Abstract
Recent studies have shown that HDAC inhibitors act synergistically with camptothecin derivatives in combination therapies. To exploit this synergy, new hybrid molecules targeting simultaneously topoisomerase I and HDAC were designed. In particular, a selected multivalent agent containing a camptothecin and a SAHA-like template showed a broad spectrum of antiproliferative activity, with IC50 values in the nanomolar range. Preliminary in vivo results indicated a strong antitumor activity on human mesothelioma primary cell line MM473 orthotopically xenografted in CD-1 nude mice and very high tolerability.
Introduction
Over the last two decades, drug discovery has been primarily focused on the development of single-target drugs with high potency and selectivity. However, single target agents can perform excellently during the treatment of diseases with linear pathways. Multi-factorial diseases with complex signalling networks, such as cancer, are robust and can be efficiently eradicated only by the simultaneous attack of multiple targets.
A well-established strategy for anticancer therapy is the rational design of drug combinations aimed at achieving synergistic effects and overcoming drug resistance. However, the combination of agents that exhibit synergistic interaction in cell culture can be less effective than expected. This is mainly due to differential pharmacokinetic behaviour of each drug and to the difficulty to afford optimal concentrations for the required time [1]. Other shortcomings of drug combinations are unpredictable drug-drug interactions and possible enhancement of adverse effects. To overcome these limitations, an increase of interest has been made recently in hybrid bifunctional agents designed to inhibit simultaneously multiple cellular targets relevant to tumour growth/survival [2–5]. The benefits of drugs with multiple targets could be (a) improvement of the efficacy by exploiting synergistic interactions; (b) enhancement of the selectivity resulting in a better tolerability; (c) modulation of drug resistance. Additionally, multivalent molecules are expected to provide pharmacokinetic and pharmacodynamic advantages over the separate administration of the two drug components [4, 5].
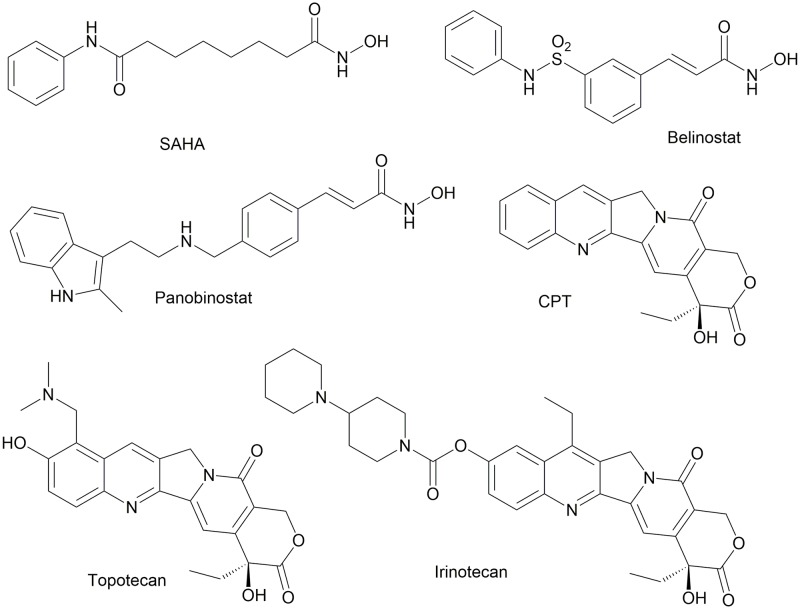
One attractive pathway for tumour growth inhibition is the protein acetylation state modulation by histone deacetylases (HDACs). To date, 18 human HDACs have been identified and grouped into four classes on the basis of their homology with yeast proteins [6]. Class I HDACs (HDAC1, 2, 3, and 8) share high homology with the yeast transcriptional regulator RPD3; class II HDACs are closely related to HDAC1 (HDAC4, 5, 6, 7, 9, and 10); class III HDACs, also called sirtuins, are homologous with Sir2 (SIRT1, 2, 3, 4, 5, 6, and 7), and class IV HDAC (HDAC11) is homologous with class I and II enzymes. HDAC inhibitors have already surpassed the clinical trial stage, culminating with the approval of Vorinostat, originally known as SAHA (suberoylanilide hydroxamic acid), romidepsin (FK228), panobinostat (LBH-589), and belinostat (formerly PXD-101) [6, 7] (Fig 1).
Fig 1. Structures of representative HDAC and topoisomerase I inhibitors.
Another class of therapeutic agents acting at a nuclear level are topoisomerase I inhibitors [8, 9]. Topoisomerases relieve the torsional stress associated with the separation of DNA strands during transcription and DNA replication. Eukaryotic Topoisomerase I (Top1) is a Type IB enzyme that nicks and rejoins only one strand of duplex DNA, and it is critically important for maintaining genome integrity [10]. Many small molecule inhibitors of Top1 have proven clinically effective, the most potent being the camptothecin (CPT) derivatives [11, 12] (Fig 1).
Two semisynthetic camptothecins have been approved by US FDA for cancer chemotherapy: Irinotecan (CPT-11) and Topotecan. Belotecan (Camtobell) has been approved in South Korea for treatment of NSCLC (Fig 1). Other derivatives are in different stages of clinical development, such as Lipotecan [13] and BAY 56–3722 [14], together with a number of macromolecular delivery agents and supramolecular assemblies [15].
The cancer-promoting mechanism of HDAC involves transcriptional silencing of tumor suppressors via deacetylation of nucleosomes containing tumor suppressor genes. HDAC inhibition has been reported to affect cancer cells mainly by a global relaxation of chromatin structure, which contributes to a recovered cell response to chemotherapeutic agents targeting DNA [16]. In this respect, there is a lot of evidence showing that co-treatment with HDAC inhibitors and camptothecin derivatives (such as Topotecan, Irinotecan) synergistically block cell proliferation [17–24]. We decided to exploit this synergistic effects by designing hybrid inhibitors which could, in principle, interact with both targets. In particular, we chose to incorporate into a prototypical HDACi pharmacophore a properly substituted CPT. Classical medicinal chemistry approaches coupled with computational analysis and biological evaluation led to the definition of key molecular insights for dual inhibition.
Materials and methods
Chemistry
General information
All reagents and solvents were reagent grade or were purified by standard methods before use. Melting points were determined in open capillaries and are uncorrected. NMR spectra were recorded in CDCl3 (where not otherwise stated) at 300 MHz. Chemical shifts (δ values) and coupling constants (J values) are given in ppm and Hz, respectively. Solvents were routinely distilled prior to use; anhydrous tetrahydrofuran (THF) and ether (Et2O) were obtained by distillation from sodium-benzophenone ketyl; dry methylenechloride was obtained by distillation from phosphorus pentoxide. All reactions requiring anhydrous conditions were performed under a positive nitrogen flow, and all glassware were oven dried and/or flame dried. Isolation and purification of the compounds were performed by flash column chromatography on silica gel 60 (230–400 mesh). Analytical thin-layer chromatography (TLC) was conducted on Fluka TLC plates (silica gel 60 F254, aluminum foil). Analyses indicated by the symbols of the elements or functions were within ±0.4% of the theoretical values. Compounds 2a [25], 1a, 2b, 2c, [3], 5 [26], 7-formylcamptothecin and 10-hydroxy-7-formylcamptothecin (1b) [27] were prepared according to literature procedures.
Synthesis of compounds 3a-d, 10
7-(camptothecin-7-ylmethyleneaminooxy)-heptanoic acid (2c). To a solution of 1a (200 mg, 0.47 mmol) in acetic acid (15 ml) with 7-aminooxyheptanoic acid hydrobromide (250 mg, 1.04 mmol) was added and the mixture heated 3 h at 80 °C. The solvent was evaporated and the residue was purified by flash chromatography with CH2Cl2:CH3OH 93:7 to give 171 mg (70%) of compound 2c. mp:124–125 °C. 1H-NMR (300 MHz, CDCl3)δ: 10.50 (1H, brs); 8.84 (1H, s); 8.09 (1H, d, J = 8.5 Hz); 8.01 (1H, d, J = 8.5 Hz); 7.98 (1H, s); 7.69 (1H, dd, J = 8.5, 8.5 Hz); 7.58 (1H, s); 7.53 (1H, dd, J = 8.5, 8.5 Hz); 5.70–5.60 (1H, m); 5.28–5.15 (3H, m); 4.34 (2H, t, J = 6.3 Hz); 2.32 (2H, t, J = 6.9 Hz); 1.91–1.77 (4H, m); 1.71–1.57 (2H, m); 1.51–1.35 (4H, m); 0.89 (3H, t, J = 7.1 Hz). Anal. calcd for C28H29N3O7: C,64.73; H, 5.63; N, 8.09; Found: C, 64.65; H, 5.61; N, 8.10.
Camptothecin-7-yl(methyleneaminooxy)-acetic acid hydroxyamide (3a). To a solution of 2a (50 mg, 0.11 mmol) in dry DMF (1 mL), DCC (35 mg, 0.17 mmol), and N-hydroxysuccinimide (20 mg, 0.17 mmol) were added under nitrogen atmosphere. The mixture was stirred 48 h at rt, then DCC (12 mg 0.06 mmol) and N-hydroxysuccinimide (7 mg, 0.06 mmol) were added. The reaction was stirred 24 h at rt, then hydroxylamine hydrochloride (23 mg, 0.33 mmol) and TEA (44 μl, 0.33 mmol) were added and stirring was continued for 24 h at room temperature. The solvent was evaporated and the crude was purified by flash chromatography with CH2Cl2:CH3OH from 195:5 to 190:10 to give 5 mg of product (10%). mp: 169 °C. 1H-NMR (300 MHz, DMSO-d6) δ: 10.82 (1H, brs); 9.37 (1H, s); 9.00 (1H, brs); 8.62 (1H, d, J = 8.5 Hz); 8.24 (1H, d, J = 8.5 Hz); 7.94 (1H, dd, J = 8.5, 8.5 Hz); 7.80 (1H, dd, J = 8.5, 8.5 Hz); 7.37 (1H, s); 6.55 (1H, s); 5.44 (2H, s); 5.34 (2H, s); 4.78 (2H, s); 1.99–1.77 (2H, m); 0.89 (3H, t, J = 7.0 Hz). Anal. calcd for C23H20N4O7: C,59.48; H, 3.34; N, 12.06; Found: C, 59.63; H, 3.35; N, 12.04.
6-(camptothecin-7-ylmethyleneaminooxy)-hexanoic acid hydroxyamide (3b). To a solution of 2b (350 mg, 0.69 mmol) in dry DMF (6 ml), WSC (172 mg, 0.9 mmol) and HOBt (112 mg, 0.83 mmol) were added under nitrogen atmosphere. The mixture was stirred 3 h at rt. cooled to 0 °C, and hydroxylamine hydrochloride (240 mg, 3.45 mmol) and TEA (0.48 ml, 3.45 mmol) were added. The reaction was stirred 1 h at room temperature. The solvent was evaporated and the residue was purified by flash chromatography with CH2Cl2:CH3OH 91: 9 to give 220 mg of product (61%). mp: 158–159°C. 1H-NMR (300 MHz, DMSO-d6) δ: 10.36 (1H, s); 9.33 (1H, s); 8.68 (1H, s); 8.60 (1H, d, J = 8.3 Hz); 8.21 (1H, d, J = 8.3 Hz); 7.91 (1H, dd, J = 8.3, 8.3 Hz); 7.76 (1H, dd, J = 8.3, 8.3 Hz); 7.36 (1H, s); 6.55 (1H, s); 5.44 (1H, s); 5.32 (1H, s); 4.35 (2H, t, J = 6.5 Hz); 2.00 (2H, t, J = 7.3 Hz); 1.94–1.75 (4H, m); 1.65–1.52 (2H, m); 1.51–1.38 (2H, m); 0.89 (3H, t, J = 7.0 Hz). Anal. calcd for C27H28N4O7: C,62.30; H, 5.42; N, 10.76; Found: C, 62.24; H, 5.43; N, 10.74.
7-(camptothecin-7-ylmethyleneaminooxy)-heptanoic acid hydroxyamide (3c). To a solution of 2c (150 mg, 0.29 mmol) in dry DMF (2.5 ml), WSC (72 mg, 0.38 mmol) and HOBt (46 mg, 0.35 mmol) were added under nitrogen atmosphere. The mixture was stirred overnight at room temperature. The reaction was cooled to 0 °C and hydroxylamine hydrochloride (101 mg, 1.45 mmol), TEA (0.2 mL, 1.45 mmol) were added. The mixture was stirred 1 h at rt. The solvent was evaporated and the residue purified by flash chromatography with CH2Cl2:CH3OH 91:9 to give 70 mg of product (45%). mp: 164–165°C. 1H-NMR (300 MHz, DMSO-d6) δ: 10.33 (1H, s); 9.31 (1H, s); 8.65 (1H, s); 8.58 (1H, d, J = 8.5 Hz); 8.21 (1H, d, J = 8.5 Hz); 7.90 (1H, dd, J = 8.5, 8.5 Hz); 7.75 (1H, dd, J = 8.5, 8.5 Hz); 7.35 (1H, s); 6.53 (1H, s); 5.43 (2H, s); 5.30 (2H, s); 4.41–4.27 (2H, m); 2.56–2.38 (2H, m); 2.03–1.68 (4H, m); 1.64–1.23 (6H, m); 0.90 (3H, t, J = 7.0 Hz). Anal. calcd for C28H30N4O7: C,62.91; H, 5.66; N, 10.48; Found: C, 62.98; H, 5.64; N, 10.50.
6-(10-hydroxycamptothecin-7-ylmethyleneaminooxy)-hexanoic acid hydroxyamide (3d). To a solution of 2d (287 mg, 0.56 mmol) in dry DMF (20 ml), WSC (140 mg, 0.73 mmol) and HOBt (140 mg, 0.73 mmol) were added. The mixture was cooled to 0 °C and hydroxylamine hydrochloride (82 mg, 1.18 mmol) and TEA (111 μl, 1.18 mmol) were added. The reaction was stirred 8 h at room temperature, the solvent was evaporated and the residue was diluted with water (40 mL). The resulting solid was filtered to obtain 220 mg of product (73%). mp: 202–203 °C. 1H-NMR (300 MHz, DMSO-d6) δ: 10.49 (1H, brs); 10.36 (1H, s); 9.02 (1H, s); 8.66 (1H, brs); 8.06 (1H, d, J = 9.2 Hz); 7.72 (1H, d, J = 1.8 Hz); 7.47 (1H, dd, J = 1.8, 9.2 Hz); 7.26 (1H, s); 6.50 (1H, s); 5.41 (2H, s); 5.27 (2H, s); 4.33 (2H, t, J = 5.8 Hz); 2.04–1.93 (2H, m); 1.92–1.72 (4H, m); 1.66–1.52 (2H, m); 1.51–1.35 (2H, m); 0.89 (3H, t, J = 7.0 Hz). Anal. calcd for C27H28N4O8: C,60.44; H, 5.26; N, 10.44; Found: C, 60.55; H, 5.27; N, 10.41.
2-(6-aminohexyloxy)-isoindole-1,3-dione) trifluoroacetate (6). To a solution of compound 5 (835 mg, 2.3 mmol) in CH2Cl2 (5 mL) trifluoroacetic acid (2 mL) and was added and the mixture was stirred 40 min at room temperature. Trifluoroacetic acid and solvent were removed by evaporation under vacuo and the residue was grounded with ethyl ether, to give 810 mg (94%) of product. mp:99 °C. 1H-NMR (300 MHz, DMSO-d6) δ: 7.89–7.51 (4H, m); 4.13 (2H, t, J = 6.4 Hz); 2.86–2.72 (2H, m); 1.73–1.29 (8H, m).
N-[6-(1,3-dioxo-1,3-dihydro-isoindol-2-yloxy)-hexyl]-2-tritylsulfanylacetamide (7). To a solution of 6 (300 mg, 0.80 mmol) in CH2Cl2 (3 ml), tritylsulfonylacetic acid (261 mg, 0.78 mmol), DMAP (5 mg, 0.04 mmol), TEA (167 μl, 1.2 mmol), and WSC (199 mg, 1.04 mmol) were added under nitrogen atmosphere. The mixture was stirred 1.5 h at room temperature. After dilution with dichloromethane, the mixture was washed with brine, dried and evaporated. Purification by flash chromatography with hexane:EtOAc 6:4, gave 238 mg (53%) of product. mp: 133 °C. 1H-NMR (300 MHz, DMSO-d6) δ: 7.88–7.70 (4H, m); 7.48–7.17 (15H, m); 6.06 (1H, brs); 4.20 (2H, t, J = 6.4 Hz); 3.14 (2H, s); 3.02–2.91 (2H, m); 1.85–1.68 (2H, m); 1.56–1.44 (2H, m); 1.43–1.23 (4H, m). Anal. calcd for C35H34N2O4S: C,72.64; H, 5.92; N, 4.84; Found: C, 72.48; H, 5.93; N, 4.85.
N-(6-Aminooxyhexyl)-2-tritylsulfanyl-acetamide (8). To a solution of 7 (94 mg, 0.16 mmol) in methanol (2 ml), NH2NH2.H2O (25 μL, 0.48 mmol) was added and the solution was heated to reflux for 1h. After evaporation of the solvent, the residue was diluted with ethyl acetate. The solid was filtered and the solvent evaporated under reduced pressure. Purification by flash chromatography with hexane:EtOAc 2:1 gave 60 mg of an oil (83%). 1H-NMR (CDCl3) δ: 7.48–7.12 (15H, m); 6.01 (1H, brs); 5.35 (1H, brs); 3.65 (2H, t, J = 6.7 Hz); 3.14 (2H, s); 2.99–2.89 (2H, m); 1.44–1.08 (8H, m). Anal. calcd for C27H32N2O2S: C,72.29; H, 7.19; N, 6.24; Found: C, 72.09; H, 7.20; N, 6.23.
N-[6-(camptothecin-7-ylmethyleneaminooxy)-hexyl]-2-tritylsulfanylacetamide (9). To a solution of 7-formylcamptothecin (25 mg, 0.07 mmol) in ethanol (2 mL) N-(6-aminooxyhexyl)-2-tritylsulfanylacetamide (8) (25 mg, 0.11 mmol) was added and the mixture was heated 5h to reflux. The solvent was evaporated and the crude was purified by chromatography with CH2Cl2:CH3OH 98:2 to obtain 51 mg of a yellow solid (91%). mp: 108 °C. 1H-NMR (300 MHz, DMSO-d6) δ: 9.32 (1H, s); 8.59 (1H, d, J = 8.5 Hz); 8.22 (1H, dd, J = 8.5, 8.5 Hz); 7.90–7.69 (2H, m); 7.40–7.15 (16H, m); 6.56 (1H, s); 5.42 (2H, s); 5.30 (2H, s); 4.34 (2H, t, J = 6.4 Hz); 3.02–2.89 (2H, m); 2.75 (2H, s); 1.94–1.69 (4H, m); 1.49–1.07 (6H, m); 0.89 (3H, t, J = 7.0 Hz). Anal. calcd for C48H46N4O6S: C,71.44; H, 5.75; N, 6.94; Found: C, 71.38; H, 5.73; N, 6.95.
N-[camptothecin-7-yl-(methyleneaminooxy)-hexyl]-2-mercaptoacetamide (10). To a solution of 9 (63 mg, 0.08 mmol) in CH2Cl2 (2.6 mL) trifluoroacetic acid (78 μl, 1.02 mmol) and triethylsilane (30 μl, 0.19 mmol) were added. The solution was stirred 2.5 h at room temperature, the solvent was evaporated and the residue was purified by flash chromatography with CH2Cl2:CH3OH to give 36 mg of compound 10 (80%). mp:120 °C. 1H-NMR (300 MHz, DMSO-d6) δ: 9.32 (1H, s); 8.60 (1H, d, J = 8.5 Hz); 8.20 (1H, d, J = 8.1 Hz); 7.96 (1H, t, J = 5.5 Hz); 7.92–7.85 (1H, m); 7.79–7.70 (1H, m); 7.33 (1H, s); 6.53 (1H, s); 5.47–5.24 (4H, m); 4.34 (2H, t, J = 6.4 Hz); 3.10–2.99 (4H, m); 1.95–1.68 (4H, m); 1.51–1.29 (6H, m); 0.87 (3H, t, J = 7.3 Hz). Anal. calcd for C29H32N4O6S: C,61.69; H, 5.71; N, 9.92; Found: C, 61.75; H, 5.72; N, 9.94.
Cell lines
Human primary epithelioid MM288, MM317, MM404, MM473, MM473-Luc; biphasic MM487 and MM491; sarcomatoid MM432 and MM472 mesothelioma cell lines (all from our cell bank collection).
Human K562 erythromyeloblastoid leukemia (ATCC # CCL-243), ARH-77 plasma leukemia (ATCC # CRL-1621), NB4 acute promyelocytic leukemia (ATCC # ACC-207), THP1 and MV4-11 monocytic leukemia (ATCC # TIB-202, CRL-9591), Jurkat acute T-cell leukemia (ECACC # 90112119), U937 histiocytic lymphoma (ATCC # CRL-1593), RAJI, DG-75, and RAMOS Burkitt lymphoma (DSMZ # ACC-319, ACC-83, ACC-603); MAVER, MINO, REC-1 (DSMZ # ACC-717, ACC-687, ACC-584), JECO-1, Z-138 (ATCC # CRL-3006, CRL-3001) mantle cell lymphoma; KM-H2 and L-428 Hodgkin lymphoma (DSMZ # ACC-8, ACC-197), OCI-LY3 and U-2932 diffuse large B-cell lymphoma (DSMZ # ACC-731, ACC-633), RPMI-8226 and NCI-H929 multiple myeloma (ATCC # CCL-155, CRL-9068).
Human NCI-H460 NSCLC (ATCC # HTB-177), Capan-1 pancreatic carcinoma (ATCC # HTB-79), A431 epidermoid carcinoma (ATCC # CRL-1555), HeLa cervix carcinoma (ATCC # CCL-2), A2780 and A2780-Dx multidrug-resistant ovarian carcinoma (ECACC # 93112519, 93112520), HT29 colorectal carcinoma (ECACC # 91072201), HepG2 hepatocellular carcinoma (ATCC # HB-8065), DU145 prostate carcinoma (ATCC # HTB-81).
Cell cultures
All primary mesothelioma cell lines (MM288, MM317, MM404, MM473, MM487, MM491, MM432, MM472) were cultured in F-10 Nutrient Mixture medium supplemented with 10% Foetal Bovine Serum (FBS), 2 mM L-glutamine and gentamicin sulfate. MM473-Luc cells were cultured in Ham’s F-10 Nutrient Mixture supplemented with 10% FBS, 2 mM L-glutamine, and G418 antibiotic. A2780, A2780-Dx, ARH-77, DG-75, Jurkat, KM-H2, HeLa, L-428, NB4, NCI-H60, U937, RAJI, REC-1, RPMI-8226, and U-2932 cells were cultured in RPMI-1640 medium supplemented with 10% FBS, 2 mM L-glutamine and gentamicin sulfate. JECO-1, MAVER-2, MINO, OCI-LY3 and RAMOS cells were cultured in RPMI-1640 medium supplemented with 20% FBS, 2 mM L-glutamine and gentamicin sulfate. H929 and THP1 cells were cultured in RPMI-1640 medium supplemented with 10% FBS, 0.05 mM mercaptoethanol, L-glutamine and gentamicin sulfate. K562, MV4-11, Z-138 cells were cultured in IMDM medium supplemented with 10% FBS, L-glutamine and gentamicin sulfate. Capan-1 cells were cultured in IMDM medium supplemented with 10% FBS, 2 mM L-glutamine and gentamicin sulfate. A431, DU145, and HepG2 cells were cultured in EMEM medium supplemented with 10% FBS, 2 mM L-glutamine and gentamicin sulfate. HT29 cells were cultured in McCoy’s medium supplemented with 10% FBS, 2 mM L-glutamine and gentamicin sulfate.
Cells were maintained in an incubator at 37°C with saturated humidity and an atmosphere of 95% air and 5% CO2, and were sub-cultured every 2–3 days.
Cytotoxicity assay
Human tumor cells were seeded in 96-well cell culture plastic plates, allowed to attach, and exposed to scalar concentrations of the test compounds. Cell survival was evaluated upon 72 h by the sulphorhodamine B (SRB) or MTT tetrazolium reduction assay and the IC50 value (drug concentration inhibiting 50% of cell growth) calculated by the ALLFIT program.
FACS analysis
Tumor cells were seeded in 10-cm tissue culture plates, allowed to attach and incubated for 72 h with the test compounds (IC80 dose). After this, cells were harvested with trypsin/EDTA, rinsed with cold PBS, and fixed in 70% ice-cold ethanol at 4 °C. The day of analysis, cells were rinsed twice with cold PBS, incubated with RNase A (50 KU/ml) for 30 min. at 37°C, stained with propidium iodide (PI, 50 μg/mL) and processed on a FACScan flow cytometer (Becton Dickinson). The CellQuest software (BD Biosciences) was used to acquire data, while cell cycle analysis was performed using the ModFit software (Verity Software House).
HDAC’s profiling
The experiments were carried out in RBC facility (http://www.reactionbiology.com/) by using in triplicate 10 concentrations of each compound starting from 10 μM. The substrate for HDAC1,2,6, and 10 was Fluorogenic peptide from p53 residues 379–382 (RHKK(Ac)AMC). The Substrate for HDAC4 was Fluorogenic HDAC Class2a Substrate (Trifluoroacetyllysine) IC50 values were calculated using the GraphPad Prism 4 program based on a sigmoidal dose-response equation.
Protein analysis
NCI-H460 cells in logarithmic phase of growth were collected in Dulbecco’s 1x PBS buffer and lysed in RIPA lysis buffer (50mM Tris-HCl, 150 mM NaCl, 0,5% Na-Deoxycolate, 1% NP-40, 0.15% SDS) containing protease and phosphatase inhibitors and then centrifuged at 12000 rpm for 10 min at 4 °C. The supernatant, containing the total proteins extracts, were transferred in new vials. The protein concentration was measured by Bradford Protein Assay. Equal amounts of proteins for each sample were then resolved by SDS-PAGE and loaded on 10–12% polyacrilamide gels. The proteins were then transferred on nitrocellulose membranes. Molecular weights were estimated based upon the relative migration with molecular weight protein markers.
The non-specific binding was blocked by incubation of membranes in TBS-tween (25 mM Tris, 150 mM NaCl, 0.1% Tween 20) with 5% nonfat dry milk (BIORAD, Milan) for 60 minutes. Subsequently, the membrane was incubated overnight with primary antibodies: anti-Histone H4 (Millipore, 1:500), anti-acetyl-H4 (Santa Cruz, 1:1000) anti-p21 (Santa Cruz, 1:1000), anti-acetyl-tubulin (abcam, 1:1000), anti-α-tubulin (Sigma, 1:5000), anti β-actin (Sigma, 1:5000) and anti-Topoisomerase 1 (Santa Cruz, 1:500). After three washes with TBST blots were incubated with HRP-coniugated secondary anti-mouse and anti-rabbit (Santa Cruz, 1:5000) 1 hour at room temperature. After three washes with TBST the blots were developed with ECL substrate (GE Healthcare) and chemiluminescence detected by ChemiDoc Imaging System (Biorad). The optical density of the protein bands detected by western blotting was normalized on α -tubulin levels and analyzed by ChemiDoc software (Biorad).
Animal model
Female 4–6 weeks old nude mice (CD1 nu/nu) were purchased from Charles River Laboratories (Calco (Lecco) Italy) and kept in polisulfone IVC cages (33.2 x 15 x 13 cm, Tecniplast) with 75 complete changes of filtered air per hour (HEPA H 14 filter); autoclaved dust-free bedding and mouse house enrichment device (Tecniplast) were used.
Animals were housed under a light-dark cycle, keeping temperature and humidity constant. Parameters of the animal rooms were assessed as follows: 22 ± 2 °C temperature, 55 ± 10% relative humidity, about 15–20 filtered air changes/hour and 12 hour circadian cycle of artificial light (7 a.m., 7 p.m.). Mice were fed with a complete irradiated pellet diet (GLP 4RF21, Mucedola) ad libitum throughout the entire duration of the experiment, and had unrestricted access to drinking water. All animal studies were conducted in accordance with Ethics approval obtained from the Italian Ministry of Health and all experiments were in accordance with the European guidelines for the care and use of laboratory animals. All procedures were performed under deep isoflurane anesthesia and at the end of experiments were sacrified by cervical dislocation.
In vivo antitumor activity
Human epithelioid mesothelioma MM473-Luc (luciferase-expressing) cells were cultured in complete medium, checked with Mycoplasma Detection Kit (Lonza) to exclude mycoplasma contamination, counted by trypan blue dye exclusion test to assess cell viability (> 95%), re-suspended in M199 medium, and orthotopically inoculated intrapleurally (i.pl.) in CD-1 nude mice (Charles River) at a concentration of 1x106/200μl M199/mouse. Compound 3d was diluted in 10% 1:1 absolute ethanol-Cremophor solution in saline and was administered i.v. at 45 mg/kg using q4dx3w schedule. Irinotecan was administered iv at 35 mg/kg, using the schedule q4dx3. Tumor growth was evaluated weekly by using the IVIS Spectrum (PerkinElmer) in vivo technology system, through intraperitoneal injection of D-Luciferin potassium salt (100μL/10gr, PerkinElmer). After the IP D-luciferin injection, mice were anesthetized in an oxygen-rich induction chamber with 2% isofluorane, and images were captured 30 minutes after D-Luciferin injection in order to allow substrate distribution. Tumor volume was measured as average radiance that is the total flux of photons or radiance (photons/second from the surface) in each pixel, summed or integrated over the ROI area, in a square centimeter (cm2) of the tissue. (photons/sec/cm2/sr). The established maximum average radiance was 1x108 photons/sec/cm2/sr, used as humane end-point.
Statistical analysis
Statistical analysis was performed by using U-Test (GraphPad Prism 6). Outliers were removed by using the Rout test (Q = 10%, GraphPad Prism 6).
Molecular modeling
The ligand molecules were obtained using a systematic conformer search followed by geometry optimisation of the lowest energy structure with MOPAC7 (PM3 Method, RMS gradient 0.0100). The DNA-Topoisomerase-I and HDAC-II models were derived from the deposited X-ray structures (Protein Data Bank entry 1T8I and 5IWG, respectively) as previously described [3].
Energy minimisations and molecular modeling calculations were performed by using the GROMACS package [28] and the AMBER-03 [29] force field, a variant of the AMBER-99 [30] potential in which charges and main-chain torsion potentials have been derived based on QM+continuum solvent calculations and each amino acid is allowed unique main-chain charges (nucleic acids have not been modified from AMBER-99).
Molecular docking experiments were performed with Autodock 4.0 [31]. We used the Lamarckian Genetic Algorithm which combines global search (Genetic Algorithm alone) to local search (Solis and Wets algorithm) [32]. Ligands and receptors were further processed using the Autodock Tool Kit (ADT) [33]. Gasteiger–Marsili charges [34] were loaded on the ligands in ADT and Cornell parameters were used for the phosphorous atoms in the DNA. Solvation parameters were added to the final structure using the Addsol utility of Autodock. Each docking consisted of an initial population of 100 randomly placed individuals, a maximum number of 200 energy evaluations, a mutation rate of 0.02, a crossover rate of 0.80, and an elitism value of 1. For the local search, the so-called pseudo-Solis and Wets algorithm was applied using a maximum of 250 iterations per local search. 250 independent docking runs were carried out for each ligand. The grid maps representing the system in the actual docking process were calculated with Autogrid. The dimensions of the grids were 80×80×80, with a spacing of 0.1 Å between the grid points and the center close to the cavity left by the ligand after its removal. The simpler inter-molecular energy function based on the Weiner force field in Autodock was used to score the docking results. Results differing by less than 1.0 Å in positional root-mean-square deviation (rmsd) were clustered together and were represented by the result with the most favorable free energy of binding. The poses were equilibrated by a 10.0 ns of partially restrained molecular dynamics simulation using the GROMACS package.
Results and discussion
Chemistry
We envisaged that the appropriate combination and outdistancing between the CPT scaffold and a SAHA-like portion could lead to compounds characterized by suitable size and geometrical shape for fitting both enzymes' binding pockets. In previous papers we reported that CPT- functionalization at C-7 is highly tolerable. Particularly, we synthesized 7-oxyiminomethyl derivatives, showing potent in vitro and in vivo activity [25–27, 35, 36]. For this reason, (E)-7-oxyminomethyl-CPTs were selected for conjugation to a SAHA-like pharmacophore via a suitable chain. The CPT ring system should act as a surface recognition cap group for HDAC inhibition, at the same time maintaining the Top1 inhibition activity. To investigate the optimal linker length, we designed a series of compounds containing a SAHA-like HDAC inhibitor connected to the CPT scaffold by linear alkyl chains.
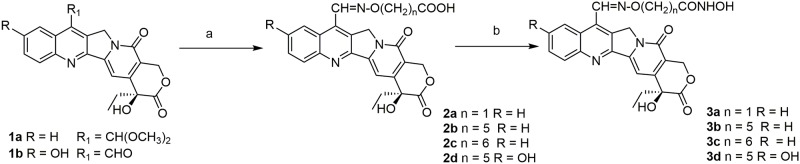
The reaction route to CPT-HDAC hybrid compounds 3a-c is shown in Scheme 1. The compounds were prepared reacting the dimethylacetal of 7-formylcamptothecin (1a) with suitable hydroxylamines to obtain the oximes 2a-c [3]. The carboxylic acid group was then transformed into a hydroxamic acid by reaction with hydroxylamine hydrochloride (Fig 2).
Fig 2. Synthesis of compounds 3a-d.
Reagents and conditions: a) for 2a-c: HCl·H2NO(CH2)nCOOH, acetic acid, 80 °C, 3h; for 2d: HCl·H2NO(CH2)nCOOH, EtOH, pyridine, reflux, 4h; 2a: 90%, 2b: 73%, 2c: 70%, 2d:75%; b) for 3a: DCC, NHS, HCl·NH2OH, rt, 48h, 10%; for 3b-d, WSC, HOBt, DMF, NH2OH·HCl, TEA, rt, 1-8h, 3b: 61%, 3c: 45%, 3d: 73%.
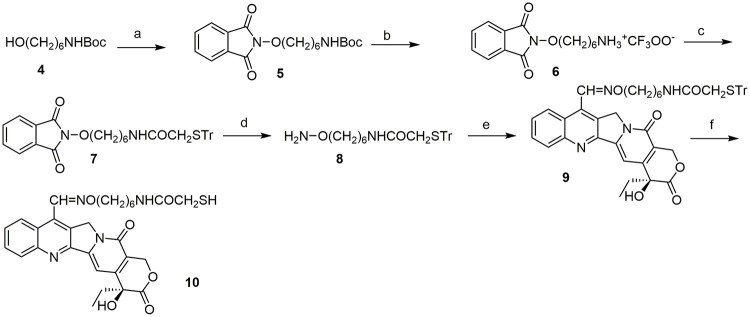
The CPT derivatives in clinical practice, Topotecan and Irinotecan, have a hydroxy group at C-10. In order to elucidate the contribution of this group, we prepared the 10-OH substituted analogue of compound 3b (e.g. compound 3d) starting from 7-formyl-10-hydroxy-CPT (1b) [27] (Fig 2). Furthermore, we decided to investigate the effect of the replacement of the hydroxamic acid moiety with a different HDAC Zn-binding group. Accordingly, we prepared compound 10, carrying a mercaptoacetamide group in place of the hydroxamic acid [37]. The synthetic route is described in Fig 3. N-Boc protected 6-aminohexanol 4 was reacted with N-hydroxyphthalimide in Mitsunobu conditions to give compound 5, which was deprotected and subsequently coupled with S-tritylmercaptoacetic acid [38]. The obtained hydroxylamine 8 was reacted with 7-formylcamptothecin [27] to give the oxime 9, which after removal of trityl group by TFA, gave mercaptoacetamide 10.
Fig 3. Synthesis of compound 10.
Reagents and conditions: a) N-hydroxyphthalimide, PPh3, DIAD, THF, 63%; b) TFA, CH2Cl2, 94%; c) TrSCH2COOH, EDCI, DMAP, TEA, CH2Cl2, 53%; d) NH2NH2, CH3OH, 83%; e) 7-formylcamptothecin, EtOH, 91%; f) TFA, Et3SiH, CH2Cl2, 80%.
A series of similar compounds, having however the SAHA-like chain attached to the C-10 of CPT via an ether bond, has been recently reported by Oyelere and coworkers [39]. Another series of 10-O-ether derivatives have been described in a patent [40]. The most active of these compounds, CY-700, has been prepared by us for comparison (see below).
Molecular modeling studies
DNA-Top1 complexes
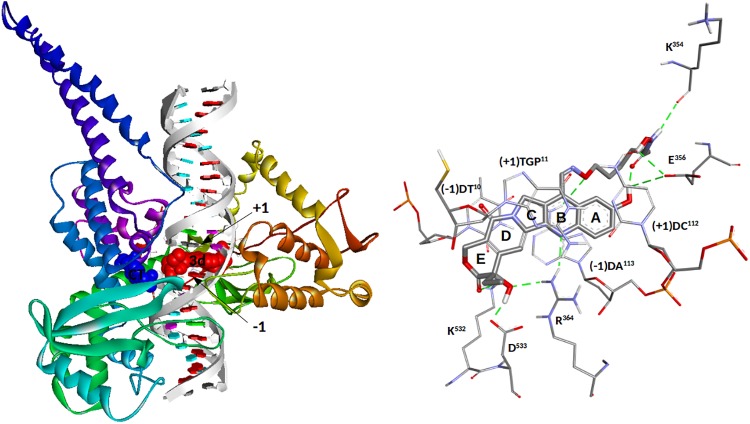
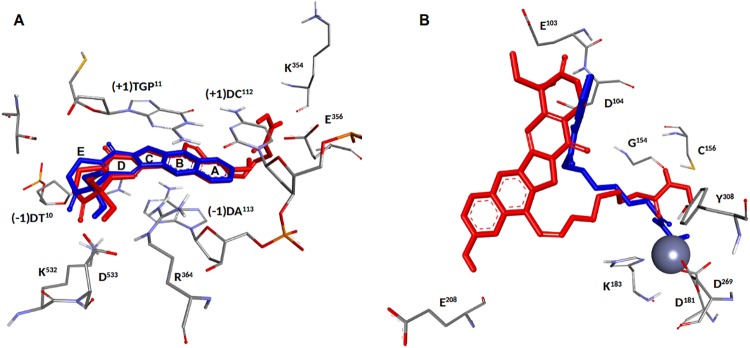
Molecular modelling studies were performed to explore the binding mode for compounds 3a-d and 10 to the DNA-Top1 complex. The compounds are properly positioned inside the intercalation binding site, created by conformational changes of the phosphodiester bond between the +1 (upstream) and -1 (downstream) base pairs of the uncleaved strand, which effectively ‘‘open” the DNA duplex, with multiple strong π-π stacking interactions between the aromatic A, B and D rings of the camptothecin and both the -1 (T10-A113) and +1 (G11-C112) base pairs (Fig 4).
Fig 4.
(Left)—Schematic representation of the proposed top-score binding mode for compound 3d in the DNA-Topoisomerase-I complex. Topoisomerase-I is represented as Backbone Ribbon colored from blue (N-term) to red (C-term), while nucleic acids are represented as an arrow along the backbone pointing toward the C3'-end, and sugar groups and bases as boxes (A in red, C in violet, G in green and T in blue). The 3d ligand and the catalytic tyrosine (CT) are rendered in CPK (3d in red and CT in blue). (Right)—Hypothetical binding mode for the Top-score conformation of 3d in complex with DNA-Topoisomerase-I, with the aminoacids side chains relevant to the discussion rendered in line and the ligands in stick. Hydrogen bonds are shown as green dotted lines.
Compounds 3b and 3d show a hydrogen bond between the nitrogen atom N1 of ring B and R364NH2 residue, which also forms an additional hydrogen bond between R364NH1 and the OH group of the lactone ring E of all five compounds (see Table 1). The lactone ring is involved in other two interactions with the DNA-Top1 target. The first hydrogen bond, common to all five compounds, still involves the OH group of the lactone ring and D533Oδ2. The other interaction is represented by a bidentate hydrogen bond formed between K532NζH2/3 and the C = O group of 3a, 3c, 3d and 10, while 3b is able to form a single hydrogen bond with K532NζH2 only. The interaction pattern of the compound 3d within the binding site is completed by a hydrogen bond between the camptothecin phenolic 10-hydroxy and E356Oε2, an interaction clearly impossible for the other compounds. Moreover, the phenolic 10-hydroxy of 3d forms a strong intramolecular hydrogen bond with the carboxyl group of the side chain (1.66Å), increasing the stability of the complex which is the best among those studied in this paper.
Table 1. Binding free energies (kcal/mol) and selected distances (in Å) between the five ligands and the residues of the DNA-Top1 system.
| Interactions occouring within the binding site | |||||||||||
| ΔGbinding | K532NζH2 | K532NζH3 | D533Oδ2 | R364NH1 | R364NH2 | E356Oε2 | (-1)T10 | (-1)A113 | (+1)G11 | (+1)C112 | |
| 3a | -9.21 | 2.66 | 2.68 | 2.32 | 2.43 | - | - | π-π | π-π | π-π | π-π |
| 3b | -10.47 | - | 2.37 | 2.05 | 2.61 | 2.83 | - | π-π | π-π | π-π | π-π |
| 3c | -9.24 | 2.70 | 2.82 | 2.44 | 2.43 | - | - | π-π | π-π | π-π | π-π |
| 3d | -11.82 | 2.70 | 2.61 | 1.64 | 2.30 | 2.67 | 2.95 | π-π | π-π | π-π | π-π |
| 10 | -9.08 | 2.85 | 2.83 | 2.60 | 2.32 | - | - | π-π | π-π | π-π | π-π |
| Interactions involving the rear region of the binding site | |||||||||||
| K425NζH1 | K425NζH2 | Y426O | E356Oε2 | E356N | K354O | W416Nε1 | (-1)A113N7 | (-1)A113H61 | (-1)A113H62 | (-1)A114H61 | |
| 3a | 2.29 | - | 1.68 | - | - | - | - | - | - | 2.86 | - |
| 3b | 2.82 | 1.75 | - | - | - | - | - | 2.18 | 2.93 | 2.92 | 1.89 |
| 3c | - | - | - | 1.63 | 1.98 | 1.56 | - | - | - | 2.44 | - |
| 3d | - | - | - | 2.55 | - | 2.57 | - | - | - | 2.88 | - |
| 10 | - | - | - | - | - | - | 1.94 | - | - | 2.68 | - |
Stacking interactions between the aromatic rings of camptothecin and the -1 (T10-A113) and +1 (G11-C112) base pairs are indicated with π-π.
The side chains of the five compounds are located in the rear region of the binding site, resting on the DNA duplex and forming a series of interactions that contribute to the stability of the complexes (Table 1). The only interaction common to all the studied compounds is the hydrogen bond between (-1)A113H62 and the oxygen of the oxime group, that only in 3b forms a second hydrogen bond with (-1)A113H61. The C = O group of the 3a side chain interact with K425NζH1, while 3b forms a bidentate bond with both K425NζH1 and K425NζH2. The interaction with K425 is absent in 3c-d and 10, where the C = O group is rotated to form a hydrogen bond with W416Nε1.
The interaction of the hydroxyamic NH group with the target is present only for compounds 3a, 3c and 3d, involving however different residues: 3a with Y426O, 3c with E356Oε2, 3d with K354O. Finally, the interaction pattern of the side chains is completed by the interactions of the hydroxamic OH group with the DNA-topoisomerase-I system, present only in 3b, 3c and 3d, but also in this case with different residues: 3b with (-1)A113N7, (-1)A114H61 and K425NζH2, 3c with E356N and K354O, 3d with E356Oε2.
HDAC-II complexes
The complexes are obtained by molecular docking of 3a-d and 10 using the Class I Rpd3-like protein HDAC2 as a model.
While the 3a-d series shows minor structural modification in the linker length, 10 is rather different, due to the presence of the SH group, which might change the interactions with the catalytic zinc ion that is essential for enacting the enzyme. Apart from these differences, all the ligands enter the binding site in a very similar way, with the functionalized side chain inside the cavity and the camptothecin system that interacts with the external surface near the entrance to the cavity (Fig 5).
Fig 5.
(Left)—Solvent Accessible Surface (SAS) representation of HDAC bond to 3d (in stick). The portions of molecule interacting with the binding site is delimited by the oxime group, located near the entrance of the cavity. (Right)—Binding mode of the top-score conformation of 3d to HDAC-II, with the aminoacids side chains relevant to the discussion rendered in line and the ligand in stick. Hydrogen bonds are shown as green dotted lines.
As expected, the docking results of 3a-d were very similar and their side chains are highly superimposed. Their common interactions include a hydrogen bond from the hydroxamic NH towards G154O and two interaction involving the hydroxamic C = O group; a hydrogen bond with H183Hδ1 and the chelation of the catalytic Zn2+ ion (Table 2). The residue Y308 is directly involved only in the binding of the compounds having the spacer with n = 5 (3b and 3d), through a hydrogen bond involving Y308O and the C = O group of both compounds, and a further hydrogen bond between Y308O and the hydroxamic OH of the 3d compound alone, which further stabilizes its conformation. Compound 10 still exhibits the interactions with G154 and H146, but there is no hydrogen bond with Y308. The loss of the interaction with Y308 is almost certainly attributable to the non-optimal spacer length, considering that the same effect is also found in compounds 3a and 3c. Furthermore, unlike 3a-d compounds, in 10 the zinc ion is complexed by the sulfur atom, which also interacts with rings of H145, H146 and Y308, forming three Pi-Sulfur interactions.
Table 2. Binding free energies (kcal/mol) and distances (in Å) between the five ligands and HDAC II residues forming the catalytic binding site.
| Interactions occouring within the binding site | Interactions involving Camptothecin | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ΔGbinding | G154O | H146Hε2 | Y308OH | Zn2+ | D104Oδ1 | E208O | Y209N | Y209 | F210 | |
| 3a | -7.53 | 2.01 | 2.57 | - | 2.86 | - | - | - | π-π | π-π |
| 3b | -7.66 | 2.03 | 2.91 | 1.97 | 2.53 | - | - | - | π-π | - |
| 3c | -7.36 | 2.00 | 2.32 | - | 2.90 | - | - | - | π-π | - |
| 3d | -7.89 | 1.90 | 2.76 | 1.91 (OH) 2.14 (C = O) |
2.74 | 1.56 | 1.98 | - | - | - |
| 10 | -7.78 | 2.35 | 2.51 | - | 2.54 | - | - | 3.01 | - | - |
Stacking interactions are indicated with π-π.
The portions of 3a-d and 10 interacting with the binding site are delimited by the oxime group, which in fact lies in all cases at the entrance of the cavity. The camptothecin chain leans to rest on the outer surface of the receptor to form interactions with residues at the rim of the pocket. For 3a-c the only observed π-π stacking interactions are with the residue Y209 (3a-c), while only 3a is able to form a pi-pi stacking with F210, evidently aided by the shorter linker that forces the camptothecin skeleton to approach the surface. In contrast, compound 10 does not show π-π stacking interactions with residues at the rim of the pocket, but is still anchored to the target surface by a hydrogen bond between the camptothecin carbonyl oxigen in 17 and Y209N.
In compound 3d, the presence of the 10-hydroxy group and the optimal spacer length makes it once again unique among the studied compounds. These two structural features provide 3d with the possibility of tightly interacting with the external surface through two strong hydrogen bonds, one between the OH of the lactone E ring and D104Oδ1 and the other between the phenolic 10-hydroxy and E208O.
Finally, compound 3d was superimposed to the reference ligands CPT and SAHA in the catalytic sites of Top1 and HDAC II, respectively (Fig 6A and 6B). Fig 6A shows the superimposition of 3d and CPT in the catalytic site of Top1. The two conformations are almost perfectly superimposed, with the exception of the D-E rings, which in 3d move close to K532 and D533 to obtain a better interaction. The overlap of 3d and SAHA within the HDAC-II catalytic site is reported in Fig 6B. Although the functional groups are different, the two molecules behave similarly within the cavity, both interacting with the same residues and chelating the Zn++ ion. The two structures diverge as one moves away from the centre of the cavity, as a result of the different interaction with the surface of the enzyme. This behaviour can be explained considering the different dimensions and properties of the 3d and SAHA terminal groups.
Fig 6.
(A)–Superimposition of the 3d (in red) and CPT (blue) structures in the catalytic site of Top1. (B)—Overlap of 3d (red) and SAHA (blue) within the HDAC-II catalytic site.
Overall, the results obtained showed that hybrids with a 5/6 carbon chain are capable of forming stable complexes with both HDAC and Top1. Compound 3d has the most favourable interactions and its promising binding mode provides a computational validation to our plan to combine the CPT and SAHA-like motifs in a single molecule.
Biological activity evaluation
The antiproliferative activity was evaluated on a series of human tumor cell lines (NCI-H460: non small cell lung cancer; CAPAN 1: human pancreatic ductal adenocarcinoma; A431: human epidermoid carcinoma; HeLa: human epithelioid cervix carcinoma; HT29: human colon adenocarcinoma; DU145: human prostate carcinoma; HepG2: human liver hepatocellular carcinoma; A2780: human ovarian carcinoma; A2780-Dox: human doxorubicin-resistant ovarian carcinoma). The activity was assessed after 72 h with the SRB assay.
Compounds 3a-c, having different linker length, showed significant antiproliferative activity, suggesting that the chain was flexible enough to effectively position the molecule within the enzymes’ active sites. Compound 3d, the 10-hydroxy substituted derivative of 3b, showed a very potent antiproliferative activity as well. In fact, it displayed a cytotoxic activity in the nanomolar range (0.03 μM < IC50 < 0.4 μM), while SAHA (IC50 > 0.9 μM), Irinotecan (IC50 > 1.5 μM), and the reference compound dual inhibitor CY-700 (0.3 μM < IC50 < 3.3 μM) were all less effective (Table 3). Compound 10, carrying a Zn-binding group different from the hydroxamic acid, maintained a significant antiproliferative activity.
Table 3. Human tumor cell lines exposed to Top1, HDAC, and dual Top1/HDAC inhibitors.
| NCI-H460 | Capan 1 | A431 | HeLa | HT29 | DU145 | HepG2 | A2780 | A2780-Dx | RI | |
|---|---|---|---|---|---|---|---|---|---|---|
| Cpda | IC50 ± SD (μM) | |||||||||
| Irinotecan | 2.4 ± 0.1 | 1.5 ± 0.1 | 4.1 ± 0.1 | 5.8± 0.6 | 4.8 ± 0.4 | 2.8 ± 0.07 | 4.0 ± 0.8 | 3.8 ± 0.5 | 20.1± 2.9 | 5.3 |
| SAHA | 5.0 ± 0.2 | 1.8 ± 0.2 | 3.5 ± 0.1 | 2.7 ± 0.2 | 2.2 ± 0.3 | 0.87 ± 0.03 | 1.54 ± 0.1 | 2.1 ± 0.2 | 3.3 ± 0.6 | 1.6 |
| CY700 | 0.36 ± 0.02 | 0.96±0.08 | 0.79± 0.1 | 3.3± 0.9 | 1.2 ± 0.2 | 0.87±0.04 | 1.5 ± 0.4 | 2.5 ± 0.3 | 5.9 ± 1.0 | 2.4 |
| 3a | 0.36 ± 0.10 | 0.05± 0.006 | n.e. | n.e. | n.e. | n.e. | n.e. | n.e. | n.e. | |
| 3b | 0.023±0.004 | 0.006±0.0007 | 0.004±0.001 | 0.16±0.04 | n.e. | n.e. | n.e. | 0.008±0.002 | 0.27±0.070 | 31.7 |
| 3c | 0.198±0.18 | 0.043± 0.003 | 0.026±0.001 | 2.4 ± 0.5 | 0.045±0.01 | 0.029±0.001 | 0.68±0.2 | 0.038±0.008 | 0.065±0.01 | 1.7 |
| 3d | 0.27 ± 0.02 | 0.035 ± 0.002 | 0.14 ± 0.02 | 0.18±0.05 | 0.39±0.1 | 0.07±0.003 | 0.11±0.03 | 0.27 ± 0.08 | 1.8 ± 0.4 | 6.6 |
| 10 | 0.1 ± 0.01 | n.e. | n.e. | n.e. | n.e. | n.e. | n.e. | 0.21 ± 0.05 | 0.35 ± 0.1 | 1.7 |
aHuman tumor cell lines were treated with various Top1, HDAC, and dual Top1/HDAC inhibitors and the anti-proliferative activity assessed after 72 h with the SRB assay. RI = Resistance Index. Ne = not evaluated.
Compounds 3b and 3d were selected for a deeper characterization of their target modulation and specificity.
Initially, a comparative study on different purified HDAC isoforms was carried out (Table 4).
Table 4. Inhibition of HDAC isoforms by compounds 3b, 3d.
| Compounda | HDAC1 | HDAC2 | HDAC4 | HDAC6 | HDAC10 |
|---|---|---|---|---|---|
| 3d | 5,26E-08 | 9,33E-08 | n.a | 1,56E-09 | 2,24E-07 |
| 3b | 4,30E-08 | 7,62E-08 | n.a. | 3,87E-09 | 9,99E-08 |
| Trichostatin A | 1,244E-08 | 2,36E-08 | - | 2,09E-09 | 3,20E-08 |
| TMP269 | -- | -- | 3.53E-07 | -- | -- |
aIC50 values were calculated using the GraphPad Prism 4 program based on a sigmoidal dose-response equation.
n.a. = not active. Trichostatin A and TMP269 were used as reference standards
Both compounds showed a significant activity against four purified HDAC isoforms, representative of class I (HDAC 1, 2) and Class IIb (HDAC 6, 10) HDACs, with IC50 comparable to those of the reference compound Trichostatin A, without substantial selectivity. Conversely, both compounds were inactive against HDAC 4 (representative of Class IIa HDACs) (Table 4). This result was not unexpected, as it is well known that SAHA inhibits class IIa enzymes at concentrations that are not clinically significant [41].
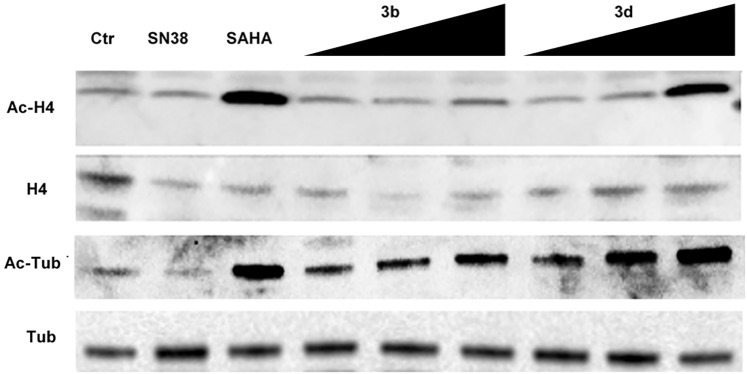
Subsequent analyses were conducted to verify whether 3b and 3d effectively targeted HDACs and Top1. To explore the HDAC inhibitory activity of the selected compounds, NCI-H460 cells were exposed to 3b and 3d (at different concentrations: 0.125, 0.2, 0.5 μM) and to the reference compounds SN38 (the active metabolite of Irinotecan) and SAHA (0.05 and 5 μM, respectively, corresponding approximately to their own IC80). The accumulation of both acetylated histone H4 and acetylated tubulin was investigated (Fig 7). Compound 3d, similarly to SAHA, showed the characteristics of a Pan-HDAC inhibitor, as revealed by H4 and tubulin acetylation in a dose dependent manner. Compound 3b, at a concentration corresponding to its IC80 (0.5 μM), showed to be less potent than 3d on both targets, particularly on H4 (Fig 7).
Fig 7. Effects of compounds 3b, 3d, SN38 and SAHA (reference compounds) on acetylation of tubulin and histone H4 in NCI-H460 cells.
Cells were treated for 24 h with SN38 (0.05 μM), SAHA (5 μM), 3b and 3d (0.125–0.2–0.5 μM). Cells were lysed in 0.5% NP-40 and 40 μg of total proteins were loaded on NuPAGE 4–12% (Invitrogen), transferred to a nitrocellulose paper and probed with specific antibodies. Acetylated Histone H4 (Ac-H4); Histone H4; Acetylated Tubulin (Ac-H4); Tubulin.
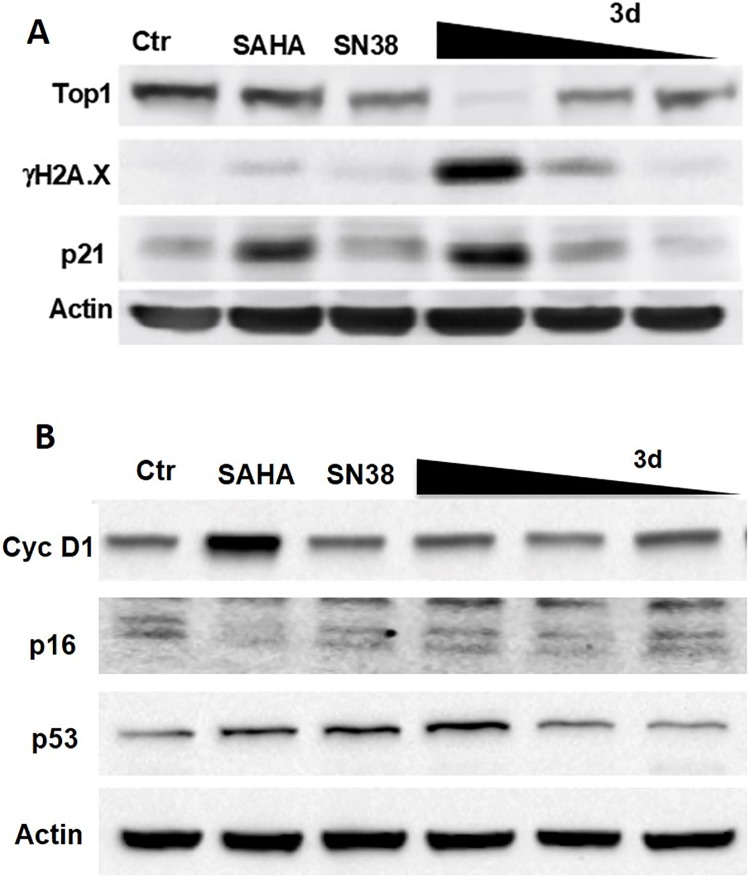
Following on from these results, 3d was further investigated to assess its ability to inhibit Top1. Interestingly, the compound drastically reduced the expression of Top1 (comparable to SN38) (Fig 8A). This finding is consistent with recent literature, where there is evidence that treatment with topoisomerase inhibitors significantly decreases the Top I expression [42]. In addition, 3d induced p53 activation (Fig 8B) and DNA damage at a dose corresponding to its IC80, confirmed by H2A.X induction (Fig 8A). Interestingly, SN38 and SAHA at equivalent doses appeared less effective in p53 up-regulation and completely ineffective in H2A.X induction (Fig 8A and 8B), thus suggesting that the 3d dual targeting activity cooperates in the DNA damage induction.
Fig 8. Effects of compound 3d, SN38 and SAHA (reference compounds) on Top1, H2A.X, p21 (A), CycD1, p16, p53 (B) in NCI-H460 cells.
H460 cells were treated for 24 h with SAHA (5 μM), SN38 (0.05 μM), and 3d (0.125–0.2–0.5μM). Cells were lysed in 0.5% NP-40 and 40 μg of total proteins were loaded on NuPAGE 4–12% (Invitrogen), transferred to a nitrocellulose paper and probed with specific antibodies.
Moreover, P16 and CycD1 were down and up-regulated, respectively, by SAHA, while 3d and SN38 didn’t show any effect. Interestingly, SAHA-induced block of G2/M progression observed in our experiment (see below) and its induction of p21, p53 and Cyclin D1 suggest an effect on senescence pathways activation, as recently reported [43].
Furthermore, similarly to SAHA, 3d activated the expression of p21, probably as a consequence of its HDAC inhibitory activity (Fig 8). Taken together, these results suggest that 3d possesses a dual-inhibitor characteristic, acting at cellular level both as HDAC and Top1 inhibitor.
To assess the antitumor potential of compound 3d, a large screening on hematologic cancer cell lines was carried out (Tables 5 and 6). The compound showed a cytotoxic potency mostly in the nanomolar range (0.006 μM < IC50 < 4 μM), superior to SAHA (0.6 μM < IC50 < 9 μM), and Irinotecan (IC50 > 10 μM) in most cell lines. Interestingly, 3d showed an activity superior to that of CY-700 (0.3 μM < IC50 < 10 μM), the most active compound described by Chen [40], taken as a reference standard of dual CPT-HDAC inhibitors. These data confirm the correctness of the choice of linking the HDAC-active chain to the position 7 of camptothecin.
Table 5. Human lymphoma and leukemia cell lines exposed to 3d and Top1, HDAC, and dual Top1/HDAC inhibitor CY700.
| Cpd | MINO | MAVER-2 | JECO-1 | U-2932 | OCI-LY3 | L-428 | KM-H2 | DG-75 | NB4 |
|---|---|---|---|---|---|---|---|---|---|
| IC50 ± SD (μM) | |||||||||
| 3d | 0.031±0.01 | 0.012±0.001 | 0.023±0.006 | 0.044±0.01 | 0.018±0.005 | 0.43±0.07 | 0.39±0.1 | 1.9±0.2 | n.e. |
| Irinotecan | >10(36%) | 0.74 ± 0.1 | >10 (48%) | 4.4 ± 0.8 | > 10 (0%) | >10(0%) | >10(12%) | >10(0%) | 1.0±0.3 |
| CY700 | 0.37± 0.1 | 0.4 ± 0.08 | 1.0 ± 0.4 | 0.9 ± 0.3 | 0.34±0.04 | 1.7 ± 0.2 | 1.5 ± 0.5 | >10(18%) | 0.24±0.06 |
| SAHA | 1.1 ± 0.3 | 5.4 ± 1.2 | 2.6 ± 0.7 | 1.1 ± 0.2 | 0.99 ± 0.3 | 2.3± 0.5 | 1.2± 0.2 | 2.1 ± 0.2 | 3.3± 0.6 |
Human hematologic cancer cells were exposed to various Top1, HDAC, and dual Top1/HDAC inhibitors and the anti-proliferative activity assessed after 72 h with the MTT assay. Ne = not evaluated.
Table 6. Human myeloma, leukemia and lymphoma cell lines exposed to 3d and SAHA.
| Cell line | SAHA | 3d |
|---|---|---|
| IC50 ± SD (μM) | ||
| K 562 | 1.53 ± 0.4 | 0.94 ± 0.2 |
| ARH-77 | 3.9 ± 0.5 | 0.21 ± 0.06 |
| THP1 | 3.9 ± 1.4 | 0.22 ± 0.09 |
| MV4-11 | 3.84 ± 0.8 | 2.73 ± 0.3 |
| Jurkat-cp | 1.99 ± 0.2 | 0.23 ± 0.05 |
| U937 | 1.62 ± 0.3 | 0.18 ± 0.05 |
| RAJI | 1.73 ± 0.6 | 0.077 ± 0.02 |
| RAMOS | 3.9 ± 0.7 | 0.99 ± 0.1 |
| REC-1 | 0.94 ± 0.2 | 0.18 ± 0.04 |
| Z-138 | 0.7 ± 0.1 | 0.006 ± 0.001 |
| RPMI-8226 | 1.98 ± 0.5 | 4.1 ± 1.4 |
| H929 | 0.56 ± 0.07 | 1.1 ± 0.4 |
Human hematologic cancer cells were exposed to 3d and SAHA and the anti-proliferative activity assessed upon 72h with the MTT assay.
Lastly, we tested 3d against a series of primary malignant pleural mesothelioma (MPM) cell lines (from our cell bank collection).
We compared 3d to drugs in clinical use for the treatment of MPM, such as Cisplatin, and drugs such as Irinotecan, SAHA, Gefitinib, 5-FU, which failed to increase the survival of MPM patients when investigated in clinical trials [44].
Gratifyingly, 3d showed a potent antiproliferative effect, with IC50 ranging from 0.05 to 11 μM, which was much higher than SAHA (4 μM < IC50 < 12 μM) and Irinotecan (IC50 > 7 μM). Moreover, 3d resulted more effective than Gefitinib (14 μM < IC50 < 38 μM), cisplatin (1.2 μM < IC50 < 8.0 μM), and 5-FU (18 μM < IC50 < 200 μM), as well (Table 7). These results are remarkable, considering that mesothelioma still remains an incurable cancer, and new therapeutic approaches against the disease are urgently needed [45].
Table 7. Human primary mesothelioma cell lines exposed to 3d and reference antitumor drugs.
| Cpd | EPITHELIOID | BIPHASIC | SARCOMATOID | ||||||
|---|---|---|---|---|---|---|---|---|---|
| IC50 ± SD (μM) | |||||||||
| MM288 | MM317 | MM404 | MM473 | MM481 | MM487 | MM491 | MM432 | MM472 | |
| 3d | 0.48±0.1 | 0.85±0.2 | 1.4 ±0.3 | 2.1±0.5 | 11.7±1.2 | 0.2±0.05 | 0.87±0.3 | 0.16±0.03 | 0.048±0.01 |
| Irinotecan | 14.9±2.0 | 7.0 ± 1.0 | 9.4 ± 0.7 | 10.0±2.0 | |||||
| SAHA | 3.8 ±0.2 | 11.2±0.9 | 8.8± 1.2 | 8.5± 0.4 | 11.5±0.4 | 7.4 ± 0.4 | 10.7±0.4 | 6.9 ± 1.0 | 4.6 ± 0.8 |
| Gefitinib | 38.7±1.4 | 26.1±2.9 | 28.4±2.4 | 18.4±0.7 | 31.2±1.0 | 15.0±0.5 | 14.0±0.7 | 17.1 ± 1.0 | 19.1 ± 0.5 |
| Cisplatin | 8.1 ± 0.3 | 4.2 ± 0.6 | 2.4 ± 0.5 | 2.4 ± 0.2 | 7.0 ± 1.5 | 3.0 ± 0.5 | 6.5 ± 0.9 | 5.0 ± 0.8 | 1.26 ± 0.1 |
| 5-FU | >200(35%) | 17.9±3.1 | 52.2±8.7 | 35.2±8.4 | 27.4±2.2 | >200(48%) | 44.0±10.3 | >200(34%) | 40. ± 10.5 |
Human primary mesothelioma cells were exposed to 3d and different anti-tumor compounds. Anti-proliferative activity was assessed upon 72 h with the SBR assay.
Compound 3d was also tested, comparing it with Irinotecan and SAHA, on NCI-H460 NSCLC cell line to assess the effect on cell cycle progression and induction of apoptosis. FACS analysis revealed a comparable effect of 3d and Irinotecan on cell cycle distribution, with a steady block of treated-cells in the S phase, while SAHA preferably arrested cells in G2/M. Interestingly, 3d also had cytotoxic activity, with induction of apoptosis (sub-G0/1 population) that was lacking in cells treated with Irinotecan and SAHA (Table 8).
Table 8. Flow cytometric evaluation of human NCI- H460 NSCLC cells exposed to 3d and reference antitumor drugs.
| NCI-H460a | G0/1 (%) | S (%) | G2/M (%) | Sub-G0/1 (%) |
|---|---|---|---|---|
| Ctr | 71.0 | 23.0 | 5.9 | 2.4 |
| 3d | 12.1 | 76.0 | 11.9 | 15.2 |
| Irinotecan | 7.9 | 89.0 | 3.2 | 2.7 |
| SAHA | 65.2 | 11.7 | 23.1 | 1.8 |
aTumor cells were exposed for 72 h to 3d and reference anti-tumor drugs (at their own IC80 dose), stained with PI, and processed on a FACScan flow cytometer. The CellQuest software was used to acquire data and assess the sub-G0/1 population, while cell cycle analysis was performed with the ModFit software.
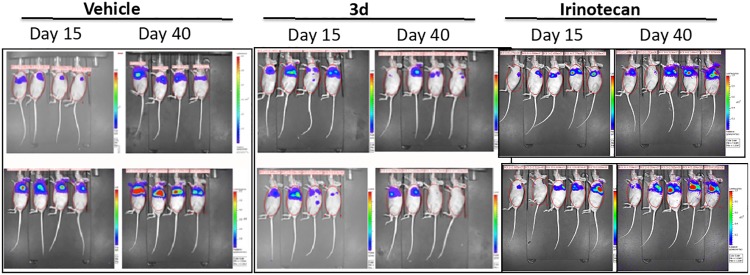
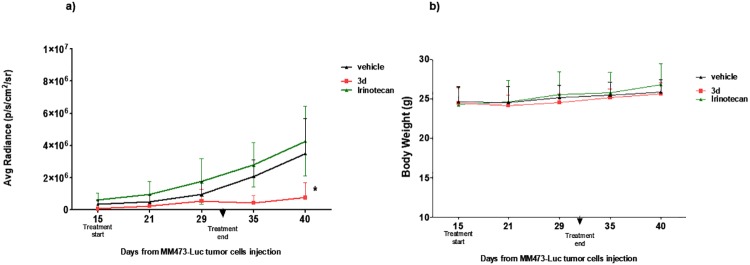
The efficacy of the new dual inhibitor 3d was evaluated on human mesothelioma primary cell line MM473-Luc orthotopically xenografted in CD-1 nude mice. This cell line was stably transfected with luciferase and was suited for the observation of tumor growth by chemoluminescence analysis. After tumor cells injection and with tumors in the growth phase (day 15), mice were intravenously treated with 3d (45 mg/kg, q4dx3w), Irinotecan (35 mg/kg, q4dx3) or vehicle as negative control (treatments ended at day 32). The tumor volume was measured as BLI signal and expressed as average radiance.
The molecule administered had a strong antitumor activity as shown in Figs 9 and 10. Moreover, the tumor chemiluminescence in animals treated with 3d indicated that the compound acts by both reducing the tumor growth progression and, in some cases, by reducing the tumor size until the disappearance of the bioluminescent signal (8 days after the end of treatments) (Figs 9 and 10). Conversely, Irinotecan had only a marginal antitumor effect (not significant, p>0.5) when administered at 35 mg/kg. Interestingly, the antitumor activity was observed at doses that appear well tolerated (no reduction in body weight was observed, data not shown).
Fig 9. Efficacy of 3d in an orthotopic xenograft model of human epithelioid mesothelioma.
Bioluminescence signal from mice injected with MM473 expressing luciferase at day 15 (first treatments) and day 40 (8 days after the last treatment).
Fig 10. Efficacy of 3d in an orthotopic xenograft model of human epithelioid mesothelioma.
a) bioluminescence signal of experimental groups throughout the experimental period; b) body weight curve. Statistical analysis performed using non parametric Mann-Whitney test (U) 3d groups versus Vehicle *p<0,05.
Conclusion
Synergistic effects have been reported on the use of HDAC and topoisomerase I inhibitors as a co-treatment on various cancer cells. In this study, new hydroxamic acid derivatives of camptothecin were evaluated as dual-action CPT-HDAC inhibitors. In particular, compound 3d displayed a broad spectrum of antiproliferative activity, with IC50 values in the nanomolar range, on a series of human solid tumor, hematologic, and mesothelioma cell lines. The new hybrid molecule showed a potency superior to both HDAC inhibitor SAHA and Top1 inhibitor Irinotecan, increased antitumor activity and very high tolerability in in vivo human tumor models with respect to SAHA and Topotecan.
In molecular modelling studies, compound 3d showed the most favourable interactions and its promising binding mode provided structural support to the molecular design and development of analogues. Overall, the docking interaction profiles of synthesized compounds were in accordance with the biological results, especially for selected compound 3d, which was found to have significantly better interactions with both target proteins. This finding can be explained considering the simultaneous presence in 3d of the phenolic -OH on ring A of camptothecin (10-hydroxy) and the right length of the linker (n = 5). These two structural features represent the driving force that makes 3d a better ligand for both the DNA-Top1 binary complex and the HDAC-II active site.
The single dual-function molecule not only attacks the cancer cells from two distinct pathways simultaneously but also improves drug efficacy of the conventional CPT without increasing its dose-limiting toxicity. Therefore, it can be used for treating tumors susceptible to camptothecin drugs and/or to HDAC inhibitors, showing at the same time a good tolerability and low toxicity.
Data Availability
All relevant data are within the paper.
Funding Statement
The authors received no specific funding for this work. Scientia Advice provided support in the form of salaries for author RA, but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific role of this author is articulated in the ‘author contributions’ section.
References
- 1.Sundar R, Valeri N, Harrington KJ, Yap TA. Clin. Combining Molecularly Targeted Agents: Is More Always Better? Cancer Res. 2017; 23: 1123–1125. [DOI] [PubMed] [Google Scholar]
- 2.Musso L, Dallavalle S, Zunino F. Perspectives in the development of hybrid bifunctional antitumour agents. Biochem. Pharmacol. 2015; 96: 297–305. 10.1016/j.bcp.2015.06.006 [DOI] [PubMed] [Google Scholar]
- 3.Cincinelli R, Musso L, Artali R, Guglielmi M, Bianchino E, Cardile F, et al. Camptothecin-psammaplin A hybrids as topoisomerase I and HDAC dual-action inhibitors. Eur. J. Med. Chem. 2018; 143: 2005–2014. 10.1016/j.ejmech.2017.11.021 [DOI] [PubMed] [Google Scholar]
- 4.Bérubé G. An overview of molecular hybrids in drug discovery. Expert Opin. Drug Discov. 2016; 11: 281–305. 10.1517/17460441.2016.1135125 [DOI] [PubMed] [Google Scholar]
- 5.Abbot V, Sharma P, Dhiman S, Noolvi MN, Patel HM, Bhardwaj V. Small hybrid heteroaromatics: resourceful biological tools in cancer research. RSC Advances 2017; 7: 28313–28349. [Google Scholar]
- 6.Roche J, Bertrand P. Inside HDACs with more selective HDAC inhibitors. Eur. J. Med. Chem. 2016; 121: 451–483. 10.1016/j.ejmech.2016.05.047 [DOI] [PubMed] [Google Scholar]
- 7.Chun P. Histone deacetylase inhibitors in hematological malignancies and solid tumors. Arch. Pharm. Res. 2015; 38: 933–949. 10.1007/s12272-015-0571-1 [DOI] [PubMed] [Google Scholar]
- 8.Pommier Y. Drugging Topoisomerases: Lessons and Challenges. ACS Chem Biol. 2013; 8: 82–95. 10.1021/cb300648v [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu G, Zekria D, Cai X, Ni X. Current status of CPT and its analogues in the treatment of malignancies. Phytochem. Rev. 2015; 14: 429–441. [Google Scholar]
- 10.Kim N, Sue Jinks-Robertson S. The Top1 paradox: Friend and foe of the eukaryotic genome. DNA Repair 2017; 56:33–41. 10.1016/j.dnarep.2017.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y.-Q, Li W-Q, Morris-Natschke SL, Qian K, Yang L, Zhu G-X, et al. Perspectives on Biologically Active Camptothecin Derivatives. Med. Res. Rev. 2015; 35: 753–789. 10.1002/med.21342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martino E, Della Volpe S, Terribile E, Benetti E, Sakaj M, Centamore A, et al. The long story of camptothecin: From traditional medicine to drugs. Bioorg. Med. Chem. Lett. 2017; 27: 701–707. 10.1016/j.bmcl.2016.12.085 [DOI] [PubMed] [Google Scholar]
- 13.Ghamande S, Lin CC, Cho DC, Shapiro GI, Kwak EL, Silverman MH, et al. A phase 1 open-label, sequential dose-escalation study investigating the safety, tolerability, and pharmacokinetics of intravenous TLC388 administered to patients with advanced solid tumors. Invest New Drugs. 2014; 32:445–451 10.1007/s10637-013-0044-7 [DOI] [PubMed] [Google Scholar]
- 14.Slingerland M, Gelderblom H. The fate of camptothecin glycoconjugate: report of a clinical hold during a phase II study of BAY 56–3722 (formerly BAY 38–3441), in patients with recurrent or metastatic colorectal cancer resistant/refractory to irinotecan. Invest New Drugs. 2012; 30: 1208–1210. 10.1007/s10637-011-9679-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Botella P, Rivero-Buceta E. Safe approaches for camptothecin delivery: Structural analogues and nanomedicines. J. Control. Release 2017; 247: 28–54. 10.1016/j.jconrel.2016.12.023 [DOI] [PubMed] [Google Scholar]
- 16.Pasini A, Marchetti C, Sissi C, Cortesi M, Giordano E, Minarini A, et al. Novel Polyamine–Naphthalene Diimide Conjugates Targeting Histone Deacetylases and DNA for Cancer Phenotype Reprogramming. ACS Med. Chem. Lett. 2017; 8: 1218–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bruzzese F, Rocco M, Castelli S, Di Gennaro E, Desideri A, Budillon A. Synergistic antitumor effect between vorinostat and topotecan in small cell lung cancer cells is mediated by generation of reactive oxygen species and DNA damage-induced apoptosis. Mol Cancer Ther. 2009, 8:3075–3087. 10.1158/1535-7163.MCT-09-0254 [DOI] [PubMed] [Google Scholar]
- 18.Sarcar B, Kahali S, Chinnaiyan P. Vorinostat enhances the cytotoxic effects of the topoisomerase I inhibitor SN38 in glioblastoma cell lines. J Neurooncol. 2010; 99: 201–207. 10.1007/s11060-010-0127-7 [DOI] [PubMed] [Google Scholar]
- 19.Sato A, Asano T, Horiguchi A, Ito K, Sumitomo M, Asano T. Antitumor Effect of Suberoylanilide Hydroxamic Acid and Topotecan in Renal Cancer Cells. Oncol Res. 2011; 19: 217–223. [DOI] [PubMed] [Google Scholar]
- 20.Munster PN, Daud AI. Preclinical and clinical activity of the topoisomerase I inhibitor, karenitecin, in melanoma. Expert Opin. Investig. Drugs. 2011; 20:1565–1574. 10.1517/13543784.2011.617740 [DOI] [PubMed] [Google Scholar]
- 21.Na YS, Kim SM, Jung KA, Yang SJ, Hong YS, Ryu MH, et al. Effects of the HDAC inhibitor CG2 in combination with irinotecan, 5-fluorouracil, or oxaliplatin on HCT116 colon cancer cells and xenografts. Oncol Rep. 2010; 24: 1509–1514. [DOI] [PubMed] [Google Scholar]
- 22.Wasim L, Chopra M. Panobinostat induces apoptosis via production of reactive oxygen species and synergizes with topoisomerase inhibitors in cervical cancer cells. Biomed. Pharmacother. 2016; 84: 1393–1405. 10.1016/j.biopha.2016.10.057 [DOI] [PubMed] [Google Scholar]
- 23.Sampson VB, Vetter NS, Kamara DF, Collier AB, Gresh RC, Kolb EA. Vorinostat Enhances Cytotoxicity of SN-38 and Temozolomide in Ewing Sarcoma Cells and Activates STAT3/AKT/MAPK Pathways. PLoS One. 2015; 10: e0142704 10.1371/journal.pone.0142704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gray J, Cubitt CL, Zhang S, Chiappori A. Combination of HDAC and topoisomerase inhibitors in small cell lung cancer. Cancer Biol Ther. 2012; 13: 614–622. 10.4161/cbt.19848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dallavalle S, Ferrari A, Biasotti B, Merlini L, Penco S, Gallo G, Marzi M, Tinti MO, Martinelli R, Pisano C, Carminati P, Carenini N, Beretta G, Perego P, De Cesare M, Pratesi G, Zunino F. Novel 7-oxyiminomethyl derivatives of camptothecin with potent in vitro and in vivo antitumor activity. Novel 7-Oxyiminomethyl Derivatives of Camptothecin with Potent in Vitro and in Vivo Antitumor Activity. J. Med. Chem 2001; 44: 3264–3274. [DOI] [PubMed] [Google Scholar]
- 26.Cincinelli R, Musso L, Dallavalle S, Artali R, Tinelli S, Colangelo D, Zunino F, De Cesare M, Beretta GL, Zaffaroni N. Design, modeling, synthesis and biological activity evaluation of camptothecin-linked platinum anticancer agents. Eur. J. Med. Chem 2013; 63: 387–400. 10.1016/j.ejmech.2013.02.022 [DOI] [PubMed] [Google Scholar]
- 27.Dallavalle S, Delsoldato T, Ferrari A, Merlini L, Penco S, Carenini N, Perego P, De Cesare M, Pratesi G, Zunino F. Novel 7-Substituted Camptothecins with Potent Antitumor Activity. J Med. Chem. 2000; 43: 3963–3969. [DOI] [PubMed] [Google Scholar]
- 28.Lindahj E, Hess B, van der Spoel D. GROMACS 3.0: a package for molecular simulation and trajectory analysis. Mol. Model. Annu. 2001; 7: 306–317. [Google Scholar]
- 29.Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, Caldwell J, Wang J, Kollman P. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comp. Chem. 2003; 24:1999–2012. [DOI] [PubMed] [Google Scholar]
- 30.Wang J, Cieplak P, Kollman PA. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J. Comp. Chem. 2000; 21: 1049–1074. [Google Scholar]
- 31.Huey R, Morris GM, Olson AJ, Goodsell DSS. A semiempirical free energy force field with charge-based desolvation.J. Comput. Chem. 2007; 28: 1145–1152. 10.1002/jcc.20634 [DOI] [PubMed] [Google Scholar]
- 32.Solis FJ, Wets RJB. Minimization by Random Search Techniques. Math. Oper. Res. 1981; 6: 19–30. [Google Scholar]
- 33.Sanner MF. Python: a programming language for software integration and development. J. Mol. Graph. Model. 1999; 17: 57–61. [PubMed] [Google Scholar]
- 34.Gasteiger J, Marsili MM. Iterative partial equalization of orbital electronegativity—a rapid access to atomic charges. Tetrahedron 2008; 36: 3219–3228. [Google Scholar]
- 35.De Cesare M, Beretta GL, Tinelli S, Benedetti V, Pratesi G, Penco S, Dallavalle S, Merlini L, Pisano C, Carminati P, Zunino F. Preclinical efficacy of ST1976, a novel camptothecin analog of the 7-oxyiminomethyl series. Biochem. Pharmacol., 2007; 73: 656–664. 10.1016/j.bcp.2006.11.004 [DOI] [PubMed] [Google Scholar]
- 36.Dallavalle S, Giannini G, Alloatti D, Casati A, Marastoni E, Musso L, Merlini L, Morini G, Penco S, Pisano C, Tinelli S, De Cesare M, Beretta G. Zunino F. Synthesis and Cytotoxic Activity of Polyamine Analogues of Camptothecin. J. Med. Chem. 2006; 49: 5177–5186. 10.1021/jm060285b [DOI] [PubMed] [Google Scholar]
- 37.Kozikowski AP, Chen Y, Gaysin A, Chen B, D'Annibale MA, Suto CM, Langley BC. Functional Differences in Epigenetic ModulatorsSuperiority of Mercaptoacetamide-Based Histone Deacetylase Inhibitors Relative to Hydroxamates in Cortical Neuron Neuroprotection Studies. J. Med. Chem. 2007; 50: 3054–3061. 10.1021/jm070178x [DOI] [PubMed] [Google Scholar]
- 38.Chen B, Petukhov PA, Jung M, Velena A, Eliseeva E, Dritschilo A, Kozikowski AP. Chemistry and biology of mercaptoacetamidesas novel histone deacetylase inhibitors. Chemistry and biology of mercaptoacetamides as novel histone deacetylase inhibitors. Bioorg. Med. Chem. Lett. 2005; 15: 1389–1392. 10.1016/j.bmcl.2005.01.006 [DOI] [PubMed] [Google Scholar]
- 39.Guerrant G, Patil V, Canzoneri JC, Yao L.P, Hood R, Oyelere AK. Dual-acting histone deacetylase-topoisomerase I inhibitors. Bioorg. Med. Chem. Lett. 2013; 23: 3283–3287. 10.1016/j.bmcl.2013.03.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Y, Chen Y. Preparation of HDAC inhibiting derivatives of camptothecin. WO2012096832 (2012).
- 41.Hsu KC, Liu CY, Eight Lin T, Hsieh JH, Sung TY, Tseng HJ, Yang JM, Huang WJ. Novel Class IIa-Selective Histone Deacetylase Inhibitors Discovered Using an in Silico Virtual Screening Approach. Scientific Reports 2017; 7: 3228 10.1038/s41598-017-03417-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zou J, Li S, Chen Z, Lu Z, Gao J, Zou J, Lin X, Li Y, Zhang C, Shen L. Cell Death Dis 2018; 9:661 10.1038/s41419-018-0700-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gire V, Dulić V. Senescence from G2 arrest, revisited. Cell Cycle 2015; 14: 297–304. 10.1080/15384101.2014.1000134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patel SC, Dowell JE. Modern management of malignant pleural mesothelioma. Lung Cancer: Targets and Therapy 2016; 7: 63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yap TA, Aerts JG, Popat S, Fennell DA. Novel insights into mesothelioma biology and implications for therapy. Nature Reviews Cancer. 2017; 17: 475–488. 10.1038/nrc.2017.42 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.