into the NheI and NdeI sites of pDsRed-RA. The two plasmids were co-injected by Genetivision, Houston, TX; two independent integrations were identified by red fluorescence in the eye and verified by PCR and sequencing.
Cell culture and western blotting
S2R+ cells [originally obtained from Ruth Lehmann (Skirball Institute, NYU School of Medicine, NY) and used regularly in our lab] were maintained in Schneider's medium (Gibco, Thermo Fisher Scientific, Waltham, MA) supplemented with 10% fetal bovine serum and 150 µg/ml of penicillin-streptomycin. His-tagged sSpiCS was purified from medium conditioned by stably transfected S2 cells as described previously (Miura et al., 2006). Its activity was confirmed by blotting lysates from treated EGFR-expressing cells for phosphorylated MAPK (Rolled). Cells were transfected via Effectene (Qiagen, Hilden, Germany) according to the manufacturer's instructions. All UAS plasmids were co-transfected with actin-GAL4. Cells were lysed in RIPA buffer as described previously (Miura et al., 2006). To extract protein from adult flies, 20 individuals were lysed in 100 µl RIPA buffer supplemented with EDTA-free cOmplete protease inhibitor (Roche, Basel, Switzerland), 5 mM NaF and 1 mM Na3VO4. The lysate was then cleared by centrifugation (11,000 g for 5 min) and an equal volume of 2× Laemmli buffer was added. For western blots, the samples were boiled for 5 min at 95°C and loaded on 8% SDS-PAGE gels. The gels were transferred onto nitrocellulose membranes (Biorad, Hercules, CA), which were blocked for 30 min in TBST (20 mM Tris-HCl pH 7.6, 137 mM NaCl and 0.2% Tween-20) supplemented with 5% low-fat milk, prior to incubation with primary antibodies overnight at 4°C in TBST with 5% milk. Blots were washed with TBST for 30 min and incubated with horseradish peroxidase-conjugated secondary antibodies (1:10,000; Jackson Immunoresearch, West Grove, PA) for 1 h in TBST plus 5% milk. Blots were developed with enhanced chemiluminescence (Thermo SuperSignal WestPico, Thermo Fisher Scientific, Waltham, MA). Primary antibodies used were mouse anti-LexADBD (Santa Cruz Biotechnology, Dallas, TX sc-7544; 1 :10,000), mouse anti-GFP (Santa Cruz Biotechnology, Dallas, TX sc-9996; 1:10,000), rabbit anti-β-gal (MPBio 08559761; 1:5000), rabbit anti-EGFR (Miura et al., 2008; 1:10,000), mouse anti-β-tubulin (Covance, Princeton, NJ MMS-410P; 1:10,000), and mouse anti-dpERK (Sigma-Aldrich, St Louis, MO M8159; 1:2500). Each experiment was replicated at least three times.
Supplementary Material
Acknowledgements
We thank Diego Alvarado, Nick Baker, Welcome Bender, Mariann Bienz, Felix Karim, Aloma Rodrigues, Trudi Schupbach, Gary Struhl, Michael Weir, the Bloomington Drosophila Stock Center, Addgene and the Developmental Studies Hybridoma Bank for reagents, and Flybase for essential information. Justine Marsolier and Jean-Yves Roignant contributed to the early stages of this project. The manuscript was improved by the critical comments of Jessica Douthit and Cheuk Hei Ho.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: M.C., K.L., J.E.T.; Investigation: M.C., D.Q.H., H.H.L., J.E.T.; Writing - original draft: J.E.T.; Writing - review & editing: M.C., D.Q.H., H.H.L., K.L.; Supervision: K.L., J.E.T.; Project administration: J.E.T.; Funding acquisition: J.E.T.
Funding
This work was supported by National Institutes of Health (grant EY013777). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.220251.supplemental
References
- Adilakshmi T., Ness-Myers J., Madrid-Aliste C., Fiser A. and Tapinos N. (2011). A nuclear variant of ErbB3 receptor tyrosine kinase regulates ezrin distribution and Schwann cell myelination. J. Neurosci. 31, 5106-5119. 10.1523/JNEUROSCI.5635-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beguelin W., Diaz Flaque M. C., Proietti C. J., Cayrol F., Rivas M. A., Tkach M., Rosemblit C., Tocci J. M., Charreau E. H., Schillaci R. et al. (2010). Progesterone receptor induces ErbB-2 nuclear translocation to promote breast cancer growth via a novel transcriptional effect: ErbB-2 function as a coactivator of Stat3. Mol. Cell. Biol. 30, 5456-5472. 10.1128/MCB.00012-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitler B. G., Goverdhan A. and Schroeder J. A. (2010). MUC1 regulates nuclear localization and function of the epidermal growth factor receptor. J. Cell Sci. 123, 1716-1723. 10.1242/jcs.062661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand T. M., Iida M., Luthar N., Starr M. M., Huppert E. J. and Wheeler D. L. (2013). Nuclear EGFR as a molecular target in cancer. Radiother. Oncol. 108, 370-377. 10.1016/j.radonc.2013.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Carpenter G. and Liao H.-J. (2013). Receptor tyrosine kinases in the nucleus. Cold Spring Harb. Perspect. Biol. 5, a008979 10.1101/cshperspect.a008979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M.-K. and Hung M.-C. (2015). Proteolytic cleavage, trafficking, and functions of nuclear receptor tyrosine kinases. FEBS J. 282, 3693-3721. 10.1111/febs.13342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M. S., Obar R. A., Schroeder C. C., Austin T. W., Poodry C. A., Wadsworth S. C. and Vallee R. B. (1991). Multiple forms of dynamin are encoded by shibire, a Drosophila gene involved in endocytosis. Nature 351, 583-586. 10.1038/351583a0 [DOI] [PubMed] [Google Scholar]
- Chou R.-H., Wang Y.-N., Hsieh Y.-H., Li L.-Y., Xia W., Chang W.-C., Chang L.-C., Cheng C.-C., Lai C.-C., Hsu J. L. et al. (2014). EGFR modulates DNA synthesis and repair through Tyr phosphorylation of histone H4. Dev. Cell 30, 224-237. 10.1016/j.devcel.2014.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark D. E., Williams C. C., Duplessis T. T., Moring K. L., Notwick A. R., Long W., Lane W. S., Beuvink I., Hynes N. E. and Jones F. E. (2005). ERBB4/HER4 potentiates STAT5A transcriptional activity by regulating novel STAT5A serine phosphorylation events. J. Biol. Chem. 280, 24175-24180. 10.1074/jbc.M414044200 [DOI] [PubMed] [Google Scholar]
- Collins R. T., Furukawa T., Tanese N. and Treisman J. E. (1999). Osa associates with the Brahma chromatin remodeling complex and promotes the activation of some target genes. EMBO J. 18, 7029-7040. 10.1093/emboj/18.24.7029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis I., Girdham C. H. and O'Farrell P. H. (1995). A nuclear GFP that marks nuclei in living Drosophila embryos; maternal supply overcomes a delay in the appearance of zygotic fluorescence. Dev. Biol. 170, 726-729. 10.1006/dbio.1995.1251 [DOI] [PubMed] [Google Scholar]
- Ehebauer M., Hayward P. and Arias A. M. (2006). Notch signaling pathway. Science STKE 364, cm7 10.1126/stke.3642006cm7 [DOI] [PubMed] [Google Scholar]
- Fey D., Matallanas D., Rauch J., Rukhlenko O. S. and Kholodenko B. N. (2016). The complexities and versatility of the RAS-to-ERK signalling system in normal and cancer cells. Semin. Cell Dev. Biol. 58, 96-107. 10.1016/j.semcdb.2016.06.011 [DOI] [PubMed] [Google Scholar]
- Fukuda M., Gotoh I., Gotoh Y. and Nishida E. (1996). Cytoplasmic localization of mitogen-activated protein kinase kinase directed by its NH2-terminal, leucine-rich short amino acid sequence, which acts as a nuclear export signal. J. Biol. Chem. 271, 20024-20028. 10.1074/jbc.271.33.20024 [DOI] [PubMed] [Google Scholar]
- Galindo M. I., Bishop S. A. and Couso J. P. (2005). Dynamic EGFR-Ras signalling in Drosophila leg development. Dev. Dyn. 233, 1496-1508. 10.1002/dvdy.20452 [DOI] [PubMed] [Google Scholar]
- Giri D. K., Ali-Seyed M., Li L.-Y., Lee D.-F., Ling P., Bartholomeusz G., Wang S.-C. and Hung M.-C. (2005). Endosomal transport of ErbB-2: mechanism for nuclear entry of the cell surface receptor. Mol. Cell. Biol. 25, 11005-11018. 10.1128/MCB.25.24.11005-11018.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golembo M., Schweitzer R., Freeman M. and Shilo B. Z. (1996). argos transcription is induced by the Drosophila EGF receptor pathway to form an inhibitory feedback loop. Development 122, 223-230. [DOI] [PubMed] [Google Scholar]
- Golemis E. A., Serebriiskii I., Finley R. L. Jr, Kolonin M. G., Gyuris J. and Brent R. (2001). Interaction trap/two-hybrid system to identify interacting proteins. Curr. Protoc. Cell Biol. Chapter 17, Unit 17 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz S. J., Ukken F. P., Rubinstein C. D., Thiede G., Donohue L. K., Cummings A. M. and O'Connor-Giles K. M. (2014). Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics 196, 961-971. 10.1534/genetics.113.160713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada N., Lo H.-W., Day C.-P., Pan Y., Nakajima Y. and Hung M.-C. (2006). Co-regulation of B-Myb expression by E2F1 and EGF receptor. Mol. Carcinog. 45, 10-17. 10.1002/mc.20147 [DOI] [PubMed] [Google Scholar]
- Hazelett D. J., Bourouis M., Walldorf U. and Treisman J. E. (1998). decapentaplegic and wingless are regulated by eyes absent and eyegone and interact to direct the pattern of retinal differentiation in the eye disc. Development 125, 3741-3751. [DOI] [PubMed] [Google Scholar]
- Hsu S.-C. and Hung M.-C. (2007). Characterization of a novel tripartite nuclear localization sequence in the EGFR family. J. Biol. Chem. 282, 10432-10440. 10.1074/jbc.M610014200 [DOI] [PubMed] [Google Scholar]
- Huang W.-C., Chen Y.-J., Li L.-Y., Wei Y.-L., Hsu S.-C., Tsai S.-L., Chiu P.-C., Huang W.-P., Wang Y.-N., Chen C.-H. et al. (2011). Nuclear translocation of epidermal growth factor receptor by Akt-dependent phosphorylation enhances breast cancer-resistant protein expression in gefitinib-resistant cells. J. Biol. Chem. 286, 20558-20568. 10.1074/jbc.M111.240796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo L., Wang Y.-N., Xia W., Hsu S.-C., Lai C.-C., Li L.-Y., Chang W.-C., Wang Y., Hsu M.-C., Yu Y.-L. et al. (2010). RNA helicase A is a DNA-binding partner for EGFR-mediated transcriptional activation in the nucleus. Proc. Natl. Acad. Sci. USA 107, 16125-16130. 10.1073/pnas.1000743107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida M., Brand T. M., Campbell D. A., Li C. and Wheeler D. L. (2013). Yes and Lyn play a role in nuclear translocation of the epidermal growth factor receptor. Oncogene 32, 759-767. 10.1038/onc.2012.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Grenley M. O., Bravo M.-J., Blumhagen R. Z. and Edgar B. A. (2011). EGFR/Ras/MAPK signaling mediates adult midgut epithelial homeostasis and regeneration in Drosophila. Cell Stem Cell 8, 84-95. 10.1016/j.stem.2010.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim F. D. and Rubin G. M. (1998). Ectopic expression of activated Ras1 induces hyperplastic growth and increased cell death in Drosophila imaginal tissues. Development 125, 1-9. [DOI] [PubMed] [Google Scholar]
- Kidd S., Struhl G. and Lieber T. (2015). Notch is required in adult Drosophila sensory neurons for morphological and functional plasticity of the olfactory circuit. PLoS Genet. 11, e1005244 10.1371/journal.pgen.1005244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai S.-L. and Lee T. (2006). Genetic mosaic with dual binary transcriptional systems in Drosophila. Nat. Neurosci. 9, 703-709. 10.1038/nn1681 [DOI] [PubMed] [Google Scholar]
- Lee H.-J., Jung K.-M., Huang Y. Z., Bennett L. B., Lee J. S., Mei L. and Kim T.-W. (2002). Presenilin-dependent gamma-secretase-like intramembrane cleavage of ErbB4. J. Biol. Chem. 277, 6318-6323. 10.1074/jbc.M110371200 [DOI] [PubMed] [Google Scholar]
- Legent K., Steinhauer J., Richard M. and Treisman J. E. (2012). A screen for X-linked mutations affecting Drosophila photoreceptor differentiation identifies Casein kinase 1alpha as an essential negative regulator of Wingless signaling. Genetics 190, 601-616. 10.1534/genetics.111.133827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Iida M., Dunn E. F., Ghia A. J. and Wheeler D. L. (2009). Nuclear EGFR contributes to acquired resistance to cetuximab. Oncogene 28, 3801-3813. 10.1038/onc.2009.234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo H.-W., Hsu S.-C., Ali-Seyed M., Gunduz M., Xia W., Wei Y., Bartholomeusz G., Shih J.-Y. and Hung M.-C. (2005). Nuclear interaction of EGFR and STAT3 in the activation of the iNOS/NO pathway. Cancer Cell 7, 575-589. 10.1016/j.ccr.2005.05.007 [DOI] [PubMed] [Google Scholar]
- Lo H.-W., Ali-Seyed M., Wu Y., Bartholomeusz G., Hsu S.-C. and Hung M.-C. (2006). Nuclear-cytoplasmic transport of EGFR involves receptor endocytosis, importin beta1 and CRM1. J. Cell. Biochem. 98, 1570-1583. 10.1002/jcb.20876 [DOI] [PubMed] [Google Scholar]
- Masuyama K., Zhang Y., Rao Y. and Wang J. W. (2012). Mapping neural circuits with activity-dependent nuclear import of a transcription factor. J. Neurogenet. 26, 89-102. 10.3109/01677063.2011.642910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurel-Zaffran C., Suzuki T., Gahmon G., Treisman J. E. and Dickson B. J. (2001). Cell-autonomous and -nonautonomous functions of LAR in R7 photoreceptor axon targeting. Neuron 32, 225-235. 10.1016/S0896-6273(01)00471-8 [DOI] [PubMed] [Google Scholar]
- Miura G. I., Buglino J., Alvarado D., Lemmon M. A., Resh M. D. and Treisman J. E. (2006). Palmitoylation of the EGFR ligand Spitz by Rasp increases Spitz activity by restricting its diffusion. Dev. Cell 10, 167-176. 10.1016/j.devcel.2005.11.017 [DOI] [PubMed] [Google Scholar]
- Miura G. I., Roignant J.-Y., Wassef M. and Treisman J. E. (2008). Myopic acts in the endocytic pathway to enhance signaling by the Drosophila EGF receptor. Development 135, 1913-1922. 10.1242/dev.017202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni C.-Y., Murphy M. P., Golde T. E. and Carpenter G. (2001). gamma -Secretase cleavage and nuclear localization of ErbB-4 receptor tyrosine kinase. Science 294, 2179-2181. 10.1126/science.1065412 [DOI] [PubMed] [Google Scholar]
- Pagliarini R. A. and Xu T. (2003). A genetic screen in Drosophila for metastatic behavior. Science 302, 1227-1231. 10.1126/science.1088474 [DOI] [PubMed] [Google Scholar]
- Pfeiffer B. D., Ngo T.-T. B., Hibbard K. L., Murphy C., Jenett A., Truman J. W. and Rubin G. M. (2010). Refinement of tools for targeted gene expression in Drosophila. Genetics 186, 735-755. 10.1534/genetics.110.119917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Port F., Chen H.-M., Lee T. and Bullock S. L. (2014). Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc. Natl. Acad. Sci. USA 111, E2967-E2976. 10.1073/pnas.1405500111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price J. V., Clifford R. J. and Schupbach T. (1989). The maternal ventralizing locus torpedo is allelic to faint little ball, an embryonic lethal, and encodes the Drosophila EGF receptor homolog. Cell 56, 1085-1092. 10.1016/0092-8674(89)90641-7 [DOI] [PubMed] [Google Scholar]
- Queenan A. M., Ghabrial A. and Schupbach T. (1997). Ectopic activation of Torpedo/Egfr, a Drosophila receptor tyrosine kinase, dorsalizes both the eggshell and the embryo. Development 124, 3871-3880. [DOI] [PubMed] [Google Scholar]
- Roch F., Jimenez G. and Casanova J. (2002). EGFR signalling inhibits Capicua-dependent repression during specification of Drosophila wing veins. Development 129, 993-1002. [DOI] [PubMed] [Google Scholar]
- Rodrigues A. B., Werner E. and Moses K. (2005). Genetic and biochemical analysis of the role of Egfr in the morphogenetic furrow of the developing Drosophila eye. Development 132, 4697-4707. 10.1242/dev.02058 [DOI] [PubMed] [Google Scholar]
- Ronchi E., Treisman J., Dostatni N., Struhl G. and Desplan C. (1993). Down-regulation of the Drosophila morphogen Bicoid by the Torso receptor-mediated signal transduction cascade. Cell 74, 347-355. 10.1016/0092-8674(93)90425-P [DOI] [PubMed] [Google Scholar]
- Sardi S. P., Murtie J., Koirala S., Patten B. A. and Corfas G. (2006). Presenilin-dependent ErbB4 nuclear signaling regulates the timing of astrogenesis in the developing brain. Cell 127, 185-197. 10.1016/j.cell.2006.07.037 [DOI] [PubMed] [Google Scholar]
- Schweitzer R., Shaharabany M., Seger R. and Shilo B. Z. (1995). Secreted Spitz triggers the DER signaling pathway and is a limiting component in embryonic ventral ectoderm determination. Genes Dev. 9, 1518-1529. 10.1101/gad.9.12.1518 [DOI] [PubMed] [Google Scholar]
- Shearin H. K., Dvarishkis A. R., Kozeluh C. D. and Stowers R. S. (2013). Expansion of the gateway multisite recombination cloning toolkit. PLoS ONE 8, e77724 10.1371/journal.pone.0077724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearin H. K., Macdonald I. S., Spector L. P. and Stowers R. S. (2014). Hexameric GFP and mCherry reporters for the Drosophila GAL4, Q, and LexA transcription systems. Genetics 196, 951-960. 10.1534/genetics.113.161141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhauer J., Gijon M. A., Riekhof W. R., Voelker D. R., Murphy R. C. and Treisman J. E. (2009). Drosophila lysophospholipid acyltransferases are specifically required for germ cell development. Mol. Biol. Cell 20, 5224-5235. 10.1091/mbc.e09-05-0382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker H., Andjelkovic M., Oldham S., Laffargue M., Wymann M. P., Hemmings B. A. and Hafen E. (2002). Living with lethal PIP3 levels: viability of flies lacking PTEN restored by a PH domain mutation in Akt/PKB. Science 295, 2088-2091. 10.1126/science.1068094 [DOI] [PubMed] [Google Scholar]
- Struhl G. and Adachi A. (1998). Nuclear access and action of Notch in vivo. Cell 93, 649-660. 10.1016/S0092-8674(00)81193-9 [DOI] [PubMed] [Google Scholar]
- Struhl G. and Basler K. (1993). Organizing activity of Wingless protein in Drosophila. Cell 72, 527-540. 10.1016/0092-8674(93)90072-X [DOI] [PubMed] [Google Scholar]
- Szuts D. and Bienz M. (2000). LexA chimeras reveal the function of Drosophila Fos as a context-dependent transcriptional activator. Proc. Natl. Acad. Sci. USA 97, 5351-5356. 10.1073/pnas.97.10.5351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateno M., Nishida Y. and Adachi-Yamada T. (2000). Regulation of JNK by Src during Drosophila development. Science 287, 324-327. 10.1126/science.287.5451.324 [DOI] [PubMed] [Google Scholar]
- Traynor A. M., Weigel T. L., Oettel K. R., Yang D. T., Zhang C., Kim K. M., Salgia R., Iida M., Brand T. M., Hoang T. et al. (2013). Nuclear EGFR protein expression predicts poor survival in early stage non-small cell lung cancer. Lung Cancer 81, 138-141. 10.1016/j.lungcan.2013.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.-C., Lien H.-C., Xia W., Chen I.-F., Lo H.-W., Wang Z., Ali-Seyed M., Lee D.-F., Bartholomeusz G., Ou-Yang F. et al. (2004). Binding at and transactivation of the COX-2 promoter by nuclear tyrosine kinase receptor ErbB-2. Cancer Cell 6, 251-261. 10.1016/j.ccr.2004.07.012 [DOI] [PubMed] [Google Scholar]
- Wang S.-C., Nakajima Y., Yu Y.-L., Xia W., Chen C.-T., Yang C.-C., McIntush E. W., Li L.-Y., Hawke D. H., Kobayashi R. et al. (2006). Tyrosine phosphorylation controls PCNA function through protein stability. Nat. Cell Biol. 8, 1359-1368. 10.1038/ncb1501 [DOI] [PubMed] [Google Scholar]
- Wang Y.-N., Yamaguchi H., Huo L., Du Y., Lee H.-J., Lee H.-H., Wang H., Hsu J.-M. and Hung M.-C. (2010). The translocon Sec61beta localized in the inner nuclear membrane transports membrane-embedded EGF receptor to the nucleus. J. Biol. Chem. 285, 38720-38729. 10.1074/jbc.M110.158659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L. and Baker N. E. (2001). Role of the EGFR/Ras/Raf pathway in specification of photoreceptor cells in the Drosophila retina. Development 128, 1183-1191. [DOI] [PubMed] [Google Scholar]
- Yang L. and Baker N. E. (2003). Cell cycle withdrawal, progression, and cell survival regulation by EGFR and its effectors in the differentiating Drosophila eye. Dev. Cell 4, 359-369. 10.1016/S1534-5807(03)00059-5 [DOI] [PubMed] [Google Scholar]
- Zscheppang K., Dörk T., Schmiedl A., Jones F. E. and Dammann C. E. L. (2011). Neuregulin receptor ErbB4 functions as a transcriptional cofactor for the expression of surfactant protein B in the fetal lung. Am. J. Respir. Cell Mol. Biol. 45, 761-767. 10.1165/rcmb.2010-0179OC [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.