ABSTRACT
Transgenic animals are invaluable for modeling cancer genomics, but often require complex crosses of multiple germline alleles to obtain the desired combinations. Zebrafish models have advantages in that transgenes can be rapidly tested by mosaic expression, but typically lack spatial and temporal control of tumor onset, which limits their utility for the study of tumor progression and metastasis. To overcome these limitations, we have developed a method referred to as Transgene Electroporation in Adult Zebrafish (TEAZ). TEAZ can deliver DNA constructs with promoter elements of interest to drive fluorophores, oncogenes or CRISPR-Cas9-based mutagenic cassettes in specific cell types. Using TEAZ, we created a highly aggressive melanoma model via Cas9-mediated inactivation of Rb1 in the context of BRAFV600E in spatially constrained melanocytes. Unlike prior models that take ∼4 months to develop, we found that TEAZ leads to tumor onset in ∼7 weeks, and these tumors develop in fully immunocompetent animals. As the resulting tumors initiated at highly defined locations, we could track their progression via fluorescence, and documented deep invasion into tissues and metastatic deposits. TEAZ can be deployed to other tissues and cell types, such as the heart, with the use of suitable transgenic promoters. The versatility of TEAZ makes it widely accessible for rapid modeling of somatic gene alterations and cancer progression at a scale not achievable in other in vivo systems.
KEY WORDS: Cancer, Electroporation, Melanoma, Zebrafish, Metastasis
Summary: Here, we developed Transgene Electroporation in Adult Zebrafish (TEAZ), a method that enables researchers to study cancer and metastasis by introducing DNA elements focally into somatic adult tissue.
INTRODUCTION
The zebrafish has become an increasingly applied model in cancer biology at the interface of basic discovery and preclinical animal experimentation. The high fecundity and relatively simple husbandry of zebrafish enable large experimental series in vivo. Early cancer models in zebrafish were largely developed using mutagens, such as N-methyl-N'-nitro-N-nitrosoguanidine (MNNG) or 7,12-dimethylbenz (α) anthracene (DMBA), which were later supplanted by transgenic technologies (Beckwith et al., 2000; Pliss et al., 1982; Spitsbergen et al., 2000). The initial transgenic cancer models were developed by injecting one-cell zebrafish embryos with DNA constructs containing a promoter and oncogene. For example, T-cell ALL was modeled by transgene-driven expression of the MYC oncogene using the rag2 promoter, and melanomas generated by expressing BRAFV600E under the mitfa promoter in a tp53−/− germline mutant background (Berghmans et al., 2005; Langenau et al., 2003; Patton et al., 2005).
Despite the documented power of transgenic tumor models for mechanism discovery and drug testing, current models have several significant drawbacks: (1) the majority of established models do not exhibit spatiotemporal control, such that the timing and anatomical location of tumor onset remains variable (Kaufman et al., 2016; Patton et al., 2005; White et al., 2011); (2) they generally do not enable the introduction of serial somatic oncogenic events for modeling second and third hit mutations after onset (Mione and Trede, 2010; White et al., 2013); and (3) discerning multifocal primary tumors versus true metastatic spread of a single tumor is challenging. These issues all impose significant limitations for investigating tumor progression and metastasis.
Transplantation-based methods address some of these issues: tumors can be dissected from a transgenic tumor-bearing animal or from patient-derived xenografts (PDXs), and then serially transplanted into recipient animals such as the casper recipient strain to allow detailed in vivo imaging (Fior et al., 2017; Heilmann et al., 2015; Hoffman, 2015; Siolas and Hannon, 2013; Tang et al., 2016; White et al., 2008; Zeng et al., 2017). Alternatively, stable cell lines can be generated from a transgenic animal, such as the ZMEL1 melanoma line, which can similarly be used for transplantation studies (Heilmann et al., 2015). Although these transplantation approaches allow for precise spatiotemporal control and are amenable to imaging of metastasis, the experiments often require immunosuppression of the recipients either through irradiation or genetic manipulation of immune cells (Tang et al., 2016), in addition to the initial generation of the suitable cell line. Recent work from the Langenau laboratory has shown that syngeneic fish can be used as transplant recipients, but these require that the tumors be developed in that particular genetic background, somewhat limiting their broad use across cancer (Blackburn et al., 2011). Furthermore, transplantation cancer models implant foreign tumors into inherently artificial microenvironments.
A variety of Cre/Lox-based approaches have been used in the zebrafish to control the cells that undergo initiation, including T-cell leukemia that can be controlled by mRNA injection (Langenau et al., 2005). A further modification uses CreERT2 drivers, such that both the cell type and timing of gene expression can be controlled (Hans et al., 2009), and inducible expression has also been achieved with heat shock Cre constructs (Hans et al., 2011). Despite these advances, there is still a paucity of verified Cre/Lox-based approaches to cancer in the zebrafish as the transgenic animals have proven time-consuming to create, and require complex breeding schemes to generate the final required genotypes.
Based on this, we wished to develop an approach that would enable introduction of oncogenic elements directly into adult somatic tissue in a spatiotemporally controlled manner. In this paper, we report oncogenesis via Transgene Electroporation in Adult Zebrafish (TEAZ), which models how tumors natively form in somatic tissues in a fully immunocompetent adult zebrafish. Electroporation applies electrical pulses to generate pores within the cell membrane, enabling extracellular biomolecules (including DNA) to enter the cell (Neumann et al., 1982; Wong and Neumann, 1982). Electroporation is widely used for stable introduction of DNA elements into cells in tissue culture and into chick and mouse embryos. Electroporation has occasionally been utilized in adult zebrafish, but these studies have been limited to cell tracking and transient morpholino knockdowns, and the technique has never been applied to cancer modeling (Hoegler et al., 2011; Münch et al., 2013; Rambabu et al., 2005; Thummel et al., 2011). Several studies in mice have harnessed electroporation to introduce transgenes into select adult tissues, including retina, muscle, brain and prostate (Maresch et al., 2016; Nomura et al., 2016; Swartz et al., 2001; Yarmush et al., 2014), and used it to model tumors such as pancreatic cancer (Jung et al., 2014; Maresch et al., 2016; Park et al., 2014). However, these approaches require surgery of the mice and can only be limited to a small number of animals at a time, limiting the number of subjects that can reasonably be studied in each experiment. Based on these prior observations and the very large cohorts we can generate, we reasoned that direct electroporation of oncogenic transgenic constructs into the zebrafish would be a straightforward, highly scalable approach to model tumor formation in cells of interest. Because electrodes and DNA solutions can be placed at defined locations, TEAZ allows for delivery of multiple transgenes specifically to the anatomical locations of interest. We find that TEAZ allows for the development of complex, aggressive melanomas driven by expression of oncogenic BRAFV600E in concert with loss of the tumor suppressors tp53 and rb1. These tumors are highly invasive and eventually metastasize to distant locations, unlike previous transgenic zebrafish melanoma models which do not generally metastasize (Patton et al., 2005). Given the wealth of functionally uncharted genetic lesions discovered from sequencing human tumors, TEAZ allows for testing of candidate mutations in a rapid, scalable in vivo system. More broadly, TEAZ can also be used to study somatic alteration of gene function in any adult tissue, which will have applications for diseases outside of cancer as well.
RESULTS
Introduction of genetic elements into adult zebrafish via electroporation
The TEAZ method is designed to introduce genetic elements into specific locations within the adult zebrafish (Fig. 1A). The method has been optimized for the use of plasmids generated in Escherichia coli and purified using standard plasmid purification protocols (midi preps). To test this protocol, we anesthetized adult zebrafish, and then injected 1.0 μl purified plasmid DNA (1000 ng/μl) directly under the dorsal fin. The injected zebrafish was then quickly placed into an agarose mold to position the animal upright. Using paddle-shaped electrodes, we directed electrical pulses across the injected region (electroporator set to LV mode, 45 V, 5 pulses, 60 ms pulse length, 1 s pulse interval). To maximize expression in the skin as opposed to deeper tissues, the cathode paddle can be placed just above the surface where the DNA was injected as this will pull the negatively charged DNA towards the surface adjacent to the injection site. After electroporation, we placed the anesthetized zebrafish into fresh water for recovery and maintenance through standard husbandry. Including anesthetization, DNA injection and electroporation, the entire protocol takes ∼45-60 s per animal.
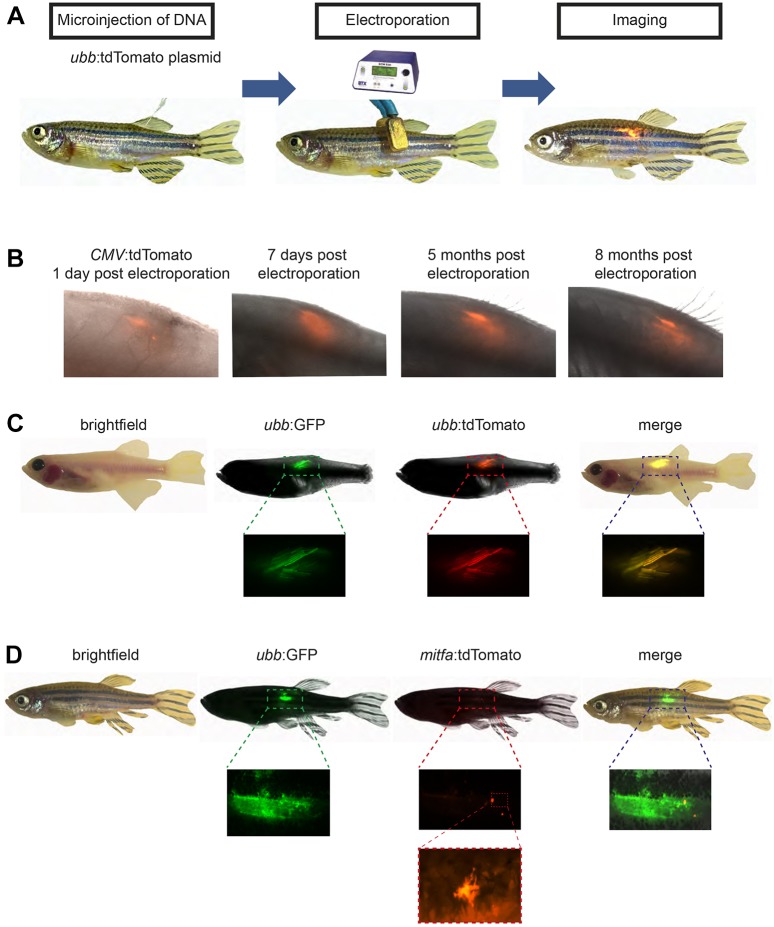
Fig. 1.
TEAZ. (A) Schematic representation of the method applied for the introduction of ubb:tdTomato directly under the dorsal fin of adult zebrafish. The purified plasmid DNA (1 μl of a 1000 ng/μl solution of ubb:tdTomato) is injected into anesthetized zebrafish using a pulled glass micropipette. Electrical pulses are directed across the injected region (settings: LV mode, 45 V, 5 pulses, 60 ms pulse length and 1 s pulse interval). Reporter expression can be visualized by fluorescent microscopy (n=2/2). (B) Electroporation of a CMV:tdTomato plasmid was performed and the animal followed for a period of 8 months (n=2/2). The fluorescent signal can be visualized as early as 1 dpe, with intensity peaking at ∼1 week and maintaining for at least 8 months. (C) Multiple plasmids will co-integrate in TEAZ. casper zebrafish were electroporated with a total volume of 1.0 μl (0.5 μl of 1000 ng/μl ubb:GFP and 0.5 μl of 1000 ng/μl ubb:tdTomato) and imaged using BF, GFP and tdTomato (n=3/3), revealing co-expression of the plasmids. (D) Promoter specificity is maintained following TEAZ. AB fish were electroporated with 1.0 μl total volume (0.5 μl of 1000 ng/μl ubb:GFP and 0.5 μl of 1000 ng/μl mitfa:tdTomato) and displayed highly restricted expression of the mitfa reporter plasmid, but widespread expression of the ubb plasmid (n=9/9). High-resolution imaging of the tdTomato+ cells reveals a dendritic phenotype, consistent with the melanocytic lineage.
To establish TEAZ, we injected wild-type zebrafish (AB strain) with a plasmid in which the zebrafish ubiquitin B (ubb) promoter (Mosimann et al., 2011) drives tdTomato expression (ubb:tdTomato). This vector was created using the Tol2 transposon system that is commonly deployed in the zebrafish (Balciunas et al., 2006; Kawakami, 2007; Kawakami et al., 2000; Kwan et al., 2007; Villefranc et al., 2007). In our studies, we did not use transposase mRNA because in preliminary tests we were unable to drive fluorescence from mRNA (either GFP or tdTomato) following electroporation (data not shown). After TEAZ, we imaged the electroporated zebrafish starting at 1 day postelectroporation (dpe) and over the course of several months. Fig. 1B shows an example animal, in which stable expression of tdTomato (under the CMV promoter) was detectable for up to 8 months (Fig. 1B). Similarly, the stable expression of ubb:GFP-p2a-tdTomato was also detectable after 8 months postelectroporation (n=4/4). We also tested whether the same ubb promoter and TEAZ would express in other parts of the animal by injecting it directly into the head. We observed robust expression of ubb:GFP in the head in four of the five animals electroporated (Fig. S1), indicating that TEAZ-mediated transgene activity is not restricted to a particular location on the adult zebrafish body. At the site of electroporation, there is initially a small area of tissue damage, which is rapidly healed within 1 week. We have never seen ectopic expression of the transgene away from the site of electroporation, nor have we observed any systemic toxicity or obvious procedure-caused death in several hundred similarly electroporated animals. In addition, to ensure lack of germline transmission, we electroporated several different constructs (Table S1), waited for adult expression, and then bred those animals to wild-type adults. We then screened the resultant embryos at both 1 day postfertilization (dpf) and 4 dpf and saw no animals with fluorescence (n=947 embryos). This indicates that the TEAZ method allows for highly focal, somatic transgenesis at the site of electroporation.
TEAZ allows for simultaneous expression of multiple plasmids
Electroporation of multiple plasmids into cultured cells in vitro generally leads to joint uptake of the plasmids by cells and subsequent co-expression of the transgenes. To test this in the zebrafish, we performed TEAZ with two plasmids in a single injection. We co-injected the ubb:tdTomato plasmid along with a ubb:GFP plasmid (each at 0.5 μl of 1000 ng/μl plasmid stock) and then monitored fluorescence. High-magnification views demonstrated that 100% of the transgene-expressing cells were double positive for both GFP and tdTomato (n=3/3 fish) (Fig. 1C). Consequently, TEAZ can be expanded to express and combine multiple transgenes in the adult zebrafish skin.
Maintenance of promoter specificity following electroporation
We next sought to determine whether TEAZ enables cell type-specific transgene expression. We co-electroporated mitfa:tdTomato (Lister et al., 1999) (which drives in melanocytes) and ubb:GFP (which drives ubiquitously) plasmids and then imaged the zebrafish using both tdTomato and GFP channels. A representative animal is shown in Fig. 1D (n=9/9). As anticipated, we detected broad and strong expression of GFP from the ubb promoter (Mosimann et al., 2011). In contrast, we found highly limited expression of the mitfa:tdTomato plasmid. High-resolution imaging of the tdTomato+ cells revealed a dendritic appearance that is consistent with the appearance of mature melanocytes (Fig. 1D). We next tested expression in the heart using the cardiomyocyte-specific myl7 promoter driving GFP (myl7:GFP; formerly referred to as cmlc:GFP) (Huang et al., 2003). We injected and electroporated myl7:GFP plasmid directly into the beating heart muscle of an anesthetized adult zebrafish. We found strong and specific expression of GFP in the beating heart (n=2/4) (Fig. S1C, Movie 1). Importantly, when myl7:GFP was electroporated below the dorsal fin (n=5) and mitfa:tdTomato was electroporated into the heart (n=5) fluorescence was not detected, showing that expression is highly cell type specific and driven by promoter specificity. We conclude that TEAZ-mediated vector delivery maintains promoter specificity following electroporation, enabling us to target specific somatic cell types within specified regions of adult zebrafish.
Melanoma initiation requires multiple transgenes
We next sought to apply TEAZ to directly model melanoma formation in adult zebrafish, circumventing embryonic manipulations. We and others have previously used a traditional germline melanoma transgenic in which the mitfa promoter drives oncogenic BRAFV600E (Patton et al., 2005; White et al., 2011). In a tp53−/− deficient background, these transgenic animals develop a 100% penetrant melanoma at 4-12 months of age, without any additional transgenes (Patton et al., 2005; White et al., 2011). This original transgenic was further extended using the miniCoopR system (Ceol et al., 2011), in which the mitfa gene itself is knocked out, creating a strain with the genotype mitfa:BRAFV600E;tp53−/−;mitfa−/− (herein referred to as the ‘triple’ strain). When this triple strain is injected at the one-cell embryo stage with a ‘rescue’ plasmid containing an mitfa:mitfa and mitfa:GFP cassette in cis, the resultant animals have rescued GFP+ melanocytes that all go on to develop GFP+ melanomas as adults (Ceol et al., 2011).
To test whether TEAZ is adaptable to this approach and could enable circumvention of initiating transgene expression at embryonic stages, we electroporated the miniCoopR:GFP rescue cassette under the dorsal fin of triple strain adult zebrafish. We found that 8/10 injected animals developed GFP fluorescence at the site of injection, and, remarkably, one of the animals developed rescued melanocytes. This indicates that it is possible to ‘rescue’ melanophore development in a germline genetic defect (i.e. mitfa−/−) by directly electroporating a minigene cassette into adult somatic tissues. However, none of these animals went on to develop melanoma over a period of 4 months, a duration that leads to melanoma in embryo injection-based experiments. This observation suggests that in TEAZ, additional genetic hits are necessary above and beyond BRAF and tp53−/−.
TEAZ-mediated CRISPR of Rb1 stimulates melanoma in adults
In the mitfa−/− mutant background, there are no mature melanocytes (Lister et al., 1999) because mitfa is required for expression of melanocytic genes, such as the Pmel genes and tyr. We suspected that there might be melanocytic precursor cells in the mitfa:BRAFV600E;tp53−/−;mitfa−/− background that are largely quiescent and not actively cycling. We therefore aimed to knock out the function of the tumor suppressor Rb1 using CRISPR-Cas9, because TP53 and RB1 mutations have a tendency to be concurrent in human melanoma patients, as seen in the cBIOPortal (P=0.017) (Berger et al., 2012; Cerami et al., 2012; Gao et al., 2013; Hodis et al., 2012; Hugo et al., 2016; Krauthammer et al., 2012). Rb1 normally acts to keep cells arrested in G1, and we therefore reasoned that loss of its function might provoke the cells to proceed through the cell cycle and be more amenable to full malignant transformation (Yu et al., 2009). To test this hypothesis, we employed the triple strain, and electroporated three plasmids: (1) miniCoopR:GFP, (2) ubb:Cas9 and (3) zU6:sgRNA against rb1 (see Materials and Methods for details). We found that of the nine electroporated animals, eight developed rescued melanocytes and went on to establish aggressively growing GFP+ lesions with the phenotypic appearance of frank melanomas (Fig. 2A-C). The tumors appeared within 3-7 weeks, in striking contrast to the 3-6 months typically required for standard embryo-injection transgenics (Fig. 2D). To confirm that the effect was due to introduced rb1 mutations, we dissected the dorsal fin (tumor) and tail fin (control normal tissue) from the same adult zebrafish for sequence analysis. Deep sequencing of the two fins and CrispRVariants-based allele analysis validated that the tumor contained two independent frameshift mutations in rb1 at the protospacer adjacent motif site that are characteristic of CRISPR mutations and were not present in the control tail fin (Fig. S2) (Burger et al., 2016; Lindsay et al., 2016). Although the percentage of mutant reads was low in this analysis, it is likely because we sequenced surrounding normal tissue as well as tumor. We previously performed whole-genome and exome sequencing on a series (n=53) of embryonic-transgenic zebrafish tumors and did not see any Rb1 mutations (Kansler et al., 2017; Yen et al., 2013). Consistent with the loss of Rb1 in TEAZ tumors, when we stained for phospho-Rb1 in both a TEAZ tumor and standard embryo injection F0 tumor (i.e. mitfa:BRAF injected into a tp53−/− background without any Cas9/sgRNA), we found that most of the cells in the TEAZ tumor are phospho-Rb1 negative, whereas most of the embryo injection tumor cells are Rb1+ (Fig. S3). The immunohistochemistry detected phospho-Rb1 but not total Rb1 protein, as we do not have an antibody that effectively stains for this. The results revealed that TEAZ-mediated transformation of adult tissue can result in rapid melanoma onset using tumor-relevant genetic lesions.
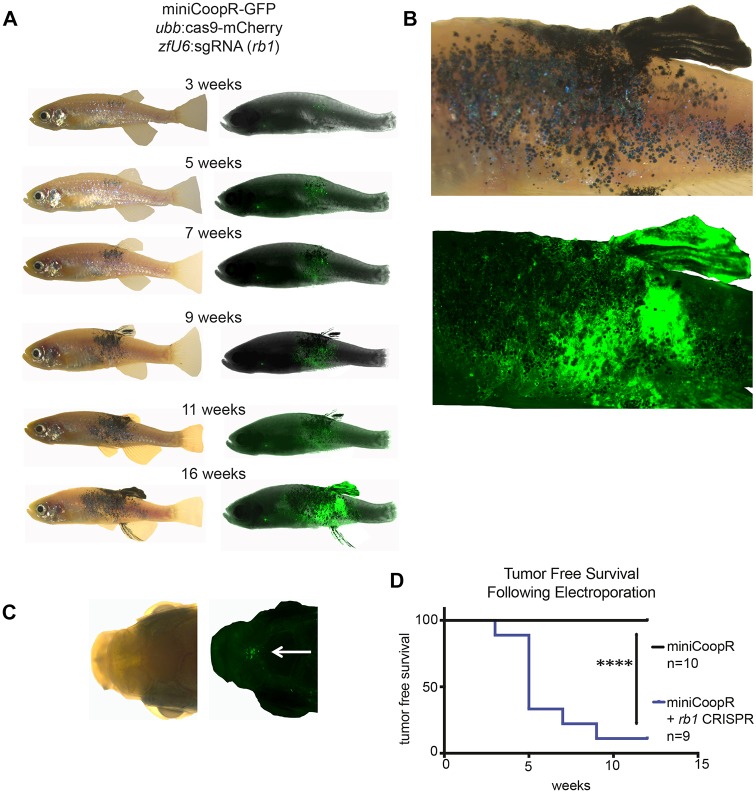
Fig. 2.
Generation of a novel melanoma model with TEAZ. (A) mitfa:BRAFV600E;tp53−/−;mitfa−/− zebrafish (triple strain) were electroporated with the miniCoopR:GFP plasmid that both rescues melanocytes and expresses GFP under the mitfa promoter, with (n=10) or without (n=9) two additional plasmids to genetically knockout rb1 (ubb:Cas9 and zfU6:sgRNA against rb1). The electroporated zebrafish were then imaged over time by both fluorescence and brightfield to monitor tumor development. Overall, 17/20 electroporated zebrafish had GFP+ cells. Tumor development in a representative zebrafish from the melanoma model including rb1 knockout is shown. (B) Higher-magnification view of the tumor-bearing animal shown in A at 16 weeks postelectroporation. (C) At 9 weeks postelectroporation, 4/8 zebrafish had evidence of GFP+ distant micrometastases in the head. (D) The loss of rb1 is essential for tumor initiation as visualized by the Kaplan–Meier curve comparing zebrafish electroporated with miniCoopR:GFP with or without rb1 sgRNA. Log-rank (Mantel–Cox) test was used for statistical analysis (****P<0.0001).
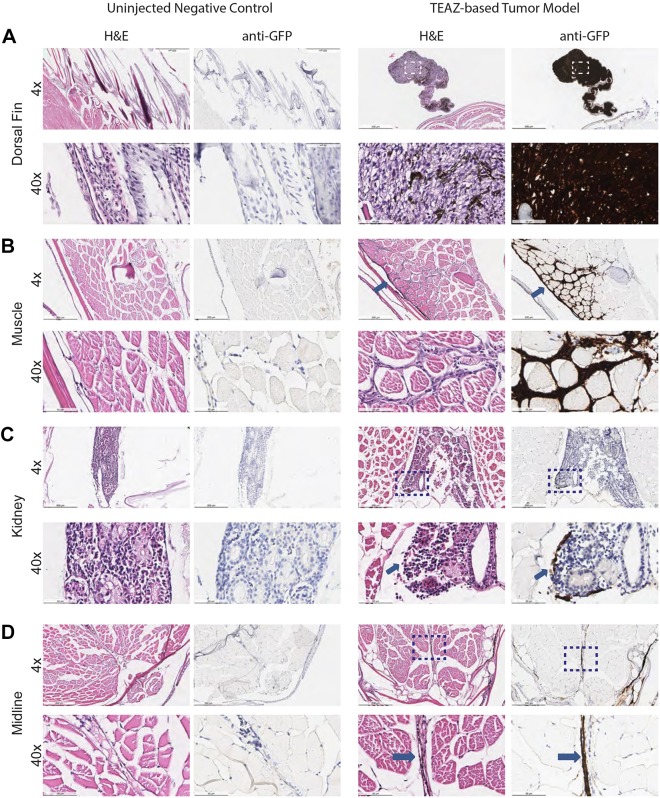
To confirm that the lesions were truly tumorigenic, we followed a cohort of these TEAZ-treated zebrafish for a period of 4 months. By 5 weeks, primary melanomas could be visualized by both fluorescence and brightfield imaging. By 9 weeks, four of eight remaining zebrafish had tumors that had rapidly progressed, and traversed the midline to the opposite side of the body. In four of eight zebrafish, we noted evidence of GFP+ distant micrometastases in the head (Fig. 2C). To investigate further, we sacrificed two tumor-bearing animals along with two control animals and performed routine histology and anti-GFP staining. The first fish had a large protruding primary tumor with uniform GFP expression by fluorescence imaging. Histology of this tumor confirmed this, identifying uniform anti-GFP staining, and Hematoxylin and Eosin (H&E) staining showed cells highly consistent with high-grade melanoma, as determined by pathologist assessment (i.e. nuclear atypia and presence of melanin) (Fig. 3A). We noted extensive invasion into the muscle (Fig. 3B), which had not previously been seen in transgenic zebrafish melanoma modeling using BRAFV600E with tp53−/−. Along with this invasion phenotype, we identified micrometastases within the kidney (Fig. 3C) and attached to blood vessels (Fig. 3D). The second fish had an atypical tumor with variable GFP expression by fluorescence imaging, and histology showed a tumor of mixed histology in the vicinity of the injection needle: surface GFP+ tumor cells consistent with a low-grade melanoma, and a deeper tumor in the muscle that was GFP–, consistent with a sarcoma (Fig. S4A,B). This second nonmelanoma tumor is likely caused by inactivation of both rb1 and tp53 in the muscle, as we used ubiquitous Cas9 in our studies, and this combination is commonly found in sarcomas (Cerami et al., 2012; Gao et al., 2013; Gonin-Laurent et al., 2007; Pérot et al., 2010; Stratton et al., 1990). We did not find any GFP staining or abnormal H&E staining in either of the control animals (Fig. 3; Fig. S4C). Taken together, our findings demonstrate that the TEAZ method can rapidly and robustly give rise to tumors in a highly defined spatiotemporal manner.
Fig. 3.
Melanoma model using TEAZ show evidence of rapid progression. (A) Pathology of tumor-bearing zebrafish (n=1) (along with control zebrafish, n=2) at 16 weeks postelectroporation, stained with H&E or anti-GFP immunohistochemistry, demonstrates a large primary tumor that is uniformly GFP+. (B-D) Histology reveals evidence of extensive invasion into the muscle (arrows) (B) along with micrometastatic sites within the kidney (C) or along blood vessels (arrows) (D). Images are visualized at 4× and 40×. Scale bars: 500 μm (4×) and 50 μm (40×). Dashed line boxes indicate the area enlarged at 40×.
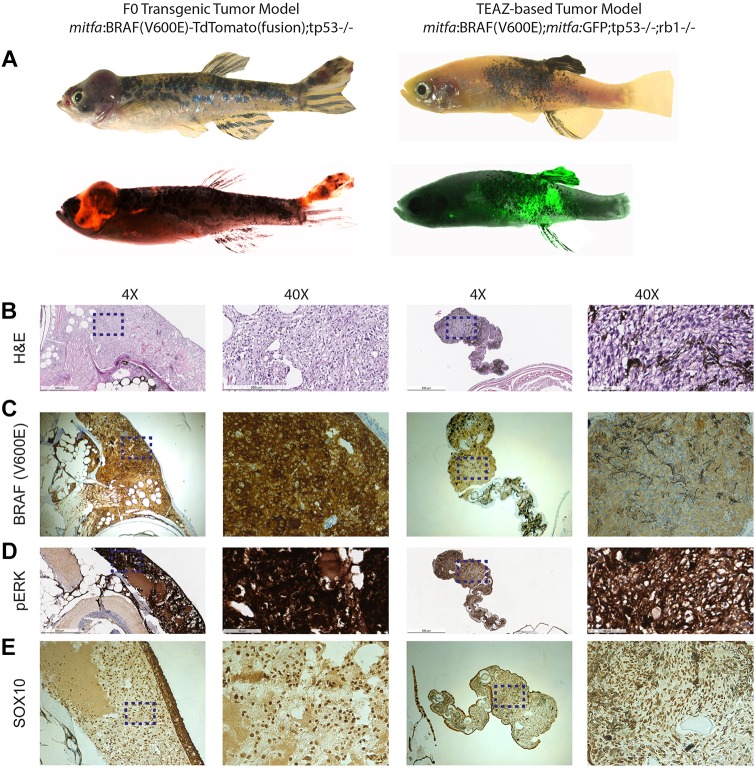
We previously showed that most melanomas, of both fish and human origin, overexpress a neural crest transcriptional program typified by crestin and sox10, making the latter gene particularly relevant for human melanoma. To compare our TEAZ-based tumors to the more traditional embryo injection models, we performed immunohistochemistry of the melanomas using antibodies against SOX10, BRAFV600E and phospho-ERK (Fig. 4). In agreement with previous models, we find that both the TEAZ melanomas and embryo injection transgenics have high levels of SOX10 protein expression. Additionally, both tumor types ubiquitously express both BRAFV600E and phospho-ERK, albeit to a lesser degree in the TEAZ tumor. These data suggest that the TEAZ melanomas are functionally similar to the embryo injection transgenics. One key difference is that we also document evidence of progression and distant metastasis using TEAZ.
Fig. 4.
Histological comparison of the embryo injection melanoma model and TEAZ melanoma model. (A) The left images show a melanoma created by injection of an mitfa:BRAFV600E-tdTomato (fusion) transgene into a tp53−/− background (n=1). Right images show a TEAZ-based melanoma created by electroporation of miniCoopR:GFP plus ubb:Cas9 plus zfU6:sgRNA against Rb1 (n=1) (example shown is fish at 16 weeks also shown in Fig. 2A). (B) H&E staining of both tumors shows similar histology, although with increased melanin pigmentation in the TEAZ tumor (also shown in Fig. 3A). (C,D) Antibody staining against BRAFV600E shows that both tumors are widely BRAFV600E positive, which correlates with high levels of phospho-ERK staining. (E) Reflecting the neural crest origin of melanocytes, both tumors show strong nuclear expression of SOX10. Images are visualized at 4× and 40×. Scale bars: 500 μm (4×) and 50 μm (40×). Dashed line boxes indicate the area enlarged at 40×.
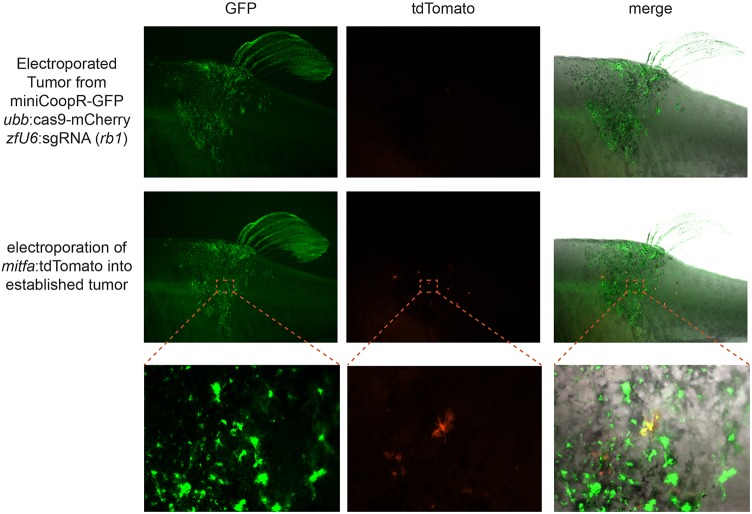
Somatic tumors are amenable to sequential transgenic manipulation
One of the major limitations of available genetic models is the inability to modify genes in a sequential order, mimicking in vivo tumor progression from malignant clones (McGranahan and Swanton, 2017). This limitation precludes the investigation into whether certain oncogenic events are driving initiation (which occur early) versus metastasis (when they may occur later). We therefore sought to determine whether we could sequentially perform TEAZ to introduce new DNA elements into an already existing tumor. We selected a TEAZ melanoma from the cohort above (4 months postinitial electroporation), and electroporated an mitfa:tdTomato plasmid directly into the tumor (Fig. 5). Within 1 week after this second electroporation, we identified tdTomato+ cells within the TEAZ-treated tumor (n=2/2). The tdTomato+ cells have a dendritic appearance typical of a melanocytic cell. We also tested whether this sequential transgene electroporation into an existing tumor was of different efficiency to de novo electroporation into unperturbed tissue. To do this, we compared electroporation of mitfa:tdTomato into an existing TEAZ tumor versus electroporation of mitfa:tdTomato plus ubb:GFP into an AB fish (the GFP was added here to control for the fact that TEAZ tumors are GFP+). We then counted the number of tdTomato+ cells in both situations, and noted a greater number of positive cells when electroporating into the AB fish compared with the established tumor (Fig. S5). However, although it does appear that sequential electroporation into tumors might be slightly less efficient than de novo electroporation, it is still efficient enough for routine use in existing tumors and will allow for sequential modeling of genetic lesions.
Fig. 5.
Cancer modeling with TEAZ enables sequential electroporation of transgenes. A tumor-bearing fish (created with rb1 sgRNA as in Fig. 2) was imaged using GFP and tdTomato. As expected, only GFP+ tumor cells were seen, with no expression in the tdTomato channel. This tumor was then electroporated with an mitfa:tdTomato plasmid and re-imaged 5 days later, showing areas that are now both GFP+ and tdTomato+ (n=2).
DISCUSSION
We have developed TEAZ, an electroporation-based approach for expressing transgenes and creating mutations in somatic cell types of interest within a region of interest in the adult zebrafish. We successfully applied TEAZ to the generation of malignant melanoma, and our results show that TEAZ can be used for sequential electroporation, which could be used to make increasingly complex tumor models. This model is amenable to initiating a tumor at a defined time and place, and will allow for a detailed analysis of tumor progression and metastasis in a fully immunocompetent zebrafish.
One major limitation of current transgenic cancer modeling in the zebrafish is the challenge of controlling both the timing and location of tumor initiation. Although both transplantation and Cre/Lox approaches can address some of these issues, neither fully solves the problem. Transplantation generally requires immune modulation of the recipient, and introduces relatively high cell burdens into tissue contexts that would not occur during natural tumor formation. Cre/Lox is extremely powerful but requires multiple genetic crosses, and there are very few verified lines that exist for cancer modeling in the fish. In contrast, TEAZ rapidly allows for introduction of the required genetic elements in a multiplexed, complex manner.
Electroporation has been used as a mechanism for tumor initiation in mouse models of cancer. Both glioblastoma and oligoastrocytomas (Chen et al., 2016) have been induced into the developing fetus using in utero electroporation and the piggyBAC transposon system. In these studies, the transposase was included on the plasmid as a helper, although in our study we did not find Tol2 transposase to be necessary for highly stable transgene expression. Recent work (Jung et al., 2014; Maresch et al., 2016; Park et al., 2014) showed that plasmid delivery and electroporation could be used to initiate KRAS-driven pancreatic cancer in the adult mouse and, similar to our findings, can be complemented with CRISPR-Cas9-mediated genome editing. One exciting area that we believe TEAZ will open up is the possibility of new epithelial cancer models in the zebrafish, because that has not been developed on a very large scale.
One of the major advantages of our TEAZ system is the high efficiency we have observed. Once trained, we find that the rate of success of transgene expression approaches 100% of injected animals with 100% survival when performed in the dorsal skin. Injections into other adult tissues, such as the heart or head, are technically more challenging, requiring more practice, and also result in higher death rates, as expected (75% survival for heart, 62.5% survival for head). In our study of the melanomas, we found that, overall, 88% of the fish developed GFP+ cells by 3 weeks, and all of those fish subsequently went on to develop tumors by 7 weeks. This timing is an advance over the previous standard transgenic melanoma models, even with the more rapid miniCoopR mosaic approaches that speed up tumors with the addition of oncogenes such as SETDB1 (Chen et al., 2016). In addition, embryo injection remains a relatively laborious process, whereas we find that the adult electroporation is simple and fast and can be easily taught to inexperienced users.
One possibility that our TEAZ system opens up in the future is the ability to initiate adult-stage tumors in virtually any genetic background (i.e. the casper strain) or other existing transgenic line. In our studies using the miniCoopR background, we needed at least three genes to obtain efficient tumors: BRAFV600E, tp53−/− and rb1−/−. This might or might not be related to the specifics of the miniCoopR system as the melanocyte progenitors likely to have to re-enter the cell cycle. We also noted that our tumors formed faster than typical miniCoopR tumors, even with the addition of genes such as SETDB1. This could relate to the specifics of the TEAZ approach or could simply be due to the accelerating effect of rb1 loss. It was surprising that the allelic fraction of rb1 mutations was low in our MiSeq analysis, but this could be due to the fact that, in isolating genomic DNA for sequencing, we included large margins of surrounding normal tissue along with the tumor. Our staining for phospho-Rb1 is suggestive that the majority of the tumor cells are deficient for this tumor suppressor. The extent of rb1 loss required for TEAZ tumors will need further analysis.
It will be important for future studies to establish the minimum number of genetic elements necessary to drive tumor formation in wild-type backgrounds, which will allow TEAZ to be applied to existing transgenic lines that label a variety of interesting cell types, such as T-cells (Dee et al., 2016), macrophages (Ellett et al., 2011) or endothelial cells (Lawson and Weinstein, 2002). Related to this, we have not yet attempted TEAZ on larval-age fish because it will require specialized, smaller electroporation paddles, but this could be useful for modeling tumors that occur in young adults. Previously, creating transgenic tumors in a strain of interest required time-consuming breeding and genotyping to obtain the final genotype of interest. In contrast, with TEAZ, one can electroporate a combination of oncogenes and sgRNAs against tumor suppressors into any given genetic background directly, saving months of breeding and unused animals. To ensure specificity of the tumor types, it will be important to use tissue-specific promoters to drive Cas9, to avoid mixed-histology tumors induced by ubiquitous Cas9 expression.
Metastasis and tumor progression have remained challenging to study using the zebrafish model; our results presented here suggest that TEAZ-mediated tumor modeling is amenable to studying metastasis in a high-throughput, immunocompetent model. Although transplantation-based approaches are powerful, they require immune system manipulation, such as irradiation or transplantation in genetically immunocompromised zebrafish to counteract rejection (Heilmann et al., 2015; Tang et al., 2016; White et al., 2008, 2013). In contrast, TEAZ allows tumor formation in fully immunocompetent animals. These features render the TEAZ model well positioned to (1) screen metastatic modulators to test rate, propensity and latency; (2) selectively alter genes within specific cell types within the tumor microenvironment; (3) image the interplay between tumor cells and specific microenvironmental cell-types using widely available transgenic lines; and (4) introduce serial mutations to study order of progression or induce competition studies within a tumor.
MATERIALS AND METHODS
Zebrafish husbandry
Zebrafish were bred and maintained in the Zuckerman fish facility, in temperature- (28°C), pH- (7.4) and salinity-controlled conditions. All fish were maintained on a 14 h on/10 h off light cycle. The animal protocols described in this paper were approved by the Memorial Sloan Kettering Cancer Center Institutional Animal Care and Use Committee (12-05-008).
Zebrafish mutant lines
Transgenic lines used in these studies included wild-type (AB), casper (D'Agati et al., 2017; White et al., 2008) and the triple line (Ceol et al., 2011) (mitfa:BRAFV600E;tp53−/−;mitfa−/−) (provided by the Houvras laboratory at Weill Cornell Medical College, New York, USA). TEAZ was equally successful in both male and female zebrafish. TEAZ was performed on zebrafish ranging from 4 to 12 months postfertilization.
Molecular biology
Purified plasmids were generated using the Gateway system and isolated from E. coli using the Qiagen HiSpeed Plasmid Maxi Kit. The miniCoopR vector was provided by the Houvras laboratory. The zebrafish U6 promoter was cloned out of genomic DNA as previously described (Heilmann et al., 2015). The sgRNA against rb1 was designed using the CHOPCHOP software and has the 20 bp sequence 5′-GGCTCAGTGAGTTTGAACGG-3′ (Labun et al., 2016; Montague et al., 2014). The ubb promoter was as described previously (Mosimann et al., 2011), and the Cas9-mCherry fusion plasmid was subcloned from Addgene plasmid number 78313 (Burger et al., 2016). All final plasmids were constructed using Gateway technology and the Tol2kit as previously described (Kawakami, 2007; Kwan et al., 2007).
Plasmids used in the study were as follows: ubb:GFP (Mosimann et al., 2011) and ubb:tdTomato (both in Tol2kit plasmid backbone #394), zfU6:Rb1gRNA (Heilmann et al., 2015), mitfa:tdTomato (Lister et al., 2001), myl7:GFP (Huang et al., 2003), miniCoopR (Ceol et al., 2011) and ubb:Cas9-mCherry fusion (Burger et al., 2016) (all in Tol2kit plasmid backbone #395).
Electroporation of adult zebrafish
At the time of electroporation, recipient adult zebrafish were anesthetized in 0.2% Tricaine. The plasmid of interest was resuspended at 1000 ng/μl in ddH2O, and 1.0 μl was injected into the dorsal skin, head or heart (using a pulled glass micropipette). No transposase mRNA was used in these studies. We have successfully tested a range of concentrations from 400 ng/μl to 2000 ng/μl and from 0.5 μl to 2.0 μl. Following injection, the zebrafish were immediately placed upright in an agarose mold for ease of handling, and electrodes were placed on either side of the fish surrounding the injection site (Fig. 1A). The cathode paddle was generally placed on the same side as the injection to promote the DNA entering cells closer to the surface of the fish, but the cathode and anode can be swapped to promote integration into cells deeper within the animal. We used the ECM 830 Electro Square Porator from BTX Harvard Apparatus and the Genepaddles, 3×5 mm. For all experiments described, the LV mode was used with a voltage of 45 V, 5 pulses, 60 ms pulse length and 1 s pulse interval. The electroporated zebrafish were immediately returned to flowing fresh water after electroporation. Electroporated zebrafish were imaged within 4 dpe to ensure successful TEAZ, and then serially imaged for up to 8 months using brightfield and fluorescence imaging.
For electroporation of deeper tissues, such as heart or brain, the same glass electrode was used to inject the DNA but penetrated more deeply directly into those tissues. The electroporation paddles were positioned around those organs, but still kept on the surface of the animal with the same electrical settings.
For the Rb1 miniCoopR experiments, the following concentrations of plasmids were used: miniCoopR:GFP (370 ng), ubb:Cas9 (205 ng) and zfU6:sgRNA against rb1 (285 ng).
Imaging and image processing
Adult zebrafish were imaged using an upright Zeiss Discovery V16 equipped with a motorized stage, brightfield, and GFP and tdTomato filter sets. To acquire images, zebrafish were lightly anesthetized with 0.2% Tricaine. Images were acquired with the Zeiss Zen software v1, and the postimage processing was performed using Fiji (Schindelin et al., 2012).
Histology
Selected zebrafish were fixed in 4% paraformaldehyde for 48 h at 4°C and then paraffin embedded. Fish were sectioned at 5 µM and placed on Apex Adhesive slides, baked at 60°C, and then stained with H&E or antibodies against GFP (Abcam, ab183734, 1:100), BRAFV600E (Abcam, ab228461, 1:400), phospho-Rb1 (Cell Signaling Technology, 8516s, 1:400), phospho-ERK (Cell Signaling Technology, 4370, 1:100) or SOX10 (Cell Marque, 383A-76, 1:50). All histology was performed by Histowiz (http://www.histowiz.com), and staining was performed by Histowiz or the Hollmann Laboratory and reviewed by a pathologist (T.J.H.).
Kaplan–Meier analysis
All animals were followed for up to 16 weeks and tumor-free survival was measured using the Kaplan–Meier method. The differences between the miniCoopR zebrafish with and without rb1 knockout were analyzed using the log-rank statistics.
MiSeq analysis
Reads were mapped to the zebrafish genome version GRCHz10 using bwa version 0.7.13-r1126. Mutation quantification was performed using CrispRVariants version 1.7.4 (Lindsay et al., 2016). MiSeq reads can be accessed through the NCBI Sequence Read Archive (SRA) (accession code SRP147816).
Supplementary Material
Acknowledgements
We thank Histowiz (www.histowiz.com) for performing histology work.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: S.J.C., R.M.W.; Methodology: S.J.C., S.T., H.L., A.B., T.J.H., C.M., R.M.W.; Software: H.L., A.B., C.M.; Validation: S.J.C., S.T., T.J.H., C.M.; Formal analysis: S.J.C., H.L., A.B., R.M.W.; Investigation: S.J.C., S.T., Y.M.Z.; Resources: S.T., Y.M.Z., A.B., N.R.C., I.S.K., R.M.W.; Writing - original draft: S.J.C., R.M.W.; Writing - review & editing: S.J.C., R.M.W.; Visualization: S.J.C., H.L., A.B., C.M.; Supervision: A.B., T.J.H., L.S., C.M., R.M.W.; Project administration: L.S., R.M.W.; Funding acquisition: L.S., R.M.W.
Funding
This work was supported by the National Institutes of Health (DP2CA186572, P30CA008748 and K08AR055368 to R.M.W.), the National Cancer Institute (F31CA196305 to S.J.C.; 1F99CA212436-01 to Y.M.Z.), the Melanoma Research Alliance (R.M.W.), the Starr Cancer Consortium (R.M.W.), the Pershing Square Sohn Foundation (R.M.W.), the Harry J. Lloyd Foundation (R.M.W.), Consano (R.M.W.), Joanna M. Nicolay Melanoma Foundation (S.J.C.), Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung (PP00P3_139093 to C.M.), SwissBridge Foundation (A.B. and C.M.), FP7 People: Marie-Curie Actions (CIG PCIG14-GA-2013-631984 to C.M.), the National Institute of General Medical Sciences (T32GM007739 to N.R.C.), the Melanoma Research Foundation (N.R.C.), and the Alan and Sandra Gerry Metastasis and Tumor Ecosystems Center at Memorial Sloan Kettering Cancer Center (R.M.W.). S.J.C. is also supported by a Robert B. Catell Fellowship at Memorial Sloan Kettering Cancer Center.
Data availability
MiSeq reads are available at the SRA (accession code SRP147816).
Supplementary information
Supplementary information available online at http://dmm.biologists.org/lookup/doi/10.1242/dmm.034561.supplemental
References
- Balciunas D., Wangensteen K. J., Wilber A., Bell J., Geurts A., Sivasubbu S., Wang X., Hackett P. B., Largaespada D. A., McIvor R. S. et al. (2006). Harnessing a high cargo-capacity transposon for genetic applications in vertebrates. PLoS Genet. 2, e169 10.1371/journal.pgen.0020169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckwith L. G., Moore J. L., Tsao-Wu G. S., Harshbarger J. C. and Cheng K. C. (2000). Ethylnitrosourea induces neoplasia in zebrafish (Danio rerio). Lab. Invest. 80, 379-385. 10.1038/labinvest.3780042 [DOI] [PubMed] [Google Scholar]
- Berger M. F., Hodis E., Heffernan T. P., Deribe Y. L., Lawrence M. S., Protopopov A., Ivanova E., Watson I. R., Nickerson E., Ghosh P. et al. (2012). Melanoma genome sequencing reveals frequent PREX2 mutations. Nature 485, 502-506. 10.1038/nature11071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berghmans S., Murphey R. D., Wienholds E., Neuberg D., Kutok J. L., Fletcher C. D. M., Morris J. P., Liu T. X., Schulte-Merker S., Kanki J. P. et al. (2005). tp53 mutant zebrafish develop malignant peripheral nerve sheath tumors. Proc. Natl. Acad. Sci. USA 102, 407-412. 10.1073/pnas.0406252102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn J. S., Liu S. and Langenau D. M. (2011). Quantifying the frequency of tumor-propagating cells using limiting dilution cell transplantation in syngeneic zebrafish. J. Vis. Exp. e2790 10.3791/2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger A., Lindsay H., Felker A., Hess C., Anders C., Chiavacci E., Zaugg J., Weber L. M., Catena R., Jinek M. et al. (2016). Maximizing mutagenesis with solubilized CRISPR-Cas9 ribonucleoprotein complexes. Development 143, 2025-2037. 10.1242/dev.134809 [DOI] [PubMed] [Google Scholar]
- Ceol C. J., Houvras Y., Jane-Valbuena J., Bilodeau S., Orlando D. A., Battisti V., Fritsch L., Lin W. M., Hollmann T. J., Ferré F. et al. (2011). The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature 471, 513-517. 10.1038/nature09806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E., Gao J., Dogrusoz U., Gross B. E., Sumer S. O., Aksoy B. A., Jacobsen A., Byrne C. J., Heuer M. L., Larsson E. et al. (2012). The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401-404. 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Becker A. and LoTurco J. (2016). Overview of transgenic glioblastoma and oligoastrocytoma CNS models and their utility in drug discovery. Curr. Protoc. Pharmacol. 72, 14.37.1-12 10.1002/0471141755.ph1437s72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Agati G., Beltre R., Sessa A., Burger A., Zhou Y., Mosimann C. and White R. M. (2017). A defect in the mitochondrial protein Mpv17 underlies the transparent casper zebrafish. Dev. Biol. 430, 11-17. 10.1016/j.ydbio.2017.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dee C. T., Nagaraju R. T., Athanasiadis E. I., Gray C., Fernandez del Ama L., Johnston S. A., Secombes C. J., Cvejic A. and Hurlstone A. F. L. (2016). CD4-transgenic Zebrafish reveal tissue-resident Th2- and regulatory T cell-like populations and diverse mononuclear phagocytes. J. Immunol. 197, 3520-3530. 10.4049/jimmunol.1600959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellett F., Pase L., Hayman J. W., Andrianopoulos A. and Lieschke G. J. (2011). mpeg1 promoter transgenes direct macrophage-lineage expression in zebrafish. Blood 117, e49-e56. 10.1182/blood-2010-10-314120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fior R., Póvoa V., Mendes R. V., Carvalho T., Gomes A., Figueiredo N. and Ferreira M. G. (2017). Single-cell functional and chemosensitive profiling of combinatorial colorectal therapy in zebrafish xenografts. Proc. Natl. Acad. Sci. USA 114, E8234-E8243. 10.1073/pnas.1618389114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Aksoy B. A., Dogrusoz U., Dresdner G., Gross B., Sumer S. O., Sun Y., Jacobsen A., Sinha R., Larsson E. et al. (2013). Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonin-Laurent N., Hadj-Hamou N. S., Vogt N., Houdayer C., Gauthiers-Villars M., Dehainault C., Sastre-Garau X., Chevillard S. and Malfoy B. (2007). RB1 and TP53 pathways in radiation-induced sarcomas. Oncogene 26, 6106-6112. 10.1038/sj.onc.1210404 [DOI] [PubMed] [Google Scholar]
- Hans S., Kaslin J., Freudenreich D. and Brand M. (2009). Temporally-controlled site-specific recombination in zebrafish. PLoS ONE 4, e4640 10.1371/journal.pone.0004640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hans S., Freudenreich D., Geffarth M., Kaslin J., Machate A. and Brand M. (2011). Generation of a non-leaky heat shock-inducible Cre line for conditional Cre/lox strategies in zebrafish. Dev. Dyn. 240, 108-115. 10.1002/dvdy.22497 [DOI] [PubMed] [Google Scholar]
- Heilmann S., Ratnakumar K., Langdon E. M., Kansler E. R., Kim I. S., Campbell N. R., Perry E. B., McMahon A. J., Kaufman C. K., van Rooijen E. et al. (2015). A quantitative system for studying metastasis using transparent zebrafish. Cancer Res. 75, 4272-4282. 10.1158/0008-5472.CAN-14-3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodis E., Watson I. R., Kryukov G. V., Arold S. T., Imielinski M., Theurillat J.-P., Nickerson E., Auclair D., Li L., Place C. et al. (2012). A landscape of driver mutations in melanoma. Cell 150, 251-263. 10.1016/j.cell.2012.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoegler K. J., Distel M., Köster R. W. and Horne J. H. (2011). Targeting olfactory bulb neurons using combined in vivo electroporation and Gal4-based enhancer trap zebrafish lines. J. Vis. Exp. 2964 10.3791/2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman R. M. (2015). Patient-derived orthotopic xenografts: better mimic of metastasis than subcutaneous xenografts. Nat. Rev. Cancer 15, 451-452. 10.1038/nrc3972 [DOI] [PubMed] [Google Scholar]
- Huang C.-J., Tu C.-T., Hsiao C.-D., Hsieh F.-J. and Tsai H.-J. (2003). Germ-line transmission of a myocardium-specific GFP transgene reveals critical regulatory elements in the cardiac myosin light chain 2 promoter of zebrafish. Dev. Dyn. 228, 30-40. 10.1002/dvdy.10356 [DOI] [PubMed] [Google Scholar]
- Hugo W., Zaretsky J. M., Sun L., Song C., Moreno B. H., Hu-Lieskovan S., Berent-Maoz B., Pang J., Chmielowski B., Cherry G. et al. (2016). Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell 165, 35-44. 10.1016/j.cell.2016.02.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S., Choi H.-J., Park H.-K., Jo W., Jang S., Ryu J.-E., Kim W.-J., Yu E.-S. and Son W.-C. (2014). Electroporation markedly improves sleeping beauty transposon-induced tumorigenesis in mice. Cancer Gene Ther. 21, 333-339. 10.1038/cgt.2014.33 [DOI] [PubMed] [Google Scholar]
- Kansler E. R., Verma A., Langdon E. M., Simon-Vermot T., Yin A., Lee W., Attiyeh M., Elemento O. and White R. M. (2017). Melanoma genome evolution across species. BMC Genomics 18, 136 10.1186/s12864-017-3518-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman C. K., Mosimann C., Fan Z. P., Yang S., Thomas A. J., Ablain J., Tan J. L., Fogley R. D., van Rooijen E., Hagedorn E. J. et al. (2016). A zebrafish melanoma model reveals emergence of neural crest identity during melanoma initiation. Science 351, aad2197 10.1126/science.aad2197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K. (2007). Tol2: a versatile gene transfer vector in vertebrates. Genome Biol. 8 Suppl. 1, S7 10.1186/gb-2007-8-s1-s7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K., Shima A. and Kawakami N. (2000). Identification of a functional transposase of the Tol2 element, an Ac-like element from the Japanese medaka fish, and its transposition in the zebrafish germ lineage. Proc. Natl. Acad. Sci. USA 97, 11403-11408. 10.1073/pnas.97.21.11403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauthammer M., Kong Y., Ha B. H., Evans P., Bacchiocchi A., McCusker J. P., Cheng E., Davis M. J., Goh G., Choi M. et al. (2012). Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat. Genet. 44, 1006-1014. 10.1038/ng.2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan K. M., Fujimoto E., Grabher C., Mangum B. D., Hardy M. E., Campbell D. S., Parant J. M., Yost H. J., Kanki J. P. and Chien C.-B. (2007). The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn. 236, 3088-3099. 10.1002/dvdy.21343 [DOI] [PubMed] [Google Scholar]
- Labun K., Montague T. G., Gagnon J. A., Thyme S. B. and Valen E. (2016). CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res. 44, W272-W276. 10.1093/nar/gkw398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenau D. M., Traver D., Ferrando A. A., Kutok J. L., Aster J. C., Kanki J. P., Lin S., Prochownik E., Trede N. S., Zon L. I. et al. (2003). Myc-induced T cell leukemia in transgenic zebrafish. Science 299, 887-890. 10.1126/science.1080280 [DOI] [PubMed] [Google Scholar]
- Langenau D. M., Feng H., Berghmans S., Kanki J. P., Kutok J. L. and Look A. T. (2005). Cre/lox-regulated transgenic zebrafish model with conditional myc-induced T cell acute lymphoblastic leukemia. Proc. Natl. Acad. Sci. USA 102, 6068-6073. 10.1073/pnas.0408708102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson N. D. and Weinstein B. M. (2002). In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev. Biol. 248, 307-318. 10.1006/dbio.2002.0711 [DOI] [PubMed] [Google Scholar]
- Lindsay H., Burger A., Biyong B., Felker A., Hess C., Zaugg J., Chiavacci E., Anders C., Jinek M., Mosimann C. et al. (2016). CrispRVariants charts the mutation spectrum of genome engineering experiments. Nat. Biotechnol. 34, 701-702. 10.1038/nbt.3628 [DOI] [PubMed] [Google Scholar]
- Lister J. A., Robertson C. P., Lepage T., Johnson S. L. and Raible D. W. (1999). nacre encodes a zebrafish microphthalmia-related protein that regulates neural-crest-derived pigment cell fate. Development 126, 3757-3767. [DOI] [PubMed] [Google Scholar]
- Lister J. A., Close J. and Raible D. W. (2001). Duplicate mitf genes in zebrafish: complementary expression and conservation of melanogenic potential. Dev. Biol. 237, 333-344. 10.1006/dbio.2001.0379 [DOI] [PubMed] [Google Scholar]
- Maresch R., Mueller S., Veltkamp C., Öllinger R., Friedrich M., Heid I., Steiger K., Weber J., Engleitner T., Barenboim M. et al. (2016). Multiplexed pancreatic genome engineering and cancer induction by transfection-based CRISPR/Cas9 delivery in mice. Nat. Commun. 7, 10770 10.1038/ncomms10770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGranahan N. and Swanton C. (2017). Clonal heterogeneity and tumor evolution: past, present, and the future. Cell 168, 613-628. 10.1016/j.cell.2017.01.018 [DOI] [PubMed] [Google Scholar]
- Mione M. C. and Trede N. S. (2010). The zebrafish as a model for cancer. Dis. Model. Mech. 3, 517-523. 10.1242/dmm.004747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montague T. G., Cruz J. M., Gagnon J. A., Church G. M. and Valen E. (2014). CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 42, W401-W407. 10.1093/nar/gku410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosimann C., Kaufman C. K., Li P., Pugach E. K., Tamplin O. J. and Zon L. I. (2011). Ubiquitous transgene expression and Cre-based recombination driven by the ubiquitin promoter in zebrafish. Development 138, 169-177. 10.1242/dev.059345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münch J., González-Rajal A. and de la Pompa J. L. (2013). Notch regulates blastema proliferation and prevents differentiation during adult zebrafish fin regeneration. Development 140, 1402-1411. 10.1242/dev.087346 [DOI] [PubMed] [Google Scholar]
- Neumann E., Schaefer-Ridder M., Wang Y. and Hofschneider P. H. (1982). Gene transfer into mouse lyoma cells by electroporation in high electric fields. EMBO J. 1, 841-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T., Nishimura Y., Gotoh H. and Ono K. (2016). Rapid and efficient gene delivery into the adult mouse brain via focal electroporation. Sci. Rep. 6, 29817 10.1038/srep29817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.-S., Lim K.-M., Park S. G., Jung S. Y., Choi H.-J., Lee D. H., Kim W.-J., Hong S.-M., Yu E.-S. and Son W.-C. (2014). Pancreatic cancer induced by in vivo electroporation-enhanced sleeping beauty transposon gene delivery system in mouse. Pancreas 43, 614-618. 10.1097/MPA.0000000000000102 [DOI] [PubMed] [Google Scholar]
- Patton E. E., Widlund H. R., Kutok J. L., Kopani K. R., Amatruda J. F., Murphey R. D., Berghmans S., Mayhall E. A., Traver D., Fletcher C. D. M. et al. (2005). BRAF mutations are sufficient to promote nevi formation and cooperate with p53 in the genesis of melanoma. Curr. Biol. 15, 249-254. 10.1016/j.cub.2005.01.031 [DOI] [PubMed] [Google Scholar]
- Pérot G., Chibon F., Montero A., Lagarde P., de Thé H., Terrier P., Guillou L., Ranchère D., Coindre J.-M. and Aurias A. (2010). Constant p53 pathway inactivation in a large series of soft tissue sarcomas with complex genetics. Am. J. Pathol. 177, 2080-2090. 10.2353/ajpath.2010.100104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pliss G. B., Zabezhinski M. A., Petrov A. S. and Khudoley V. V. (1982). Peculiarities of N-nitramines carcinogenic action. Arch. Geschwulstforsch. 52, 629-634. [PubMed] [Google Scholar]
- Rambabu K. M., Rao S. H. N. and Rao N. M. (2005). Efficient expression of transgenes in adult zebrafish by electroporation. BMC Biotechnol. 5, 29 10.1186/1472-6750-5-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676-682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siolas D. and Hannon G. J. (2013). Patient-derived tumor xenografts: transforming clinical samples into mouse models. Cancer Res. 73, 5315-5319. 10.1158/0008-5472.CAN-13-1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitsbergen J. M., Tsai H.-W., Reddy A., Miller T., Arbogast D., Hendricks J. D. and Bailey G. S. (2000). Neoplasia in zebrafish (Danio rerio) treated with 7,12-dimethylbenz[a]anthracene by two exposure routes at different developmental stages. Toxicol. Pathol. 28, 705-715. 10.1177/019262330002800511 [DOI] [PubMed] [Google Scholar]
- Stratton M. R., Moss S., Warren W., Patterson H., Clark J., Fisher C., Fletcher C. D., Ball A., Thomas M. and Gusterson B. A. (1990). Mutation of the p53 gene in human soft tissue sarcomas: association with abnormalities of the RB1 gene. Oncogene 5, 1297-1301. [PubMed] [Google Scholar]
- Swartz M., Eberhart J., Mastick G. S. and Krull C. E. (2001). Sparking new frontiers: using in vivo electroporation for genetic manipulations. Dev. Biol. 233, 13-21. 10.1006/dbio.2001.0181 [DOI] [PubMed] [Google Scholar]
- Tang Q., Moore J. C., Ignatius M. S., Tenente I. M., Hayes M. N., Garcia E. G., Torres Yordán N., Bourque C., He S., Blackburn J. S. et al. (2016). Imaging tumour cell heterogeneity following cell transplantation into optically clear immune-deficient zebrafish. Nat. Commun. 7, 10358 10.1038/ncomms10358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thummel R., Bailey T. J. and Hyde D. R. (2011). In vivo electroporation of morpholinos into the adult zebrafish retina. J. Vis. Exp. e3603 10.3791/3603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villefranc J. A., Amigo J. and Lawson N. D. (2007). Gateway compatible vectors for analysis of gene function in the zebrafish. Dev. Dyn. 236, 3077-3087. 10.1002/dvdy.21354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White R. M., Sessa A., Burke C., Bowman T., LeBlanc J., Ceol C., Bourque C., Dovey M., Goessling W., Burns C. E. et al. (2008). Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell 2, 183-189. 10.1016/j.stem.2007.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White R. M., Cech J., Ratanasirintrawoot S., Lin C. Y., Rahl P. B., Burke C. J., Langdon E., Tomlinson M. L., Mosher J., Kaufman C. et al. (2011). DHODH modulates transcriptional elongation in the neural crest and melanoma. Nature 471, 518-522. 10.1038/nature09882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White R., Rose K. and Zon L. (2013). Zebrafish cancer: the state of the art and the path forward. Nat. Rev. Cancer 13, 624-636. 10.1038/nrc3589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong T.-K. and Neumann E. (1982). Electric field mediated gene transfer. Biochem. Biophys. Res. Commun. 107, 584-587. 10.1016/0006-291X(82)91531-5 [DOI] [PubMed] [Google Scholar]
- Yarmush M. L., Golberg A., Serša G., Kotnik T. and Miklavčič D. (2014). Electroporation-based technologies for medicine: principles, applications, and challenges. Annu. Rev. Biomed. Eng. 16, 295-320. 10.1146/annurev-bioeng-071813-104622 [DOI] [PubMed] [Google Scholar]
- Yen J., White R. M., Wedge D. C., Van Loo P., de Ridder J., Capper A., Richardson J., Jones D., Raine K., Watson I. R. et al. (2013). The genetic heterogeneity and mutational burden of engineered melanomas in zebrafish models. Genome Biol. 14, R113 10.1186/gb-2013-14-10-r113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., McDaid R., Lee J., Possik P., Li L., Kumar S. M., Elder D. E., Van Belle P., Gimotty P., Guerra M. et al. (2009). The role of BRAF mutation and p53 inactivation during transformation of a subpopulation of primary human melanocytes. Am. J. Pathol. 174, 2367-2377. 10.2353/ajpath.2009.081057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng A., Ye T., Cao D., Huang X., Yang Y., Chen X., Xie Y., Yao S. and Zhao C. (2017). Identify a blood-brain barrier penetrating drug-TNB using Zebrafish Orthotopic Glioblastoma Xenograft model. Sci. Rep. 7, 14372 10.1038/s41598-017-14766-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.