Abstract
UGT1A4 is primarily expressed in the liver and exhibits catalytic activities for various drugs. Amongst the few UGT1A4 polymorphisms evaluated, studies support the alteration of UGT1A4-mediated glucuronidation by a few variations including the Pro24Thr and Leu48Val variants (referred to as UGT1A4*2 and *3). We therefore investigated genetic mechanisms that might contribute to interindividual variation in UGT1A4 expression and activity. The UGT1A4 gene was sequenced from −4963 bp relative to the ATG to 2000 bp after the first exon in 184 unrelated Caucasians and African-Americans. We identified a large number of genetic variations, including 13 intronic, 39 promoter, as well as 14 exonic polymorphisms, with 10 that lead to amino-acid changes. Of the nucleotide variations found in the −5kb promoter region, 5 are located in the proximal region (first 500 bp), and positioned in putative HNF-1 and OCT-1 binding sites. Four of these variants, placed at −163, −219, −419 and −463, are in complete linkage disequilibrium with the Leu48Val coding region variant and with several variants in the upstream region of the promoter. Transient transfections of reference and variant promoter constructs (from position −500 to +1) in different cell lines with or without co-expression of HNF-1 and/or OCT-1, demonstrated limited effect of these variations. Additional functional studies on promoter variants are still required to predict their potential influence on UGT1A4 expression in vivo. Besides, several coding variants significantly modified the enzyme kinetics for tamoxifen and Z-4-hydroxytamoxifen (Val48, Asp50, Gln56, Phe176, Asn250, Leu276) and are expected to have a potential in vivo effect.
Keywords: pharmacogenetics, polymorphisms, tamoxifen, UGT, UGT1A4
INTRODUCTION
UDP-glucuronosyltransferase (UGT) enzymes comprise a superfamily of membrane proteins that catalyze the glucuronidation of a wide range of xenobiotics and endogenous compounds. As a major pathway the among phase II reactions in drug metabolism, interindividual variation in the glucuronidation pathway has received much attention and a number of genetic variations has been described, some of which have a clear pharmacological impact. To this day, nineteen functional proteins have been described, classified into three subfamilies: UGT1A, UGT2A and UGT2B [1–3]. The subfamily UGT1 includes nine active UGT isoforms encoded by a single gene UGT1 than spans 200kb. The UGT1A isoforms are encoded by four shared exons (2–5) that code for the carboxy terminal region of the protein, and variable exons 1, which corresponds to the substrate binding domain and provides differential substrate specificity [4].
UGT1A4, together with UGT1A3, is the main enzyme responsible for addition of glucuronic acid to amino groups (N-glucuronidation) [5,6]. The UGT1A4 protein is primarily expressed in the liver and in bile ducts, colon and small intestine. This enzyme exhibits catalytic activities mostly for primary and secondary amines, present in various therapeutic drugs [7]. Among these are a number of psychiatric drugs, including tricyclic antidepressants, antipsychotics and anticonvulsants. Trifluoperazine (TFP) is known to be readily and specifically glucuronidated by UGT1A4 [8]. The UGT1A4 enzyme was also reported to have the capacity to generate one of the glucuronides of the dietary carcinogen N-OH-PhIP found in cooked meat [9,10]. Tamoxifen, a nonsteroidal antiestrogen widely used for chemotherapy of hormone-dependent breast cancer, was also reported as a substrate for this enzyme [11]. Genetic variability in tamoxifen metabolism is one putative causal factor associated with interpatient variability in response to therapy [12]. Tamoxifen can be converted into several oxidative compounds [13], including 4-OH-tamoxifen, which is considered as its active metabolite, and both are shown to be metabolized by UGT1A4 into ammonium-linked glucuronides [11,12,14].
Frequency of genetic variations and haplotypes in UGT1A4 has been described for the Caucasian and Japanese populations, however, these studies included limited analysis of the promoter and intronic regions [15–17]. A careful evaluation of the function of UGT1A4 coding region polymorphisms has been limited to the P24T and L48V variants [15,18]. In contrast, only two promoter variations (−163G>A and −219C>T) were evaluated in vitro in a single study [19].
The main objective of this study was 1) to screen the UGT1A4 gene for polymorphisms in the promoter and coding region, from −4962bp relative to the transcription start site to 2000bp after the first exon, and, 2) to establish their potential effect on UGT1A4 enzymatic function and expression in vitro.
METHODS
Materials
Methanol, acetonitrile and formic acid were purchased from VWR (Montreal, Canada). Ammonium formate was purchased from Laboratoire Mat (Quebec, Canada). Tamoxifen, z-4-hydroxytamoxifen, β-glucuronidase type VII from E. Coli and UDP-glucuronic acid were obtained from Sigma-Aldrich (St. Louis, MO). N-hydroxy-2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (N-OH-PhIP), d5-tamoxifen and d5-4-hydroxytamoxifen were purchased from Toronto Research Chemical (Toronto, ON, Canada). Tamoxifen glucuronide, d5-tamoxifen glucuronide, z-4-hydroxytamoxifen glucuronide and d5-z-4-hydroxytamoxifen were obtained from enzymatic assays using liver microsomes. Human cell lines, HEK293, ACHN, HepG2 and Caco-2, were obtained from the American Type Culture Collection (Rockville, MD). All other chemicals and reagents were of the highest grade commercially available.
DNA samples from 100 unrelated Caucasians and from 36 Caucasian/African-American were described from previous studies [20,21]. Liver microsomes, DNA and RNA from American subjects were used as described previously [22,23]. All subjects provided written consent for experimental purposes, and the Institutional Review Boards approved the use of these samples.
UGT1A4 sequencing
PCR was used to amplify the UGT1A4 gene from −4962bp in the promoter region to 2000 bp after the first exon in 100 Caucasian individuals. Sequencing of the promoter (−2500bp) and exonic region was also performed for the 52 liver samples. The first exon was also sequenced in 36 African-American individuals. Amplification and sequencing primers are shown in Table 1. PCR conditions were 95°C for 1 min for initial denaturation, followed by 40 cycles at 95°C for 30 sec, 55–65°C for 30–40 sec and 72°C for 1–1.5 min, with a final extension at 72°C for 7 min. Specificity of primers was confirmed by direct sequencing of all PCR products. Amplicons were sequenced with an ABI 3700 automated sequencer using Big Dye (PerkinElmer Life Sciences, Boston, MA) dye primer chemistry. Samples were sequenced on both strands with primers listed in Table 1. Samples with ambiguous sequencing chromatograms were subjected to a second, independent amplification, followed by DNA sequencing. Sequences were analyzed with Staden preGap4 and Gap4 programs (Open Source Technology Group, CA, USA, http://staden.sourceforge.net/) and compared with the reference sequence (GenBank AF297093) to assess genetic variations. Linkage disequilibrium analysis and haplotype determination were performed with the Haploview 3.32 software (www.broad.mit.edu/mpg/haploview). Ambiguous sequencing chromatograms and samples with new SNPs were systematically re-amplified and re-sequenced.
Table 1.
Primer sequences
| Location in UGT1A4 | Sense | Primer Sequence (5’−3’) | Annealing temperature °C |
|---|---|---|---|
| Promoter | |||
| −5152 to −4024 bp | F* | TGTTTCTGCTCCTTATGCAAGCCT | 57 |
| R* | TGGAATAGCCACATTTCAAGTGCTC | ||
| −4231 to −3176 bp | F | TCTAGTGGGCATGTTAAACACCAG | 65 |
| R | TTATCCAGGTGTGATGGTGCACA | ||
| F* | CCACTAAAATTAATGTGAATA | ||
| R* | TGGAATAGCCACATTTCAAGTGCTC | ||
| F* | AGATGGAGTCTTGCTCTGTCACCCA | ||
| R* | AGGCGCATGGCACCACATCCAGC | ||
| F* | AGATGGAGTTTCATCATATTGGCCA | ||
| R* | GGTGCATGTAGGTTTGAGGTTTG | ||
| R* | GCAACATGCTGAGACTCCATCT | ||
| −3297 to −1959 bp | F | CCTTTGGGAGACCGAGGG | 58 |
| R | GCTACCACATATGCTGATGGC | ||
| F* | GCAGCATCTTCAAGTCTCTAAT | ||
| R* | ATAAAGTCTCTTCCACCTG | ||
| −2080 to −742 bp | F | GATTGGGAGAGGGGAGCTAGAT | 55 |
| R | GTGGGTCCTTGCTAGGGTTGT | ||
| F* | GCTGAGAATCCCTTTCTAGCA | ||
| R* | ATATGCCTCCCTTGAGCTGGG | ||
| −1013 to +22bp | F | TTATGCAGCCCGTTCTGTTCTGGA | 56 |
| R | GAGATGGCCAGAGGACTCCAGGTTC | ||
| F* | GACTTGGAGAAAAGCCTGG | ||
| Exon 1 | |||
| −108 to +701bp | F | CTGATTTGCTAGGTGGCTCAA | 56 |
| R | CCCTTATGCAAGTCTTGCC | ||
| F* | CTGACAGCCTATGCTGTTC | ||
| R* | GTATCTTTGGCCCTTCATAGGTG | ||
| +229 to IV1S+159bp | F | CTGACAGCCTATGCTGTTC | 56 |
| R | GTTTTCAGTGGTCACTGAGAG | ||
| F* | TCCTTCCTCCTATATTCCTAAG | ||
| R* | CCCTTATGCAAGTCTTGCC | ||
| Intron 1 | |||
| +687 to IV1S+1017bp | F | TTATGCAAGTCTTGCCTCTGAGC | 61 |
| R | AAATCCATTAAGGGAGCCATCC | ||
| F* | TATGCATCCGTGTGGCTGTTCCGA | ||
| F* | CTCAGGTGAAGCTGATCATATCA | ||
| IV1S+934bp to IV1S+1988bp | F | TCATTGAAATAGTACTCTGGGATG | 55 |
| R | GGTCATGTAAGGGTTAATCCAAT | ||
| F* | TTCTTGAGACTGAGCCTCGTTCTG | ||
| R* | CATGTGCCACTGCTCCTGGCAATT | ||
PCR primers are indicated in bold.
Indicates primers used for sequencing.
Reference sequence is AF297093. Position is relative to the first nucleotide of each primer. F, forward; R, reverse.
UGT1A4 variants and microsomal preparations
UGT1A4 variant alleles were generated by site-directed mutagenesis using the pcDNA3 vector, using a procedure described previously [24,25]. Stable HEK293 cell transfection with variant pcDNA3-UGT expression plasmids, preparation of microsomes by differential centrifugation, and assessment of UGT protein levels by Western blot have been previously described [24–26].
Transient transfections of UGT1A4 promoter constructs
Reference and variant promoter constructs of 606 bp were isolated by PCR from carriers of reference (−163G, −219C, −419G and −457C) or variant promoter (−163A, −219T, −419A and −457T). Additional variant alleles were generated by site-directed mutagenesis using the pcDNA3 vector. Promoter constructs of 500 bp were cloned upstream of the luciferase reporter gene in the PGL3 basic vector (Promega, Madison, WI) and transient transfections were performed using Lipofectamine2000 as the transfectant (Invitrogen, Burlington, ON). Reference and variant UGT1A4 promoter constructs (500ng) were transfected in HepG2, Caco-2 and ACHN cell lines in the presence or absence of 25ng HNF-1 and/or 25ng OCT-1 (in pcDNA1.1 vector) transcription factors. Addition of pRL-null (5ng) and the pBS-SK+ empty vector yielded a total of 555ng of DNA for each transfection. Luciferase activity was assessed with the Dual-Luciferase Reporter Assay kit (Promega, Madison, WI) following the manufacturer’s protocol. Luciferase activity was measured with 30 μL of cell lysate in a 96-well plate on an LB96V microplate luminometer (EG&G Berthold, Bad Wildbad, Germany). Results were obtained by comparing luciferase in transfected cells to the basal level ratio in cell lines transfected with an empty pGL3 vector (fold over pGL3).
Enzymatic assays with UGT1A4 substrates: tamoxifen, trans-4-hydroxy-tamoxifen, trifluoperazine and N-OH-PhIP
Enzymatic assays were performed according to a standard procedure [26,27]. Briefly, assays consisted of 1h incubations at 37°C with 5 to 50 μg of microsomal preparation according to the linearity of the reaction, 50 mM Tris-HCl, 10 mM MgCl2, 2 mM UDP-glucuronic acid, leupeptin, pepstatin and phosphatidylcholine. Assays were terminated by adding 100μL ACN + 0.000002% (v/v) butylated hydroxytoluene (BHT) for tamoxifen or 100μL MeOH + 0.000002% BHT for 4-hydroxytamoxifen. For glucuronidation experiments in liver samples, a fixed concentration of either N-OH-PhIP (25 μM) or trifluoperazine (200 μM) was used. In kinetic analysis experiments, varying tamoxifen (1–100μM) and Z-4-hydroxytamoxifen (1–500μM) concentrations were used. The procedure for enzymatic assays with N-OH-PhIP and trifluoperazine was described previously [8,28]. Statistical evaluation of best-fit model was used to select the enzyme kinetic model and confirmed by a visual inspection of fitted functions (V as a function of [S]) and Eadie-Hofstee plots (V as a function of V/[S]). Kinetic parameter calculations were performed with Sigma Plot 8.0 software assisted by Enzyme Kinetics 1.1 software (SPSS Inc., Chicago, IL). Values are expressed as the mean of at least two experiments performed at least in duplicate.
LC/MS analysis of glucuronide products
Detection of UGT1A4-generated N-OH-PhIP and TFP glucuronides was performed as described previously [8,10]. For the detection of tamoxifen, 4-hydroxytamoxifen glucuronides, the LC-MS/MS system consisted of a mass spectrometer (model API 3200, Concord, Canada) operated in multiple reactions monitoring mode (MRM) and equipped with an electrospray ionization interface in positive ion mode. Positive-product MRM ions pairs were m/z 548.3 → 372.1 for tamoxifen-glucuronide, m/z 553.3 → 377.1 for d5-tamoxifen-glucuronide, m/z 564.3 → 388.3 for Z-OH-tamoxifen-glucuronide and m/z 569.0 → 393.0 for d5-Z-OH-tamoxifen-glucuronide. The ion source temperature was set at 450°C. The ion spray voltage, declusting potential and entrance potential were set at 5000, 15 and 12V respectively. The collision energy for tamoxifen glucuronide and Z-OH-tamoxifen-glucuronide were –42 V. The chromatographic system consisted of an Agilent 1200 series (Agilent, Palo Alto, CA, USA). For tamoxifen glucuronide, the chromatographic separation was achieved with a Gemini C6 column with 3 μm packing material, 100 × 4.6 mm (Phenomenex). The mobiles phases were (solvent A) water 0,1% formic acid (v/v) and (solvent B) acetonitrile 0,1% acid formic (v/v). Separation was achieved using a linear gradient of 45–60% B in 2.0min at a flow rate of 0.9 ml/min. Then, the column was flushed with 95%B for 2 minutes and re-equilibrated to initial conditions over 3 min. For the Z-OH-tamoxifen glucuronide, the chromatographic separation was achieved with a ACE-3 HL column with 3 μm packing material, 100 × 4.6 mm (Life Science, Peterborough, Canada). The mobiles phases were (solvent A) water 0,4% formic acid (v/v) and (solvent B) methanol. Separation was achieved using isocratic conditions of 30% A and 70% B for 3 minutes at a flow rate of 0.9 ml/min. The column was flushed with 95%B for 2 minutes and re-equilibrated to initial conditions over 3 min.
UGT1A4 Quantitative Real-time-PCR
UGT1A4 RNA was quantified in the liver samples and expression levels were normalized for r18S content [29]. Briefly, reverse transcription reaction was performed using 200 units of Superscript II (Invitrogen, Burlington, Ontario, Canada) with 1 μg RNA, and 7.5 ng random hexamers (Roche, Laval, Québec, Canada) at 42°C for 50 minutes. UGT1A4 forward and reverse primer sequences for real-time PCR were located in the exon 1: 5’- CCT GTC CTA CAT TTG CCA TA-3’ and 5’- TCA AAT TCC TGA GAT AGT GGC TTC CC-3’, respectively. For each reaction, performed in triplicate, 50ng of cDNA was amplified in a 20μL total reaction volume, containing 10 μL SyBr Green PCR Mix (Applied Biosystems, Warrington, UK), 2 μL each primer (200 nM), and 6 μL of cDNA. Quantification was performed with an ABI Prism 7000 SDS v1.1 apparatus (Applied Biosystems, Warrington, UK). Amplification conditions were as follow: 10 minutes at 95°C followed by 40 cycles of 15 seconds at 95°C and 60 seconds at 60 °C.
Statistical analyses
All statistical analyses were performed with the JMP software v4.0.2 (SAS Institute, Cary, NC). Normality of distribution was assessed using the Shapiro-Wilk W test. Logarithmic or Box Cox Y transformation was applied to values for which distribution was not normal. P-values of less than 0.05 for ANOVA (Student’s t test) or Chi-Square pairwise comparisons were considered statistically significant.
RESULTS
Sequencing of the UGT1A4 gene
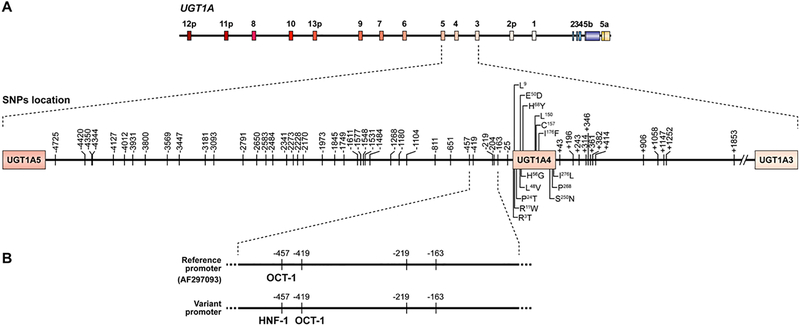
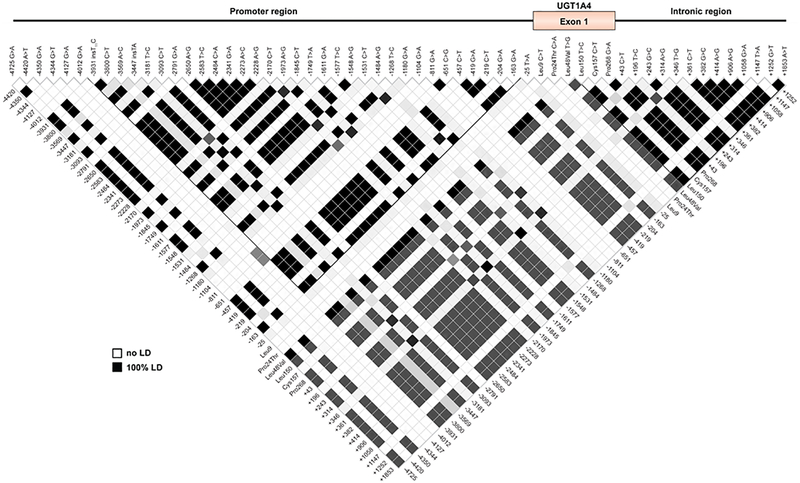
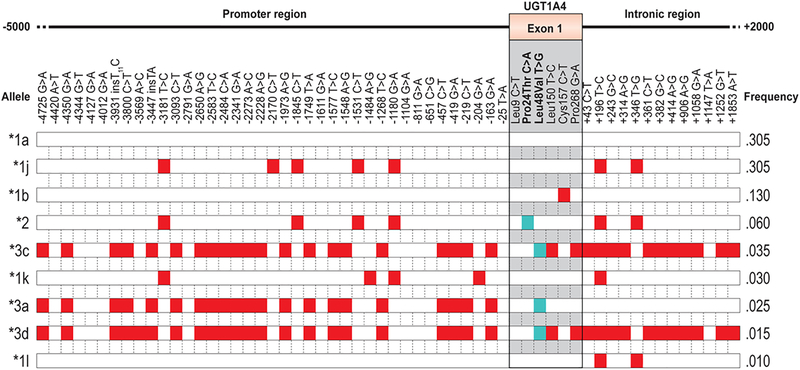
Sequencing of the UGT1A4 exonic region in Caucasian and African-American samples as well as in a set of liver samples, allowed the identification of 14 nucleotide changes in the first exon, including 10 resulting in amino acid change (Arg3Thr, Arg11Trp, Pro24Thr, Leu48Val, Glu50Asp, His56Gln, His68Tyr, Ile176Phe, Ser250Asn and Ile276Leu) (Fig. 1A). In the 100 Caucasian individuals, 13 intronic and 39 promoter polymorphisms were found. Linkage disequilibrium (LD) analyses revealed a strong linkage between most promoter SNPs and also between intronic SNPs, resulting in two block structures, which were also in relatively elevated linkage (67%) (Fig. 2). Haplotypes were inferred in the cohort of Caucasian individuals (n=100) and revealed the existence nine different haplotypes of variable frequencies (Fig. 3). A series of variations in the promoter region (haplotypes *3a, *3c and *3d) were found to be in complete LD with the previously reported L48V coding region variant, with an allelic frequency of 6%. These variations were also frequently associated with a series of intronic variations (haplotypes *3c and *3d).
Fig. 1.
(A) Sequencing of the UGT1A4 gene allowed the identification of 66 polymorphisms. (B) In silico analyses revealed that four polymorphisms located in the first 500 bp were associated with deletions or additions of putative HNF-1 and OCT-1 transcription factors binding sites.
Fig. 2.
Pairwise LD and bloc structure of UGT1A4 polymorphisms.
Fig. 3.
UGT1A4 haplotypes and their frequencies in Caucasian individuals (n=100). Variants at codons 24 and 48 were already reported.
Computational analysis of UGT1A4 promoter variations
To better assess if these variants could potentially modulate the transcription of the UGT1A4 gene, a transcription factor binding site localization analysis was performed with the MatInspector software (Genomatix, Ann Arbor, MI). A region comprising the first 500 bp of the UGT1A4 promoter was found to be associated with deletions or additions of putative HNF-1 and OCT-1 binding sites in accordance with genotype status at positions −163, −219, −419 and −457 (Fig. 1B). While the variant at positions −419 was associated the addition of an OCT-1 binding site, the variant at position −457 was associated with the simultaneous deletion of an OCT-1 and the addition of an HNF-1 binding sites. Nucleotide changes were all found to be located in the core sequences of putative binding domains. Consequently, this variant haplotype is predicted to result in the putative displacement of one OCT-1 and addition of one HNF-1 binding sites.
UGT1A4 promoter variants and gene expression
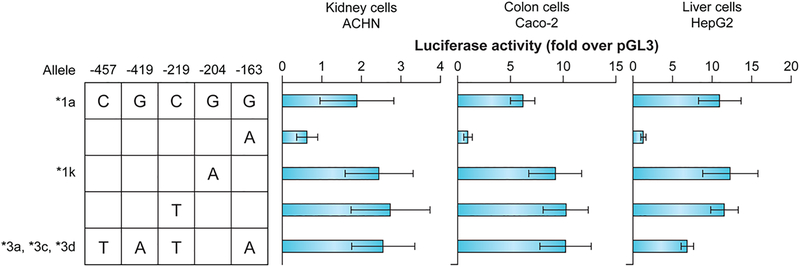
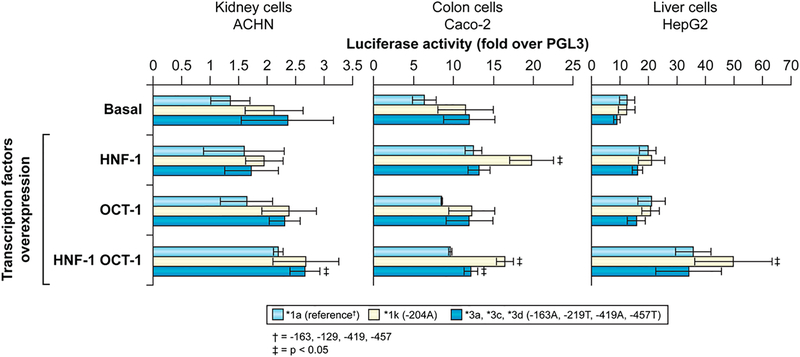
Transcriptional activity modulation by the UGT1A4 promoter variations was assessed by transient transfections in ACHN (kidney), Caco-2 (colon) and HepG2 (liver) cell lines. Although the basal activity was markedly reduced with the −163A variant in all cell lines, basal activity of the complete variant haplotype (−163A, −219T, −419A, −457T) did not differ significantly from the reference promoter (Fig. 4). The addition of HNF-1 and OCT-1 transcription factors increased the luciferase activity in HepG-2 cells while no noticeable change was detected in the other two cell lines (Fig. 5). The infrequent −204G>A polymorphism, which demonstrated no evidence of LD with other proximal variations, enhanced the positive action of co-expressed HNF-1 on UGT1A4 promoter activity, and this effect was further increased by co-expression of OCT-1 in Caco-2 and HepG2 cells. However, no significant differences were observed in luciferase activity between the reference and variant promoter (−163A, −219T, −419A, −457T) constructs with the addition of either or both the HNF-1 and OCT-1 transcription factors in two of the three cell lines (Fig. 5). These results suggest that those promoter variants, using a proximal construction around 500 bp of the UGT1A4 promoter, do not substantially modify gene expression.
Fig. 4.
Transient transfections of reference and variant UGT1A4 promoter constructs in diverse human cell lines.
Fig. 5.
Cotransfections of reference and variant UGT1A4 promoter constructs (500 bp) with HNF-1 and/or OCT-1 in ACHN, Caco-2 and HepG2 cell lines do not result in marked modification of expression. * p≤0.05
UGT1A4 coding variants modulate in vitro glucuronide formation
Several UGT1A4 coding variants have been previously shown to alter in vitro glucuronidation. To confirm and extend upon these observations, we performed enzymatic assays and included the less frequent coding variations. For these assays, tamoxifen and Z-4-hydroxytamoxifen were tested (Table 3). Kinetics for tamoxifen fitted the Michaelis-Menten model. Several variants presented an altered Km and/or Vmax compared to the UGT1A4*1 reference protein. This resulted in a 105% increased Vmax/Km for UGT1A4*3 (Val48), and a Vmax/Km for Asp50, Gln56, Phe176, Asn250, Leu276 11–38% of the reference UGT1A4*1 protein. While relative Vmax for z-4-OH-tamoxifen was reduced for all tested variants but UGT1A4*4 (Trp11), Km was increased 2–10 fold for variants Phe176, Asn250, Leu276, resulting in a significantly reduced Vmax/Km for variants Asp50, Gln56, Phe176, Asn250, Leu276. Variants Phe176 and Leu276, when present together with Thr24 or Val48, respectively, also resulted in markedly reduced the Vmax/Km for these two substrates.
Table 3.
UGT1A4 coding variants and tamoxifen and trans-4-hydroxytamoxifen glucuronidation activities.
| Apparent Km | Ki | n | Relative Vmax | Vmax / Km | |
|---|---|---|---|---|---|
| (μM) | (μM) | (pmol/min/mg protein) | (μl/min/mg) | ||
| Tamoxifen | |||||
| 1A4*1 | 17.7±2.0 | 565±35 | 32.3±5.7 | ||
| 1A4*2 (T24) | 22.2±1.1 | 568±62 | 28.2±5.3 | ||
| 1A4*3 (V48) | 6.7±0.7* | 440±23 | 66.2±3.6 | ||
| 1A4*4 (W11) | 16.8±3.4 | 416±12* | 25.4±5.8 | ||
| 1A4*9 (T3) | 14.2±2.5 | 507±58 | 36.5±10.4 | ||
| 1A4*10 (D50) | 17.0±2.3 | 200±7* | 11.8±1.2 | ||
| 1A4*11 (Q56) | 70.7±18.4 | 285±1* | 4.2±1.1 | ||
| 1A4*12 (Y68) | 11.7±0.8 | 419±115 | 36.3±12.3 | ||
| 1A4*13 (F176) | 9.7±0.5* | 117±3* | 12.1±0.9 | ||
| 1A4*14 (N250) | 14.6±5.1 | 262±1* | 19.1±6.7 | ||
| 1A4*15 (L276) | 51.9±2.0* | 177±16* | 3.4±0.2 | ||
| 1A4*16 (V48Y68) | 7.3±0.2* | 318±17* | 43.3±3.5 | ||
| 1A4*17 (T24L276) | 43.4±13.2 | 93±16* | 2.3±1.1 | ||
| 1A4*18 (V48F176) | 7.9±2.4* | 31±3* | 4.1±1.6 | ||
| z-4-OH-tamoxifen | |||||
| 1A4*1 | 15.4±1.5 | 261±24 | ± | 2193±181 | 143.4±25.4 |
| 1A4*2 (T24) | 15.2±1.1 | 242±46 | ± | 1151±162* | 80.0±11.3 |
| 1A4*3 (V48) | 10.4±1.1 | 217±17 | ± | 1528±77* | 147.8±23.3 |
| 1A4*4 (W11) | 12.9±1.2 | 214±14 | ± | 1560±241 | 122.1±30.0 |
| 1A4*9 (T3) | 15.5±1.6 | 314±27 | ± | 1025±20* | 66.6±8.0 |
| 1A4*10 (D50) | 18.3±0.6 | 252±16 | ± | 434±33* | 23.7±2.6 |
| 1A4*11 (Q56) | 20.6±1.4 | 222±0 | ± | 197±12* | 9.6±0.1 |
| 1A4*12 (Y68) | 16.6±0.3 | 319±46 | ± | 1098±45* | 66.0±3.7 |
| 1A4*13 (F176) | 99.1±9.9* | 1.70±0.13 | 357±28* | 3.6±0.6 | |
| 1A4*14 (N250) | 160.5±16.6* | 1.09±0.07 | 289±24* | 1.8±0.3 | |
| 1A4*15 (L276) | 24.3±1.3* | 247±14 | ± | 109±12* | 4.5±0.3 |
| 1A4*16 (V48Y68) | 16.3±1.1 | 357±35 | ± | 1348±48* | 83.1±8.7 |
| 1A4*17 (T24L276) | 31.5±4.2* | 256±1 | ± | 351±31* | 11.2±0.5 |
| 1A4*18 (V48F176) | 92.1±0.1* | 1.69±0.04 | 22±5* | 0.2±0.1 |
Vmax was adjusted for relative expression of each variant isoform: UGT1A4*1=1 (arbitrarily set at 1), UGT1A4*2=0.07, UGT1A4*3=0.45, UGT1A4*4=0.66, UGT1A4*9 (T3)=0.31, UGT1A4*10 (D50)=0.86, UGT1A4*11 (Q56)=0.47, UGT1A4*12 (Y68)=0.21, UGT1A4*13 (F176)=1.09, UGT1A4*14 (N250)=0.50, UGT1A4*15 (L276)=0.51, UGT1A4*16 (V48Y68)=0.94, UGT1A4*17 (T24L276)=0.44, UGT1A4*18 (V48F176)=0.16.
For 4-OH-tamoxifen, UGT1A4*13 (F176), UGT1A4*14 (N250) and UGT1A4*18 (V48F176) were characterized by a Hill’s profile.
The F176 variation was found in subjects of African origin.
Correlation between the presence of genetic variations, UGT1A4 mRNA, and glucuronidation activity in a human liver bank
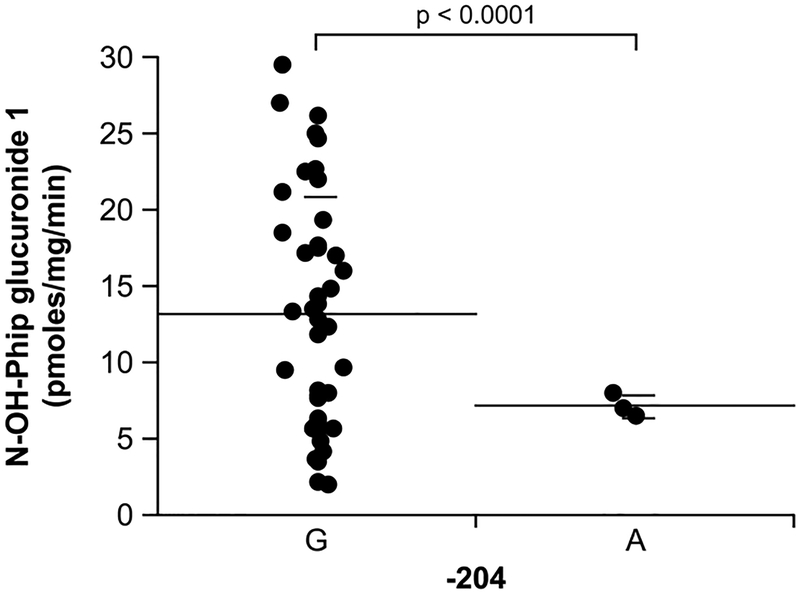
In order to study the potential influence of identified UGT1A4 variants, UGT1A4 RNA expression and formation of TFP-glucuronide, as well as glucuronide 1 of N-OH-PhIP (uncharacterized at this time), were correlated with UGT1A4 genotypes (−2497A>G, −2170C>T, −1663C>T, −1484A>G, −1180G>A, Arg3Thr, Arg11Trp, Thr14, Leu18Phe, Pro24Thr, Leu48Val) in a set of liver samples. Firstly, UGT1A4 RNA content in liver samples demonstrated a wide distribution (20±26 arbitrary units; range 1–117). Although RNA content correlated weakly with the extent of TFP-gluruconide formation (r2=0.301; p=0.047), it did not correlate with N-OH-PhIP glucuronide 1. A lower N-OH-PhIP glucuronide formation was observed for the −204A carriers (n=3; 13.2±7.6 vs 7.2±0.7nmoles/mg/min for −204G vs A carriers, respectively; p = 0.0132) (Fig. 6). Interestingly, a carrier of a silent variation at codon 14 (42G) had a significantly lower TFP glucuronidation (0.52±0.25 vs 0.03 for 42A vs G carriers, respectively; p=0.0005 rank for G carrier = 1). No significant correlation could be established between the other promoter and coding region variations and UGT1A4 RNA expression or hepatic glucuronidation activities.
Fig. 6.
Relationship between N-OH-PhIP glucuronidation rates and the UGT1A4 −204 variation.
DISCUSSION
UGT1A4 is the one of the major enzymes responsible for the N-glucuronidation of various compounds, including therapeutic drugs [5–7]. In previous studies, UGT1A4 was demonstrated to have the capacity to conjugate antihistamines (cyproheptadine), tricyclic antidepressants (imipramine) and of numerous antipsychotics (clozapine, trifluoperazine) [10,15,30]. UGT1A4 was also found to generate tamoxifen and 4-OH-tamoxifen glucuronides [11], as well as one of the glucuronides of N-OH-PhIP, a dietary carcinogen found in cooked meat [9,10]. For a number of these compounds, metabolic variations were predicted to originate from individual genetic profiles of UGT enzymes. While a number of coding region polymorphisms, including the P24T and L48V variants, have been described, the 5’-untranslated region of the UGT1A4 gene has not been as extensively studied [16,18]. Although UGT1A4 variations have previously been described in the Caucasian and Japanese population [15–17], to our knowledge, the present study is the most extensive to have been published so far.
Several coding variants in the UGT1A4 gene significantly modify the activity of recombinant UGT1A4 for tamoxifen and Z-4-hydroxytamoxifen. The frequencies of non-synonymous variants established in this study are similar to those reported previously: 8% for the T24 variant and of 9–17% for the V48 variant [15,30]. We also demonstrated a high density of promoter variations. However, according to the in vitro analysis of the first 500 base pairs of its promoter region, predicted to be involved in gene regulation according to transcription factors binding sites analyses, no polymorphisms are significantly associated with a modulation of transcriptional activity, or trifluoperazine glucuronidation activity in liver microsomes.
Of all promoter variants found herein, haplotypes *3a, *3c and *3d were of particular interest due to their tight LD with the V48 coding variation. Transient transfections in various cell lines derived from human liver, kidney or colon, did not reveal markedly modified transcriptional activity. The HNF-1 and OCT-1 transcription factors are known regulators of UGT genes. HNF-1 was found to regulate several UGT1A and 2B genes, including UGT1A4 [31]. In turn, OCT-1 was previously reported to bind to HNF-1, consequently enhancing its capacity to activate the UGT2B7 promoter [32]. The increased activity observed in the hepatic cell line HepG2 upon co-transfection with these transcription factors confirms their likely role in UGT1A4 regulation. In a previous study, the −163A variation was found to be associated with significantly reduced transcriptional activity upon transient transfection [19]. Although a similar observation was done in our study, the complete haplotype (−163A, −219A, 419T and −463A) did not show such a repression, despite its possible association with the addition and deletion of HNF-1 and OCT-1 putative binding sites. However, it is clear that transcription factors other than HNF-1 and OCT-1 may differentially affect the SNPs studied herein. Besides, since several other SNPs have also been found in the region beyond −1000 bp, additional studies will be required to carefully assess the contribution of the distal promoter in the transcriptional regulation of the UGT1A4 gene and the potential influence of variable amino acids.
Kinetic analysis with tamoxifen and 4-hydroxytamoxifen have been previously published [11,14], while only the effect of variants P24T and L48V had been assessed for these substrates. The apparent Km derived in this study were in a range similar to those observed by Kaku et al. [11], but higher than those noted by Sun et al. [14]. No significant effect was evidenced for the T24 allozyme compared to the reference UGT1A4.1 protein, but an increased glucuronidation activity for the V48 variant was reported against tamoxifen and 4-OH-tamoxifen [14]. In the present study, a significantly increased activity was observed only for tamoxifen. We also identified additional novel although rare variations in the UGT1A4 gene, including Asp50, Gln56, Phe176, Asn250, and Leu276, all of which are likely to be functionally significant since they are associated with reduced UGT1A4 activity with one or both substrates.
Despite the fact that several variants are predicted to modulate glucuronidation based on in vitro functional date, this could not be demonstrated by correlative studies in a set of 52 human liver microsomes. The haplotype comprising the V48 coding region variant in addition to the four linked nucleotide changes at positions −163T, −219A, −419T and −457A, was not associated with significantly different N-OH-PhIP or TFP glucuronidation activities. Only the −204A variant appeared to change N-OH-PhIP glucuronidation, although this variation was found in only 3 liver samples; however, it is unknown whether this effect is due to the −204A variant or to other variations found in LD in the promoter and intronic regions. The T24 variant was only found in four liver samples and this limited our capacity to determine functional significance. Additional studies are required in order to assess the in vivo impact of genetic variations in UGT1A4.
Table 2.
UGT1A4 variants frequency in the population of 100 Caucasians.
| Region | rs number | Amino acid change | Allele name | Position* | Nucleotide change | Minor allele frequency (%) |
|---|---|---|---|---|---|---|
| Promoter | rs17862870 | 130704 | −4725 g > a | 8.0 | ||
| - | 131009 | −4420 a > t | 0.5 | |||
| rs4663945 | 131079 | −4350 g > a | 8.0 | |||
| - | 131085 | −4344 g > t | 1.0 | |||
| - | 131302 | −4127 g > a | 1.0 | |||
| - | 131417 | −4012 g > a | 0.5 | |||
| - | 131498 | −3931 T11 ins | 8.0 | |||
| rs12477216 | 131629 | −3800 c > t | 8.0 | |||
| - | 131860 | −3569 a > c | 1.0 | |||
| - | 131984 | −3445 ta ins | 8.0 | |||
| rs6749496 | 132248 | −3181 t > c | 43.0 | |||
| rs17862871 | 132336 | −3093 c > t | 8.0 | |||
| - | 132638 | −2791 g > a | 0.5 | |||
| rs17868334 | 135779 | −2650 a > g | 8.0 | |||
| rs17863791 | 132846 | −2583 t > c | 8.0 | |||
| rs17868335 | 132945 | −2484 c > a | 8.0 | |||
| rs17863792 | 133088 | −2341 g > a | 8.0 | |||
| rs17864696 | 133156 | −2273 a > c | 8.0 | |||
| rs17874943 | 133201 | −2228 a > g | 8.0 | |||
| rs6744284 | 133259 | −2170 c > t | 32.5 | |||
| - | 133456 | −1973 a > g | 8.0 | |||
| rs1875263 | 133584 | −1845 c > t | 38.0 | |||
| rs3806595 | 133680 | −1749 t > a | 8.0 | |||
| - | 133818 | −1611 g > a | 0.5 | |||
| rs3806594 | 133852 | −1577 t > c | 8.0 | |||
| rs3806593 | 133881 | −1548 a > g | 8.0 | |||
| rs3806592 | 133898 | −1531 c > t | 38.0 | |||
| rs11676072 | 133945 | −1484 a > g | 5.0 | |||
| rs3806591 | 134161 | −1268 t > c | 8.0 | |||
| rs869283 | 134249 | −1180 g > a | 44.0 | |||
| - | 134324 | −1104 g > a | 0.5 | |||
| rs28898608 | 134618 | −811 g > a | 0.5 | |||
| rs28898609 | 134778 | −651 c > g | 0.5 | |||
| rs3732221 | 134972 | −457 c > t | 8.0 | |||
| rs3732220 | 135010 | −419 g > a | 8.0 | |||
| rs3732219 | 135210 | −219 c > t | 8.0 | |||
| - | 135225 | −204 g > a | 4.0 | |||
| rs3732218 | 135266 | −163 g > a | 8.0 | |||
| - | 135404 | −25 t > a | 0.5 | |||
| Exon 1 | - | Leu9 | 135453 | 25 c > t | 0.5 | |
| rs6755571 | Pro24Thr | 1A4*2 | 135498 | 70 c > a | 6.0 | |
| rs2011425 | Leu48Val | 1A4*3 | 135570 | 142 t > g | 8.0 | |
| rs12468274 | Leu150 | 135876 | 448 t > c | 5.5 | ||
| rs2011404 | Cys157 | 135899 | 471 c > t | 15.0 | ||
| rs3732217 | Pro268 | 136232 | 805 g > a | 5.5 | ||
| Intron 1 | rs2011219 | 136338 | 910 c > t | 5.5 | ||
| rs871514 | 136491 | 1063 t > c | 50.0 | |||
| rs904855 | 136538 | 1110 g > c | 5.5 | |||
| rs904856 | 136609 | 1181 a > g | 5.5 | |||
| rs13401281 | 136641 | 1213 t > g | 39.5 | |||
| rs12466779 | 136656 | 1228 c > t | 5.5 | |||
| rs12463641 | 136677 | 1249 g > c | 5.5 | |||
| rs12468356 | 136709 | 1281 a > g | 5.5 | |||
| rs12468543 | 137201 | 1773 a > g | 5.5 | |||
| rs12463910 | 137353 | 1925 g > a | 5.5 | |||
| - | 137442 | 2014 t > a | 0.5 | |||
| - | 137547 | 2119 g > t | 5.5 | |||
| - | 138148 | 2720 a > t | 5.5 |
Reference sequence: AF297093
ins: insertion
ACKNOWLEDGEMENTS
The authors thank Lyne Villeneuve for her technical assistance. We also thank Robert Millikan for DNA samples from African-Americans subjects. This work was supported by the Canadian Institutes of Health Research (MOP-42392) and Canada Research Chair Program (C.G.). M.O.B.B. is holder of a Canada Graduate Scholarship Doctoral Research Award from the CIHR and of the Canadian Federation of University Women Dr. Marion Elder Grant Fellowship, funded by CFUW Wolfville. J.P.A. and M.H.L. were supported by undergraduate studentship awards from the Fonds de la Recherche en Sauté du Quebec (FRSQ) and CIHR, respectively. O.B. was the recipient of a health professional studentship award from the FRSQ. C.G. is the holder of a Canada Research Chair in Pharmacogenomics. Dr Court was supported by grant GM-061834 from the National Institute of General Medical Sciences (NIGMS), National Institutes of Health (Bethesda, MD). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This work was supported by the Canadian Institutes of Health Research (MOP-42392) and Canada Research Chair Program (C.G.).
Contributor Information
Marie-Odile Benoit-Biancamano, Pharmacogenomics Laboratory, CHUQ Research Center and Faculty of Pharmacy, Laval University, G1V 4G2, Québec, Canada.
Jean-Philippe Adam, Pharmacogenomics Laboratory, CHUQ Research Center and Faculty of Pharmacy, Laval University, G1V 4G2, Québec, Canada.
Olivier Bernard, Pharmacogenomics Laboratory, CHUQ Research Center and Faculty of Pharmacy, Laval University, G1V 4G2, Québec, Canada.
Michael H. Court, Department of Pharmacology and Experimental Therapeutics, Tufts University School of Medicine, Boston, Massachusetts.
Marie-Hélène Leblanc, Pharmacogenomics Laboratory, CHUQ Research Center and Faculty of Pharmacy, Laval University, G1V 4G2, Québec, Canada.
Patrick Caron, Pharmacogenomics Laboratory, CHUQ Research Center and Faculty of Pharmacy, Laval University, G1V 4G2, Québec, Canada.
Chantal Guillemette, Pharmacogenomics Laboratory, CHUQ Research Center and Faculty of Pharmacy, Laval University, G1V 4G2, Québec, Canada.
REFERENCES
- 1.Guillemette C. Pharmacogenomics of human UDP-glucuronosyltransferase enzymes. Pharmacogenomics J 2003; 3:136–158. [DOI] [PubMed] [Google Scholar]
- 2.Mackenzie PI, Bock KW, Burchell B, Guillemette C, Ikushiro S, Iyanagi T, et al. Nomenclature update for the mammalian UDP glycosyltransferase (UGT) gene superfamily. Pharmacogenet Genomics 2005; 15:677–685. [DOI] [PubMed] [Google Scholar]
- 3.Sten T, Bichlmaier I, Kuuranne T, Leinonen A, Yli-Kauhaluoma J, Finel M. UDP-glucuronosyltransferases (UGTs) 2B7 and UGT2B17 display converse specificity in testosterone and epitestosterone glucuronidation, whereas UGT2A1 conjugates both androgens similarly. Drug Metab Dispos 2009; 37:417–423. [DOI] [PubMed] [Google Scholar]
- 4.Gong QH, Cho JW, Huang T, Potter C, Gholami N, Basu NK, et al. Thirteen UDPglucuronosyltransferase genes are encoded at the human UGT1 gene complex locus. Pharmacogenetics 2001; 11:357–368. [DOI] [PubMed] [Google Scholar]
- 5.Green MD, Tephly TR. Glucuronidation of amines and hydroxylated xenobiotics and endobiotics catalyzed by expressed human UGT1.4 protein. Drug Metab Dispos 1996; 24:356–363. [PubMed] [Google Scholar]
- 6.Green MD, King CD, Mojarrabi B, Mackenzie PI, Tephly TR. Glucuronidation of amines and other xenobiotics catalyzed by expressed human UDP-glucuronosyltransferase 1A3. Drug Metab Dispos 1998; 26:507–512. [PubMed] [Google Scholar]
- 7.Tukey RH, Strassburg CP. Human UDP-glucuronosyltransferases: metabolism, expression, and disease. Annu Rev Pharmacol Toxicol 2000; 40:581–616. [DOI] [PubMed] [Google Scholar]
- 8.Court MH. Isoform-selective probe substrates for in vitro studies of human UDP-glucuronosyltransferases. Methods Enzymol 2005; 400:104–116. [DOI] [PubMed] [Google Scholar]
- 9.Malfatti MA, Felton JS. N-glucuronidation of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) and N_hydroxy-PhIP by specific human UDP-glucuronosyltransferases. Carcinogenesis 2001; 22:1087–1093. [DOI] [PubMed] [Google Scholar]
- 10.Girard H, Thibaudeau J, Court MH, Fortier LC, Villeneuve L, Caron P, et al. UGT1A1 polymorphisms are important determinants of dietary carcinogen detoxification in the liver. Hepatology 2005; 42:448–457. [DOI] [PubMed] [Google Scholar]
- 11.Kaku T, Ogura K, Nishiyama T, Ohnuma T, Muro K, Hiratsuka A. Quaternary ammonium-linked glucuronidation of tamoxifen by human liver microsomes and UDP-glucuronosyltransferase 1A4. Biochem Pharmacol 2004; 67:2093–2102. [DOI] [PubMed] [Google Scholar]
- 12.Lazarus P, Blevins-Primeau AS, Zheng Y, Sun D. Potential role of UGT pharmacogenetics in cancer treatment and prevention: focus on tamoxifen. Ann N Y Acad Sci 2009; 1155:99–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poon GK, Chui YC, McCague R, Llnning PE, Feng R, Rowlands MG, et al. Analysis of phase I and phase II metabolites of tamoxifen in breast cancer patients. Drug Metab Dispos 1993; 21:1119–1124. [PubMed] [Google Scholar]
- 14.Sun D, Chen G, Dellinger RW, Duncan K, Fang JL, Lazarus P. Characterization of tamoxifen and 4-hydroxytamoxifen glucuronidation by human UGT1A4 variants. Breast Cancer Res 2006; 8:R50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ehmer U, Vogel A, Schutte JK, Krone B, Manns MP, Strassburg CP. Variation of hepatic glucuronidation: Novel functional polymorphisms of the UDP-glucuronosyltransferase UGT1A4. Hepatology 2004; 39:970–977. [DOI] [PubMed] [Google Scholar]
- 16.Saeki M, Saito Y, Jinno H, Sai K, Hachisuka A, Kaniwa N, et al. Genetic variations and haplotypes of UGT1A4 in a Japanese population. Drug Metab Pharmacokinet 2005; 20:144–151. [DOI] [PubMed] [Google Scholar]
- 17.Menard V, Girard H, Harvey M, Perusse L, Guillemette C. Analysis of inherited genetic variations at the UGT1 locus in the French-Canadian population. Hum Mutat 2009; 30:677–687. [DOI] [PubMed] [Google Scholar]
- 18.Wiener D, Fang JL, Dossett N, Lazarus P. Correlation between UDP-glucuronosyltransferase genotypes and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone glucuronidation phenotype in human liver microsomes. Cancer Res 2004; 64:1190–1196. [DOI] [PubMed] [Google Scholar]
- 19.Erichsen TJ, Ehmer U, Kalthoff S, Lankisch TO, Muller TM, Munzel PA, et al. Genetic variability of aryl hydrocarbon receptor (AhR)-mediated regulation of the human UDP glucuronosyltransferase (UGT) 1A4 gene. Toxicol Appl Pharmacol 2008; 230:252–260. [DOI] [PubMed] [Google Scholar]
- 20.Levesque E, Delage R, Benoit-Biancamano MO, Caron P, Bernard O, Couture F, et al. The impact of UGT1A8, UGT1A9, and UGT2B7 genetic polymorphisms on the pharmacokinetic profile of mycophenolic acid after a single oral dose in healthy volunteers. Clin Pharmacol Ther 2007; 81:392–400. [DOI] [PubMed] [Google Scholar]
- 21.Butler LM, Sinha R, Millikan RC, Martin CF, Newman B, Gammon MD, et al. Heterocyclic amines, meat intake, and association with colon cancer in a population-based study. Am J Epidemiol 2003; 157:434–445. [DOI] [PubMed] [Google Scholar]
- 22.Girard H, Court MH, Bernard O, Fortier LC, Villeneuve L, Hao Q, et al. Identification of common polymorphisms in the promoter of the UGT1A9 gene: evidence that UGT1A9 protein and activity levels are strongly genetically controlled in the liver. Pharmacogenetics 2004; 14:501–515. [DOI] [PubMed] [Google Scholar]
- 23.Court MH, Duan SX, Guillemette C, Journault K, Krishnaswamy S, Von Moltke LL, et al. Stereoselective conjugation of oxazepam by human UDP-glucuronosyltransferases (UGTs): S-oxazepam is glucuronidated by UGT2B15, while R-oxazepam is glucuronidated by UGT2B7 and UGT1A9. Drug Metab Dispos 2002; 30:1257–1265. [DOI] [PubMed] [Google Scholar]
- 24.Guillemette C, Ritter JK, Auyeung DJ, Kessler FK, Housman DE. Structural heterogeneity at the UDP-glucuronosyltransferase 1 locus: functional consequences of three novel missense mutations in the human UGT1A7 gene. Pharmacogenetics 2000; 10:629–644. [DOI] [PubMed] [Google Scholar]
- 25.Villeneuve L, Girard H, Fortier LC, Gagne JF, Guillemette C. Novel functional polymorphisms in the UGT1A7 and UGT1A9 glucuronidating enzymes in Caucasian and African-American subjects and their impact on the metabolism of 7-ethyl-10-hydroxycamptothecin and flavopiridol anticancer drugs. J Pharmacol Exp Ther 2003; 307:117–128. Epub 2003 Aug 2027. [DOI] [PubMed] [Google Scholar]
- 26.Gagne JF, Montminy V, Belanger P, Journault K, Gaucher G, Guillemette C. Common human UGT1A polymorphisms and the altered metabolism of irinotecan active metabolite 7-ethyl-10-hydroxycamptothecin (SN-38). Mol Pharmacol 2002; 62:608–617. [DOI] [PubMed] [Google Scholar]
- 27.Bernard O, Guillemette C. The main role of UGT1A9 in the hepatic metabolism of mycophenolic acid and the effects of naturally occurring variants. Drug Metab Dispos 2004; 32:775–778. [DOI] [PubMed] [Google Scholar]
- 28.Girard H, Thibaudeau J, Court MH, Fortier LC, Villeneuve L, Caron P, et al. UGT1A1 polymorphisms are important determinants of dietary carcinogen detoxification in the liver. Hepatology 2005; 42:448–457. [DOI] [PubMed] [Google Scholar]
- 29.Verreault M, Senekeo-Effenberger K, Trottier J, Bonzo JA, Belanger J, Kaeding J, et al. The liver X-receptor alpha controls hepatic expression of the human bile acid-glucuronidating UGT1A3 enzyme in human cells and transgenic mice. Hepatology 2006; 44:368–378. [DOI] [PubMed] [Google Scholar]
- 30.Mori A, Maruo Y, Iwai M, Sato H, Takeuchi Y. UDP-glucuronosyltransferase 1A4 polymorphisms in a Japanese population and kinetics of clozapine glucuronidation. Drug Metab Dispos 2005; 33:672–675. Epub 2005 Feb 2011. [DOI] [PubMed] [Google Scholar]
- 31.Gardner-Stephen DA, Mackenzie PI. Isolation of the UDP-glucuronosyltransferase 1A3 and 1A4 proximal promoters and characterization of their dependence on the transcription factor hepatocyte nuclear factor 1alpha. Drug Metab Dispos 2007; 35:116–120. [DOI] [PubMed] [Google Scholar]
- 32.Ishii Y, Hansen AJ, Mackenzie PI. Octamer transcription factor-1 enhances hepatic nuclear factor-1alpha-mediated activation of the human UDP glucuronosyltransferase 2B7 promoter. Mol Pharmacol 2000; 57:940–947. [PubMed] [Google Scholar]