Abstract
Staphylococcus aureus bacteremia is one of the most frequent severe bacterial infections worldwide, with an associated mortality of about 20–40% in developed countries. In 2013, we noted an increase in this infection in the teaching hospital in Grenoble, France, compared to 2012. The mean incidence of S. aureus bacteremia was 0.28 per 1,000 patient-days in 2012 and 0.35 per 1,000 patient-days in 2013. This trend was confirmed in 2014 (0.35 per 1,000 patient-days). In the present work we aimed to study the population of patients presenting with S. aureus bacteremia in 2013 and to genotype the corresponding S. aureus strains in order to identify a successful and/or virulent genotype to design a specific infection control program. One hundred ninety-one S. aureus isolates (including 9 methicillin-resistant) out of 199 corresponding cases of bacteremia were characterized with the spa typing method. Among 108 spa types, t571, t002, t008 and t084 were the most prevalent. Although not widely prevalent, t571 was the most frequently identified clone (8.4% of all isolates). Spa type t571 has been described in previous studies as belonging to the clonal complex CC398, which is consistent with the recent emergence of methicillin-susceptible S. aureus CC398 reported in blood cultures in Europe.
Introduction
Staphylococcus aureus represents the second leading cause of community- and healthcare-associated bacteremia in developed countries with an overall annual incidence rate of 15–40 per 100 000 population [1–4]. A 30-day mortality of 20% has been reported in developed countries [5]; therefore, the identification of the factors influencing its rise, both genetic and/or environmental, is of major importance for the prevention of this infection. The distribution of methicillin-resistant S. aureus (MRSA) clonal types has been well characterized across different geographic regions and care settings. Although methicillin-susceptible S. aureus (MSSA) accounts for a large proportion of staphylococcal infections, comparatively little is known about its molecular epidemiology, in particular in France. S. aureus bacteremia, regardless of methicillin-susceptibility, is a worldwide problem associated with increased morbidity and mortality among hospitalized patients. An epidemiological study conducted in a cohort of French hospitals allowed the identification of first cases of blood infections due to S. aureus ST398 [6]. Initially, the ST398 was a MRSA clone associated with livestock carriage in pig farms. Subsequently, these clones spread to human with a worldwide dissemination [7, 8]. Several human infections of MRSA CC398, including bacteremia, have been documented, sometimes without livestock contact [9, 10]. Methicillin-susceptible variants of CC398 have been occasionally described and phylogenetic studies suggest that livestock-associated MRSA CC398 emerged from human MSSA CC398 [11]. Several studies have shown that MSSA CC398 isolated in humans was not livestock associated [6, 12]. Many spa types have been associated with the CC398 background [13], but the assignation of spa t571 to CC398 has been repeatedly observed [13, 14, 15].
In the Grenoble-Alpes University Hospital, France, the mean incidence of S. aureus bacteremia cases rose from 0.28 per 1,000 patient-days in 2012 to 0.35 per 1,000 patient-days in 2013. This trend was confirmed in 2014 (0.35 per 1,000 patient-days). To investigate this alarming observation, clinical data on all cases of S. aureus bacteremia in 2013 were collected and corresponding isolates were characterized by spa typing, as this molecular typing method is accurate, simple, cost-effective, rapid and easily interpretable [16, 17].
Material and methods
Study population
All consecutive patients hospitalized in Grenoble-Alpes University Hospital (2000 beds) with S. aureus bacteremia diagnosed between 1st January 2013 and 31st December 2013 were retrospectively enrolled. S. aureus bacteremia was defined as patients with at least one blood culture bottle, performed with the Bactec system (Becton Dickinson, USA) that was positive for S. aureus. Study ethics approval was obtained from Center for Ethics Committee (CECIC) in Rhône-Alpes-Auvergne District (agreement n°IRB 5891).
Clinical and therapeutic data were retrospectively collected from the patient’s medical records. Clinical data included demographics, background characteristics (comorbidities, immunocompromised status), and healthcare contacts before hospitalisation. Characteristics of S. aureus bacteremia comprised duration, and associated infection site (endocardium, lung, bones and joints, muscles, urine). Therapeutic data consisted of antibiotics, catheter removal, surgery, and admission to an intensive care unit. A healthcare-associated infection could be either nosocomial or non-nosocomial. An infection was considered nosocomial if S. aureus bacteremia developed in a patient hospitalized at least 48 hours prior to the onset of signs/symptoms indicating bacteremia. A non-nosocomial healthcare–associated infection was defined as S. aureus bacteremia diagnosed within 48 hours of admission in a patient with extensive healthcare contact as reflected by any of the following criteria: (a) received intravenous therapy, wound care, or specialized nursing care at home within the 30 days prior to the onset of S. aureus bacteremia; (b) attended a hospital or hemodialysis clinic or received intravenous chemotherapy within the 30 days before the onset of S. aureus bacteremia; (c) was hospitalized in an acute care hospital for 2 or more days in the 90 days before the beginning of S. aureus bacteremia; or (d) lived in a nursing home or long-term care facility. Community-acquired infection was identified as S. aureus bacteremia within 48 hours of admission with no criteria for healthcare-associated infection.
The diagnosis of infective endocarditis (IE) was classified as definite, possible or was excluded according to modified Duke’s criteria [18].
S. aureus isolates
Bacteremia-associated S. aureus strains were routinely identified on the basis of their colony morphology, Gram staining, catalase test and slide test for clumping factor (Staphytect Plus; Oxoid, UK). All identifications were confirmed by MALDI-TOF MS, using a Microflex LT instrument, Flexcontrol 3.0 software and the Biotyper 2.0 database (Bruker Daltonics, USA). Following antimicrobial susceptibility testing (see below), all isolates were stored at room temperature in agar tubes (BioRad, USA).
Antimicrobial susceptibility-testing
Susceptibility-testing was performed on Mueller-Hinton agar plates by the disk diffusion method (antibiotic disks; Bio-Rad, USA), according to French guidelines for antibiogram (CA-SFM, 2015). Antibiotics tested were cefoxitin, kanamycin, tobramycin, gentamicin, netilmycin, amikacin, doxycycline, erythromycin, lincomycin, pristinamycin, linezolide, rifampicin, cotrimoxazole, ofloxacin, fusidic acid, fosfomycin, vancomycin, and teicoplanin. The detection of methicillin resistance has been determined with the cefoxitin disk screen test in accordance with EUCAST recommendations.
Spa sequencing
Spa typing was performed with the Ridom Staph Type standard protocol [19], by using an in house procedure with an ABI 3130xl sequencer (Applied, USA) for double stranded DNA sequencing and by using Ridom Staph Type software (version 1.5) [20] (http://spaserver.ridom.de/), which automatically analyses spa repeats and assigns spa types. Spa types were analysed using the integrated based upon repeat pattern (BURP) algorithm [21].
Characterization of spa t571 strains
For all spa t571 strains, a PCR assay was performed to detect the CC398-specific sau1-hsdS1 variant [22]. Erythromycin-resistant isolates were screened with primers specific for the ermT gene as previously described [23].
Results
Strain characteristics
Over the study period, 199 patients with S. aureus bacteremia were included. Among the 199 corresponding strains, two were not stored, six did not recover after storage, leaving 191 strains for further characterization. Of the 191 strains that we typed, the overall MRSA rate was 4.7% (n = 9) and the major resistances of the 191 isolates were to erythromycin (n = 39, 20.4%) and ofloxacin (n = 12, 6.3%) (Table 1). All the isolates were susceptible to vancomycin. Among the 191 characterized isolates, MRSA were detected in 5.3% (n = 8) of the healthcare-associated (HCA) infections (n = 151) and in 2.5% (n = 1) of the community-acquired (CA) infections (n = 40). The resistance of MRSA towards fluoroquinolones was 88.9%.
Table 1. Antibiotic resistance profiles of 191 isolates of S. aureus collected in 2013 in Grenoble-Alpes University Hospital, France.
| MSSA (n = 182) |
MRSA (n = 9) |
t571 (n = 16) |
||||
|---|---|---|---|---|---|---|
| S | R (%) | S | R (%) | S | R (%) | |
| Antibiotic | ||||||
| Oxacillin | 182 | 0 | 0 | 9 (100) | 16 | 0 |
| Gentamicin | 181 | 1 (0.5) | 9 | 0 | 16 | 0 |
| Tobramycin | 181 | 1 (0.5) | 5 | 4 (44.4) | 16 | 0 |
| Kanamycin | 179 | 3 (1.6) | 4 | 5 (55.6) | 16 | 0 |
| Amikacin | 180 | 2 (1.1) | 6 | 3 (33.3) | 16 | 0 |
| Netilmycin | 181 | 1 (0.5) | 9 | 0 | 16 | 0 |
| Doxycycline | 178 | 4 (2.2) | 9 | 0 | 16 | 0 |
| Erythromycin | 145 | 37 (20.3) | 6 | 3 (33.3) | 4 | 12 (75) |
| Lincomycin | 178 | 4 (2.2) | 7 | 2 (22.2) | 16 | 0 |
| Cotrimoxazole | 181 | 1 (0.5) | 9 | 0 | 16 | 0 |
| Fosfomycin | 182 | 0 | 9 | 0 | 16 | 0 |
| Fusidic acid | 182 | 0 | 9 | 0 | 16 | 0 |
| Pristinamycin | 182 | 0 | 9 | 0 | 16 | 0 |
| Rifampicin | 182 | 0 | 8 | 1 (11.1) | 16 | 0 |
| Nitrofurantoin | 182 | 0 | 9 | 0 | 16 | 0 |
| Ofloxacin | 178 | 4 (2.2) | 1 | 8 (88.9) | 16 | 0 |
| Linezolid | 182 | 0 | 9 | 0 | 16 | 0 |
| Teicoplanin | 182 | 0 | 9 | 0 | 16 | 0 |
| Vancomycin | 182 | 0 | 9 | 0 | 16 | 0 |
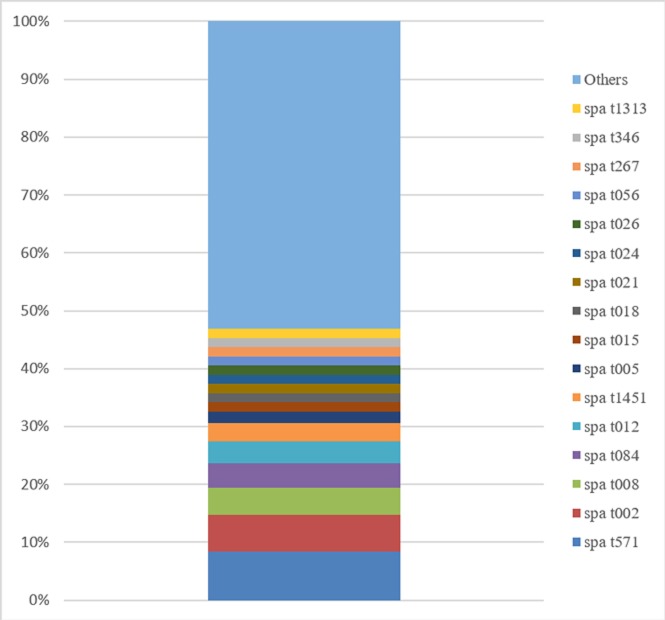
Among the 191 S. aureus isolates examined, 108 spa types were identified, two of which were newly assigned during this study. One of the 191 isolates could not be assigned to a spa type through the protocol described above and was consequently excluded from clustering analyses. For two patients, two S. aureus isolates were identified corresponding to two different spa types. The frequency of each spa type was variable, with 1 to 16 isolates per spa group type. Eighty-one spa types were represented by a single isolate, while 27 were represented by at least two isolates. The most prevalent spa type was t571, which accounted for 16 isolates (8.4% of all isolates). The 16 isolates were assigned to CC398 using the PCR targeting the CC398-specific variant of sau1-hsdS1. Twelve of these isolates were resistant to erythromycin (75%) (Table 1). We showed that ermT gene was the most common mechanism responsible for erythromycin resistance in these isolates, with 11 of the 12 strains positive for the ermT specific PCR (92%). The other most prevalent spa types were t002 (n = 12, 6.3%), t008 (n = 9, 4.7%) and t084 (n = 8, 4.2%). Among MRSA isolates, t008 was the most frequently detected (n = 5). The other spa types detected in MRSA isolates were t024 (n = 2), t080 (attributed most likely to the European-ST80 CA MRSA lineage) (n = 1) and t15255 (n = 1). Fig 1 shows the prevalence of the spa types within our collection. Using the integrated BURP algorithm, the 108 spa types were divided into 15 spa clusters and 19 singleton spa types. The prevalence of each spa cluster is noted in Table 2. Eleven strains were excluded because the number of repeats was less than five. The four major clusters were spa CC t015, CC t012, CC t024 and CC t002 and accounted for 42.6% (n = 46) of the spa types and 53.4% (n = 102) of the strains. Spa CC t015 was the most frequently detected for strains associated with infective endocarditis. Spa CC t015 was most prevalent for strains isolated in bone and joint infections, in skin and soft tissue infections and in infections of intravascular devices. Spa CC t012 was mostly detected for strains associated with respiratory infections and urinary tract infections.
Fig 1. Overall frequency of spa types.
Types found in ≤2 isolates are categorized as “others” (n = 92 of 108 total spa types).
Table 2. Frequencies of spa clusters.
CA: Community-acquired infections; HCA: Healthcare-associated infections; IE: Infective endocarditis; severe infections: intensive care unit admission, septic shock and death.
| Cluster (most common spa types) | No. of spa types (%) (n = 108) |
No. of strains (%) (n = 191) |
No. of CA strains (%) (n = 40) |
No. of HCA strains (%) (n = 151) |
No. of IE strains (%) (n = 8) |
No. of strains associated with severe infections (%) (n = 64) |
|---|---|---|---|---|---|---|
| t015 (t015, t050, t230, t571, t1451, t4896) | 22 (20.4) | 46 (24.1) | 12 (30) | 35 (23.2) | 3 (37.5) | 15 (23.4) |
| t012 (t012, t018, t021) | 10 (9.3) | 20 (10.5) | 4 (10) | 16 (10.6) | 0 | 11 (17.2) |
| t024 (t008, t024, t1171) | 7 (6.5) | 18 (9.4) | 1 (2.5) | 17 (11.3) | 1 (12.5) | 3 (4.7) |
| t002 (t002) | 7 (6.5) | 18 (9.4) | 4 (10) | 14 (9.3) | 1 (12.5) | 5 (7.8) |
| t189/3380 (t267) | 6 (5.6) | 8 (4.2) | 1 (2.5) | 7 (4.6) | 0 | 3 (4.7) |
| t216 (t216) | 5 (4.6) | 6 (3.1) | 0 | 4 (2.6) | 0 | 2 (3.1) |
| t084 (t084, t279, t346) | 5 (4.6) | 15 (7.9) | 5 (12.5) | 10 (6.6) | 0 | 5 (7.8) |
| t005 (t005) | 4 (3.7) | 7 (3.7) | 2 (5) | 5 (3.3) | 1 (12.5) | 3 (4.7) |
| t167 | 3 (2.8) | 3 (1.6) | 0 | 2 (1.3) | 0 | 2 (3.1) |
| t127 (t177) | 3 (2.8) | 4 (2.1) | 1 (2.5) | 3 (2) | 0 | 2 (3.1) |
| Other spa types | 36 (33.3) | 46 (24.1) | 10 (25) | 38 (25.2) | 2 (25) | 13 (20.3) |
Patient characteristics
As mentioned above, 199 patients with S. aureus bacteremia were included. The main characteristics of patients are shown in Table 3.
Table 3. Main characteristics of the 199 patients with S. aureus bacteremia and of the 16 patients infected with spa t571 strains.
| n (%) | t571 (n = 16) | ||
|---|---|---|---|
| Median age (years) | 48 (range 0–95) | 44 (range 0–87) | |
| Male gender | 131 (65.8) | 10 (62.5) | |
| Care unit |
Medical | 145 (72.9) | 12 (75) |
| Surgical | 28 (14.1) | 2 (12.5) | |
| Intensive care | 26 (13.1) | 2 (12.5) | |
| Renal failure | 49 (24.6) | 3 (18.8) | |
| Renal failure with hemodialysis | 14 (7.0) | 1 (6.3) | |
| Diabetes mellitus | 50 (25.1) | 5 (31.3) | |
| Immunosuppression | 56 (28.1) | 5 (31.3) | |
| Associated infection site | Vascular foreign body | 63 (31.7) | 9 (56.3) |
| Skin and soft tissue | 36 (18.1) | 1 (6.3) | |
| Bone and joint | 32 (16.1) | 3 (18.8) | |
| Lung | 19 (9.5) | 1 (6.3) | |
| Urinary tract | 5 (2.5) | 1 (6.3) | |
| Infective endocarditis | 8 (4) | 0 | |
| Severe infectionsa | 68 (34.2) | 4 (25) | |
| Presumed setting of acquired infection | Community | 41 (20.6) | 2 (12.5) |
| Healthcare-associated | 158 (79.4) | 14 (87.5) | |
| Healthcare-associated nosocomial | 105 (52.8) | 6 (37.5) | |
| Healthcare-associated non nosocomial | 53 (26.6) | 8 (50) | |
aIntensive care unit admission, septic shock and death.
Most patients had severe comorbidities. Renal failure was reported in 24.6% of patients (n = 49). Immunosuppression was present in 28.1% of patients (n = 56). Only 4 patients (2%) were past or current injecting drug users. A deep focus of infection was detected in 86.9% of patients (n = 173). The most frequent deep focus of infection was an invasive vascular device, followed by skin and soft tissue and bone and joint localizations. Echocardiography (transthoracic or transoesophageal) was performed in 67.3% of patients and definite IE was diagnosed in 4% of patients (n = 8). Severe infections involving septic shock, admission to the intensive care unit and/or death concerned 68 patients (34.2%). The mortality rate was 21.6%.
Spa t571 bacteremia were associated with an infected vascular foreign body with 56.3% of the cases, which explain why 87.5% of these bacteremia are healthcare-associated infections.
Discussion
In 2013, a large increase in S. aureus, mainly methicillin-susceptible, bacteremia was observed in our University Hospital compared to previous years (2012). Here we aimed to characterize all S. aureus isolates associated with bacteremia in 2013 in order to determine whether this increased prevalence could be attributed to the diffusion of an epidemic strain. Clinical data were also collected so as to determine any unknown risk factors for S. aureus bacteremia and interpret spa types according to clinical status.
The incidence of healthcare-associated S. aureus bacteremia was high (79.4%), as observed by Le Moing et al. [24]. Therefore, it seems essential to improve the prevention of these infections. In Australia the implementation of non-specific infection prevention and control initiatives coincided with a significant reduction in the incidence of healthcare-associated S. aureus bacteremia [25]. Several studies have shown that universal decolonization may also be appropriate, especially in the intensive care units [26, 27].
Only 8 cases of IE were noted in our population (4%). As echocardiography (transthoracic or transoesophageal) was not performed in 32.7% of patients, this estimation is probably conservative. In studies with more systematic use of echocardiography, the proportion of IE was higher with 15.6%, 22% and 19% reported respectively by Le Moing et al., Rasmussen et al. and Vos et al. [24, 28, 29]. This observation suggests the importance of actively searching for IE by performing transthoracic or transoesophageal echocardiography in cases of S. aureus bacteremia, as it is recommended in the 2009 European guidelines on IE. As no change in the population profile was clearly manifest compared to similar cohort studies conducted before 2013 [30–32], it seemed useful to study the genotype of S. aureus strains by spa typing. In our study, spa t571 accounts for 8.4% of our isolates and is the predominant spa type. The assignation of spa t571 to CC398 has been repeatedly observed [13, 14, 15, 33]. In our study, 100% of spa t571 strains belong to the CC398. Recently, Valentin-Domelier et al. highlighted the emergence of MSSA ST398 in bacteremia in the “Centre” region of France (2.5 million inhabitants), as has been reported worldwide [6, 34]. The first cases of ST398 bacteremia in French patients living in animal free environments were reported in 2007. The ST398 clone accounted for 15% of MSSA isolates responsible for bacteremia in 2015 whereas it had never been detected in bacteremia cases before 2007 [35]. MSSA CC398 isolates, harboring distinct characteristics from the livestock-associated multiresistant highly pathogenic clone, seem to be associated with high mortality in bacteremia and easy transmissibility [14, 36]. We note that ST398-spa t571 strains described so far exhibited the macrolide-lincosamide-streptogramin B resistance gene ermT [13]. In our study, 75% of the spa t571 strains were erythromycin-resistant and we demonstrated that the ermT gene conferred this resistance for 92% of the strains. This marker of the human clade [15] suggests that all CC398 MSSA likely correspond to the ancestral human population. Two other evidences argue in favor of this conclusion: theses strains are all MSSA and they are not resistant to tetracycline (a marker of LA-MRSA). The predominance of the clone spa t571 implies that our University Hospital is probably concerned by the spread of this clone. This is not the case for all European countries, e.g. in Germany where MSSA spa t571 constituted only 0.14% of German isolates from infections in human between 2006 and 2012 [15].
In contrast to the clustering usually noted with MRSA strains, the majority of our isolates, which were methicillin susceptible, showed broad genetic diversity. Similar MSSA heterogeneity has been reported across Europe and in the USA [37, 38]. Two of the most frequent MSSA clones observed in our study (spa t002 and t008) were equivalent to MRSA strains and also to other commonly circulating ones in Europe [37]. These data suggest that the spread of S. aureus through hospitals and the community is related more to the characteristics of some clones than to methicillin resistance [39].
Spa types were equally distributed irrespective of the seriousness of the infection. Likewise, no specific spa type or clonal complex was implicated when cases of IE were considered. In the study by Nienaber et al., MSSA IE isolates were significantly more likely to be CC30 [40]. This result was not verified in our study, since none of the 8 CC30 were from IE cases. Tristan et al. also described a large diversity of S. aureus clonal complexes implicated in IE [41]. Feil et al. reported the molecular characteristics of 334 S. aureus and noted no link between MLST type and capability of triggering an infection [42]. Feil et al. explained their results by a weak link between clonality and virulence factors. In other words, isolates within the same lineage may be distinct due to their virulence factors and therefore their capability of infecting the patient.
Our study was not without limitations. First, it would have been interesting to characterize S. aureus strains found in 2012 in order to compare them to those of 2013 and distinguish any genetic differences particularly as the rate of S. aureus bacteremia was lower in 2012; however, they were not available. Furthermore, given that the type of molecular characterisation of strains concerns only a small part of the genome, our results give only a general overview of the genetic structure of our strain collection, albeit with a well-validated method [19]. Despite spa typing has been shown to be highly concordant with MLST [16, 17, 43, 44], spa CC t015 groups two distinct STs; spa type t015 has been related to ST45 (according to RIDOM database), while t571 is associated with ST398. One explanation given by Nübel et al. is that the sequence identity between spa types in different STs probably reflects the result of repeated convergent evolution of spa sequences on multiple occasions [45]. Due to the limitation on spa typing, the question of t571 attributed to CC398 has been solved by performing a specific PCR. Finally, the impact of the host’s genetic background has not yet been studied.
In conclusion, our results indicated that S. aureus spa t571 might be growing cause of bloodstream infections in our hospital. These results are concordant with other findings showing that S. aureus CC398, especially spa type t571 can cause invasive infections in humans. These strains appear to be highly receptive to horizontal gene transfer and acquisition of genetic elements leading to virulence and antibiotic resistance could occur. Therefore, careful monitoring of the evolution and epidemiology of spa t571 bacteremia is necessary. Further to this study, an infection control program of bacteremia was set up in Grenoble-Alpes University Hospital. Catheter related bacteremia are specifically targeted during weekly meeting gathering infectious disease specialists, bacteriologists and infection control specialists. The supervision is organized from the results of blood culture, in a prospective and continuous way over all year. The interpretation of these data constitutes the first stage of the follow-up of this infection, allowing the discussion of prevention measures.
Acknowledgments
We thank Alison Foote PhD (Grenoble-Alpes University Hospital) for critically editing the manuscript and Patricia Martins Simões for her expertise in spa typing.
Data Availability
All relevant data are within the paper.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Laupland KB, Lyytikäinen O, Søgaard M, Kennedy KJ, Knudsen JD, Ostergaard C, et al. The changing epidemiology of Staphylococcus aureus bloodstream infection: a multinational population-based surveillance study. Clin Microbiol Infect Dis. 2013;19:465–71. [DOI] [PubMed] [Google Scholar]
- 2.Asgeirsson H, Gudlaugsson O, Kristinsson KG, Heiddal S, Kristjansson M. Staphylococcus aureus bacteraemia in Iceland, 1995–2008: changing incidence and mortality. Clin Microbiol Infect Dis. 2011;17:513–8. [DOI] [PubMed] [Google Scholar]
- 3.Lyytikäinen O, Ruotsalainen E, Järvinen A, Valtonen V, Ruutu P. Trends and outcome of nosocomial and community-acquired bloodstream infections due to Staphylococcus aureus in Finland, 1995–2001. Eur J Clin Microbiol Infect Dis. 2005;24:399–404. 10.1007/s10096-005-1345-3 [DOI] [PubMed] [Google Scholar]
- 4.Uslan DZ, Crane SJ, Steckelberg JM, Cockerill FR, St Sauver JL, Wilson WR, et al. Age- and sex-associated trends in bloodstream infection: a population-based study in Olmsted County, Minnesota. Arch Intern Med. 2007;167:834–9. 10.1001/archinte.167.8.834 [DOI] [PubMed] [Google Scholar]
- 5.Vogel M, Schmitz RPH, Hagel S, Pletz MW, Gagelmann N, Scherag A, et al. Infectious disease consultation for Staphylococcus aureus bacteremia—A systematic review and meta-analysis. J Infect. 2016;72:19–28. 10.1016/j.jinf.2015.09.037 [DOI] [PubMed] [Google Scholar]
- 6.Valentin-Domelier A-S, Girard M, Bertrand X, Violette J, François P, Donnio P-Y, et al. Methicillin-susceptible ST398 Staphylococcus aureus responsible for bloodstream infections: an emerging human-adapted subclone? PloS One. 2011;6:e28369 10.1371/journal.pone.0028369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharma M, Nunez-Garcia J, Kearns AM, Doumith M, Butaye PR, Argudín MA, et al. Livestock-Associated Methicillin Resistant Staphylococcus aureus (LA-MRSA) Clonal Complex (CC) 398 Isolated from UK Animals belong to European Lineages. Front Microbiol. 2016;7:1741 10.3389/fmicb.2016.01741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grøntvedt CA, Elstrøm P, Stegger M, Skov RL, Skytt Andersen P, Larssen KW, et al. Methicillin-Resistant Staphylococcus aureus CC398 in Humans and Pigs in Norway: A “One Health” Perspective on Introduction and Transmission. Clin Infect Dis. 2016;63:1431–8. 10.1093/cid/ciw552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Becker K, Ballhausen B, Kahl BC, Köck R. The clinical impact of livestock-associated methicillin-resistant Staphylococcus aureus of the clonal complex 398 for humans. Vet Microbiol. 2017;200:33–8. 10.1016/j.vetmic.2015.11.013 [DOI] [PubMed] [Google Scholar]
- 10.Salmenlinna S, Lyytikäinen O, Vainio A, Myllyniemi AL, Raulo S, Kanerva M, et al. Human cases of methicillin-resistant Staphylococcus aureus CC398, Finland. Emerg Infect Dis. 2010;16:1626–9. 10.3201/eid1610.091571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Price LB, Stegger M, Hasman H, Aziz M, Larsen J, Andersen PS, et al. Staphylococcus aureus CC398: host adaptation and emergence of methicillin resistance in livestock. mBio. 2012;3(1). pii: e00305-11. 10.1128/mBio.00305-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Mee-Marquet N, Corvaglia AR, Valentin AS, Hernandez D, Bertrand X, Girard M, et al. Analysis of prophages harbored by the human-adapted subpopulation of Staphylococcus aureus CC398. Infect Genet Evol. 2013;18:299–308. 10.1016/j.meegid.2013.06.009 [DOI] [PubMed] [Google Scholar]
- 13.Vandendriessche S, Kadlec K, Schwarz S, Denis O. Methicillin-susceptible Staphylococcus aureus ST398-t571 harbouring the macrolide-lincosamide-streptogramin B resistance gene erm(T) in Belgian hospitals. J Antimicrob Chemother. 2011;66:2455–9. 10.1093/jac/dkr348 [DOI] [PubMed] [Google Scholar]
- 14.David MZ, Siegel J, Lowy FD, Zychowski D, Taylor A, Lee CJ, et al. Asymptomatic carriage of sequence type 398, spa type t571 methicillin-susceptible Staphylococcus aureus in an urban jail: a newly emerging, transmissible pathogenic strain. J Clin Microbiol. 2013;51:2443–7. 10.1128/JCM.01057-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cuny C, Layer F, Köck R, Werner G, Witte W. Methicillin susceptible Staphylococcus aureus (MSSA) of clonal complex CC398, t571 from infections in humans are still rare in Germany. PloS One. 2013;8(12):e83165 15. 10.1371/journal.pone.0083165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koreen L, Ramaswamy SV, Graviss EA, Naidich S, Musser JM, Kreiswirth BN. spa typing method for discriminating among Staphylococcus aureus isolates: implications for use of a single marker to detect genetic micro- and macrovariation. J Clin Microbiol. 2004;42:792–9. 10.1128/JCM.42.2.792-799.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strommenger B, Braulke C, Heuck D, Schmidt C, Pasemann B, Nübel U, et al. spa Typing of Staphylococcus aureus as a frontline tool in epidemiological typing. J Clin Microbiol. 2008;46:574–81. 10.1128/JCM.01599-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li JS, Sexton DJ, Mick N, Nettles R, Fowler VG, Ryan T, et al. 2000. Proposed modifications to the Duke criteria for the diagnosis of infective endocarditis. Clin Infect Dis. 2000;30:633–8. 10.1086/313753 [DOI] [PubMed] [Google Scholar]
- 19.Shopsin B, Gomez M, Montgomery SO, Smith DH, Waddington M, Dodge DE, et al. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J Clin Microbiol. 1999;37:3556–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mellmann A, Weniger T, Berssenbrügge C, Keckevoet U, Friedrich AW, Harmsen D, et al. Characterization of clonal relatedness among the natural population of Staphylococcus aureus strains by using spa sequence typing and the BURP (based upon repeat patterns) algorithm. J Clin Microbiol. 2008;46:2805–8. 10.1128/JCM.00071-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harmsen D, Claus H, Witte W, Rothgänger J, Claus H, Turnwald D, et al. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol. 2003;41:5442–8. 10.1128/JCM.41.12.5442-5448.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stegger M, Lindsay JA, Moodley A, Skov R, Broens EM, Guardabassi L. Rapid PCR detection of Staphylococcus aureus clonal complex CC398 by targeting the restriction-modification system carrying sau1-hsdS1. J Clin Microbiol. 2011;49:732–4. 10.1128/JCM.01970-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chroboczek T, Boisset S, Rasigade JP, Tristan A, Bes M, Meugnier H, et al. Clonal complex 398 methicillin susceptible Staphylococcus aureus: a frequent unspecialized human pathogen with specific phenotypic and genotypic characteristics. PloS One. 2013;8:e68462 10.1371/journal.pone.0068462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le Moing V, Alla F, Doco-Lecompte T, Delahaye F, Piroth L, Chirouze C, et al. Staphylococcus aureus Bloodstream Infection and Endocarditis—A Prospective Cohort Study. PloS One. 2015;10:e0127385 10.1371/journal.pone.0127385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitchell BG, Collignon PJ, McCann R, Wilkinson IJ, Wells A. A major reduction in hospital-onset Staphylococcus aureus bacteremia in Australia-12 years of progress: an observational study. Clin Infect Dis. 2014;59:969–75. 10.1093/cid/ciu508 [DOI] [PubMed] [Google Scholar]
- 26.Huang SS, Septimus E, Kleinman K, Moody J, Hickok J, Avery TR, et al. Targeted versus universal decolonization to prevent ICU infection. N Engl J Med. 2013;368:2255–65. 10.1056/NEJMoa1207290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Climo MW, Yokoe DS, Warren DK, Perl TM, Bolon M, Herwaldt LA, et al. Effect of daily chlorhexidine bathing on hospital-acquired infection. N Engl J Med. 2013;368:533–42. 10.1056/NEJMoa1113849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rasmussen RV, Høst U, Arpi M, Hassager C, Johansen HK, Korup E, et al. Prevalence of infective endocarditis in patients with Staphylococcus aureus bacteraemia: the value of screening with echocardiography. Eur J Echocardiogr. 2011;12:414–20. 10.1093/ejechocard/jer023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vos FJ, Bleeker-Rovers CP, Sturm PD, Krabbe PFM, van Dijk APJ, Oyen WJG, et al. Endocarditis: effects of routine echocardiography during Gram-positive bacteraemia. Neth J Med. 2011;69:335–40. [PubMed] [Google Scholar]
- 30.Forsblom E, Ruotsalainen E, Mölkänen T, Ollgren J, Lyytikäinen O, Järvinen A. Predisposing factors, disease progression and outcome in 430 prospectively followed patients of healthcare- and community-associated Staphylococcus aureus bacteraemia. J Hosp Infect. 2011;78:102–7. 10.1016/j.jhin.2011.03.010 [DOI] [PubMed] [Google Scholar]
- 31.Jacobsson G, Dashti S, Wahlberg T, Andersson R. The epidemiology of and risk factors for invasive Staphylococcus aureus infections in western Sweden. Scand J Infect Dis. 2007;39:6–13. 10.1080/00365540600810026 [DOI] [PubMed] [Google Scholar]
- 32.Bassetti M, Trecarichi EM, Mesini A, Spanu T, Giacobbe DR, Rossi M, et al. Risk factors and mortality of healthcare-associated and community-acquired Staphylococcus aureus bacteraemia. Clin Microbiol Infect. 2012;18:862–9. 10.1111/j.1469-0691.2011.03679.x [DOI] [PubMed] [Google Scholar]
- 33.van Wamel WJB, Hansenová Manásková S, Fluit AC, Verbrugh H, de Neeling AJ, van Duijkeren E, et al. Short term micro-evolution and PCR-detection of methicillin-resistant and -susceptible Staphylococcus aureus sequence type 398. Eur J Clin Microbiol Infect Dis. 2010;29:119–22. 10.1007/s10096-009-0816-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Verkade E, Bergmans AMC, Budding AE, van Belkum A, Savelkoul P, Buiting AG, et al. Recent emergence of Staphylococcus aureus clonal complex 398 in human blood cultures. PloS One. 2012;7:e41855 10.1371/journal.pone.0041855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Diene SM, Corvaglia AR, François P, van der Mee-Marquet N, Regional Infection Control Group of the Center Region. Prophages and adaptation of Staphylococcus aureus ST398 to the human clinic. BMC Genomics. 2017;18:133 10.1186/s12864-017-3516-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bouiller K, Gbaguidi-Haore H, Hocquet D, Cholley P, Bertrand X, Chirouze C. Clonal complex 398 methicillin-susceptible Staphylococcus aureus bloodstream infections are associated with high mortality. Clin Microbiol Infect. 2016;22:451–5. 10.1016/j.cmi.2016.01.018 [DOI] [PubMed] [Google Scholar]
- 37.Grundmann H, Aanensen DM, van den Wijngaard CC, Spratt BG, Harmsen D, Friedrich AW, et al. Geographic distribution of Staphylococcus aureus causing invasive infections in Europe: a molecular-epidemiological analysis. PLoS Med. 2010;7:e1000215 10.1371/journal.pmed.1000215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miko BA, Hafer CA, Lee CJ, Sullivan SB, Hackel MAM, Johnson BM, et al. Molecular characterization of methicillin-susceptible Staphylococcus aureus clinical isolates in the United States, 2004 to 2010. J Clin Microbiol. 2013;51:874–9. 10.1128/JCM.00923-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Orscheln RC, Hunstad DA, Fritz SA, Loughman JA, Mitchell K, Storch EK, et al. Contribution of genetically restricted, methicillin-susceptible strains to the ongoing epidemic of community-acquired Staphylococcus aureus infections. Clin Infect Dis. 2009;49:536–42. 10.1086/600881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nienaber JJC, Sharma Kuinkel BK, Clarke-Pearson M, Lamlertthon S, Park L, Rude TH, et al. Methicillin-susceptible Staphylococcus aureus endocarditis isolates are associated with clonal complex 30 genotype and a distinct repertoire of enterotoxins and adhesins. J Infect Dis. 2011;204:704–13. 10.1093/infdis/jir389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tristan A, Rasigade J-P, Ruizendaal E, Laurent F, Bes M, Meugnier H, et al. Rise of CC398 lineage of Staphylococcus aureus among Infective endocarditis isolates revealed by two consecutive population-based studies in France. PloS One. 2012;7:e51172 10.1371/journal.pone.0051172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feil EJ, Cooper JE, Grundmann H, Robinson DA, Enright MC, Berendt T, et al. How clonal is Staphylococcus aureus? J Bacteriol. 2003;185:3307–16. 10.1128/JB.185.11.3307-3316.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hallin M, Deplano A, Denis O, De Mendonça R, De Ryck R, Struelens MJ. Validation of pulsed-field gel electrophoresis and spa typing for long-term, nationwide epidemiological surveillance studies of Staphylococcus aureus infections. J Clin Microbiol. 2007;45:127–33. 10.1128/JCM.01866-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O'Hara FP, Suaya JA, Ray GT, Baxter R, Brown ML, Mera RM, et al. spa Typing and Multilocus Sequence Typing Show Comparable Performance in a Macroepidemiologic Study of Staphylococcus aureus in the United States. PloS One. 2012;7:e51172 10.1371/journal.pone.0051172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nübel U, Roumagnac P, Feldkamp M, Song JH, Ko KS, Huang YC, et al. Frequent emergence and limited geographic dispersal of methicillin-resistant Staphylococcus aureus. Proc Natl Acad Sci U S A. 2008;105:14130–5. 10.1073/pnas.0804178105 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.