Abstract
Background
A large portion of intronic and intergenic space in our genome consists of repeated sequences. One of the most prevalent is the Long INterspersed Element-1 (LINE-1, L1) mobile DNA. LINE-1 is rightly receiving increasing interest as a cancer biomarker.
Content
Intact LINE-1 elements are self-propagating. They code for RNA and proteins which function to make more copies of the genomic element. Our current understanding is that this process is repressed in most normal cells, but that LINE-1 expression is a hallmark of many types of malignancy. Here, we will consider features of cancer cells when cellular defense mechanisms repressing LINE-1 go awry. We will review evidence that genomic LINE-1 methylation, LINE-1-encoded RNAs, and LINE-1 open reading frame 1 protein (ORF1p) may be useful in cancer diagnosis.
Summary
The repetitive and variable nature of LINE-1 DNA sequences pose unique challenges to studying them, but recent advances in reagents and next generation sequencing present opportunities to characterize LINE-1 expression and activity in cancers, and identify clinical applications.
Introduction
A very small portion - about one percent - of our DNA is recognizable as protein-coding gene exons. Much of the intervening sequence is intronic and intergenic space littered with the remains of mobile DNAs. These are known as transposable elements (TEs), and their ability to copy themselves over time has shaped much of the modern human genome. We carry hundreds of thousands of copies of these sequences scattered as interspersed repeats. Collectively, they make up half of our DNA (1, 2).
TEs exist as transposons, which operate via a “cut-and-paste” mechanism, or retrotransposons, which propagate by a “copy-and-paste” mechanism known as retrotransposition. Retrotransposons use an RNA intermediate expressed from a genomic locus which is then reverse transcribed by retrotransposon-encoded proteins to make a new genomic insertion. Retrotransposons are classified as long terminal repeat (LTR) or non-LTR elements. They can furthermore be described as autonomous or non-autonomous depending on whether they encode the protein machinery necessary for retrotransposition. The only autonomous, active elements in humans are non-LTR retrotransposons known as Long INterspersed Elements (LINEs). LINEs have an evolutionary history that predates humans by hundreds of millions of years. In aggregate, the human genome is 17% LINE-1 sequence and 5–6% LINE-2 and LINE-3 sequences (1, 3). All retrotransposition today is driven by LINE-1 (L1), the only autonomous element in humans, which remains the focus of this review (4–7).
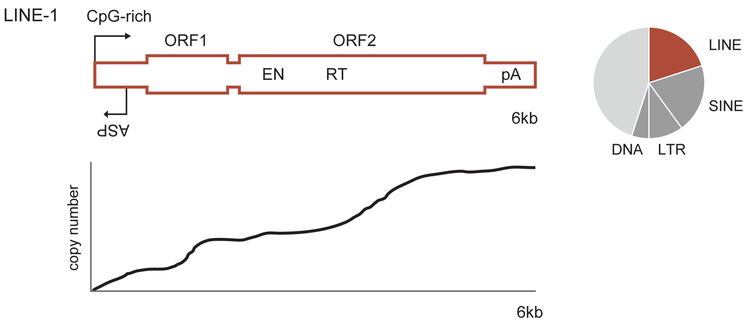
An intact LINE-1 sequence measures approximately 6 kilobases in length and encodes two well-recognized proteins, open reading frame 1 protein (ORF1p) and open reading frame 2 protein (ORF2p)(Figure 1). LINE-1 also has an antisense promoter (ASP) activity that can initiate fusion transcripts (8–10) and aberrant coding sequence (ORF0) (11) in the opposing direction.
Figure 1.
A schematic of a LINE-1 element. A full-length LINE-1 is 6 kilobases (kb) in length. It includes a CpG-rich bidirectional promoter and two open reading frames for ORF1p and ORF2p proteins. The element is illustrated as a block with widened open reading frames. ASP=antisense promoter; EN=endonuclease, RT=reverse transcriptase, pA=polyA tail. Beneath this schematic is a plot showing the relative genomic copy number of L1Hs sequences in the human genome as a function of position along the length of the 6kb consensus sequence. There are relatively more copies of the 3’ end of the element because many copies are 5’ truncated at the time of their integration. The length of a LINE-1 is stable after insertion with the exception of the polyA portion. The pie chart to the right illustrates the percentage of the human genome comprised of repetitive elements. LINE=Long INterspersed Element; SINE=Short INterspersed Element; LTR=Long Terminal Repeat; DNA=DNA transposons (‘cut-and-paste’ transposons).
ORF1p trimerizes to form an RNA binding complex required for LINE-1 transposition (12–14). ORF2p encodes two enzymatic activities also essential for retrotransposition, an endonuclease and a reverse transcriptase (15–17). ORF2p reverse transcribes new genomic DNA copies of LINE-1 from its RNA and is co-opted to copy other repeats, namely the Alu Short INterspered Element (SINE)(18), and the SVA (SINE, VNTR, Alu) composite elements (19, 20).
Each individual inherits a small complement of full-length, retrotransposition-active or ‘hot’ LINE-1 loci (21–24). The specific loci vary from person to person. The prevailing hypothesis in the field is that these potentially protein-coding LINE-1 are then kept in check by a series of host defenses to maintain genome integrity. In this article, we will briefly reference aspects of normal LINE-1 control and contrast this with LINE-1 expression in malignancy.
Mechanisms of LINE-1 Repression
Expression of LINE-1 sequences and subsequent steps in retrotransposition are repressed by host factors. In the germ line, the piRNA pathway is critical in establishing repressive DNA methylation patterns (reviewed in (25)). In somatic cells, the SWI/SNF2-related, matrix-associated, actin-dependent regulator of chromatin, subfamily A, member 6 (SMARCA6) helicase (26, 27) and p53 (28) feature prominently.
Once expressed, LINE-1 transcripts and proteins can be altered in their activities. Mechanisms include RNAi pathways (29, 30), nonsense mediated decay via up-frame shift 1 (UPF1) (31), and antiviral proteins like apolipoprotein B mRNA editing enzyme catalytic polypeptide 3 (APOBEC3) cytidine deaminase (32), Moloney leukemia virus 10 (MOV10) (33, 34), and zinc-finger antiviral protein (ZAP) (35, 36).
LINE-1 Hypomethylation in Cancer
Full-length LINE-1 transcription is driven by a CpG dinucleotide-rich internal promoter. Many genomic LINE-1 sequences are 5’ truncated at the time of genomic integration, and so have lost their promoter. Many intact promoter sequences are found throughout the genome, however, and CpG methylation of LINE-1 is used by many as a surrogate marker of whole genome methylation levels (37). It is important to note that while many tumor suppressor gene promoters are methylated in tumors, whole genome methylation and LINE-1 methylation specifically tend to be reduced in malignancies.
LINE-1 methylation studies have been conducted in many of the most common lethal cancers, including (in order of mortality) lung cancer, colon and rectal cancers, breast cancer, prostate cancer, liver cancer, ovarian cancer, and esophageal cancer (Figure 2). In non-small cell lung cancer, LINE-1 promoter hypomethylation is common (38) and is associated with genomic instability (39) and poor prognosis (40). In colon cancer, LINE-1 hypomethylation appears to be an early event (41) also associated with poor outcomes (42, 43). It appears inversely correlated with microsatellite instability (44, 45). Interestingly, colon cancer patients whose tumors exhibit extremely low levels of LINE-1 methylation may be a clinically distinct group, with a tendency to present at a younger age (46). LINE-1 hypomethylation is more pronounced in colon cancer liver metastases compared to matched primary tumors (47). In breast cancer, LINE-1 hypomethylation has been reported in preneoplastic phases of epithelial atypia with persistently low LINE-1 promoter methylation seen in in situ and invasive lesions (48). It has also been associated with decreased overall survival and drug resistance in younger patients (49). In prostate cancers, LINE-1 hypomethylation is also reported, particularly in association with chromosome 8 abnormalities (50); it appears more pronounced in metastatic lesions than in primary tumors (51). In hepatocellular carcinoma, several groups have associated LINE-1 hypomethylation with poor clinical outcomes, including disease recurrence after resection (52–54). In epithelial ovarian cancers, LINE-1 hypomethylation is correlated with more aggressive histology, poorer progression-free intervals, and poorer survival (55). Finally, in esophageal squamous cell carcinomas, LINE-1 hypomethylation is also recognized and associated with poorer survival (56). Evidence of aberrant LINE-1 hypomethylation is less frequently reported for hematolymphoid neoplasias.
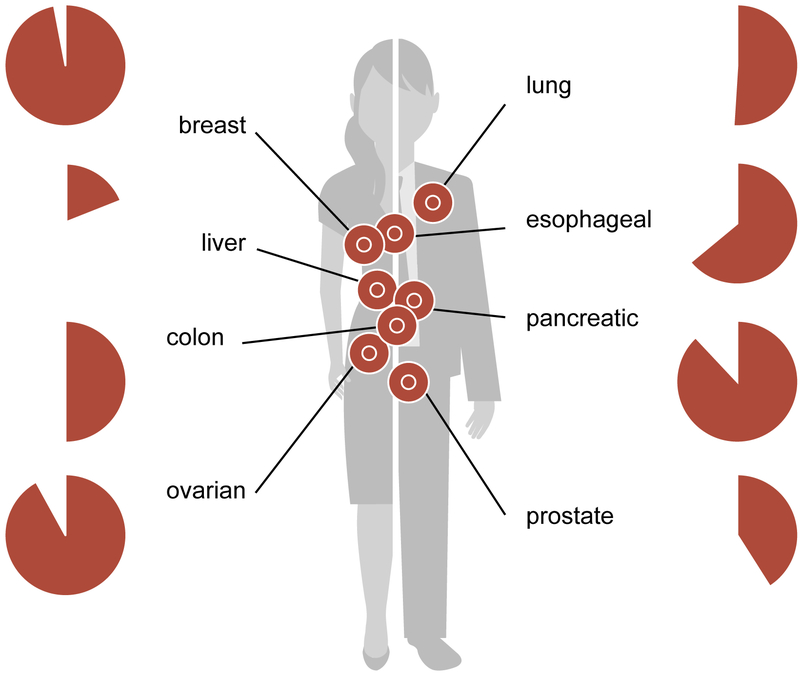
Figure 2.
LINE-1 ORF1p expression in cancer. Diagram of tissue types with known LINE-1 positive cancers. Hypomethylation of LINE-1 promoters has been described for tumors originating from all of these sites. The proportion of cases with LINE-1 ORF1p expression detectable by sensitive immunohistochemistry (IHC) is shown next to each tissue of origin.
A meta-analysis of LINE-1 hypomethylation as a marker for cancer risk revealed that tissue-based DNA assays fairly consistently reveal LINE-1 hypomethylation in cancers compared to controls (57). In contrast, LINE-1 methylation status in blood is apparently not a marker of cancer risk across 19 studies included in the meta-analysis, suggesting that direct assays of malignant tissues are more sensitive to these changes (57).
Many of the studies cited above compare aggregate normal and tumor LINE-1 methylation levels without delineating the specific LINE-1 loci driving these changes or accounting for differences in the complement of inherited LINE-1 sequences between individuals. However, very recent work suggests that more personalized approaches are likely to yield new insight, and add nuance to the overly simplistic model that relating cancerous histopathology or poor outcomes with LINE-1 hypomethylation. Emma Scott and her colleagues in Scott Devine’s laboratory recently traced somatic retrotransposition events in a case of colon cancer to three inherited, full-length LINE-1 elements. One of these hot elements, a LINE-1 on chromosome 17, appears responsible for the cancer as it generated a 1.4 kb insertion in the adenomatous polyposis coli (APC) gene (58). This is a classic ‘driver mutation’ positioned to inactivate the tumor suppressor. The ‘parent’ or ‘source’ element is a polymorphic variant in human populations, which can be methylated appropriately – it is methylated in reference DNA from lymphoblastoid cell lines. However, its promoter sequence was largely unmethylated in both normal colon and cancerous tissue from this patient. So, while some LINE-1 may acquire hypomethylation in the permissive environment of a cancer cell, others appear to escape silencing in normal tissues and be poised to play important inciting roles in tumor pathogenesis.
LINE-1 RNA
LINE-1 and other interspersed repeat RNAs have been less well characterized than gene messenger RNAs (mRNAs) in cancer. They pose a challenge because there are hundreds of thousands of LINE-1 genomic loci that have the potential to be incorporated into larger transcripts. Indeed, most of these genomic copies are 5’ truncated, and it follows that their transcription will not be directed by the LINE-1 promoter, but rather exclusively by ‘read-through’ transcription. Most LINE-1 do not have intact ORFs, and roles of their RNAs are not well understood. LINE-1 RNA has long been recognized as a component of heterogeneous nuclear RNAs (59), and recent in situ hybridization studies demonstrate that repetitive RNAs, and 3’ LINE-1 RNA in particular, are long-lived components of chromatin (60). The abundant expression of these fragments of LINE-1 RNA has been seen across cancers (61).
LINE-1 RNA that is the intermediate for retrotransposition is encoded by the LINE-1 promoter and is the same length as a full-length genomic element, 6 kilobases (kb) (i.e., the so-called unit LINE-1 transcript (62)). Before advances in next generation sequencing, RNA expression directed specifically from the LINE-1 promoter could be most reliably detected by Northern blots so that the size of the resulting RNA could be assessed. This was how LINE-1 RNA was first identified in cytoplasmic fractions of Ntera2D1 teratocarcinoma cells (63), and it remains a valuable approach for experimentalists today (64). Despite caveats for interpreting their results, RNAse protection assays, RT-PCRs, and in situ hybridizations have also been used to infer unit LINE-1 expression. When these assays target the 5’ end of LINE-1, they are expected to be relatively more specific than when 3’ positioned probes and primers are used. Similarly, selecting for polyadenylated, cytoplasmic RNA and/or using assays specific for the sense strand of LINE-1 can further promote specificity for unit transcripts.
There is recent evidence reported by Claude Philippe and colleagues with Gaël Cristofari’s laboratory that active LINE-1 RNAs [L1Hs or L1(Ta)] can be traced to specific templating loci using next generation sequencing data. Their analysis involves integrating a combination of data types, including a genomic LINE-1 insertion map, RNA-seq reads, and chromatin immunoprecipitation (ChIP)-seq data (65). Actively transcribed LINE-1 loci have a two-part ‘signature’ : (i.) RNA-seq reads corresponding to ‘read-through’ transcription downstream (3’) of the polyA tail, as well as (ii.) histone H3K4 trimethylation, H3K27 acetylation, and RNA polymerase II ChIP-seq reads extending into upstream (5’) sequence. With these as proxies for LINE-1 expression, the group was able to identify a small number of transcribed LINE-1 loci in human cancer cell lines – about 5 to 15 elements appear responsible for most unit LINE-1 RNA expression in cells. These data are consistent with a model wherein cancer cells maintain LINE-1 repression at most full length loci, with only a handful of escaping elements with the capability of retrotransposition.
Long read sequencing may be useful for detecting unit LINE-1 RNAs as well as resolving RNA species transcribed from other genomic repeats. These long reads may incorporate unique flanking sequence, may also enable unequivocal mapping of full-length transcripts. Being able to accurately phase internal sequence variants of unit LINE-1 RNAs and relate these variants back to individual genomic loci (58) will represent an important advance.
ORF1p expression
Intact, full length LINE-1 sequences code for two proteins, ORF1p and ORF2p. Of these, expression of the first has been best characterized in human cancers. LINE-1 expression constructs in in vitro transfected cells produce ORF1p at 1,000- to 10,000-fold higher levels than ORF2p (31).
ORF1p (p40) is an RNA binding protein essential for retrotransposition (12–14, 66). Its crystal structure has been solved (67). Three ORF1p protein molecules intertwine throughout the length of their N-terminal coiled coil to form a homotrimeric complex. The central RNA recognition motifs (RRMs) (68) and C-terminal domains (CTDs) project outward from the coiled coil axis to form deep intervening clefts. These clefts are highly positively charged surfaces that likely interact with the backbone of single-stranded RNA.
At least two studies have sequenced the RNAs that interact with LINE-1 protein in HEK293T cells overexpressing exogenous LINE-1. Taylor et al. immunoprecipitated LINE-1 ribonucleoproteins using FLAG-tagged ORF1p or ORF2p and found that L1 RNA represented 8.3–10.3% and 18.0–28.2% of reads associated with ORF1p pulldown or ORF2p pulldown, respectively (31). There was also enrichment of U6 snRNA. Mandal and colleagues used photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP) followed by sequencing to discover that 22% of all ORF1p-associated RNAs were mRNAs with known pseudogenes (69). ORF1p also associated with small structured RNAs included spliceosomal and hY RNAs, in addition to LINE-1, Alu, and SVA RNA. It is not known whether ORF1p sequesters cellular RNAs within tumors or what impacts this may have on cancer cell biology.
The first antibody developed against ORF1p was a rabbit polyclonal reagent described in 1990 by Debra Leibold and colleagues in Thomas Fanning’s laboratory (70). The group reported detecting LINE-1 ORF1p in embryonal carcinoma cells (Ntera2), teratocarcinoma cells (2102Ep), and choriocarcinoma cells (JEG-3). Many of the first studies of ORF1p expression then focused on these highly expressing germ cell tumors. Gary Bratthauer also with Thomas Fanning surveyed primary adult testicular germ cell tumors and pediatric germ cell tumors by immunohistochemistry to find ~10% positive for L1Hs expression. All were epithelial with typical embryonal carcinoma or yolk sac tumor appearances (71, 72).
A larger study of pediatric malignant germ cell tumors (MGCT) was next conducted by the Children’s Oncology Group (COG), also using the same Fanning laboratory reagent, but with a more sensitive immunohistochemistry protocol. Using this method, the group, led by Xiao-Ou Shu found evidence for expression in all of the 162 MGCT cases assessed. They stratified these into cases that were strongly, moderately, and weakly immunoreactive for LINE-1 ORF1p and reported that strong expression of ORF1p was associated with poor differentiation, extragonadal sites of disease, and yolk sac tumor histologies (73).
Breast malignancies also received early recognition as LINE-1 ORF1p-expressing cancers. Bonnie and Harold Asch in collaboration with the Fanning laboratory reported that ORF1p expression could be detected by Western blot in both malignant and nonmalignant breast epithelium. Most (4/5) normal tissue samples recovered from reduction mammoplasties had low levels as compared with tumors. However, their work suggested that malignant cells produce more of the protein. They suggest that immunostaining intensity can serve as an indicator of malignancy; they report ORF1p reactivity in all cases of invasive cancer examined (12), whereas benign proliferative disease and normal tissues were weakly reactive and negative (74).
In 2010, Chris Harris and colleagues prepared rabbit polyclonal antibody against ORF1p and described a broader expression pattern in human tumors (75). The group reported positivity in 99% of breast cancers, but as well, expression in a significant proportion of bladder cancers, prostate cancers, colorectal cancers, ileal carcinoids, and pancreatic neuroendocrine tumors. Much of their work focused on breast cancers, where they reported nuclear localization of ORF1p in a subset of cases. Nuclear immunoreactivity was associated with increased incidence of local recurrence, distant metastases, and poorer overall survival.
We have since developed a mouse monoclonal antibody against ORF1p (76, 77). The reagent was raised against amino acids 35–44 of ORF1p (AAB60344.1, MENDFDELRE). This is a region close to the N-terminus of the protein, before the coiled-coil domain begins and where mouse and human LINE-1 sequences diverge. Using a combination of rabbit polyclonal antibody from Chris Harris and this mouse monoclonal antibody, Nemanja Rodić surveyed human cancers using tissue microarray immunostaining. We did not see nuclear staining in this study, but cytoplasmic immunoreactivity was very common, and either specific for malignant tissue or overexpressed in malignant cells as compared to adjacent normal. LINE-1 ORF1p immunoreactivity was seen in significant proportions of lung cancers (51%), esophageal cancers (64%), breast cancers (97%), liver cancers (19%), colon cancers (50%), ovarian cancers (92%), and prostate cancers (41%). These data are summarized in Figure 2.
There is no direct way to trace ORF1p protein expression back to individual LINE-1 genomic loci. Within an individual, multiple LINE-1 loci may contribute to protein expression, and between individuals, the LINE-1 loci contributing to protein expression need not be shared.
Several types of tumors have been shown to have genomic evidence of LINE-1 retrotransposition, implying that in addition to ORF1p, LINE-1 unit-length RNA and ORF2p are also expressed. In partnership with the Kazazian laboratory, we have demonstrated expression of ORF1p by immunohistochemistry in specific cases of pancreatic ductal adenocarcinomas (78), esophageal adenocarcinoma (79) and esophageal squamous cell carcinoma (80) with somatically-acquired genomic LINE-1 insertions.
Concluding Remarks
Once dismissed as obscure junk DNA, LINE-1 is now the focus of increasing studies in cancer biology and in the search for cancer markers. Genomic LINE-1 hypomethylation, increases in LINE-1 RNA transcription, and ORF1p accumulation seem to be features of malignant cells more so than adjacent normal tissues. Going forward, a better understanding of consequences of LINE-1 expression in cancer biology is critical in our view.
We note that the phenomenon is not uniform across tumor types or individual patients. This may reflect features inherent to the pathogenesis of each disease, p53 function for example, raising the possibility that LINE-1 expression will be useful for subclassification or prognosis. For instance, ORF1p expression positively correlates with TP53 deficiency and higher-grade lesions (76). Tumor cell type also matters. Aberrant LINE-1 ORF1p expression is a hallmark of epithelial tumors more than, for example, hematolymphoid malignancies. Finally, personalized approaches that interpret LINE-1 expression markers in light of the inherited complement of LINE-1 loci may become important.
Due to the complexity of LINE-1 genomic sequences and RNA species, we remind readers that biomarkers related to these should be regarded as uncoupled from LINE-1 protein expression. Depending on assay design, LINE-1 methylation or RNA expression may serve as surrogates of global or local chromatin status, whereas protein may be a more specific indicator of bona fide expression of the unit LINE-1 element.
Acknowledgments
The authors thank Alex Forrest-Hay for discussions. This work has been supported by the Burroughs Wellcome Trust (D.T.T.), K12CA087723 (D.T.T.), Affymetrix, Inc. (D.T.T.), the Warshaw Institute for Pancreatic Cancer Research (D.T.T.), and the Verville Family Pancreatic Cancer Research Fund (D.T.T.) as well as the Burroughs Wellcome Fund (K.H.B.), R01CA163705 (K.H.B.), R01GM103999 (K.H.B.), and the Systems Biology of Retrotransposition P50GM107632 (K.H.B.).
Footnotes
Conflict of Interest
D.T.T. receives sponsored research from Affymetrix, Inc. related to RNA in situ hybridization technologies. M.S.T. and K.H.B. license a mouse monoclonal antibody against LINE-1 ORF1p through the Johns Hopkins University School of Medicine Technology Transfer Office. This is sold by EMD Millipore (Billerica, Massachusetts, catalog #MABC1152).
References
- 1.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature 2001;409:860–921. [DOI] [PubMed] [Google Scholar]
- 2.de Koning AP, Gu W, Castoe TA, Batzer MA, Pollock DD. Repetitive elements may comprise over two-thirds of the human genome. PLoS genetics 2011;7:e1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nature reviews Genetics 2009;10:691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ostertag EM, Kazazian HH Jr. Biology of mammalian l1 retrotransposons. Annu Rev Genet 2001;35:501–38. [DOI] [PubMed] [Google Scholar]
- 5.Hancks DC, Kazazian HH Jr. Active human retrotransposons: Variation and disease. Curr Opin Genet Dev 2012;22:191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levin HL, Moran JV. Dynamic interactions between transposable elements and their hosts. Nature reviews Genetics 2011;12:615–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burns KH, Boeke JD. Human transposon tectonics. Cell 2012;149:740–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Criscione SW, Theodosakis N, Micevic G, Cornish TC, Burns KH, Neretti N, Rodic N. Genome-wide characterization of human l1 antisense promoter-driven transcripts. BMC genomics 2016;17:463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roman-Gomez J, Jimenez-Velasco A, Agirre X, Cervantes F, Sanchez J, Garate L, et al. Promoter hypomethylation of the line-1 retrotransposable elements activates sense/antisense transcription and marks the progression of chronic myeloid leukemia. Oncogene 2005;24:7213–23. [DOI] [PubMed] [Google Scholar]
- 10.Weber B, Kimhi S, Howard G, Eden A, Lyko F. Demethylation of a line-1 antisense promoter in the cmet locus impairs met signalling through induction of illegitimate transcription. Oncogene 2010;29:5775–84. [DOI] [PubMed] [Google Scholar]
- 11.Denli AM, Narvaiza I, Kerman BE, Pena M, Benner C, Marchetto MC, et al. Primate-specific orf0 contributes to retrotransposon-mediated diversity. Cell 2015;163:583–93. [DOI] [PubMed] [Google Scholar]
- 12.Hohjoh H, Singer MF. Cytoplasmic ribonucleoprotein complexes containing human line-1 protein and rna. The EMBO journal 1996;15:630–9. [PMC free article] [PubMed] [Google Scholar]
- 13.Martin SL, Branciforte D, Keller D, Bain DL. Trimeric structure for an essential protein in l1 retrotransposition. Proceedings of the National Academy of Sciences of the United States of America 2003;100:13815–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin SL, Cruceanu M, Branciforte D, Wai-Lun Li P, Kwok SC, Hodges RS, Williams MC. Line-1 retrotransposition requires the nucleic acid chaperone activity of the orf1 protein. Journal of molecular biology 2005;348:549–61. [DOI] [PubMed] [Google Scholar]
- 15.Feng Q, Moran JV, Kazazian HH Jr, Boeke JD. Human l1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell 1996;87:905–16. [DOI] [PubMed] [Google Scholar]
- 16.Cost GJ, Boeke JD. Targeting of human retrotransposon integration is directed by the specificity of the l1 endonuclease for regions of unusual DNA structure. Biochemistry 1998;37:18081–93. [DOI] [PubMed] [Google Scholar]
- 17.Mathias SL, Scott AF, Kazazian HH Jr., Boeke JD, Gabriel A. Reverse transcriptase encoded by a human transposable element. Science 1991;254:1808–10. [DOI] [PubMed] [Google Scholar]
- 18.Dewannieux M, Esnault C, Heidmann T. Line-mediated retrotransposition of marked alu sequences. Nat Genet 2003;35:41–8. [DOI] [PubMed] [Google Scholar]
- 19.Raiz J, Damert A, Chira S, Held U, Klawitter S, Hamdorf M, et al. The non-autonomous retrotransposon sva is trans-mobilized by the human line-1 protein machinery. Nucleic acids research 2012;40:1666–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hancks DC, Goodier JL, Mandal PK, Cheung LE, Kazazian HH Jr. Retrotransposition of marked sva elements by human l1s in cultured cells. Human molecular genetics 2011;20:3386–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Streva VA, Jordan VE, Linker S, Hedges DJ, Batzer MA, Deininger PL. Sequencing, identification and mapping of primed l1 elements (simple) reveals significant variation in full length l1 elements between individuals. BMC genomics 2015;16:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Macfarlane CM, Collier P, Rahbari R, Beck CR, Wagstaff JF, Igoe S, et al. Transduction-specific atlas reveals a cohort of highly active l1 retrotransposons in human populations. Human mutation 2013;34:974–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beck CR, Collier P, Macfarlane C, Malig M, Kidd JM, Eichler EE, et al. Line-1 retrotransposition activity in human genomes. Cell 2010;141:1159–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, Kazazian HH Jr. Hot l1s account for the bulk of retrotransposition in the human population. Proceedings of the National Academy of Sciences of the United States of America 2003;100:5280–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O’Donnell KA, Burns KH, Boeke JD. A descent into the nuage: The maelstrom of transposon control. Developmental cell 2008;15:179–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang J, Fan T, Yan Q, Zhu H, Fox S, Issaq HJ, et al. Lsh, an epigenetic guardian of repetitive elements. Nucleic acids research 2004;32:5019–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu W, McIntosh C, Lister R, Zhu I, Han Y, Ren J, et al. Genome-wide DNA methylation patterns in lsh mutant reveals de-repression of repeat elements and redundant epigenetic silencing pathways. Genome research 2014;24:1613–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wylie A, Jones AE, D’Brot A, Lu WJ, Kurtz P, Moran JV, et al. P53 genes function to restrain mobile elements. Genes & development 2016;30:64–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soifer HS, Zaragoza A, Peyvan M, Behlke MA, Rossi JJ. A potential role for rna interference in controlling the activity of the human line-1 retrotransposon. Nucleic acids research 2005;33:846–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang N, Kazazian HH Jr. L1 retrotransposition is suppressed by endogenously encoded small interfering rnas in human cultured cells. Nature structural & molecular biology 2006;13:763–71. [DOI] [PubMed] [Google Scholar]
- 31.Taylor MS, LaCava J, Mita P, Molloy KR, Huang CR, Li D, et al. Affinity proteomics reveals human host factors implicated in discrete stages of line-1 retrotransposition. Cell 2013;155:1034–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niewiadomska AM, Tian C, Tan L, Wang T, Sarkis PT, Yu XF. Differential inhibition of long interspersed element 1 by apobec3 does not correlate with high-molecular-mass-complex formation or p-body association. Journal of virology 2007;81:9577–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arjan-Odedra S, Swanson CM, Sherer NM, Wolinsky SM, Malim MH. Endogenous mov10 inhibits the retrotransposition of endogenous retroelements but not the replication of exogenous retroviruses. Retrovirology 2012;9:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li X, Zhang J, Jia R, Cheng V, Xu X, Qiao W, et al. The mov10 helicase inhibits line-1 mobility. The Journal of biological chemistry 2013;288:21148–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moldovan JB, Moran JV. The zinc-finger antiviral protein zap inhibits line and alu retrotransposition. PLoS genetics 2015;11:e1005121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goodier JL, Pereira GC, Cheung LE, Rose RJ, Kazazian HH Jr. The broad-spectrum antiviral protein zap restricts human retrotransposition. PLoS genetics 2015;11:e1005252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang AS, Estecio MR, Doshi K, Kondo Y, Tajara EH, Issa JP. A simple method for estimating global DNA methylation using bisulfite pcr of repetitive DNA elements. Nucleic acids research 2004;32:e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rauch TA, Zhong X, Wu X, Wang M, Kernstine KH, Wang Z, et al. High-resolution mapping of DNA hypermethylation and hypomethylation in lung cancer. Proceedings of the National Academy of Sciences of the United States of America 2008;105:252–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Daskalos A, Nikolaidis G, Xinarianos G, Savvari P, Cassidy A, Zakopoulou R, et al. Hypomethylation of retrotransposable elements correlates with genomic instability in non-small cell lung cancer. Int J Cancer 2009;124:81–7. [DOI] [PubMed] [Google Scholar]
- 40.Saito K, Kawakami K, Matsumoto I, Oda M, Watanabe G, Minamoto T. Long interspersed nuclear element 1 hypomethylation is a marker of poor prognosis in stage ia non-small cell lung cancer. Clin Cancer Res 2010;16:2418–26. [DOI] [PubMed] [Google Scholar]
- 41.Sunami E, de Maat M, Vu A, Turner RR, Hoon DS. Line-1 hypomethylation during primary colon cancer progression. PloS one 2011;6:e18884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ogino S, Nosho K, Kirkner GJ, Kawasaki T, Chan AT, Schernhammer ES, et al. A cohort study of tumoral line-1 hypomethylation and prognosis in colon cancer. J Natl Cancer Inst 2008;100:1734–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mima K, Nowak JA, Qian ZR, Cao Y, Song M, Masugi Y, et al. Tumor line-1 methylation level and colorectal cancer location in relation to patient survival. Oncotarget 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ogino S, Kawasaki T, Nosho K, Ohnishi M, Suemoto Y, Kirkner GJ, Fuchs CS. Line-1 hypomethylation is inversely associated with microsatellite instability and cpg island methylator phenotype in colorectal cancer. Int J Cancer 2008;122:2767–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Estecio MR, Gharibyan V, Shen L, Ibrahim AE, Doshi K, He R, et al. Line-1 hypomethylation in cancer is highly variable and inversely correlated with microsatellite instability. PloS one 2007;2:e399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baba Y, Huttenhower C, Nosho K, Tanaka N, Shima K, Hazra A, et al. Epigenomic diversity of colorectal cancer indicated by line-1 methylation in a database of 869 tumors. Mol Cancer 2010;9:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hur K, Cejas P, Feliu J, Moreno-Rubio J, Burgos E, Boland CR, Goel A. Hypomethylation of long interspersed nuclear element-1 (line-1) leads to activation of proto-oncogenes in human colorectal cancer metastasis. Gut 2014;63:635–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park SY, Seo AN, Jung HY, Gwak JM, Jung N, Cho NY, Kang GH. Alu and line-1 hypomethylation is associated with her2 enriched subtype of breast cancer. PloS one 2014;9:e100429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Hoesel AQ, van de Velde CJ, Kuppen PJ, Liefers GJ, Putter H, Sato Y, et al. Hypomethylation of line-1 in primary tumor has poor prognosis in young breast cancer patients: A retrospective cohort study. Breast Cancer Res Treat 2012;134:1103–14. [DOI] [PubMed] [Google Scholar]
- 50.Schulz WA, Elo JP, Florl AR, Pennanen S, Santourlidis S, Engers R, et al. Genomewide DNA hypomethylation is associated with alterations on chromosome 8 in prostate carcinoma. Genes Chromosomes Cancer 2002;35:58–65. [DOI] [PubMed] [Google Scholar]
- 51.Yegnasubramanian S, Haffner MC, Zhang Y, Gurel B, Cornish TC, Wu Z, et al. DNA hypomethylation arises later in prostate cancer progression than cpg island hypermethylation and contributes to metastatic tumor heterogeneity. Cancer Res 2008;68:8954–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harada K, Baba Y, Ishimoto T, Chikamoto A, Kosumi K, Hayashi H, et al. Line-1 methylation level and patient prognosis in a database of 208 hepatocellular carcinomas. Ann Surg Oncol 2015;22:1280–7. [DOI] [PubMed] [Google Scholar]
- 53.Zhu C, Utsunomiya T, Ikemoto T, Yamada S, Morine Y, Imura S, et al. Hypomethylation of long interspersed nuclear element-1 (line-1) is associated with poor prognosis via activation of c-met in hepatocellular carcinoma. Ann Surg Oncol 2014;21 Suppl 4:S729–35. [DOI] [PubMed] [Google Scholar]
- 54.Gao XD, Qu JH, Chang XJ, Lu YY, Bai WL, Wang H, et al. Hypomethylation of long interspersed nuclear element-1 promoter is associated with poor outcomes for curative resected hepatocellular carcinoma. Liver Int 2014;34:136–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pattamadilok J, Huapai N, Rattanatanyong P, Vasurattana A, Triratanachat S, Tresukosol D, Mutirangura A. Line-1 hypomethylation level as a potential prognostic factor for epithelial ovarian cancer. Int J Gynecol Cancer 2008;18:711–7. [DOI] [PubMed] [Google Scholar]
- 56.Iwagami S, Baba Y, Watanabe M, Shigaki H, Miyake K, Ishimoto T, et al. Line-1 hypomethylation is associated with a poor prognosis among patients with curatively resected esophageal squamous cell carcinoma. Ann Surg 2013;257:449–55. [DOI] [PubMed] [Google Scholar]
- 57.Barchitta M, Quattrocchi A, Maugeri A, Vinciguerra M, Agodi A. Line-1 hypomethylation in blood and tissue samples as an epigenetic marker for cancer risk: A systematic review and meta-analysis. PloS one 2014;9:e109478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scott EC, Gardner EJ, Masood A, Chuang NT, Vertino PM, Devine SE. A hot l1 retrotransposon evades somatic repression and initiates human colorectal cancer. Genome research 2016;26:745–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shafit-Zagardo B, Brown FL, Zavodny PJ, Maio JJ. Transcription of the kpni families of long interspersed dnas in human cells. Nature 1983;304:277–80. [DOI] [PubMed] [Google Scholar]
- 60.Hall LL, Carone DM, Gomez AV, Kolpa HJ, Byron M, Mehta N, et al. Stable c0t-1 repeat rna is abundant and is associated with euchromatic interphase chromosomes. Cell 2014;156:907–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ting DT, Lipson D, Paul S, Brannigan BW, Akhavanfard S, Coffman EJ, et al. Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science 2011;331:593–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Skowronski J, Fanning TG, Singer MF. Unit-length line-1 transcripts in human teratocarcinoma cells. Mol Cell Biol 1988;8:1385–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Skowronski J, Singer MF. Expression of a cytoplasmic line-1 transcript is regulated in a human teratocarcinoma cell line. Proceedings of the National Academy of Sciences of the United States of America 1985;82:6050–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Deininger P, Belancio VP. Detection of line-1 rnas by northern blot. Methods in molecular biology 2016;1400:223–36. [DOI] [PubMed] [Google Scholar]
- 65.Philippe C, Vargas-Landin DB, Doucet AJ, van Essen D, Vera-Otarola J, Kuciak M, et al. Activation of individual l1 retrotransposon instances is restricted to cell-type dependent permissive loci. Elife 2016;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hohjoh H, Singer MF. Sequence-specific single-strand rna binding protein encoded by the human line-1 retrotransposon. The EMBO journal 1997;16:6034–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Khazina E, Truffault V, Buttner R, Schmidt S, Coles M, Weichenrieder O. Trimeric structure and flexibility of the l1orf1 protein in human l1 retrotransposition. Nature structural & molecular biology 2011;18:1006–14. [DOI] [PubMed] [Google Scholar]
- 68.Khazina E, Weichenrieder O. Non-ltr retrotransposons encode noncanonical rrm domains in their first open reading frame. Proceedings of the National Academy of Sciences of the United States of America 2009;106:731–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mandal PK, Ewing AD, Hancks DC, Kazazian HH Jr. Enrichment of processed pseudogene transcripts in l1-ribonucleoprotein particles. Human molecular genetics 2013;22:3730–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Leibold DM, Swergold GD, Singer MF, Thayer RE, Dombroski BA, Fanning TG. Translation of line-1 DNA elements in vitro and in human cells. Proceedings of the National Academy of Sciences of the United States of America 1990;87:6990–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bratthauer GL, Fanning TG. Active line-1 retrotransposons in human testicular cancer. Oncogene 1992;7:507–10. [PubMed] [Google Scholar]
- 72.Bratthauer GL, Fanning TG. Line-1 retrotransposon expression in pediatric germ cell tumors. Cancer 1993;71:2383–6. [DOI] [PubMed] [Google Scholar]
- 73.Su Y, Davies S, Davis M, Lu H, Giller R, Krailo M, et al. Expression of line-1 p40 protein in pediatric malignant germ cell tumors and its association with clinicopathological parameters: A report from the children’s oncology group. Cancer letters 2007;247:204–12. [DOI] [PubMed] [Google Scholar]
- 74.Asch HL, Eliacin E, Fanning TG, Connolly JL, Bratthauer G, Asch BB. Comparative expression of the line-1 p40 protein in human breast carcinomas and normal breast tissues. Oncology research 1996;8:239–47. [PubMed] [Google Scholar]
- 75.Harris CR, Normart R, Yang Q, Stevenson E, Haffty BG, Ganesan S, et al. Association of nuclear localization of a long interspersed nuclear element-1 protein in breast tumors with poor prognostic outcomes. Genes & cancer 2010;1:115–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rodic N, Sharma R, Sharma R, Zampella J, Dai L, Taylor MS, et al. Long interspersed element-1 protein expression is a hallmark of many human cancers. The American journal of pathology 2014;184:1280–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sharma R, Rodic N, Burns KH, Taylor MS. Immunodetection of human line-1 expression in cultured cells and human tissues. Methods in molecular biology 2016;1400:261–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rodic N, Steranka JP, Makohon-Moore A, Moyer A, Shen P, Sharma R, et al. Retrotransposon insertions in the clonal evolution of pancreatic ductal adenocarcinoma. Nature medicine 2015;21:1060–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Doucet-O’Hare TT, Rodic N, Sharma R, Darbari I, Abril G, Choi JA, et al. Line-1 expression and retrotransposition in barrett’s esophagus and esophageal carcinoma. Proceedings of the National Academy of Sciences of the United States of America 2015;112:E4894–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Doucet-O’Hare TT, Sharma R, Rodic N, Anders RA, Burns KH, Kazazian HH Jr. Somatically acquired line-1 insertions in normal esophagus undergo clonal expansion in esophageal squamous cell carcinoma. Human mutation 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]