ABSTRACT
Loss of the paired-like homeodomain transcription factor 2 (Pitx2) in cardiomyocytes predisposes mice to atrial fibrillation and compromises neonatal regenerative capacity. In addition, Pitx2 gain-of-function protects mature cardiomyocytes from ischemic injury and promotes heart repair. Here, we characterized the long-term myocardial phenotype following myocardial infarction (MI) in Pitx2 conditional-knockout (Pitx2 CKO) mice. We found adipose-like tissue in Pitx2 CKO hearts 60 days after MI induced surgically at postnatal day 2 but not at day 8. Molecular and cellular analyses showed the onset of adipogenic signaling in mutant hearts after MI. Lineage tracing experiments showed a non-cardiomyocyte origin of the de novo adipose-like tissue. Interestingly, we found that Pitx2 promotes mitochondrial function through its gene regulatory network, and that the knockdown of a key mitochondrial Pitx2 target gene, Cox7c, also leads to the accumulation of myocardial fat tissue. Single-nuclei RNA-seq revealed that Pitx2-deficient hearts were oxidatively stressed. Our findings reveal a role for Pitx2 in maintaining proper cardiac cellular composition during heart regeneration via the maintenance of proper mitochondrial structure and function.
KEY WORDS: Mouse, Myocardial infarction, Cardiac regeneration, Mitochondria, Adipogenesis
Highlighted Article: Pitx2 is important in the maintenance of the proper cellular composition of the regenerating myocardium by reducing mitochondrial dysfunction and preventing fat accumulation.
INTRODUCTION
Heart failure is a common and devastating consequence of heart disease. The hallmark of a failing heart is degenerative cardiac muscle that loses contractility (Xin et al., 2013). Currently, there is no cure for heart failure; however, extensive efforts have been made to identify the biological principles underlying heart failure caused by major heart diseases such as myocardial infarction (MI) and arrhythmia and to develop effective treatment for these diseases. One strategy to repopulate an infarcted area caused by MI is to induce endogenous cardiomyocytes to re-enter the cell cycle. This strategy was inspired by the observation that the mammalian heart has regenerative capacity in the neonatal period (Porrello et al., 2011). Although neonatal mouse heart can fully regenerate after injury, this ability wanes quickly and disappears completely by 7 days after birth in the mouse (Porrello et al., 2011).
Previous work has revealed that modifications of molecular pathways, such as the Hippo pathway, can improve cardiac function and structural repair (Heallen et al., 2013). Recently, we have reported a protective role of the paired-like homeodomain transcription factor 2 (Pitx2) in heart injury (Tao et al., 2016). PITX2 is a gene mutated in Rieger Syndrome (Semina et al., 1996). Importantly, PITX2 is crucial for regulating cardiac left-right asymmetry during development (Lu et al., 1999). Moreover, mice heterozygous for a Pitx2 null allele, Pitx2nu/+, are prone to atrial fibrillation when paced (Wang et al., 2010). We have also shown that knocking out Pitx2 in cardiomyocytes using a muscle cell-specific MCKcre (Brüning et al., 1998) caused increased reactive oxygen species (ROS) in the myocardium. These defects cause a larger scar area and reduced heart function in mutant hearts at 3 weeks after neonatal MI when compared with control. Thus, Pitx2 functions to maintain appropriate myocardial redox balance and metabolism during regeneration (Tao et al., 2016). Interestingly, we also found that Pitx2 interacts with the Hippo signaling effector Yap to activate downstream antioxidant scavengers, which protects the myocardium from ROS damage (Tao et al., 2016).
To study the long-term effects of the loss of Pitx2 in the injured myocardium, we performed a histological analysis of mouse hearts 2 months after MI at postnatal day (P) 2 and observed adipose-like tissue within the scar zone. Gene expression analysis indicated activation of the adipogenic program and lineage-tracing experiments showed a non-cardiomyocyte origin of the adipose-like cells. Further studies using an embryonic stem cell- (ESC) to-cardiomyocyte differentiation system showed that loss of Pitx2 in ESCs promotes the expression of adipogenic markers. In addition, defective mitochondrial function may contribute to the adipose-like phenotype. We suggest that, within the regenerative window, Pitx2 activity in cardiomyocytes protects against the deleterious accumulation of fatty tissue by reducing mitochondrial dysfunction.
RESULTS
Pitx2 knockdown in cardiomyocytes during the regenerative window leads to changes in cardiac tissue composition
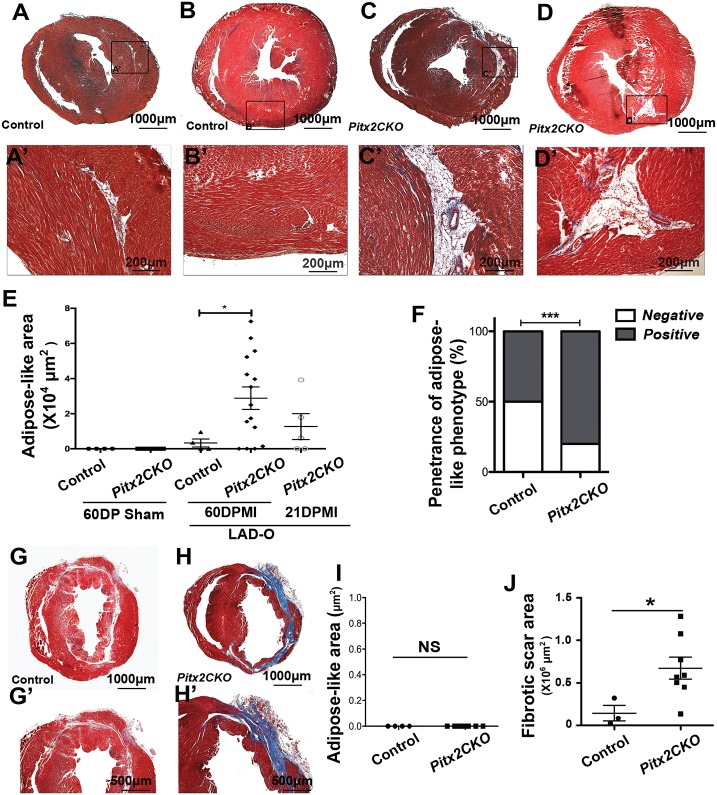
We used the neonatal heart model in which the left anterior descending artery (LAD) was occluded surgically at P2 to induce MI (Tao et al., 2016). Neonatal hearts have been shown to fully repair damaged myocardium by 3 weeks after P2 LAD occlusion, whereas conditional knockdown of Pitx2 in cardiomyocyte lineage using MCKcre/+; Pitx2f/f (Pitx2 CKO) mice compromises this reparative ability (Tao et al., 2016). To investigate the long-term effects of the loss of Pitx2 in damaged myocardium, we performed histological examination of Pitx2 CKO hearts at 21 and 60 days post myocardial infarction (DPMI). Histological analysis of trichrome-stained heart sections at 60 DPMI showed large areas of scarring mixed with the presence of adipose-like tissue in Pitx2 CKO hearts (Fig. 1C-D′), whereas minimal scarring was seen in control hearts that maintained regenerative capacity (Fig. 1A-B′). Quantification of cardiac tissue sections showed a significantly larger area and phenotypic penetrance of adipose-like tissue in Pitx2 CKO hearts than in control hearts (Fig. 1E,F). Overall, control hearts and 60 DPMI Pitx2 CKO hearts represented the phenotypic extremes, whereas 21 DPMI Pitx2 CKO hearts displayed an intermediate phenotype (Fig. 1E and Fig. S1). We also examined the effect of surgery on myocardial repair at 8 days after birth in a separate cohort of mice, a time point beyond the neonatal regenerative window. Although severe scarring was observed in both control and Pitx2 CKO groups, no adipose-like tissue was observed in either group (Fig. 1G-I). Consistent with the findings from our previous study, our results indicate that Pitx2 CKO hearts are more vulnerable to ischemic injury (Fig. 1J). These results indicate that the observed adipogenic injury response occurred only during the regenerative postnatal stage.
Fig. 1.
Adipose-like tissue in injured hearts with loss of Pitx2 in cardiomyocytes. (A-E) Trichrome staining showing scarring and an area of adipose-like tissue in the left ventricle of control (A,A′,B,B′) and Pitx2 CKO (C,C′,D,D′) hearts at 60 days after LAD occlusion performed at P2. Red, muscle; blue, collagen; white, adipose-like tissue. This adipose-like area is quantified in E. (F) Penetrance of adipose phenotype in Pitx2 CKO and control hearts. (G,G′,H,H′) Trichrome staining showing scarring in control (G,G′) and Pitx2 CKO (H,H′) hearts at 60 days after LAD occlusion performed at P8. (I,J) Quantification of the adipose-like (I) and scar areas (J) in hearts in which the LAD occlusion was performed at P8. *P<0.05, ***P<0.001; non-parametric (Mann–Whitney) test. NS, not significant.
Pitx2 CKO mice displayed a significant increase in adipose composition after injury
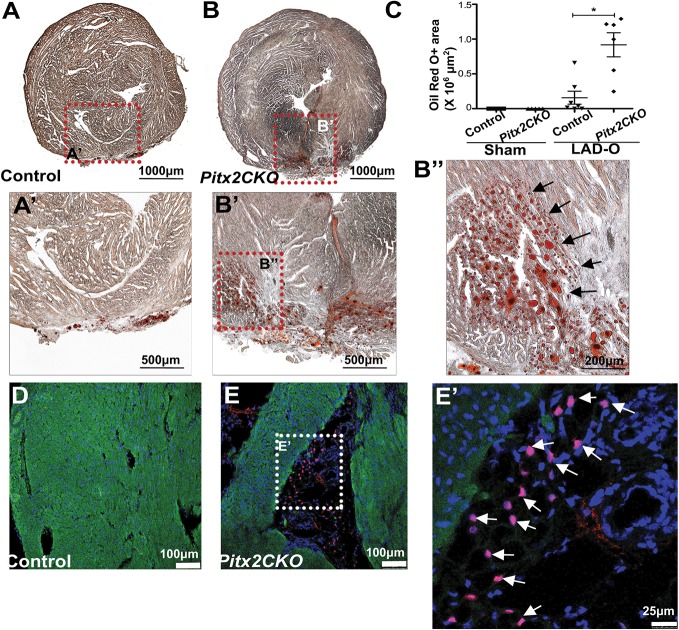
Standard Mason's trichrome histological staining lacks the specificity to definitively determine the presence of adipocytes within a complex tissue. Therefore, to determine the identity of the adipose-like tissue in injured Pitx2 CKO hearts, we performed Oil Red O staining of histological sections. We observed an accumulation of oil droplets within the damaged Pitx2 CKO myocardium (Fig. 2B-B″) and in an area of potential epicardial adipose tissue in control hearts (Fig. 2A,A′) (Yamaguchi et al., 2015; Zangi et al., 2017); the oil droplet accumulation was significantly increased in the mutant hearts compared with control hearts (Fig. 2C). To further characterize the adipose-like cells, we performed immunofluorescence staining with an antibody against the transcription factor CCAAT/enhancer-binding protein alpha (C/EBPα), a marker for adipogenesis and mature adipocytes (Fig. 2D,E) (Tontonoz et al., 1994). Pitx2 CKO heart sections stained positive for more C/EBPα+ cells, which were located within the scar tissue (Fig. 2E,E′), than did control hearts (Fig. 2D). These findings indicate the activation of the adipose gene program during myocardial repair. Strikingly, we also observed the coexistence of the adipose-like tissue with major coronary vessels in Pitx2 CKO hearts (Fig. 3).
Fig. 2.
Lipid accumulation and adipose marker expression in injured Pitx2 CKO hearts. (A-C) Oil Red O staining showing lipid droplets in Pitx2 CKO (B-B″) and control (A,A′) hearts at 60 days after LAD occlusion performed at P2. Black arrows in B″ indicate lipids. The positive-staining area is quantified in C. *P<0.05; non-parametric (Mann–Whitney) test. (D-E′) Immunostaining of cTnT (green) and C/EBPα (red; white arrows in E′) antibodies in control (D) and Pitx2 CKO (E,E′) hearts.
Fig. 3.
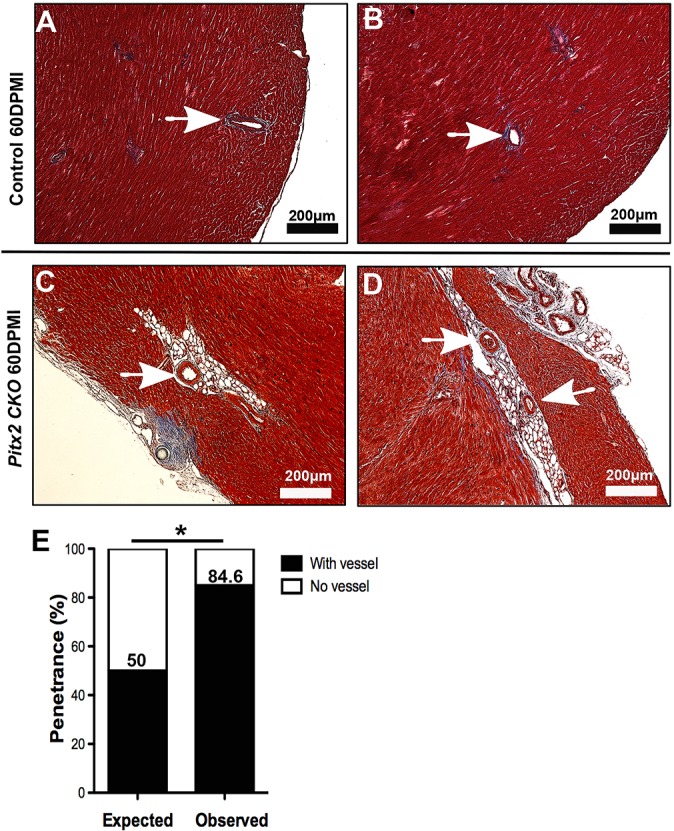
Perivascular localization of adipose-like tissue in regenerative Pitx2 CKO myocardium. (A,B) Representative images of normal coronary arteries in a regenerating control heart at 60 DPMI. (C,D) Representative images of adipose-like tissue located in close proximity to coronary vessels in regenerating Pitx2 CKO hearts. Arrow, coronary artery. (E) Phenotypic penetrance of the coexistence of adipose-like tissue and coronary arteries in Pitx2 CKO hearts compared with expected penetrance. *P<0.05; two-sided χ2 test.
Adipose-like cells have a non-cardiomyocyte origin
To elucidate the origin of the adipose-like cells in regenerating Pitx2 CKO hearts in which Pitx2 was specifically knocked out in cardiomyocytes, we performed lineage tracing in MCKCre; Pitx2f/f; mTmG mice that underwent LAD occlusion at P2. Hearts were harvested and studied at 60 DPMI. With the mTmG reporter, the MCKcre allele marks and traces the cardiomyocyte lineage as GFP+, whereas non-myocytes are labeled as TdTomato+ (Fig. S2A) (Brüning et al., 1998). C/EBPα immunofluorescence combined with GFP fluorescence showed no double-positive cells within the scar area in control and mutant hearts (Fig. S2B-C‴). These results indicate a non-cardiomyocyte origin of the adipose-like cells and a non-cell-autonomous switch of cell identity during the regenerative stage in the mutant heart.
Pitx2 expression inhibits adipogenic gene program deployment in heterogeneous in vitro differentiation assay
Mature cardiomyocytes are considered to be terminally differentiated and to have exited the cell cycle and, therefore, have a limited turnover rate (Bergmann et al., 2015); however, neonatal cardiomyocytes show increased cell division in response to damage (Heallen et al., 2013; Porrello et al., 2011). Although there is evidence that direct cardiomyocyte proliferation contributes to the repopulation of damaged myocardium (Tao et al., 2016), the cellular events involving other cardiac lineages and their interaction with cardiomyocytes are unknown. To assess the role of Pitx2 in maintaining an appropriate composition of different cardiac lineage cells in the heart, we used an in vitro assay examining the differentiation of ESCs into cardiomyocytes, which allowed us to specifically study the requirement of Pitx2 in cardiomyocyte lineage commitment (Hartman et al., 2014). Wild-type and Pitx2nu/nu ESCs were induced to differentiate into cardiomyocytes (Fig. 4A) (Lu et al., 1999). Importantly, in this system the differentiation of mouse ESCs to cardiomyocytes is less than 100%, and it therefore represents a heterogeneous cell environment of both cardiomyocyte-like cells and non-myocyte populations. Quantitative PCR (qPCR) showed increased expression of adipogenic markers in Pitx2nu/nu-derived cells compared with control cells (Fig. 4B). Flow cytometry studies, using C/EBPα as a sorting marker, also identified an increase in the percentage of C/EBPα+ cells in the plates differentiated from Pitx2nu/nu ESCs, compared with control non-mutant ESCs (Fig. 4C,D). These in vitro results suggest that Pitx2 deficiency in a pro-cardiogenic cellular environment promotes adipogenesis. To further characterize the putative pro-adipogenic gene program in Pitx2 CKO hearts in vivo, we used a previously published RNA-seq dataset that was generated from injured control and Pitx2 CKO murine ventricular tissue to examine the change in transcription levels of adipogenic genes (Fig. 4E). Indeed, many adipogenic genes, including Acacb, Klf9, Ppara and Ppard, were upregulated in Pitx2 CKO hearts. Therefore, we see evidence for adipogenesis in vitro as well as in vivo.
Fig. 4.
Loss of Pitx2 in ESCs promotes cell commitment to adipose lineage. (A) Flow chart showing the procedure carried out to assess the differentiation of control and Pitx2nu/nu ESCs into cardiomyocytes. (B) qPCR was carried out to detect adipose marker expression in differentiated Pitx2nu/nu and control cells. *P<0.05; non-parametric (Mann–Whitney) test. (C) Flow cytometry showing differentiated control and Pitx2nu/nu ESCs stained with antibody against the adipose marker C/EBPα. (D) Quantification of the adipose fraction from C. Data are mean±s.d. (E) RNA-seq datasets from injured control and Pitx2 CKO myocardium were used to generate a heat map showing the expression of adipogenesis-relevant genes.
Defective mitochondrial function contributes to the development of intramyocardial adipose tissue
In the damaged myocardium, the majority of ROS are believed to be derived from the leaky electron transport chain (ETC) in the inner mitochondrial membrane (Kirkinezos and Moraes, 2001). Oxidative stress caused by dramatic increases in ROS levels has also been shown to promote adipogenesis (Lee et al., 2009). Importantly, Pitx2 CKO hearts have higher levels of ROS after neonatal injury than the control (Tao et al., 2016). To confirm these findings in a more tractable system, we deployed our previously reported Pitx2 null P19 cell line (P19-Pitx2 null; Tao et al., 2016) in a culture model in which we mimicked increased ROS with H2O2 treatments. Using this in vitro ROS assay, we found that H2O2 treatment caused a significant increase in adipogenic gene activity, as measured by the expression of the adipocyte marker peroxisome proliferator activated receptor gamma (Pparg), of P19-Pitx2 null cells when compared with non-mutant P19 cells (Fig. 5A). These results suggest that Pitx2 may prevent ROS-induced adipogenesis. Importantly, we have previously shown that ultrastructural mitochondrial defects and mis-expression of key mitochondrial components were key features of adult Pitx2 CKO hearts (Tao et al., 2014). Moreover, in a desmoglein 2 (Dsg2)-deficient mouse model of arrhythmogenic right ventricular cardiomyopathy (ARVC), mitochondrial dysfunction has also been reported; mitochondrial swelling and abnormal cristae structures were found to be characteristic of mutant cardiomyocytes (Kant et al., 2012). Therefore, we next sought to delineate the dysfunctional mitochondrial phenotype that is common between models of ARVC and Pitx2 CKO hearts. Firstly, we examined mitochondrial activity by measuring the cellular oxygen consumption rate (OCR) and the mitochondrial gene expression in P19-Pitx2 null and P19 control cells. Seahorse studies showed a decrease in OCR in P19-Pitx2 null cells (Fig. 5B). In addition, P19-Pitx2 null cells showed disrupted expression of genes that encode ETC components including Ndufb8 and mt-Co1 (Fig. 5C). These findings suggest that the disruption of the Pitx2 gene program alters mitochondrial ETC composition, resulting in mitochondrial dysfunction and ultimately adipogenesis.
Fig. 5.
Compromised mitochondrial function contributes to adipose-like phenotype in injured neonatal mouse heart. (A) qPCR of Pparg in control and Pitx2 null P19 cell lines with or without 8 h of H2O2 treatment. (B) Seahorse assay shows oxygen consumption rate (OCR) of control and P19-Pitx2 null cell lines with readout normalized to the cell number in each assay. The x-axis indicates measurements 1-11. The dashed vertical lines indicate injections into media of the specific stressors oligomycin, FCCP and antimycin A/rotenone (A/R). (C) Western blot shows expression of ETC components in control and Pitx2 null P19 cell lines. (D,E) Trichrome staining of Cox7clacZ/+ and control mouse hearts that were subjected to LAD occlusion at P2 and analyzed at 60 days after occlusion. (F) Quantification of the amount of scarring seen in D,E. (G) Echocardiography shows the ejection fraction of control and Cox7clacZ/+ mice at 60 days after sham or LAD occlusion surgery. (H-J) Immunostaining for cTnT (green), C/EBPα (red) and DAPI (blue) in control (H) and Cox7clacZ/+ (I) hearts at 60 days after LAD occlusion. Asterisks indicate scar area. Arrows indicate C/EBPα positive cells. The percentage of C/EBPα+ scar cells is quantified in J. *P<0.05; non-parametric (Mann–Whitney) test. NS, not significant.
Pitx2 directly regulates the expression of several mitochondrial genes, including members of the terminal oxidase of the ETC, known as complex IV or cytochrome c oxidase (CcO) (Tao et al., 2016). Importantly, the vast majority of CcO deficiencies are known to induce mitochondrial ROS production (Srinivasan and Avadhani, 2012). Pitx2 may promote, among other things, CcO expression during regeneration to maintain mitochondrial function and electron flux through the ETC to reduce deleterious ROS production and ectopic myocardial adipogenesis. Cytochrome c oxidase 7c (Cox7c) is a nuclear-encoded component of CcO, and a direct Pitx2 target (Tao et al., 2016). To better decipher the contributions of mitochondrial Pitx2 target genes, we performed LAD occlusion at P2 in heterozygous Cox7ctm1b(KOMP)Mbp animals, which carry a Cox7c-targeted lacZ reporter-tagged deletion allele. Trichrome staining showed no significant increase in scarring in Cox7clacZ/+ hearts 60 DPMI, compared with non-mutant controls (Fig. 5D-F); however, cardiac function was significantly compromised in both sham and LAD-occluded groups (Fig. 5G). Similar to Pitx2 CKO animals injured at P2, Cox7clacZ/+ possessed adipose tissue within the myocardium 60 days after P2 surgery. Immunostaining confirmed the presence of a significantly higher percentage of C/EBPα+ cells in the scar tissue of Cox7clacZ/+ hearts than in control hearts (Fig. 5H-J). Thus, with endogenous Pitx2 still present, Cox7clacZ/+ animals can repair scar tissue, but are unable to prevent the accumulation of fat-like tissue within the myocardium. These findings suggest that Pitx2 is, at least in part, cardioprotective during regeneration, and that the expression of its mitochondrial gene targets work to maintain proper cardiac mitochondrial structure and function in the presence of ischemic stress. It is also suggested that defective ETC in Pitx2 CKO heart contributes to its compromised regeneration and formation of the adipose-like tissue.
Single-nuclei RNA-seq in injured control and mutant hearts
To better characterize transcriptional distinctions between control and Pitx2 CKO hearts, we performed single-nuclei RNA-seq (snRNA-seq) on control (Pitx2f/f) and Pitx2 CKO heart left ventricles 3 weeks after LAD occlusion on P2 (Fig. 6A). We chose 3 weeks post-MI as an intermediate timepoint between regeneration and the onset of the obvious phenotype, in an effort to capture the events leading to the progression of the fat deposition. Unsupervised graph-based clustering of the combined snRNA-seq dataset mapped to both introns and exons, comprising 7849 cells, was visualized using t-distributed stochastic neighbor embedding (tSNE) (van der Maaten and Hinton, 2008) (Fig. 6B). We identified nine transcriptionally distinct clusters representing all the major cardiac cell types, including cardiac fibroblasts, cardiomyocytes, endothelial cells, vascular smooth muscle cells, macrophages, epicardial cells, endocardial cells, lymphatic endothelial cells, and mural cells or pericytes (Fig. 6B-D). Interestingly, three distinct populations of fibroblasts, designated FB-1, FB-2, and FB-3 were also identified (Fig. 6B-D).
Fig. 6.
snRNA-seq on control and Pitx2 CKO cardiac tissue. (A) Diagram of snRNA-seq protocol. Samples were collected at 3 weeks after LAD occlusion on P2. Nuclei were isolated via density gradient centrifugation and nuclear snRNA-seq was performed. (B) Left, tSNE plot of 7849 cells showing the contribution of control (black) and Pitx2 CKO (red) samples. Right, the same tSNE as on the left but labeled by cell/cluster identity. (C) Average differential expression heat map for the top 1619 genes, with genes as rows and clusters as columns. Colors for each column match those in panel B, right. (D) Dot plot showing the average expression for each indicated gene (column) across all clusters (rows). FB, fibroblast; CM, cardiomyocyte; EC, endothelial cell; LEC, lymphatic endothelial cell; EndC, endocardial cell; EpiC, epicardial cell; SMC, (vascular) smooth muscle cell; Mφ, macrophage; PeC, pericyte.
Among the cardiac cell types identified, the cardiomyocytes showed significant heterogeneity, with at least three subpopulations seen in the dataset (Fig. 6B). The CM-1 population expressed high levels of fibroblast growth factor 14 (Fgf14), vascular endothelial growth factor a (Vegfa) and several long non-coding RNAs (lncRNAs) including Neat1, Mhrt and Malat1 (Fig. S3A-B), whereas CM-2 cells expressed higher levels of Wnt5b. Finally, the CM-3 population expressed low levels of the lncRNAs but high levels of cardiac genes such as Jph2 (Fig. 6C,D). Despite the heterogeneity, no single subpopulation of cardiomyocytes was unique to the Pitx2 CKO heart (Fig. 6B, left panel); however, a higher proportion of Pitx2 CKO cell nuclei was seen in CM-2 than in CM-1 (Fig. 6B). The CM clusters were computationally interrogated further. Subclustering of the CM-1, CM-2 and CM-3 populations uncovered transcriptional distinctions; unsupervised graph-based clustering and tSNE visualization of the 4136 CM nuclei identified four clusters of cardiomyocytes, arbitrarily designated c1-c4 (Fig. 7A). Examination of the contributions from control and Pitx2 CKO mice showed that c3 had significantly more nuclei from the mutant mice than nuclei from the control, with a χ2 P-value <0.05 (Fig. 7A,B). To determine the molecular characteristics of the c3 population, we performed differential expression analysis using Seurat's implementation of the likelihood ratio test for single-cell gene expression of the top 200 non-overlapping genes between clusters; our findings identified a total of 598 significantly differentially expressed genes (Fig. 7B, bottom). The genes distinguishing c3 were enriched for gene ontology terms including oxidative phosphorylation, response to oxidative stress, cristae formation and antioxidant activity (Fig. 7B,C). Among the genes enriched in this cluster were Dsp, Emd and Jph2, which are directly transcriptionally repressed by Pitx2 (Tao et al., 2014) (Fig. 7D). Furthermore, Dsp has been identified as a genetic cause of ARVC (Rampazzo et al., 2002). Increased expression of ROS scavengers, such as Sod1, Sod2 and Prdx5, indicated a higher oxidative stress level present in the Pitx2 CKO myocardium at this timepoint (Fig. 7D). Therefore, Pitx2 CKO cardiomyocytes do not possess a radically altered nuclear transcriptome 3 weeks post-MI, however, they do tend to exist in a more oxidatively stressed cellular state than control cardiomyocytes.
Fig. 7.
Subclustering of cardiomyocytes shows differential subpopulations and gene expression between control and Pitx2 CKO groups. (A) Top, tSNE of 4136 subclustered cardiomyocytes (CM-1, CM-2 and CM-3 from Fig. 6) showing clusters c1-c4 in different colors. Bottom, χ2 cluster composition analysis of CMs in Pitx2 CKO hearts compared with controls. Blue, cluster that significantly decreases in Pitx2 CKO hearts. Red, cluster that significantly increases in Pitx2 CKO hearts. Gray, no significant change in cluster composition between control and Pitx2 CKO conditions. (B) Differential expression of genes for clusters c1-c4 shown on a heat map, with the top candidate genes as rows and different cell clusters as columns. Expression of each gene was averaged across all cells in the indicated cluster. Low expression is indicated by cyan and high expression is shown in pink. (C) Gene ontology analysis of c3-enriched genes. (D) Expression plots for each gene projected across the tSNE. Pink indicates gene expression and white represents no expression. c3 is highlighted in green.
DISCUSSION
The role of Pitx2 in cardiac development, homeostasis, conduction and regeneration are not yet fully appreciated. In this study, we found that Pitx2 is required in cardiomyocytes for the prevention of intramyocardial fat tissue accumulation following P2 injury. After 60 DPMI, Pitx2 CKO hearts possessed fatty areas with high densities of lipid droplets and cells of non-cardiomyocyte origin expressing adipocyte markers. Pitx2-deficient cells had severe mitochondrial dysfunction. Indeed, Pitx2 regulates ETC gene expression, and among these gene targets is Cox7c. Genetic disruption of a single copy of Cox7c recapitulated the intramyocardial fat accumulation phenotype observed in Pitx2 CKO animals after P2 MI. Thus, Pitx2 deficiency in neonatal cardiomyocytes during heart regeneration leads to an ARVC-like phenotype in a cell non-autonomous fashion associated with mitochondrial dysfunction.
Notably, when the LAD occlusion procedure was performed after the regenerative window closed (at P8), no noticeable intramyocardial fat tissue was observed at 60 DPMI. Postnatal ventricular Pitx2 expression is very low, and by P8 is virtually undetectable (Tao et al., 2016). Coincidentally, immediately following birth, cardiomyocyte metabolism switches from glycolysis to fatty acid oxidation to meet the demanding functional requirements of a maturing animal. Moreover, both ROS and DNA damage increase dramatically in the heart after birth (Puente et al., 2014). Thus, Pitx2 is not required to drive the expression of metabolic genes in mature ventricular cardiomyocytes, including hypoxia-inducible factor 1a (Hif1a) and Pparg (Lopaschuk and Jaswal, 2010). However, we found increased expression of the Pparg co-activator Ppargc1b in Pitx2 CKO hearts, which may suggest that the two gene regulatory networks of these metabolic transcriptional regulators overlap in cardiomyocytes during the regenerative window. Interestingly, Ppargc1b has been found to modulate mitochondrial gene expression in adipose tissue (Enguix et al., 2013), similar to the role played by Pitx2 in cardiomyocytes. Future studies aimed at profiling the cistromes of these two gene regulatory networks are necessary. Based on the above findings, we propose that Pitx2 is required during the regenerative window to maintain proper mitochondrial function. Meanwhile in the mature heart, the regulatory role of Pitx2 in maintaining mitochondrial function is replaced by other transcription factors. Our research highlights the temporal importance of Pitx2 expression during the regenerative window, an ontological oxidative and metabolic transition.
Pitx2 directly regulates the expression of genes encoding ROS scavengers and ETC components. Structural defects in mitochondria are a major source of ROS (Kirkinezos and Moraes, 2001). As Pitx2 CKO adult cardiomyocytes display ultrastructural mitochondrial defects, we examined the OCR in a Pitx2-null P19 cell line and found a decreased OCR and disrupted expression of key ETC components. Consistent with these results, the removal of Pitx2 in the heart has been shown to increase cardiac susceptibility to oxidative stress (Tao et al., 2016). Moreover, increased ROS levels cause lipid droplet accumulation in glia (Liu et al., 2015). We validated this finding with a Cox7c null allele. Cox7c is a target of Pitx2 and a CcO component. It is known that CcO deficiencies lead to increased ROS. We found that heterozygous disruption of Cox7c partially phenocopied the Pitx2 fatty myocardial phenotype. As the main fuel for beating cardiomyocytes, fatty acids may easily accumulate in the myocardium when metabolism is compromised and their consumption is reduced, which would explain the increased Oil Red O staining in injured Pitx2 CKO hearts. Oncological studies have shown that CcO defects can cause metabolic reprogramming and Warburg effect induction, therefore global metabolism in Pitx2 CKO cardiomyocytes may shift towards glycolysis (Srinivasan et al., 2016). Interestingly, although the loss of Pitx2 in the injured neonatal mouse heart resulted in a lower expression of antioxidant scavengers and ETC components at 5 days after injury (Tao et al., 2016), our snRNA-seq performed at 3 weeks after injury showed a higher contribution of c3 from Pitx2 CKO cardiomyocytes, likely due to the adaptive compensation via the long-term injury response. Moreover, a c3-enriched gene, ROMO1, has been linked to high levels of DNA damage and dysfunction in endocrine cells of older patients (Enge et al., 2017). Whereas the metabolic phenotype observed in vivo and in vitro support a role for Pitx2 in mitochondrial function maintenance, P19 embryonal carcinoma cell lines that lack Pitx2 may suffer altered fate potential or differentiation status, which could have affected data interpretation. Future improvements in cardiomyocyte primary culture procedures or in vivo mitochondrial functional assessment will allow for a higher resolution view of Pitx2-dependent mitochondrial output. Altogether, the loss of Pitx2 combined with injury burden may cause high oxidative stress in a subpopulation of cardiomyocytes that contributes to a pro-adipogenic environment (Lee et al., 2009).
Inactivating Pitx2 in postnatal cardiomyocytes that are regenerating caused accumulation of adipose-like cells within the myocardium, similar to the phenotype that is observed in ARVC patients. The pathological adipose tissue that is found in ARVC patients is thought to be derived from cardiomyocytes of the second heart field and/or bi-potent Isl1+Wt1+ cell populations (Dorn et al., 2018; Lombardi et al., 2009). In contrast, genetic lineage tracing with the mTmG double-fluorescent Cre reporter of Pitx2 CKO animals injured at P2 showed that the fat cells were of non-myocyte origin. As such, the etiology of ARVC is distinct from that of Pitx2 CKO animals in terms of cellular origin. Moreover, this indicates that Pitx2 prevents injury-induced adipogenesis in a non-autonomous manner within the regenerative window. Interestingly, we also found that the majority of fat tissue was localized to large cardiac vessels (Fig. 3A,B). The vascular localization of the intramyocardial fat tissue suggests two likely cellular origins: non-myocytes residing within the perivascular microenvironment that surrounds large vessels or coronary arteries, or circulating fat cells or fat cell precursors that migrate to the injury sites. Regarding the first potential source, cardiac resident mesenchymal stem cells (MSCs) possess trilineage potential to differentiate into chondrocytes, osteoblasts and adipocytes, and are localized to the perivascular niche. Indeed, recent lineage tracing analysis found that cardiac MSCs express the hedgehog (Hh) signaling component Gli1 and respond robustly to injury (Kramann et al., 2015). Coincidentally, it has also been reported that, in skeletal muscle, the differentiation of adipogenic precursor cells into adipocytes is regulated by ciliary Hh signaling (Kopinke et al., 2017). Thus, the perivascular Gli1+ MSCs are a potential cellular source for intramyocardial fat and deserve future investigation. Secondly, in Drosophila, circulating fat body cells, the vertebrate adipocyte equivalent, are mobilized following injury and home to the wound to promote tissue repair in concert with macrophages (Franz et al., 2018). Mammalian adipocytes have been observed populating skin wounds immediately following immune cell occupation and, as a result, the mobilization and expansion of adipocytes or adipogenic precursors has been suggested (Schmidt and Horsley, 2013). However, evidence for long-distance fat cell migration has yet to be shown definitively. Nevertheless, direct infiltration of fat cells to the perivascular cardiac environment remains a distinct possibility: inflammation-induced endothelial-cell-mediated monocyte recruitment has been shown to exacerbate the secretion of pro-inflammatory cytokines and adipokines in the liver (Neels and Olefsky, 2006). Another potential adipocyte source is the epicardium, which has been shown through lineage tracing experiments to contribute to epicardial adipose tissue (Yamaguchi et al., 2015). Thus, it is possible that the Pitx2 CKO cardiomyocytes localized near large oxygen-rich coronary arteries are more inflamed or prone to apoptosis and/or necrosis, which activates a perivascular-localized immune response that brings in immune cells that, in turn, recruit adipocytes or adipocyte precursors.
The differentiation dynamics of pre-adipocytes or other adipogenic cellular sources to mature adipocytes in the context of the heart is not well characterized. Here, we found evidence of fat tissue accumulation as early as 21 days following P2 MI in Pitx2 CKO animals (Fig. S1), with the most obvious accumulation of fat tissue found at 60 DPMI. We postulate that the prolonged accumulation is likely explainable by the cellular properties and population size of the elusive pre-adipocyte population. Human adipose-derived stromal cells (ASCs) take 14-21 days to differentiate into adipocytes using an adipogenic induction cocktail that consists of Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), insulin, dexamethasone, isobutylmethylxanthine and indomethacin (Ambele et al., 2016). Human bone-marrow-derived MSCs are allowed to differentiate after induction (AdipoDiff media, Mitenyi Biotec) for 28 days (Battula et al., 2013). Importantly, fluorescence-activated cell sorted Gli2+ perivascular murine MSCs are allowed to differentiate in adipogenic medium (R&D Systems) for 21 days before Oil Red O staining is performed (Kramann et al., 2015). Therefore, the appreciable accumulation of adipocytes at 21 DPMI is comparable with in vitro adipocyte differentiation. Furthermore, the observed accumulation of intramyocardial fat cells is consistent with a rare precursor cell pool (e.g. perivascular MSC). Overall, the pro-adipogenic potential of neonatal Pitx2-deficient cardiomyocytes after injury appears to be comparable with commercially formulated and highly optimized adipogenic in vitro culture protocols.
Large-scale genome-wide association studies have found that the association between PITX2 and atrial fibrillation (AF) is significant (Weng et al., 2017), and that Pitx2 CKO animals are prone to atrial arrhythmias (Tao et al., 2014). Interestingly, the amount of epicardial adipose tissue that accumulates around the atria is also highly associated with not only the risk of AF, but also the severity of AF in humans (Hatem et al., 2016). Epicardial adipose tissue also represents a source of excess ROS (Hatem et al., 2016). Given that Pitx2 expression in left atrial cardiomyocytes persists throughout life, the correlation of Pitx2 expression with both cardiac fat accumulation and oxidative damage caused by excess ROS is very intriguing, based on our findings. We propose that AF-associated mutations affecting Pitx2 expression may lead to decreased mitochondrial function and increased ROS damage. The consequent increase in adipose composition, and likely fibrotic tissue, may ultimately contribute to the pathogenesis of AF. Our data indicate that targeting Pitx2 and ROS scavenging to reduce the formation of pathogenic intracardial and epicardial fat deposition could be a viable approach to treat AF and ARVC.
MATERIALS AND METHODS
Mouse model
Pitx2 CKO mice were bred as previously described (Tao et al., 2016). Lineage tracing experiments were conducted using MCKCre; mTmG and MCKCre/+; Pitx2f/f; mTmG mice. Cox7ctm1b(KOMP)Mbp heterozygous mice were from the Knockout Mouse Project (KOMP) Repository (Project CSD41649). Genotyping and primer sequences were performed as previously described (Heallen et al., 2013; Tao et al., 2016). All animal procedures and surgeries conformed to the Institutional Animal Care and Use Committee (IACUC) of Baylor College of Medicine, Texas, USA.
LAD occlusion and echocardiography
To induce a myocardial infarction, we occluded the LAD in mice at P2 or P8 as previously described (Tao et al., 2016). Sham group refers to the mice without surgery. Mice were euthanized either at 60 days or 21 days after LAD occlusion. Echocardiography was performed using a Vevo 2100 ultrasound system (Fujifilm VisualSonics), equipped with a MS550S transducer, B-mode and M-mode datasets as previously described (Tao et al., 2016).
Trichrome staining
Mouse hearts were collected at 2 months after LAD occlusion, fixed in 10% formalin overnight and embedded in paraffin. We prepared 7 μm sections and carried out trichrome staining as previously described (Heallen et al., 2013).
Oil Red O staining
Mouse hearts were collected at 2 months after LAD occlusion, fixed in 10% formalin overnight and embedded in optimal cutting temperature compound. Cryostat sections were cut at 15 µm. Oil Red O (Sigma-Aldrich, O0625) was dissolved in 99% isopropanol to make a 0.3% stock solution, which was mixed with water before use to make a working solution. The ratio of stock solution to water was 3:2. Cryostat sections were rinsed with water and covered with 60% isopropanol for 5 min. After removing the isopropanol, we covered the sections with Oil Red O working solution for 15 min at 37°C and then rinsed them with tap water. Hematoxylin was added to the slides for 1 min. The sections were rinsed with tap water, mounted with 10% glycerol in phosphate-buffered saline and sealed.
Immunofluorescence
Paraffin sections were deparaffinized, rehydrated and boiled in antigen unmasking solution (Vector Laboratories, H-3300) for 15 min. The tissue sections were treated with 0.05% trypsin (HyClone, SH30042.01) for 20 min at 37°C, 0.1% Tween-20 in PBS for 5 min, 0.5% Tween-20 in PBS for 5 min, 0.5% Triton X-100 in PBS for 15 min and 0.3% H2O2 in PBS for 15 min, and were then blocked with 10% donkey serum in PBS with 0.1% Tween-20 for 1 h. For staining of C/EBPα, sections were incubated with rabbit monoclonal anti-C/EBPα antibody (1:200, Cell Signaling Technology, 8178) in blocking buffer overnight at 4°C, washed and incubated in horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG antibody (1:200, Vector Laboratories, PI-1000) for 30 min. After washing, sections were incubated in tyramide signal amplification working solution for 10 min (1:100, PerkinElmer, TSA Plus Fluorescence kits, NEL741001KT). For staining for cardiac troponin T (cTnT), sections were incubated in mouse on mouse diluent working solution for 5 min (Vector Laboratories, M.O.M. Basic Kit, BMK-2202) and in mouse monoclonal anti-cTnT (1:200, Thermo Fisher Scientific, MS-295-P1) diluted in M.O.M. diluent for 30 min. After washing, the sections were incubated in Alexa Fluor 546 donkey anti-mouse IgG (1:500, Life Technologies, A10036) for 1 h. The slides were counterstained with DAPI and mounted with Vectashield hard-set mounting medium (Vector Laboratories). A Zeiss LSM 780 confocal microscope was used for imaging.
Quantitative PCR
Cells were collected at day 19 of the ESC-cardiomyocyte differentiation assay (Fig. 4B). P19 control and P19-Pitx2 null cells were collected after being treated with 300 µM H2O2 for 8 h in a CO2 incubator (Fig. 5A). Tissues and cells were lysed in 200 µl TRIzol Reagent (Thermo Fisher Scientific, 15596026) and total RNA was prepared according to the manufacturer's instructions. We used 1 µg of total RNA as a template to synthesize cDNA using qScript cDNA supermix (Quanta BioSciences). We conducted qPCR using iTaq Universal SYBR Green Supermix (Bio-Rad). The following primers were used for qPCR: Pparg forward: TCCATTCACAAGAGCTGACC, reverse: ATAAGGTGGAGATGCAGGTTC; Pgc1a forward: CACCAAACCCACAGAAAACAG, reverse: GGGTCAGAGGAAGAGATAAAGTTG; Dlk1 (or Pref-1) forward: TGTCAATGGAGTCTGCAAGG, reverse: ATTCGTACTGGCCTTTCTCC; C/EBPα forward: TGGACAAGAACAGCAACGAG, reverse: GTCAACTCCAGCACCTTCTG.
Cell culture
P19 and P19-Pitx2 null cells were cultured in αMEM medium (Corning, 10-022-CV) supplemented with 10% FBS and 1% penicillin/streptomycin. Mouse wild-type AK7 ESCs (mESCs) and Pitx2nu/nu ESCs were cultured in media prepared as follows: 250 ml DMEM/F-12 Media (Gibco), 250 ml Neurobasal medium (Gibco), 2.5 ml N-2 supplement (Life Technologies), 4.5 ml B-27 supplement (Life Technologies), 1.5 mM GlutaMAX (Gibco), 0.1 mM β-mercaptoethanol, 2% FBS and 1×2i-LIF (ESGRO 2i Supplement kit, EMD Millipore). The differentiation of mESCs to cardiomyocytes was induced according to the published protocol (Hartman et al., 2014). By day 12, beating cells were observed. The cells were collected at day 19 for qPCR and flow cytometry.
Western blot
P19 cells were harvested and lysed in radioimmunoprecipitation assay buffer [150 mM sodium chloride, 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate and 50 mM Tris (pH 8.0) (all Sigma-Aldrich), with added phosphatase inhibitor (Roche), protease inhibitor (Roche) and sodium fluoride (Sigma-Aldrich)] and the protein concentration was quantified using the Pierce BCA protein assay kit (Pierce Biotechnology) as previously described (Heallen et al., 2011). We mixed the lysate with the sample buffer and heated it at 50°C for 10 min before loading. Briefly, after separation via SDS PAGE, the proteins were transferred to polyvinylidene difluoride membranes (EMD Millipore), blocked in 5% milk/TBST [50 mM Tris-Cl, 150 mM NaCl (pH 7.4) and 0.1% Tween 20], and incubated with the primary antibody, mouse anti-α-tubulin IgG (1:5000, Abcam, 110413) overnight at 4°C. The membranes were then washed 3× in TBST and incubated with goat anti-mouse HRP-conjugated secondary antibody (1:5000, Santa Cruz Biotechnology, sc-2055) for 1 h at room temperature. Protein detection was performed using SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific).
Flow cytometry
Cells were collected on day 19 of the cardiomyocyte induction procedure and were resuspended in PBS. Paraformaldehyde was added to achieve a final concentration of 4%. Cells were fixed for 15 min at room temperature and pelleted by centrifugation. We discarded the supernatant and resuspended the cells in pre-chilled methanol for 30 min on ice. We dissolved 1 g of bovine serum albumin (BSA) in 100 ml of PBS to make incubation and blocking buffer. Cells were aliquoted into tubes, washed with the incubation buffer and incubated with the blocking buffer for 30 min at room temperature. The primary antibody, anti-C/EBPα antibody (1:200, Cell Signaling Technology, 8178), was added and the solution was incubated for 1 h at room temperature. The cells were washed with the incubation buffer and incubated with Alexa Fluor 488 donkey anti-rabbit secondary antibody (1:500, Life Technologies, A-21206) for 1 h at room temperature. The cells were washed twice with incubation buffer and resuspended in 1% FBS in PBS. Cells were sorted on a BD Biosciences LSR II flow cytometer and the result was analyzed using Diva version 8.2.
Seahorse O2 consumption assay
The Seahorse program was performed on an XFe-96 analyzer (Agilent Technologies) according to the manufacturer's guidelines. The program was set up and monitored through the Wave Desktop software. Briefly, P19 control cells and P19-Pitx2 null cells were seeded into the seahorse assay plate the day before the experiment, at a density of 1×104 cells/well for control cells, and 2×104 cells/well for the P19-Pitx2 null cells. P19-Pitx2 null cells proliferate at a slower rate compared with the control cells, therefore the different seeding density assures similar cell density at the time of the assay. On the assay plate, the wells on the outer edge can easily be affected by temperature change and cause measurement error, therefore were excluded. The rest of the wells were divided into two groups, one for the control and one for Pitx2 null cells (30 wells/cell type). The standard program was used with slight modification, a total of 11 measurements were performed in the program, with each measurement cycle including a 3 min mixing step and a 3 min measuring step. OCR was recorded and later normalized to cell number, which was found by counting trypsinized cells with a hemocytometer. Data were exported using Excel and Graphpad Prism. The assay was repeated three times with comparable results, with a representative graph presented in Fig 5.
In situ hybridization
Two-month-old ICR mice hearts were collected. RNA of whole heart was extracted and reverse transcription was conducted to get cDNA. cDNA was used as template for PCR in a 20 μl system. T3 sequence was added to the PCR forward primer and T7 sequence was added to the reverse primer. The following primers were used for PCR:
Malat1 forward: GCGAATTAACCCTCACTAAAGGGACCACACAGAATGCAGGTGT, reverse: GCGTAATACGACTCACTATAGGGGGGTAGTCCCCACTGCTAAT; Neat1 forward: GCGAATTAACCCTCACTAAAGGGAATCCCTGCAGGTGGCACTA, reverse: GCGTAATACGACTCACTATAGGGGGGGAGGAAAATGGTTAGTGG; Mhrt forward: GCGAATTAACCCTCACTAAAGGGGCCCTACAGTCTGATGAACAT, reverse: GCGTAATACGACTCACTATAGGGGCTCATTCCACTTTCCACCTC.
PCR product size was confirmed by electrophoresis and purified product indicated that the sequence was correct. The PCR product was used as template for large-scale amplification using the Q5 hot start high-fidelity 2× master mix (New England Biolabs). The reaction product was purified using the PCR purification kit (Qiagen). The DNA product and 2-month-old ICR mice heart paraffin sections were sent to Baylor College of Medicine RNA In Situ Hybridization Core for making RNA probes and conducting in situ hybridization.
Cardiac nuclei purification and single-cell nucleus transcriptome sequencing
Mouse left ventricles were collected, minced and mechanically homogenized before being subjected to Dounce homogenization in the presence of RNase inhibitors. Then, nuclei were isolated via density-gradient centrifugation (sucrose gradient). The purified nuclei were counted and diluted in 1× PBS with 0.04% BSA before being loaded onto the 10× Genomics Chromium instrument. The snRNA-seq libraries were generated using the 10× Chromium Single Cell 3′ v2 reagent kit according to the manufacturer's instructions and were then sequenced on an Illumina Nextseq500.
Sequencing data were handled using the 10× Genomics Cell Ranger software (www.10xgenomics.com). Fastq files were mapped to the mm10 genome and gene counts were quantified using a gtf file with merged exons and introns (pre-mRNA). Subsequently, expression matrices from each experiment were merged and were then imported into Seurat (version 2.1.0), in which log normalization was performed. We corrected for batch effects by regressing out the number of molecules per cell and the batch identity with the RegressOut function (Seurat package). Next, we performed a principal components analysis, and significant principal components were used as input for graph-based clustering. We used tSNE for two-dimensional visualization of the multi-dimensional dataset. Differential expression of the individual clusters was achieved using the likelihood ratio test for single-cell gene expression (FindMarkers, default). To account for overclustering, we merged clusters that were not transcriptionally distinct into a single cluster. Clusters that were composed of doublets (two different cell types within a single droplet) were removed from the dataset. Gene ontology analysis was performed using Metascape (www.metascape.org). The χ2 test was carried out on clusters as described previously (Xiao et al., 2018).
Statistics
We used ImageJ to measure areas of trichrome-stained and Oil Red O-stained samples. GraphPad PRISM was used to analyze data and to draw figures. Differences between groups were evaluated by the non-parametric (Mann–Whitney) test and the χ2 test. A P<0.05 was considered significant.
Supplementary Material
Acknowledgements
We thank Dr Peter C. Kahr for advice on experiment design and Dr Ross A. Poché for providing reagents. We also thank Alon R. Azares from the flow cytometry core at the Texas Heart Institute for providing technical support.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: L.L., G.T., M.C.H., J.F.M.; Methodology: L.L., G.T., M.C.H., Y.M.; Software: M.C.H., M.Z.; Formal analysis: L.L., G.T., M.C.H.; Investigation: L.L.; Resources: J.F.M.; Data curation: M.C.H.; Writing - original draft: L.L., G.T., M.C.H., J.F.M.; Writing - review & editing: L.L., G.T., M.C.H., J.F.M.; Visualization: L.L., M.C.H., M.Z., Y.M.; Supervision: J.F.M.; Project administration: J.F.M.; Funding acquisition: J.F.M.
Funding
This work was supported by a Fondation Leducq Transatlantic Networks of Excellence in Cardiovascular Research grant (14CVD01 to J.F.M.). The project was also supported in part by the RNA In Situ Hybridization Core Facility at the Baylor College of Medicine, which is supported by a Shared Instrumentation grant from the National Institutes of Health (NIH) (1S10OD016167) and the NIH Intellectual and Developmental Disability Research Centers grant U54 HD083092 from the Eunice Kennedy Shriver National Institute of Child Health and Human Development. This work also received funding from the NIH (DE 023177, HL 127717, HL 130804 and HL 118761 to J.F.M.; F31HL136065 to M.C.H.) and the Vivian L Smith Foundation and MacDonald Research Fund Award 16RDM001 (J.F.M.). Deposited in PMC for release after 12 months.
Data availability
snRNA-seq data have been deposited at Gene Expression Omnibus under accession number GSE117893.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.168609.supplemental
References
- Ambele M. A., Dessels C., Durandt C. and Pepper M. S. (2016). Genome-wide analysis of gene expression during adipogenesis in human adipose-derived stromal cells reveals novel patterns of gene expression during adipocyte differentiation. Stem Cell Res. 16, 725-734. 10.1016/j.scr.2016.04.011 [DOI] [PubMed] [Google Scholar]
- Battula V. L., Chen Y., Cabreira M. G., Ruvolo V., Wang Z., Ma W., Konoplev S., Shpall E., Lyons K., Strunk D. et al. (2013). Connective tissue growth factor regulates adipocyte differentiation of mesenchymal stromal cells and facilitates leukemia bone marrow engraftment. Blood 122, 357-366. 10.1182/blood-2012-06-437988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann O., Zdunek S., Felker A., Salehpour M., Alkass K., Bernard S., Sjostrom S. L., Szewczykowska M., Jackowska T., dos Remedios C. et al. (2015). Dynamics of cell generation and turnover in the human heart. Cell 161, 1566-1575. 10.1016/j.cell.2015.05.026 [DOI] [PubMed] [Google Scholar]
- Brüning J. C., Michael M. D., Winnay J. N., Hayashi T., Hörsch D., Accili D., Goodyear L. J. and Kahn C. R. (1998). A muscle-specific insulin receptor knockout exhibits features of the metabolic syndrome of NIDDM without altering glucose tolerance. Mol. Cell 2, 559-569. 10.1016/S1097-2765(00)80155-0 [DOI] [PubMed] [Google Scholar]
- Dorn T., Kornherr J., Parrotta E. I., Zawada D., Ayetey H., Santamaria G., Iop L., Mastantuono E., Sinnecker D., Goedel A. et al. (2018). Interplay of cell–cell contacts and RhoA/MRTF-A signaling regulates cardiomyocyte identity. EMBO J. 37, e98133 10.15252/embj.201798133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enge M., Arda H. E., Mignardi M., Beausang J., Bottino R., Kim S. K. and Quake S. R. (2017). Single-cell analysis of human pancreas reveals transcriptional signatures of aging and somatic mutation patterns. Cell 171, 321-330.e14. 10.1016/j.cell.2017.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enguix N., Pardo R., González A., López V. M., Simó R., Kralli A. and Villena J. A. (2013). Mice lacking PGC-1β in adipose tissues reveal a dissociation between mitochondrial dysfunction and insulin resistance. Mol. Metab. 2, 215-226. 10.1016/j.molmet.2013.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz A., Wood W. and Martin P. (2018). Fat body cells are motile and actively migrate to wounds to drive repair and prevent infection. Dev. Cell 44, 460-470.e3. 10.1016/j.devcel.2018.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman M. E., Librande J. R., Medvedev I. O., Ahmad R. N., Moussavi-Harami F., Gupta P. P., Chien W.-M. M. and Chin M. T. (2014). An optimized and simplified system of mouse embryonic stem cell cardiac differentiation for the assessment of differentiation modifiers. PLoS ONE 9, e93033 10.1371/journal.pone.0093033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatem S. N., Redheuil A. and Gandjbakhch E. (2016). Cardiac adipose tissue and atrial fibrillation: the perils of adiposity. Cardiovasc. Res. 109, 502-509. 10.1093/cvr/cvw001 [DOI] [PubMed] [Google Scholar]
- Heallen T., Zhang M., Wang J., Bonilla-Claudio M., Klysik E., Johnson R. L. and Martin J. F. (2011). Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science 332, 458-461. 10.1126/science.1199010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heallen T., Morikawa Y., Leach J., Tao G., Willerson J. T., Johnson R. L. and Martin J. F. (2013). Hippo signaling impedes adult heart regeneration. Development 140, 4683-4690. 10.1242/dev.102798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant S., Krull P., Eisner S., Leube R. E. and Krusche C. A. (2012). Histological and ultrastructural abnormalities in murine desmoglein 2-mutant hearts. Cell Tissue Res. 348, 249-259. 10.1007/s00441-011-1322-3 [DOI] [PubMed] [Google Scholar]
- Kirkinezos I. G. and Moraes C. T. (2001). Reactive oxygen species and mitochondrial diseases. Semin. Cell Dev. Biol. 12, 449-457. 10.1006/scdb.2001.0282 [DOI] [PubMed] [Google Scholar]
- Kopinke D., Roberson E. C. and Reiter J. F. (2017). Ciliary Hedgehog signaling restricts injury-induced adipogenesis. Cell 170, 340-351.e12. 10.1016/j.cell.2017.06.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramann R., Schneider R. K., DiRocco D. P., Machado F., Fleig S., Bondzie P. A., Henderson J. M., Ebert B. L. and Humphreys B. D. (2015). Perivascular Gli1+ progenitors are key contributors to injury-induced organ fibrosis. Cell Stem Cell 16, 51-66. 10.1016/j.stem.2014.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Lee Y. J., Choi H., Ko E. H. and Kim J.-W. (2009). Reactive oxygen species facilitate adipocyte differentiation by accelerating mitotic clonal expansion. J. Biol. Chem. 284, 10601-10609. 10.1074/jbc.M808742200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Zhang K., Sandoval H., Yamamoto S., Jaiswal M., Sanz E., Li Z., Hui J., Graham B. H., Quintana A. et al. (2015). Glial lipid droplets and ROS induced by mitochondrial defects promote neurodegeneration. Cell 160, 177-190. 10.1016/j.cell.2014.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi R., Dong J., Rodriguez G., Bell A., Leung T. K., Schwartz R. J., Willerson J. T., Brugada R. and Marian A. J. (2009). Genetic fate mapping identifies second heart field progenitor cells as a source of adipocytes in arrhythmogenic right ventricular cardiomyopathy. Circ. Res. 104, 1076-1084. 10.1161/CIRCRESAHA.109.196899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopaschuk G. D. and Jaswal J. S. (2010). Energy metabolic phenotype of the cardiomyocyte during development, differentiation, and postnatal maturation. J. Cardiovasc. Pharmacol. 56, 130-140. 10.1097/FJC.0b013e3181e74a14 [DOI] [PubMed] [Google Scholar]
- Lu M., Pressman C., Dyer R., Johnson R. L. and Martin J. F. (1999). Function of Rieger syndrome gene in left–right asymmetry and craniofacial development. Nature 401, 45797 10.1038/45797 [DOI] [PubMed] [Google Scholar]
- Neels J. G. and Olefsky J. M. (2006). Inflamed fat: what starts the fire? J. Clin. Invest. 116, 33-35. 10.1172/JCI27280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrello E. R., Mahmoud A. I., Simpson E., Hill J. A., Richardson J. A., Olson E. N. and Sadek H. A. (2011). Transient regenerative potential of the neonatal mouse heart. Science 331, 1078-1080. 10.1126/science.1200708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puente B. N., Kimura W., Muralidhar S. A., Moon J., Amatruda J. F., Phelps K. L., Grinsfelder D., Rothermel B. A., Chen R., Garcia J. A. et al. (2014). The oxygen-rich postnatal environment induces cardiomyocyte cell-cycle arrest through DNA damage response. Cell 157, 565-579. 10.1016/j.cell.2014.03.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampazzo A., Nava A., Malacrida S., Beffagna G., Bauce B., Rossi V., Zimbello R., Simionati B., Basso C., Thiene G. et al. (2002). Mutation in human desmoplakin domain binding to plakoglobin causes a dominant form of arrhythmogenic right ventricular cardiomyopathy. Am. J. Hum. Genet. 71, 1200-1206. 10.1086/344208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt B. A. and Horsley V. (2013). Intradermal adipocytes mediate fibroblast recruitment during skin wound healing. Development 140, 1517-1527. 10.1242/dev.087593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semina E. V., Reiter R., Leysens N. J., Alward W. L. M., Small K. W., Datson N. A., Siegel-Bartelt J., Bierke-Nelson D., Bitoun P., Zabel B. U. et al. (1996). Cloning and characterization of a novel bicoid-related homeobox transcription factor gene, RIEG, involved in Rieger syndrome. Nat. Genet. 14, 392-399. 10.1038/ng1296-392 [DOI] [PubMed] [Google Scholar]
- Srinivasan S. and Avadhani N. G. (2012). Cytochrome c oxidase dysfunction in oxidative stress. Free Radic. Biol. Med. 53, 1252-1263. 10.1016/j.freeradbiomed.2012.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S., Guha M., Dong D. W., Whelan K. A., Ruthel G., Uchikado Y., Natsugoe S., Nakagawa H. and Avadhani N. G. (2016). Disruption of cytochrome c oxidase function induces the Warburg effect and metabolic reprogramming. Oncogene 35, 1585-1595. 10.1038/onc.2015.227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y., Zhang M., Li L., Bai Y., Zhou Y., Moon A. M., Kaminski H. J. and Martin J. F. (2014). Pitx2, an atrial fibrillation predisposition gene, directly regulates ion transport and intercalated disc genes. Circ. Cardiovasc. Genet. 7, 23-32. 10.1161/CIRCGENETICS.113.000259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao G., Kahr P. C., Morikawa Y., Zhang M., Rahmani M., Heallen T. R., Li L., Sun Z., Olson E. N., Amendt B. A. et al. (2016). Pitx2 promotes heart repair by activating the antioxidant response after cardiac injury. Nature 534, 119-123. 10.1038/nature17959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tontonoz P., Hu E. and Spiegelman B. M. (1994). Stimulation of adipogenesis in fibroblasts by PPARγ2, a lipid-activated transcription factor. Cell 79, 1147-1156. 10.1016/0092-8674(94)90006-X [DOI] [PubMed] [Google Scholar]
- van der Maaten L. and Hinton G. (2008). Visualizing data using t-SNE. J. Mach. Learn. Res 9, 2579-2605. [Google Scholar]
- Wang J., Klysik E., Sood S., Johnson R. L., Wehrens X. H. T. and Martin J. F. (2010). Pitx2 prevents susceptibility to atrial arrhythmias by inhibiting left-sided pacemaker specification. Proc. Natl. Acad. Sci. USA 107, 9753-9758. 10.1073/pnas.0912585107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng L.-C. C., Choi S. H., Klarin D., Smith J. G., Loh P.-R. R., Chaffin M., Roselli C., Hulme O. L., Lunetta K. L., Dupuis J. et al. (2017). Heritability of atrial fibrillation. Circ. Cardiovasc. Genet. 10, e001838. 10.1161/CIRCGENETICS.117.001838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y., Hill M., Zhang M., Martin T., Morikawa Y., Wang S., Moise A., Wythe J. and Martin J. (2018). Hippo signaling plays an essential role in cell state transitions during cardiac fibroblast development. Dev. Cell 45, 153-169.e6. 10.1016/j.devcel.2018.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M., Olson E. N. and Bassel-Duby R. (2013). Mending broken hearts: cardiac development as a basis for adult heart regeneration and repair. Nat. Rev. Mol. Cell Biol. 14, 529-541. 10.1038/nrm3619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y., Cavallero S., Patterson M., Shen H., Xu J., Kumar S. R. and Sucov H. M. (2015). Adipogenesis and epicardial adipose tissue: a novel fate of the epicardium induced by mesenchymal transformation and PPARγ activation. Proc. Natl. Acad. Sci. USA 112, 2070-2075. 10.1073/pnas.1417232112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zangi L., Oliveira M. S., Ye L. Y., Ma Q., Sultana N., Hadas Y., Chepurko E., Später D., Zhou B., Chew W. L. et al. (2017). Insulin-like growth factor 1 receptor-dependent pathway drives epicardial adipose tissue formation after myocardial injury. Circulation 135, 59-72. 10.1161/CIRCULATIONAHA.116.022064 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.