SUMMARY
The actin cytoskeleton is critical to shape cells and pattern intracellular organelles, which collectively drives tissue morphogenesis. In multiciliated cells (MCCs), apical actin drives expansion of the cell surface necessary to host hundreds of cilia. The apical actin also forms a lattice to uniformly distribute basal bodies. This apical actin network is dynamically remodeled, but the molecules that regulate its architecture remain poorly understood. We identify the chromatin modifier, WDR5, as a regulator of apical F-actin in multiciliated cells. Unexpectedly in MCCs, WDR5 has a function independent of chromatin modification. We discover a scaffolding role for WDR5 between the basal body and F-actin. Specifically, WDR5 binds to basal bodies and migrates apically, where F-actin organizes around WDR5. Using a monomer trap for Gactin, we show that WDR5 stabilizes F-actin to maintain lattice architecture. In summary, we identify a non-chromatin role for WDR5 in stabilizing F-actin in multiciliated cells.
Keywords: congenital heart disease, Xenopus, H3K4, methylation, histone modifier, cilia, basal body
IN BRIEF
Kulkarni et al. uncover a chromatin modification-independent function for the H3K4 methyltransferase subunit WDR5 in multiciliated cell formation. WDR5 localizes to basal bodies at the ciliary base, where it stabilizes the actin lattice that allows multiciliated cells to expand their apical surface, pattern basal bodies, and generate hundreds of cilia.
Graphical Abstract
INTRODUCTION
Cilia play a central role in development and disease as they perform multiple sensory, motility and signaling functions (Oh and Katsanis, 2012; Bettencourt-Dias, et al., 2011; Sharma, et al., 2008). Cilia in multiciliated cells (MCCs) are specifically important for creating mechanical force to drive extracellular fluid flow, which is critical for clearing mucus in the airways, for transporting the egg through the oviduct, and for circulating cerebrospinal fluid in the cerebral ventricles (Oh and Katsanis, 2012; Bettencourt-Dias, et al., 2011; Houtmeyers, et al., 1999). Despite the importance of multiciliated cells (MCCs) in health, our understanding of ciliogenesis remains incomplete. While the formation of a single cilium is complex, in a MCC, this complexity is compounded to generate the hundreds of oriented cilia that beat in a coordinated fashion. First, a specialized structure called the deuterosome produces hundreds of centrioles that become the basal bodies necessary to seed many cilia (Klos Dehring, et al., 2013; Tang, 2013; Zhao, et al., 2013). Second, these basal bodies associate with vesicles that migrate and dock to the apical cell surface (Burke, et al., 2014; Vladar and Axelrod, 2008). Finally, ciliary axonemes protrude from the basal bodies and elongate to form functional cilia (Ishikawa and Marshall, 2011; Pedersen and Rosenbaum, 2008). To effectively drive extracellular fluid flow, the next challenge is to orient and coordinate hundreds of cilia. To do so, the cilia must be aligned within the epithelial plane, which is established by the rootlets and basal feet, which are attached to the basal body (Kunimoto, et al., 2012; Mitchell, et al., 2009).
Actin is a major driver of MCC formation (Sedzinski, et al., 2016; Werner, et al., 2011; Vladar and Axelrod, 2008). First, establishing a mucociliary epithelia such as in the Xenopus embryonic epidermis requires actin dependent radial intercalation of nascent MCCs (Walck-Shannon and Hardin, 2014). These nascent MCCs are first specified in deeper layers but migrate apically to insert in the surface (Sedzinski, et al., 2016; Werner, et al., 2014; Stubbs, et al., 2006). Then, once inserted, apical actin forms a lattice that generates the cell autonomous 2D force to expand the MCC’s apical surface, which is necessary to host hundreds of cilia (Sedzinski, et al., 2016). In addition, within this apical actin lattice, basal bodies are evenly distributed across the cell surface (Antoniades, et al., 2014; Werner, et al., 2011). Finally, the planar polarization of cilia may require this apical actin lattice which is essential for coordinated beating to generate fluid flow (Werner, et al., 2011; Park, et al., 2008). Therefore, F-actin plays a central role in the formation, patterning, and subsequent function of cilia in MCCs. However, our understanding of the molecular regulation of actin assembly in MCCs remains rudimentary (Sedzinski, et al., 2017; Sedzinski, et al., 2016). Specifically, how the actin lattice architecture is defined and maintained is not understood.
Here, we show that WDR5 is a key regulator of the apical organization of actin in MCCs. WDR5 is a core subunit of the human histone H3 Lys4 methyltransferase (H3K4MT) complexes that are essential for chromatin modification and transcriptional regulation (Trievel and Shilatifard, 2009; Patel, et al., 2008b; Dou, et al., 2006; Wysocka, et al., 2005). In particular, WDR5 is a highly conserved scaffolding protein essential for the association of RbBP5, ASH2L, and mDPY30 with MLL1 (Odho, et al., 2010; Trievel and Shilatifard, 2009; Patel, et al., 2008b). The assembly of this complex is dependent on the 7-fold β-propeller structure of WDR5 and is essential for H3K4MT activity (Dharmarajan, et al., 2012; Wysocka, et al., 2005). As will become important below, the N-terminal tail of WDR5 (1–29 amino acids) is dispensable for the formation of the β-propeller as well as the assembly of a functional H3K4MT complex (Schuetz, et al., 2006). On the other hand, WDR5 has an essential arginine-binding cavity that interacts with the arginine-containing WIN (WDR5-interacting) motif of MLL (Dharmarajan, et al., 2012; Patel, et al., 2008a). Specifically, a mutation in the WIN motif of the MLL protein (R3765A) or the arginine-binding cavity in WDR5 (S91K) disrupts assembly and activity of the H3K4MT complex (Patel, et al., 2008b). Nearly all studies of WDR5 focus on its nuclear function regulating H3K4 methylation, and while WDR5 has been localized outside the nucleus, a cytoplasmic role is not well defined (Bailey, et al., 2015; Wang, et al., 2010).
Our studies began when WDR5 was identified from a genomic analysis of congenital heart disease (CHD) and heterotaxy patients (Zaidi, et al., 2013). Heterotaxy (Htx) is a disorder of leftright patterning that can have a severe effect on cardiac patterning and function (Sutherland and Ware, 2009). A patient with a de novo missense mutation (K7Q) in WDR5 exhibited a conotruncal defect and a right aortic arch (normally the arch is on the left) (Zaidi, et al., 2013). How this chromatin modifier might affect a specific phenotype such as cardiac development was unknown. Depletion of WDR5 in Xenopus recapitulates a left-right patterning defect with abnormal ciliogenesis in the left-right organizer (Kulkarni et al in press).
Importantly, patients with CHD and Htx often suffer from postsurgical morbidity and mortality associated with respiratory complications, a result of cilia dysfunction in MCCs (Garrod, et al., 2014; Nakhleh, et al., 2012; Swisher, et al., 2011; Kennedy, et al., 2007). Since a number of genes are essential for ciliogenesis in both monociliated cells (as in the left-right organizer) and MCCs (mucociliary epithelia), here, we demonstrate that WDR5 is critical for ciliogenesis in the MCCs (Casey, et al., 2015; Nakhleh, et al., 2012; Becker-Heck, et al., 2011; Merveille, et al., 2011; Kennedy, et al., 2007; Tan, et al., 2007). Unexpectedly, in addition to its role in transcriptional regulation, we discovered a chromatin independent role in ciliogenesis in MCCs. Using mutational analysis, we show that the N-terminal tail of WDR5 is essential for ciliogenesis albeit unnecessary for chromatin modification. We discover that Wdr5 is localized near the base of cilia, and the β-propeller structure of WDR5 is necessary and sufficient for this localization. Interestingly, Wdr5 regulates the enrichment of apical actin, and MCCs depleted of Wdr5 fail to expand their apical surface and lose planar distribution and organization of basal bodies. Using high-resolution confocal imaging, we show that Wdr5 localizes between F-actin and the basal bodies suggesting an alternative scaffolding role between these two structures. Further, using live imaging, we find that Wdr5 first associates with basal bodies deep in the cytoplasm and migrates apically. Upon reaching the apical surface, Wdr5 dynamically interacts with F-actin, where F-actin appears to organize around Wdr5. Finally, using a monomer trap experiment, we show that Wdr5 is necessary to stabilize these actin polymers. Taken together, we discover a non-chromatin function for WDR5, where it stabilizes the actin lattice architecture to promote formation and function of MCCs.
RESULTS
WDR5 is essential for cilia in MCCs
In a separate report, we describe the role of WDR5 in left-right patterning via regulation of ciliogenesis in the left-right organizer (Kulkarni et al in press). Because respiratory complications are an established co-morbidity in CHD patients with Htx (Li, et al., 2015; Nakhleh, et al., 2012; Swisher, et al., 2011), we also examined the cilia in the MCCs of the Xenopus embryonic epidermis, which is the focus of this study. In Xenopus, the embryonic epidermis has an array of MCCs that provide an ideal model to study cilia assembly and function (Werner and Mitchell, 2013). Wdr5 depletion resulted in a dramatic loss of cilia, specifically fewer and shorter cilia in MCCs (Figure 1A), suggesting that WDR5 may play an important role in mucociliary clearance. To this end, we examined cilia-driven extracellular fluid flow over the surface of the Xenopus embryo as a test of cilia function in MCCs. We visualized the flow by adding latex microspheres (beads) to the culture media followed by time-lapse imaging of bead movement. In wildtype (WT) embryos, cilia-generated flow was brisk (Figure 1B,C, Video S1). Wdr5 depletion led to significant reduction in cilia generated fluid flow as expected for a dramatic loss of cilia (Figure 1B, C, Video S1). To quantitate the loss of fluid flow, we measured the maximum displacement of beads along the length of the embryo (Fig 1B, purple arrow to white dashed line) after six seconds. Depletion of Wdr5 led to a significant reduction in bead displacement (Fig 1C).
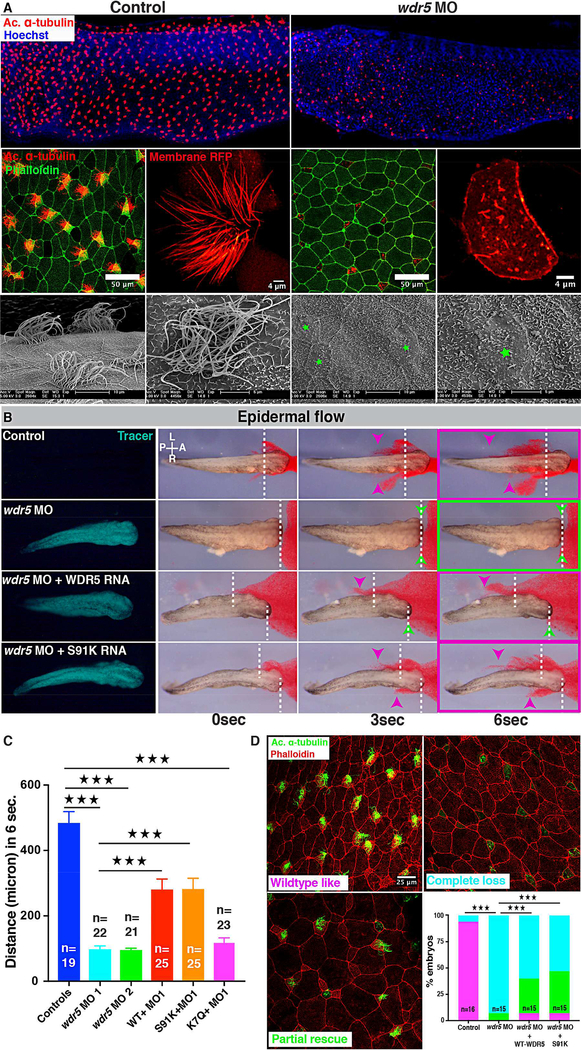
Figure 1: Wdr5 regulates ciliogenesis in MCCs independently of H3K4MT.
(A) X. tropicalis epidermal MCCs marked either by anti-acetylated α-tubulin (red) or membraneRFP, actin is labeled with phalloidin and nuclei with Hoechst. Scanning Electron Microscope images of multiciliated cells of Xenopus embryos (green stars mark MCCs). Uninjected control embryos on left and embryos injected at one cell stage with wdr5 MO on right.
(B) Cilia driven epidermal flow is visualized using red microbeads over the period of 6 seconds (see Movie S1). Magenta arrowheads indicate bead displacement from starting point (dashed lines). Green arrowheads indicate no displacement. Green fluorescent protein traces the morpholino and RNA. Experimental conditions include wdr5 MO, wdr5 MO + human WT WDR5 RNA, wdr5 MO + human mutant S91K WDR5 RNA and compared to uninjected controls.
(C) Quantification of maximum bead displacement (micron) in 6 seconds in uninjected controls, first wdr5 MO, second wdr5 MO, first wdr5 MO + WT-WDR5–3×GFP, first wdr5 MO + S91K-WDR5–3×GFP, first wdr5 MO + K7Q-WDR5–3×GFP. n = number of embryos. ★★★ indicate statistical significance at P < 0.0005.
(D) X. tropicalis epidermal MCCs marked with anti-acetylated α-tubulin (green) for cilia and phalloidin (red) for actin. Percentage of embryos with wildtype-like cilia (magenta), partial rescue (green), or complete loss (cyan) of cilia under different conditins: uninjected controls, wdr5 MO, wdr5 MO + human WT-WDR5, and wdr5 MO + human S91K-WDR5. n = number of embryos.
★★★ indicate statistical significance at P < 0.0005
To test the specificity and efficiency of our Wdr5 depletion, we employed multiple tests. First by Western Blot, we detected a reduction in Wdr5 protein in morphants at stage 20 (when MCCs are intercalating) that is partially rescued by injecting human wildtype (WT) WDR5 mRNA (Figure S1A). Second, we tested the specificity of our Wdr5 antibody by serial dilution of a blocking peptide that reduced Wdr5 signal on the Western Blot (Figure S1B). Third, injecting a scrambled MO did not result in any change in cilia driven flow compared to WT embryos (Figure S1D). Fourth, co-injecting a WT human WDR5 mRNA (tagged or untagged) in wdr5 morphants rescued the flow defect, confirming the specificity of our knockdown (Figure 1B, C, Figure S1E, I, J, Video S1). Fifth, co-injecting a WT human WDR5 mRNA in wdr5 morphants partially rescued the cilia loss (Figure 1D). Sixth, we also injected a non-overlapping second morpholino, which also showed significantly reduced cilia driven fluid flow (Figure 1C, Figure S1F). Lastly as an alternative depletion strategy, we injected a wdr5 CRISPR and examined F0 CRISPR MCCs, which also showed defective cilia in and reduced flow (Figure S1G, K) (Bhattacharya, et al., 2015). Of note, the effect of wdr5 CRISPR on cilia loss and cilia driven flow was of less magnitude than wdr5 MO. wdr5 mRNA is expressed maternally (Figure S1L) (Owens, et al., 2016), which could be manipulated by our ATG MO but not by our F0 CRISPR and may explain the difference in the degree of phenotype. In sum, we conclude that the Wdr5 depletion by MO is specific, and Wdr5 is essential for ciliogenesis in the MCCs.
WDR5 regulates foxj1 expression in MCCs
WDR5 is a core subunit of the MLL/SET1 H3K4MT complex and has been studied extensively for its role in histone methylation, chromatin modification, and transcriptional regulation (Dharmarajan, et al., 2012; Odho, et al., 2010; Trievel and Shilatifard, 2009; Patel, et al., 2008a; Patel, et al., 2008b; Dou, et al., 2006; Gori, et al., 2006; Wysocka, et al., 2005). Therefore, we began our analysis of the wdr5 cilia phenotype by examining the transcriptional regulation of genes critical for ciliogenesis. Except for foxj1, we did not detect a significant difference in the expression of a collection of genes required for proper cilia specification and function between morphants and controls (Figure S2A, D). We also examined molecular markers of non-ciliated cells in the embryonic epidermis including ionocytes (xeel) and goblet cells (atpv1a) but detected no significant difference in expression between controls and morphants (Figure S2B, D). This suggests that Wdr5 depletion specifically affects MCCs and predominately the expression of foxj1. The reduced expression of foxj1 in morphants was consistent with WDR5 localization in the nucleus during MCC specification stages (Figure S3). Also, previous genomic studies determined that RbBP5 and WDR5 bind to the FOXJ1 locus and that the FOXJ1 locus is a direct target of H3K4 tri-methylation (Mikkelsen, et al., 2007). To test the role of foxj1, we injected foxj1 mRNA into Wdr5 depleted embryos and examined cilia driven flow (Figure S2C). In WT embryos, cilia-generated flow was brisk. Wdr5 depletion led to significantly reduced fluid flow, but foxj1 mRNA did not rescue the flow defect (Figure S2C). However, we did detect monocilia in cells flanking the MCCs, an established effect of foxj1 overexpression confirming that our mRNA dose was functional (data not shown). Our results are consistent with a role for WDR5 in the transcriptional control of foxj1 but also suggest that WDR5 has a downstream or additional role in MCCs.
Mutational analysis reveals WDR5 has an H3K4 independent role in ciliogenesis in MCCs
WDR5 forms a 7-fold β-propeller structure that is essential for its scaffolding function between the catalytic subunit (SET1A, SET1B, MLL1, MLL2, MLL3, or MLL4) and the core regulatory subunits (RbBP5, ASH2L, and mDPY-30) to form a functional H3K4MT complex (Odho, et al., 2010; Trievel and Shilatifard, 2009; Patel, et al., 2008b). Interestingly, the first N-terminal 29 amino acids of WDR5 are dispensable for the β-propeller structure and are not required for H3K4MT function (Schuetz, et al., 2006). Therefore, the relevance of our patient’s allele, K7Q, was uncertain. To test the K7Q variant functionally, we attempted to rescue the loss of ciliadriven fluid flow phenotype in wdr5 morphants with the K7Q WDR5 variant. In contrast to WT WDR5, the K7Q WDR5 variant failed to rescue ciliary flow defects in wdr5 morphants (Figure 1C, Figure S1H) despite being expressed (Figure S1C) supporting the hypothesis that it is a loss of function allele (Zhu, et al., 2017).
While the N-terminal 29 amino acids of WDR5 are not required for the structure and function of the H3K4MT, it remained possible that K7Q allele could disrupt the assembly of the β-propeller in a “dominant negative” manner (although overexpression of the K7Q allele alone did affect cilia flow, Figure 1C). Alternatively, the N-terminal tail of WDR5 (1–29) could play a vital role in ciliogenesis. Therefore to distinguish between these two hypotheses, we deleted the first 25 amino acids of WDR5 (26–334-3×GFP) and attempted to rescue the cilia flow defects in morphants (Figure S1I). Again, in contrast to WT WDR5, the 26–334 WDR5 variant failed to rescue ciliary flow defects in morphants supporting the hypothesis that the N-terminal tail of WDR5 is essential for ciliogenesis.
Since the N-terminal tail is essential for cilia in MCCs but dispensable for H3K4MT activity, we postulated that WDR5 might have an additional role that is independent of chromatin modification. Previous biochemical studies have identified that the arginine-binding cavity on one face of the WDR5 β-propeller binds to the WIN motif of the MLL/SET catalytic subunit (Dharmarajan, et al., 2012; Trievel and Shilatifard, 2009; Patel, et al., 2008a). A mutation in the arginine-binding cavity (S91K WDR5) disrupts the assembly and activity of the H3K4MT complex (Patel, et al., 2008b). Therefore, to test for an H3K4 independent role, we injected the S91K WDR5 variant mRNA into wdr5 morphants and assayed cilia driven fluid flow. Co-injecting the S91K mutant RNA partially rescued the cilia as well as flow defect in wdr5 morphants strongly suggesting that Wdr5 may be playing an H3K4 independent role in cilia (Figure 1B-D, Figure S1J, Video S1). We verified that the S91K WDR5 protein is expressed at similar levels to WT and K7Q WDR5 using western blot (Figure S1C). To summarize, WDR5 mutant constructs that are sufficient for chromatin function (26–334-WDR5) are insufficient for cilia function, while mutant constructs that abrogate chromatin function (S91K-WDR5) are sufficient for cilia function. These studies in MCCs strongly suggest an H3K4 independent role for WDR5.
WDR5 is localized near the base of the cilium
To identify an alternative, H3K4 independent role for Wdr5 in MCCs, we overexpressed a human WDR5-GFP construct in Xenopus embryos. As expected, we did detect GFP signal in the nucleus but also in a punctate pattern near the apical surface of the MCCs, reminiscent of basal bodies (Figure S4A,B). Earlier, we had verified that WDR5-GFP is functional and localized appropriately by injecting WDR5-GFP mRNA in wdr5 morphants to rescue cilia driven flow defects (Figure S1E). Even then, wary of overexpression artifacts of GFP-tagged proteins, we sought to confirm this Wdr5 localization near the bases of cilia by immunofluorescence. Using an anti γ−tubulin and an anti-WDR5 antibody, we found that WDR5 localizes near the bases of cilia in the MCCs of the Xenopus epidermis (the membrane localization of the WDR-GFP may be non-specific) (Figure S4C). To see if this finding generalizes to mammals, we examined the MCCs of mouse tracheal epithelia. Again, we found that WDR5 is localized near the ciliary base in mammalian MCCs (Figure S4D), and the signal can be eliminated in the presence of a blocking peptide (ratio 1:2 antibody: blocking peptide, Figure S1B) indicating specificity (Figure S4D). Therefore, in Xenopus and mouse MCCs, WDR5 is located near the base of cilia.
The β-propeller structure is necessary and sufficient for WDR5 localization to the ciliary base in MCCs
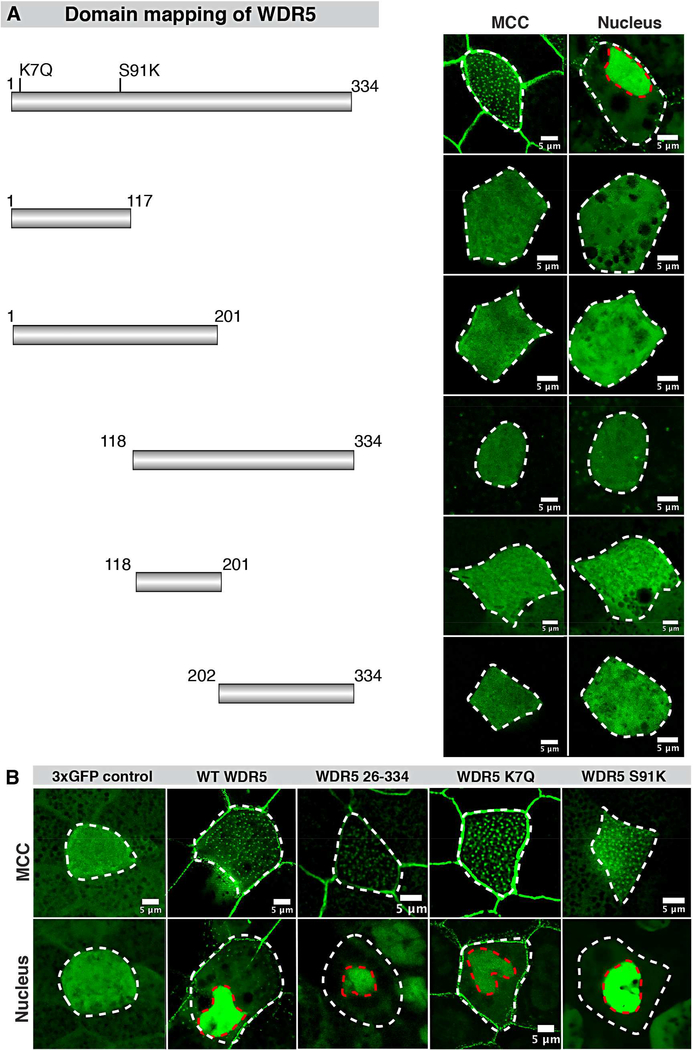
Since the N-terminal tail is essential for cilia in MCCs, we hypothesized that it may be necessary for ciliary localization. Alternatively, the β-propeller, WD repeats, or another portion of the protein may be required for ciliary localization. To test these alternatives, we made different deletion constructs tagged with 3×GFP and expressed them in Xenopus embryos. We divided WDR5 into 5 constructs 1–117 (1st and 2nd WD repeats), 118–201 (3rd and 4th WD repeats), 1201 (1–4 WD repeats), 118–334 (3–7 WD repeats), 202–334 (5–7 WD repeats) (Figure 2A). As we expected, WT WDR5–3×GFP was expressed in the nucleus as well as at the ciliary base in the MCCs (Figure 2A). However, none of the deletion constructs localized to the nucleus or bases of cilia (Figure 2A,B). These results suggest that the entire β-propeller structure is necessary for WDR5 ciliary localization in MCCs.
Figure 2: WDR5 mutants localize near the base of cilia and nucleus in MCCs.
(A) WDR5 was tagged with 3×GFP at the C-terminal end to study the localization of WDR5 mutants at the ciliary base. Wildtype WDR5 was localized to the nucleus and ciliary base in MCCs. None of the WDR5 deletion constructs (1–117, 1–201, 118–334, 118–201, and 202–334) localized either to the nucleus or ciliary base.
(B) Mutants 26–334 WDR5, patient mutation (K7Q) and methylation dead mutant (S91K) localize at the ciliary base and nucleus. Control 3×GFP only does not localize specifically to either location.
See also Figure S4
To explore the role of the N-terminal, we examined our 26–334-3×GFP construct, which contains the β-propeller structure but not the N-terminal tail of WDR5 (Schuetz, et al., 2006). Surprisingly, 26–334-3×GFP localized similarly to WT WDR5, both to the nucleus and bases of cilia, suggesting that β-propeller structure is sufficient for WDR5 ciliary localization in MCCs (Figure 2B). Consistently, the patient variant K7Q, also localized to the bases of cilia and nucleus (Figure 2B). Based on these results, we conclude that the β-propeller structure of WDR5 is necessary and sufficient for ciliary localization.
Finally, to test if the assembly of the H3K4MT complex is essential for the localization of WDR5, we examined the localization of the S91K variant. Interestingly, S91K-3×GFP also localized to the ciliary base (Figure 2B), suggesting the assembly of H3K4MT complex may not be necessary for WDR5 ciliary localization.
Wdr5 is necessary for basal body patterning and polarization
To understand how WDR5 localization translates into its function, we organized our analysis into four “steps” of ciliogenesis (Figure 3A). The four steps are: 1) MCCs are specified in the deeper epithelial layer, 2) MCCs insert themselves into the superficial epithelia. They simultaneously begin basal body biogenesis within the cytoplasm, 3) Apical enrichment of actin leads to apical expansion of MCCs. Basal bodies also start to migrate and dock to the apical surface, and 4) Basal bodies dock, distribute evenly, and orient at the apical surface, which leads to ciliogenesis(Sedzinski, et al., 2016; Zhang and Mitchell, 2015; Klos Dehring, et al., 2013; Werner, et al., 2011; Vladar and Axelrod, 2008). Cilia mediated flow then reinforces basal body polarity(Mitchell, et al., 2007). Because WDR5 is localized to basal bodies, we predicted that WDR5 might play an important function in basal body patterning during cilia assembly (step 4). Therefore, we investigated apical docking and the distribution and polarization of basal bodies in wdr5 morphants.
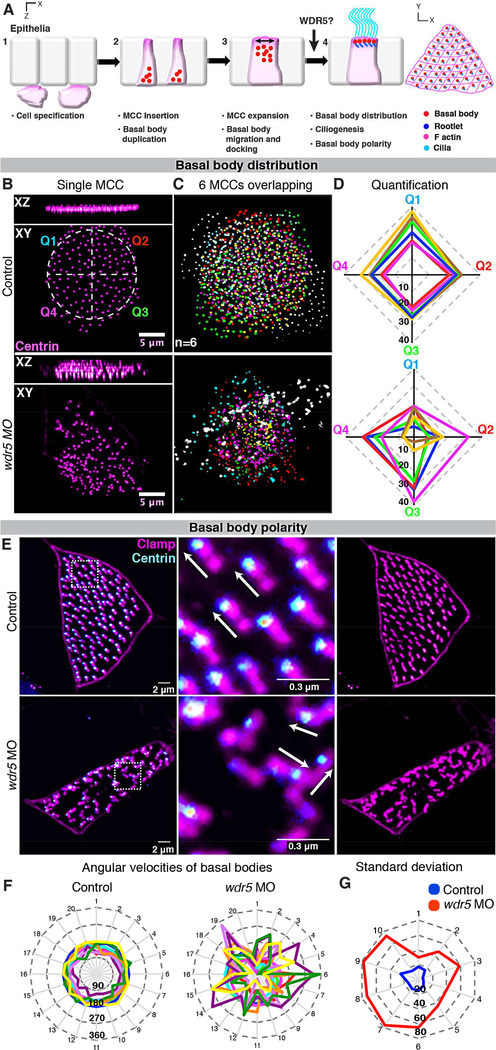
Figure 3: Wdr5 is necessary for uniform distribution and polarization of basal bodies.
(A) The four steps of ciliogenesis in MCCs: 1) MCCs are specified in the deeper epithelial layer, 2) MCCs insert themselves into the superficial epithelia. They simultaneously begin basal body biogenesis deep in the cytoplasm, 3) Apical enrichment of actin leads to apical expansion of MCCs. Basal bodies also start to migrate and dock to the apical surface, and 4) Basal bodies dock, distribute evenly, and orient at the apical surface, which leads to ciliogenesis. Cilia mediated flow then reinforces basal body polarity.
(B-D) Wdr5 is essential for uniform basal body distribution in MCCs. (B) MCC showing uniform and clumped distribution of basal bodies (Centrin-RFP) in uninjected control and wdr5 morphant respectively. Control MCC is divided into 4 quadrants labeled Q1–4 with different colors to use for quantification of distribution pattern in (D). n = number of MCCs.
(C) Basal bodies from 6 individual MCCs from different embryos are overlaid on each other to show loss of uniform distribution of basal bodies in wdr5 morphants. Basal bodies from each MCC are colored differently.
(D) Quantitative analysis of basal body distribution in uninjected controls and wdr5 MO injected embryos. Each MCC was divided into 4 quadrants and the number of basal bodies in each quadrant are plotted on their respective axes (Q1-Q4, see panel D). We measured number of basal bodies in each quadrants for 6 MCCs. In the graph, each color represents one MCC. Squareness represents uniform distribution of basal bodies in a MCC.
(E-G) Wdr5 is essential for planar organization of basal bodies in MCCs. (E) MCCs showing parallel and random orientation of rootlets (Clamp-GFP, magenta) with relation to basal bodies (Centrin-RFP – cyan) in controls and wdr5 MO injected embryos respectively.
(F) Quantitative analysis of basal body polarity with angular velocity graphs. Each color represents a MCC (10 total) and axes show orientation of rootlets for 20 basal bodies per MCC. Length of axis represents angle of orientation (0–360). Circularity depicts rootlets are parallel to each other and basal bodies are planar polarized.
(G) Angular velocity graph showing standard deviation in the orientation of rootlets. Here, each axis represents each MCC and length represents standard deviation in angle of orientation of 20 rootlets within each MCC. Standard deviation is smaller in controls, as rootlets are more parallel to each other compared to wdr5 MO injected embryos. ★★ indicate statistical significance at P < 0.005.
We first examined basal body migration and distribution at the apical surface. In WT embryos, centrin-RFP, which marks the basal bodies, is expressed apically and distributed across the cell surface as an ordered array (Figure 3B, upper panels). Depletion of Wdr5 mildly affected apical docking of basal bodies but dramatically disrupted their planar distribution across the apical cell membrane (Figure 3B, lower panels). To visualize the apical basal body distribution defects, we overlaid 6 MCCs (where each color represents a different MCC) from control and wdr5 morphants (Figure 3C). To quantify their distribution, we divided each MCC into four quadrants and counted the number of basal bodies in each quadrant (Figure 3B,D). In the graph (Figure 3D), each axis represents a different quadrant, the length from the center represents the number of basal bodies in a particular quadrant, and each color identifies a MCC where basal bodies are counted. Control MCCs show similar number of basal bodies in each quadrant (squareness of the plot), whereas wdr5 morphant MCCs show dramatically different numbers of basal bodies among quadrants (loss of squareness).
Basal body polarity helps drive directional beating and unidirectional fluid flow, which in turn reinforces the polarity of the cilia (Vladar and Axelrod, 2008; Mitchell, et al., 2007). Therefore, in Wdr5 depleted embryos, the loss of cilia and abnormal distribution of basal bodies suggests that the basal bodies may also fail to orient properly. We marked the basal bodies with centrin-RFP and the ciliary rootlets with clamp-GFP to visualize orientation (Figure 3E) (Werner and Mitchell, 2013). In control embryos, the basal bodies and rootlets are organized in an ordered, parallel array (Figure 3E, upper panels). Depletion of Wdr5 disrupts this orientation of the basal bodies and rootlets (Figure 3E, lower panels). We quantified orientation using angular velocity graphs (Figure 3F). The angular velocity measures the orientation of each rootlet, where the vector starts at the end of the rootlet, proceeds along the length of the rootlet, and terminates at the basal body. In Figure 3F, each axis represents the angular velocity of one basal body for a total of 20 basal bodies, where larger angles are plotted further from the center. Each color represents a different MCC for a total of 10 MCCs. In WT cells, the plot is nearly circular, which depicts uniformly polarized basal bodies. The circularity of the plot is clearly disrupted in morphants (Figure 3F). If we measure the standard deviation of the angular velocity in each of the 10 cells, we see that the wdr5 morphants are dramatically more randomly oriented (Figure 3G). Taken together, we conclude that Wdr5 is necessary for the planar distribution and polarization of the basal bodies, a result that may be related to its location near the ciliary base.
Wdr5 is essential for apical expansion and actin enrichment in MCCs
To dissect the timing of Wdr5 function, we examined an earlier step (step 3, Figure 4A) of MCC formation. MCCs originate in the basal layers of the epidermis and are then inserted apically to form the superficial epithelia (Stubbs, et al., 2006). Once inserted, the apical surface of the MCC must expand to generate the surface area necessary to host hundreds of cilia (Figure 4A). First, we measured the number of MCCs inserted in the superficial epithelia during the insertion process and found no difference between wdr5 morphants and controls (Figure S5A,B). Next, we measured the apical surface area and found that MCCs in wdr5 MO injected embryos fail to expand their apical surface over time despite starting at a similar apical surface area at early stages (Figure 4B,C, Figure S5A,C). Interestingly, the smaller MCC surface area is coupled to a significantly larger surface area in the non-ciliated cells (Figure 4D), suggesting compensation across the epithelial sheet. Of note, MCCs depleted of Wdr5 by F0 CRISPR also had a smaller apical area albeit to a lesser degree than MO depleted embryos (Figure S6).
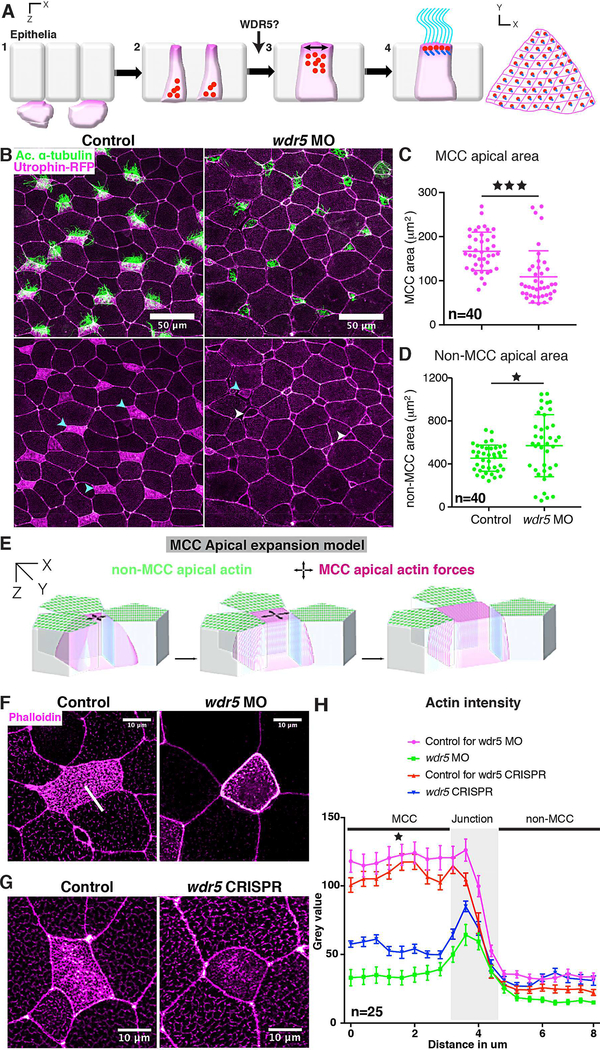
Figure 4: Wdr5 is essential for apical expansion and actin assembly in ciliated cells.
(A) The four steps of ciliogenesis in MCCs with emphasis on the role of WDR5 during the third step of MCC formation.
(B) Xenopus epidermal MCCs marked with anti-acetylated α-tubulin (green, cilia) and utrophinGFP (magenta). Cyan and white arrowheads show ciliated cells with apical enrichment and loss of actin respectively.
(C, D) Apical area of MCCs (C) and non-MCCs (D) in controls and wdr5 MO injected embryos. n = number of MCCs.
(E) Model representing actin dependent apical surface expansion in MCCs.
(F, G, H) Quantitative analysis of apical enrichment of actin in MCCs and neighboring nonMCCs in controls and (F) wdr5 MO or (G) wdr5 CRISPR injected embryos. Actin enrichment was quantified (H) using a line scan (white line) spanning MCC and neighboring non-MCC. Actin was stained using phalloidin. n = number of MCCs.
★ and ★★★ indicate statistical significance at P < 0.05 and P < 0.0005. Data are represented as mean ± SEM.
See also Figure S5 and S6
Little is known about the molecular regulators that are essential for apical expansion of MCCs although the apical actin network plays a prominent role (Sedzinski, et al., 2017; Sedzinski, et al., 2016). Previous work has shown that actin is enriched apically during MCC emergence and is critical for generating cell autonomous 2D pushing forces to expand the MCC (Figure 4E) (Sedzinski, et al., 2016). Given the central role of actin in apical expansion, we wondered if Wdr5 might regulate the enrichment of F-actin at the apical surface of the MCC. We measured F-actin (labeled using phalloidin) in the MCC and the neighboring non-ciliated cells of controls and wdr5 MO and F0 CRISPR injected embryos (Figure 4F,G, H) using a line scan. In WT embryos, we detected an apical enrichment of actin in the MCCs depicted by high fluorescence intensity; however, loss of Wdr5 led to a significant loss of apical actin (Figure 4F,G, H). On the other hand, F-actin was only mildly affected in neighboring, non-ciliated cells (Figure 4F,G, H). Our results suggest that Wdr5 plays a vital role in apical actin enrichment essential for apical expansion and basal body patterning.
WDR5 is interwoven with the actin lattice and connects basal bodies to actin
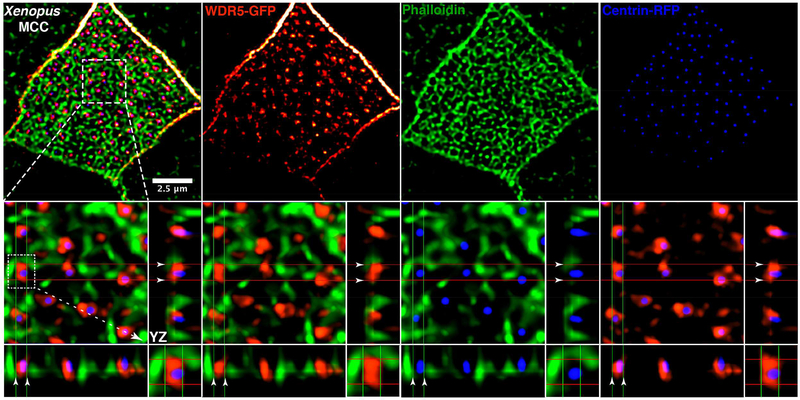
To connect the non-nuclear localization of Wdr5 (at the ciliary base), basal body patterning defects, and the loss of apical actin in MCCs depleted of Wdr5, we carefully examined their localization. In MCCs, apical actin not only creates the 2D force to expand the apical surface of the cell, it also creates a lattice in which basal bodies are embedded providing a framework for their ordered distribution across the apical cell surface (Sedzinski, et al., 2016; Werner, et al., 2011). Using high-resolution imaging in MCCs, we marked human WDR5 with WDR5-GFP, basal bodies with Centrin-RFP, and F-actin with phalloidin (Figure 5). We can readily resolve the apical actin lattice (green) in the MCCs and visualize the basal bodies (blue) docked within the lattice. WDR5-GFP (red) is interwoven within the actin lattice. Multiciliated cells in the mouse trachea also showed a similar localization pattern for WDR5 and F-actin, such that WDR5 is interwoven within the actin network (Figure S7A). Interestingly, WDR5 localizes immediately adjacent to basal bodies and the actin network and appears to bridge the gap between the two. Using optical sections (Figure 5), we can distinguish a clear connection between WDR5 and both the actin filaments and the basal bodies.
Figure 5: WDR5 is a scaffold connecting basal bodies to actin.
A Xenopus epidermal MCC expressing WDR5-GFP (red), actin (green) labeled with phalloidin, and centrin-RFP (blue). Basal bodies (blue) dock in the space within the actin lattice. WDR5 localizes between actin and centrin. XZ and YZ projections show orthogonal view. In the orthogonal view, white arrowheads indicate the space between actin and a basal body, which is occupied by WDR5.
See also Figure S7
To independently investigate a scaffolding function of WDR5 between the basal bodies and the actin lattice, we employed a co-immunoprecipitation assay. We immunoprecipitated endogenous WDR5 from human RPE cells and detected both actin and γ-tubulin, which suggests a physical interaction of WDR5 with actin and γ-tubulin (Figure S7B).
The Wdr5-basal body complex interacts dynamically with actin during apical expansion
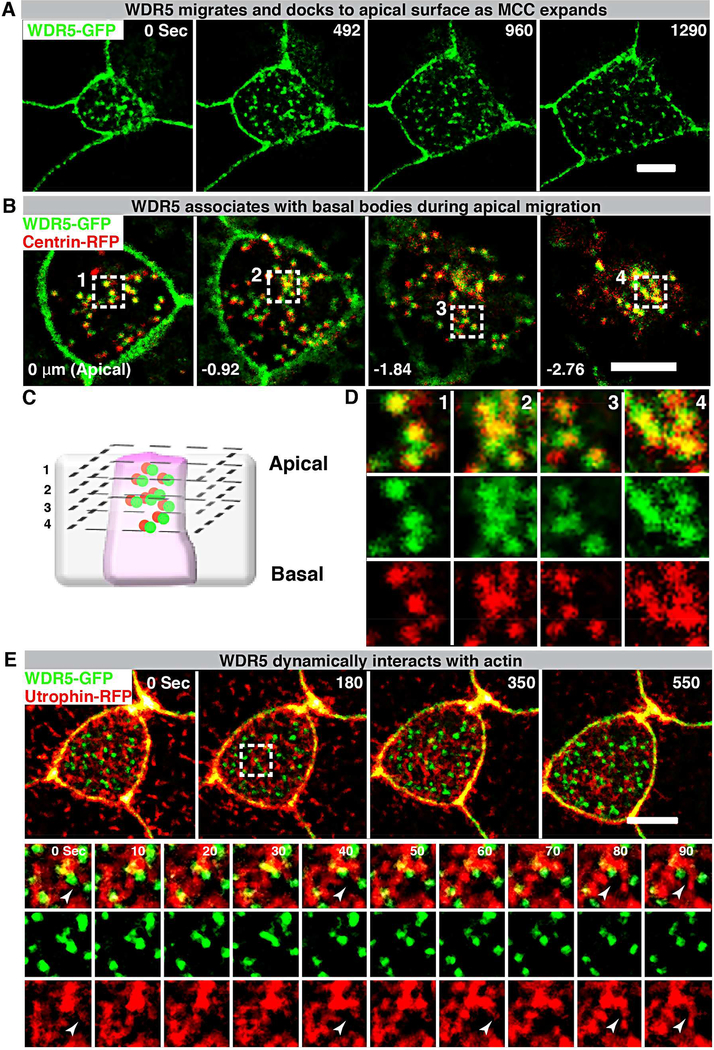
Basal bodies are each centered within the apical actin lattice. We could envision two scenarios that would involve WDR5 as a scaffold between actin and the basal body. In the first scenario, WDR5 associates with the actin network possibly organizing the apical actin lattice and then creating a “docking site” for the basal body. In the second scenario, WDR5 associates with the basal body and then dock at the apical surface to organize actin lattice. To distinguish between these two possibilities, we used live imaging to monitor these proteins during MCC apical expansion. In the case of WDR5-GFP, the punctate expression appears deep in the cell and migrates apically to the cell surface as the cell expands (Figure 6A, Video S2). This pattern resembled the appearance of basal bodies; therefore, we examined localization of basal bodies (Centrin-RFP) and WDR5-GFP concurrently (Figure 6B,C,D). We discovered that WDR5 was co-localized with basal bodies in the cytoplasm and migrated to the apical surface together. To gain insight into the relation with F-actin, we visualized F-actin with Utrophin-RFP and concurrently visualized WDR5-GFP (Figure 6E, Video S3, S4). Actin appeared to dynamically organize around WDR5 when WDR5 migrated to the apical surface (Figure 6E, Video S3, S4). From these studies, we conclude that WDR5 first localizes with the basal body within the cytoplasm and then migrates apically together. Interestingly, WDR5 appears to be a focal point around which actin organizes dynamically rather than docking into a stable actin lattice. To understand this process further, we examined the dynamic relationship between F-actin and WDR5.
Figure 6: WDR5-basal body complex interacts with F-actin during apical expansion.
(A) A montage of Xenopus epidermal MCC undergoing apical expansion over 21 minutes. WDR5 marked with WDR5-GFP localizes to the apical membrane as MCC expands.
(B,D) A montage of Xenopus epidermal MCC expressing WDR5-GFP and centrin-RFP early during MCC expansion process. Basal bodies (centrin-RFP) and WDR5 appear to co-localize deep in the cytoplasm.
(C) Schematic showing optical sections of a MCC to examine WDR5 and basal body colocalization in B and D. Optical section 1–4 correspond to the optical sections in the panels B and D.
Scale bar = 5 μM
(E) A montage of Xenopus epidermal MCC undergoing apical expansion over 9 minutes. WDR5 (WDR5-GFP) interacts with F-actin (Utrophin-RFP) at the apical surface as MCC expands. The region in a white square is magnified to show F-actin organizing around WDR5 (note white arrowheads).
WDR5 stabilizes F-actin in the MCCs
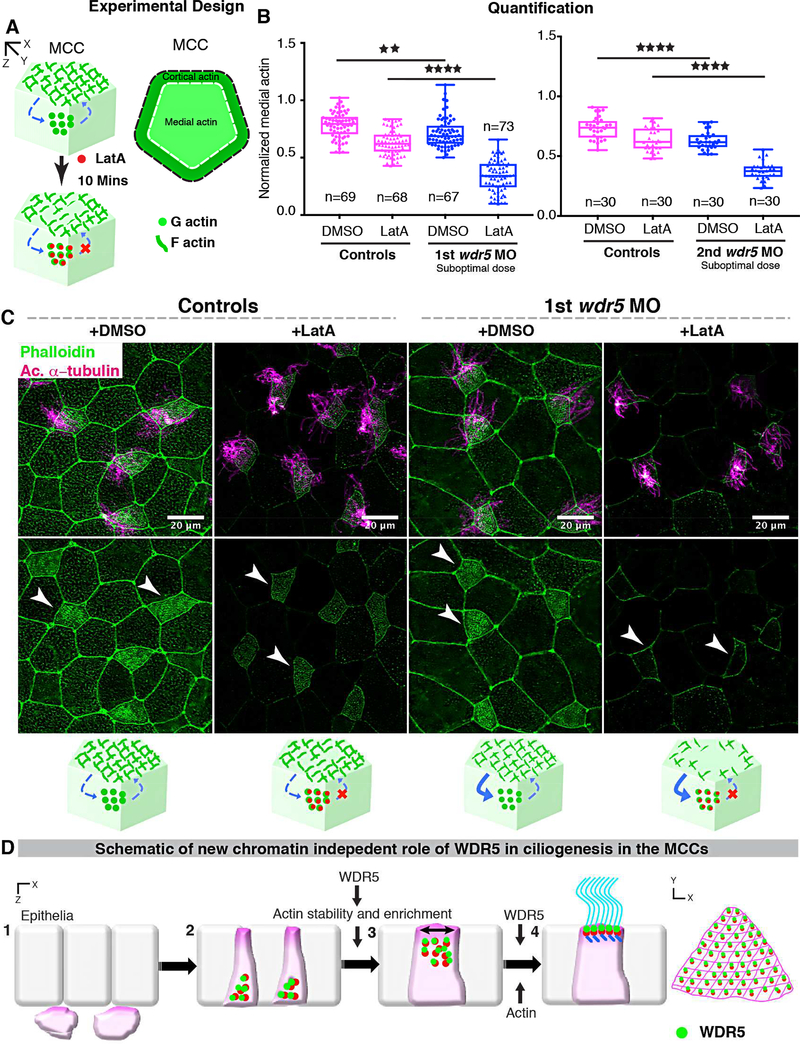
The F-actin network is a dynamic structure that is undergoing continuous assembly and disassembly from G-actin monomers (Blanchoin, et al., 2014). While key proteins like Formins and ARP2/3 are essential for actin assembly, other proteins may play a vital role in regulating the stability of this network by limiting disassembly (Pollard, 2016; Pollard, 1986). Since WDR5 depletion leads to a loss of apical actin enrichment, we hypothesized that either WDR5 promotes the assembly of actin or limits the rate of disassembly. To test these alternatives, we used a Latrunculin A (LatA) based monomer trap for G-actin, which inhibits actin polymerization by binding to G-actin in a stoichiometric 1:1 ratio (Yarmola, et al., 2000; Pollard, 1986). By exposing MCCs to LatA for a specific amount of time and measuring medial actin intensity, we can evaluate the rate of disassembly in MCCs of wildtype and Wdr5 depleted embryos (Figure 7A). For this experiment, we injected a suboptimal dose of wdr5 MO, which only partially affected actin enrichment in the MCCs (Figure 7B, C). We then exposed the embryos to either DMSO or 2 μm LatA for 10 mins (Figure 7B, C). In MCCs, actin enrichment was indistinguishable before or after DMSO treatment so we used DMSO treated embryos as a control for comparisons. We first normalized the medial actin with cortical actin and then compared the medial actin enrichment in MCCs between treatments (Figure 7B). We found that depletion of Wdr5 led to a greater loss of apical actin in response to LatA exposure, consistent with the hypothesis that Wdr5 is essential for stabilizing actin polymers and in turn maintaining the architecture of apical actin in MCCs.
Figure 7: Wdr5 is essential for stabilization of F-actin.
(A) Schematic showing the experimental design to examine the rate of F-actin disassembly in MCCs of Xenopus embryos. To isolate the effect of WDR5 in disassembly, we exposed epidermal MCCs to Latrunculin A (LatA) treatment for 10 mins. Latrunculin A specifically binds to G-actin in stoichiometric 1:1 ratio preventing the F-actin polymerization. Thus, exposure to LatA prevents the assembly but not disassembly of F-actin allowing us to evaluate the effect of WDR5 depletion on F-actin disassembly in MCCs. Based on earlier results that medial actin is necessary for apical expansion and our results that Wdr5 depletion leads to loss of medial Factin enrichment, we specifically examined the medial F-actin intensity in controls and wdr5 morphants. Medial actin intensity was normalized to cortical actin to allow us to combine results from different experiments for statistical comparison.
(B) Quantification of normalized medial actin intensity in the MCCs of controls and suboptimal dose of wdr5 morphants after exposure to DMSO (vehicle) or LatA (2 μM) for 10 mins using two non-overlapping wdr5 MOs. Our results show dramatic reduction in actin intensity in response to LatA in wdr5 morphants compared to DMSO only. n = number of MCCs.
(C) Immunofluorescence showing Xenopus epidermal MCCs labeled for F-actin (Phalloidin) and cilia (anti-acetylated α-tubulin) in controls and wdr5 morphants after exposure to DMSO (vehicle) or LatA (2uM) for 10 mins. Cartoon depicts the hypothesized rates of disassembly and the effect of LatA on medial actin enrichment in controls and wdr5 morphants.
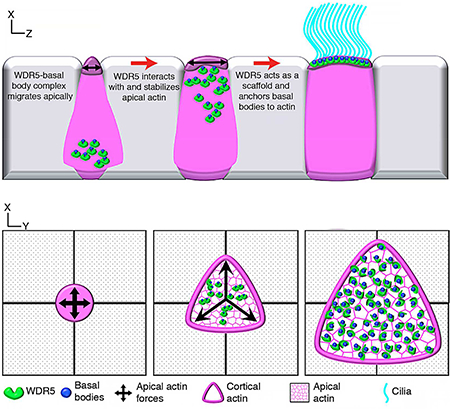
(D) Schematic of a model proposing the role of WDR5 in formation of a MCC. WDR5 binds to basal bodies as basal bodies are synthesized deep in the cytoplasm. WDR5 then migrates apically, where F-actin organizes around WDR5. WDR5 interacts with F-actin to stabilize actin network essential for apical expansion and basal body distribution. In the final stages, WDR5 anchors basal bodies to apical actin network to form functional MCCs.
DISCUSSION
Our results establish a non-chromatin role for WDR5 in F-actin stabilization that is essential for the organization of apical actin in MCCs. In addition to the traditional role for WDR5 as a scaffold for the H3K4 methyltransferase complex, our results suggest an unexpected scaffolding function between basal bodies and F-actin in MCCs. We propose the following model for WDR5 function at the ciliary base in MCCs. First, WDR5 associates with basal bodies and migrates to the apical surface. There it interacts with the developing apical actin lattice to stabilize it, which also generates the force for apical expansion. Stabilization of this actin lattice then provides the organizing framework to uniformly distribute the basal bodies across the apical surface. Finally, WDR5 is essential for anchoring basal bodies to the apical actin network for MCC function (Figure 7D).
In the nucleus, WDR5 plays a critical role in chromatin modification. Consistent with a multitude of studies emphasizing the role of WDR5 in H3K4MT activity and transcription, we found that foxj1 was downregulated in wdr5 morphants. Our results are consistent with multiple different findings: 1) the FOXJ1 locus is methylated (Mikkelsen, et al., 2007), 2) global ChIP-seq analysis identifies the FOXJ1 locus as a target of WDR5, RbBp5 and H3K4m3 (Ang, et al., 2011) and 3) Foxj1 is an established regulator of ciliogenesis (Stubbs, et al., 2008; Yu, et al., 2008). Specifically, Foxj1 is necessary and sufficient for cilia in mono-ciliated cells but not in MCCs. Interestingly, while foxj1 was downregulated in MCCs in wdr5 morphants, exogenous foxj1 did not rescue the loss of cilia suggesting that Wdr5 has another role downstream of foxj1. While foxj1 and other targets of transcriptional control by WDR5 may play a role in ciliogenesis of MCCs, our mutant analysis suggested an unexpected chromatin independent role, which became the focus of our studies.
Building a MCC is a complex biological process, which involves not only cell migration and intercalation but also dramatic intracellular patterning of organelles for effective function (Meunier and Azimzadeh, 2016; Brooks and Wallingford, 2014). MCCs first form in the basal epithelial layer and then insert into the superficial epithelia. Actin is critical for radial intercalation of cells; however, we did not detect any defects in the apical migration of the MCCs. This suggests specificity in the role of WDR5 in regulating F-actin. WDR5 affects the enrichment of F-actin locally at the apical surface of MCCs, a role that is supported by its localization. In addition, F-actin is hypothesized to bind basal bodies and play a vital role in the apical migration within a MCC (Brooks and Wallingford, 2014; Vladar and Axelrod, 2008; Boisvieux-Ulrich, et al., 1990). The original study used Cytochalasin D to disrupt actin and demonstrated that basal bodies arrest in the cytoplasm. However, whether F-actin associates with basal bodies within the cytoplasm was unknown (Boisvieux-Ulrich, et al., 1990). We did not observe any F-actin bound to apically migrating basal bodies. Moreover, Wdr5 depletion led to relatively small defects in basal body apical migration. In our experiments Wdr5 depletion is partial, whether the remaining Wdr5 is sufficient or whether Wdr5/actin does not have a role in basal body migration requires further testing. Clearly, WDR5 and actin are essential for the apical expansion, planar distribution and polarity of the basal bodies, and ciliogenesis.
F-actin organizes into a network that is essential for apical expansion (Sedzinski, et al., 2016; Stubbs, et al., 2006). Given that F-actin is inherently dynamic, the molecules that stabilize Factin are necessary to maintain the actin lattice in the MCCs. Using live imaging and an actin monomer trap, we discovered that WDR5 is essential to stabilize F-actin in MCCs. Importantly, Wdr5 first associates with basal bodies before they dock to the apical surface. Then, Wdr5 interacts with actin polymers to stabilize them. Numerous studies have found that basal body migration/docking defects are often associated with failure to enrich apical actin (Epting, et al., 2015; Antoniades, et al., 2014; Brooks and Wallingford, 2014; Ioannou, et al., 2013; Park, et al., 2008). Our results suggest that the process of basal body migration recruits molecules such as WDR5 to stabilize the apical actin network. Interestingly, molecules essential for actin remodeling are often localized to the basal bodies (Epting, et al., 2015; Antoniades, et al., 2014; Park, et al., 2008; Huang, et al., 2003) suggesting that the basal bodies are actively organizing the actin lattice.
Finally, our results emphasize the importance of patient driven mechanism discovery. Recent studies in CHD and autism clearly point to a role for chromatin modifiers in disease pathogenesis, but the molecular mechanisms remain unclear (De Rubeis, et al., 2014; Zaidi, et al., 2013). WDR5 was identified as a candidate CHD/Htx gene, and given the respiratory complications associated with these diseases, we sought to examine MCC function in our highthroughput Xenopus mucociliary disease model (Garrod, et al., 2014; Werner and Mitchell, 2013; Nakhleh, et al., 2012; Swisher, et al., 2011; Tan, et al., 2007; Houtmeyers, et al., 1999). In doing so, we identified an unexpected non-chromatin function for WDR5 in mucociliary clearance providing a plausible pathogenesis mechanism for respiratory complications in patients with WDR5 related CHD/Htx. Thus, the discovery of a cilia phenotype may have important clinical implications for patient management.
STAR METHODS
Contact for Reagent and Resource Sharing
Further information and requests for reagents may be directed to and will be fulfilled by the Lead Contact, Mustafa K. Khokha (mustafa.khokha@yale.edu)
Experimental Model and Subject Details
Xenopus:
Xenopus tropicalis were housed and cared for in our aquatics facility according to established protocols that were approved by the Yale Institutional Animal Care and Use Committee (IACUC). Embryos were produced by in vitro fertilization. First we harvested the testes of adult male in 1×MBS + 0.2%BSA. Testes were then crushed and incubated with eggs for 3 minutes and then flooded with 0.1×MBS (pH 7.8–8) for 10 minutes. Fertilized Eggs were then degellied using 3% Cysteine in 1/9MR (pH 7.8–8) for 6 minutes. Embryos were then washed using 1/9MR and used for microinjections (described below) or raised to appropriate stages in 1/9MR + gentamycin according to established protocols (del Viso and Khokha, 2012; Khokha, et al., 2002). Xenopus tadpoles were staged according to the staging table previously described in (Nieuwkoop, 1994). The developmental stages of embryos used for experiments are reported throughout the text and figures as appropriate. The embryos were collected for experimental purposes before the sex could be determined.
Mice
Mice were housed and cared for according to established protocols that were approved by the Yale IACUC.
Method Details
Microinjection of morpholino oligonucleotides and mRNA in Xenopus
Morpholino oligonucleotides or mRNA were injected using a fine glass needle and Picospritzer system into one-cell or two-cell embryos as described previously (Khokha, et al., 2002). The following morpholino oligonucleotide was injected: wdr5 translation blocking (4 ng/embryo 5’CGGGTTTCTTTTCTTCTGTTGCCAT-3’), the second wdr5 translation blocking (15 ng/ embryo 5’-TGCAAGAACAACTTGTGGCCGGATA-3’) and the scrambled MO (4ng/embryo 5’-CCTCTTACCTCAGTTACAATTTATA −3’) was injected as a negative control. The suboptimal dose was 2 and 10 ng/ embryo for first and second wdr5 MO respectively. For F0 CRISPR, we generated sgRNAs using the EnGen sgRNA synthesis kit (NEB) following the manufacturer’s instructions after creating a template using the following oligo (5’-GGATGGGGTGGATTGCGTCT-3’) as previously described (Bhattacharya, et al., 2015). We then injected 1000 ng of sgRNA and 1.2ul of Cas9 protein (PNABio) with the total volume of 3ul into one-cell stage embryos. Alexa488 (Invitrogen), mini-ruby (Invitrogen) or green fluorescent protein (100 pg/embryo) were injected as tracers. We generated in vitro capped mRNA using the mMessage machine kit (Ambion) and followed the manufacturer’s instructions. Full length human WDR5 was purchased from Thermo Scientific (IMAGE clone: 3538255) and subcloned into the pCSDest vector using Gateway recombination techniques. S91K-WDR5 was a kind gift from Dr. Cosgrove, SUNY Upstate Medical University. We generated the K7Q mutation using PCR amplification. C-3×GFP was a gift from Dr. Ann Miller, University of Michigan, Ann Arbor. Human WT-WDR5, all deletion constructs including 26–334-WDR5 for domain analysis, S91KWDR5 and K7Q-WDR5 were PCR cloned in frame into the 3×GFP vector. We injected 400pg wild type human WT-WDR5, WDR5-GFP, WDR5–3×GFP, 26–334-3×GFP, K7Q-3×GFP and S91K-WDR5 RNA for rescue of wdr5 morphants. We injected 200pg of WT WDR5–3×GFP and all mutants tagged with 3×GFP for localization. Centrin-RFP (100pg), Clamp-GFP (150pg), mem-RFP/GFP (150pg), WDR5-GFP (200pg), and utrophin-RFP/GFP (150pg) were injected into one-cell embryos to mark basal bodies, ciliary rootlets, cilia and membrane, WDR5, and Factin respectively. Latrunculin A was a gift from Dr. Thomas Pollard at Yale University. Latrunculin A was dissolved in DMSO and used at a final concentration of 2 μM.
Immunofluorescence
We harvested mouse trachea from euthanized adult mice. Mouse tracheas were fixed in 4% paraformaldehyde/PBS overnight at 40C when we assayed for acetylated α-tubulin, WDR5 and phalloidin. For immunofluorescence of multiciliated epidermal cells, we used stage 28–30 Xenopus embryos. Xenopus embryos were fixed in 100% chilled methanol overnight at −200C for γ-tubulin and WDR5. Mouse trachea and Xenopus embryos were mounted in Pro-Long Gold (Invitrogen) before imaging. WDR5 blocking peptide (Bethyl) was used to examine WDR5 specificity in 1:1 and 1:2 (antibody:blocking peptide) ratio.
RNA in situ hybridization
X. tropicalis embryos were collected at stages 27–30 for in situ hybridization. Digoxigeninlabeled antisense probes for rfx2: IMAGE:7680423, foxj1: Tneu058M03, MCIDAS: Tgas069h05, dnah9: IMAGE:7650198, xeel: IMAGE:5380579 (Dharmacon), and atpv1a: Ttpa014f09 were in vitro transcribed with T7 High Yield RNA Synthesis Kit (E2040S) from New England Biolabs.
Embryos were collected at the desired stages, fixed in MEMFA for 1 2 hours at room temperature and dehydrated into 100% ETOH. Briefly summarized, whole mount in situ hybridization of digoxigenin-labeled antisense probes w as performed overnight, the labeled embryos were then washed, incubated with anti-digoxigenin-AP Fab fragments (Roche 11093274910), and signal was detected using BM-purple (Roche 11442074001), as previously described in detail (Khokha, et al., 2002).
Image analysis
Images were captured using a Zeiss 710 Live or Leica SP8 confocal microscope. Images were processed in Fiji, Image J, or Adobe Photoshop. Figure 5 was deconvolved using Huygens Professional to increase the clarity of the signal. The 3D and orthogonal projections of Fig 5 was generated using Leica LAS X software. Basal body polarity was measured using Fiji. Final figures were made in Adobe illustrator.
Antibody concentrations
Mouse monoclonal Anti-acetylated α-tubulin (Sigma, T-6793; 1:1000), Rabbit polyclonal AntiWDR5 (Bethyl, A302–430A; 1:150 for immunofluorescence, 1:400–1:500 for immunoprecipitation and 1:500–1:1000 for western blot), Rabbit polyclonal Anti-WDR5 (Bethyl A302–429A; 1:4001:500 for immunoprecipitation), Goat polyclonal Anti-Actin (Santacruz, SC-1615; 1:250 for western blot), Mouse monoclonal Anti-γ tubulin (Sigma, T6557; 1:100 for immunofluorescence, and 1:4000 for western blot), Mouse monoclonal Anti-GAPDH (Ambion, AM4300; 1:10,000 for western blot), HRP tagged secondary antibodies (Jackson Immuno Research Laboratories, rabbit 1:12000, mouse 1:16000, goat 1:5000: for western blot) were used. Alexa 488, Texas red and Alexa-647 (all 1:500) were used as secondary antibodies for immunofluorescence. Alexa 633 and 488 phalloidin (both 1:40) were used. Blocking peptide for WDR5 was purchased from Bethyl for antibody A302–430A.
Co-immunoprecipitation
For immunoprecipitation, RPE cells were grown until 100% confluence and then were starved for 24–48 hours before lysis. Cells were lysed with NP-40 buffer (150 mM NaCl, □1.0% NP-40, 50 mM Tris, pH 8.0) with protease inhibitor on ice at a concentration of 1ml of buffer for a 10 cm plate. Cells were then centrifuged to collect supernatant. Supernatant was then pre-incubated with protein A/G beads for 1–3 hours to eliminate unspecific binding. The mixture with beads was centrifuged to collect supernatant. Supernatant was incubated with a respective antibody for 1–2 hrs, followed by overnight incubation with beads at 40C. Lysate was then centrifuged the following day and the supernatant was discarded, and the beads were collected and washed with 0.1% PBS-Tween. 2× SDS loading dye was then added to the beads and the samples were analyzed with Western blot. Western Blot was carried out using standard protocol with 4–12% gels.
Protein extraction for Western Blot analysis
To obtain total cell lysate from Xenopus, pools of 20 staged matched embryos for each treatment were placed in 200 μl of 1× RIPA buffer and crushed using a pestle. Samples were spun down twice (10,000g rpm for 20 and 10 mins at 4o degrees) to remove fat and debris. Protein samples were used immediately for western blot or stored at −80o degrees). Western Blot was carried out using standard protocol with 4–12% gels. Quantifications of changes in protein level were calculated using ImageJ software from NIH.
Quantification and Statistical analysis
Statistical significance is reported in the figures and legends. In all figures, statistical significance was defined at P<0.05. Quantifications of changes in protein level were calculated using ImageJ software from NIH. The comparisons were made either using t-tests or ANOVA after confirming the normal distribution of the data. Multiple comparisons correction was done using tukey-Kramer test. We randomly picked one cell X. tropicalis embryos from fertilization as uninjected controls or for morpholino or RNA injections. Investigators were not blind to experiments or the statistical analysis.
Supplementary Material
Video S1: Cilia driven epidermal flow visualized using red microbeads over the period of 9 seconds in X. tropicalis embryos. Related to Figure 1.
Embryos were either uninjected (controls) or injected with wdr5 MO, wdr5 MO + WT human WDR5 RNA and wdr5 MO + human WDR5 (S91K) RNA.
Video S2: WDR5 migrates and docks apically as MCC expands. Related to Figure 6.
Video S3: WDR5 and F-actin interact dynamically as WDR5 docks to apical membrane in MCCs. Related to Figure 6.
Video S4: F-actin appears to organize around WDR5 at the apical surface. Related to Figure 6.
The video shows the region marked with a white square in Figure 6.
HIGHLIGHTS.
WDR5 has a chromatin modification-independent role in multiciliated cell formation
The β-propeller structure of WDR5 is essential but not sufficient for ciliogenesis.
WDR5 controls apical cell expansion and basal body patterning
WDR5 binds actin and γ-tubulin and stabilizes the actin network
ACKNOWLEDGEMENTS
We thank the patients and their families who are the inspiration for this study. We thank Sarah Kubek and Michael Slocum for animal husbandry. Thanks to the Center for Cellular and Molecular Imaging at Yale for confocal imaging. The authors thank Dr. Ann Miller and Dr. Michael Cosgrove for the C-3×GFP construct and human WT and S91K-WDR5 constructs respectively. The authors wish to thank Prof. Thomas Pollard for helpful discussions on actin biology and Emily Legue for helpful comments on the manuscript. This work is supported by an NIH/NHLBI K99/R00 award to SSK. This work was supported by a Pilot Project as part of NIH/NHLBI 5U01HL098188 and NIH/NICHD R01HD081379 and NIH/NHLBI R33HL120783 to MKK. MKK is a Mallinckrodt Scholar. The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the NHLBI.
The authors also wish to thank the Pediatric Cardiac Genomics Consortium for providing congenital heart disease candidate genes (Zaidi. et al 2013) to our pilot project as part of their ongoing genomic analysis of the genetic causes of CHD. These investigators include: A. Roberts, J. Newburger, C. Seidman, J. Seidman (Boston Children’s Hospital); W. Chung, D. Warburton (Columbia); E. Goldmuntz (Children’s Hospital of Philadelphia); B. D. Gelb (Icahn School of Medicine at Mount Sinai); M. Brueckner, R. P. Lifton (Yale University School of Medicine); A. Romano-Adesman (Steven and Alexandra Cohen Children’s Medical Center); J. Deanfield, A. Giardini (University College London); G. Porter (University of Rochester Medical Center); R. Kim (Children’s Hospital Los Angeles); J. Kaltman (NHLBI); J. Miller, T. Hamza, K. Dandreo, S. Tennstedt (New England Research Institutes).
Footnotes
Declaration of Interests:
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ang YS, Tsai SY, Lee DF, Monk J, Su J, Ratnakumar K, Ding J, Ge Y, Darr H, Chang B, et al. (2011). Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell 145, 183–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniades I, Stylianou P, and Skourides PA (2014). Making the connection: ciliary adhesion complexes anchor basal bodies to the actin cytoskeleton. Developmental cell 28, 70–80. [DOI] [PubMed] [Google Scholar]
- Bailey JK, Fields AT, Cheng K, Lee A, Wagenaar E, Lagrois R, Schmidt B, Xia B, and Ma D (2015). WD Repeat-containing Protein 5 (WDR5) Localizes to the Midbody and Regulates Abscission. The Journal of biological chemistry 290, 89879001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker-Heck A, Zohn IE, Okabe N, Pollock A, Lenhart KB, Sullivan-Brown J, McSheene J, Loges NT, Olbrich H, Haeffner K, et al. (2011). The coiled-coil domain containing protein CCDC40 is essential for motile cilia function and left-right axis formation. Nature genetics 43, 79–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Hildebrandt F, Pellman D, Woods G, and Godinho SA (2011). Centrosomes and cilia in human disease. Trends in genetics : TIG 27, 307–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya D, Marfo CA, Li D, Lane M, and Khokha MK (2015). CRISPR/Cas9: An inexpensive, efficient loss of function tool to screen human disease genes in Xenopus. Developmental biology 408, 196–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchoin L, Boujemaa-Paterski R, Sykes C, and Plastino J (2014). Actin dynamics, architecture, and mechanics in cell motility. Physiological reviews 94, 235–63. [DOI] [PubMed] [Google Scholar]
- Boisvieux-Ulrich E, Laine MC, and Sandoz D (1990). Cytochalasin D inhibits basal body migration and ciliary elongation in quail oviduct epithelium. Cell and tissue research 259, 443–54. [DOI] [PubMed] [Google Scholar]
- Brooks ER, and Wallingford JB (2014). Multiciliated cells. Current biology : CB 24, R973–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks ER, and Wallingford JB (2015). In vivo investigation of cilia structure and function using Xenopus. Methods Cell Biol 127, 131–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke MC, Li FQ, Cyge B, Arashiro T, Brechbuhl HM, Chen X, Siller SS, Weiss MA, O’Connell CB, Love D, et al. (2014). Chibby promotes ciliary vesicle formation and basal body docking during airway cell differentiation. The Journal of cell biology 207, 123–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey JP, Goggin P, McDaid J, White M, Ennis S, Betts DR, Lucas JS, Elnazir B, and Lynch SA (2015). A case report of primary ciliary dyskinesia, laterality defects and developmental delay caused by the co-existence of a single gene and chromosome disorder. BMC Med Genet 16, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, Kou Y, Liu L, Fromer M, Walker S, et al. (2014). Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 515, 209–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Viso F, and Khokha M (2012). Generating diploid embryos from Xenopus tropicalis. Methods Mol Biol 917, 33–41. [DOI] [PubMed] [Google Scholar]
- Dharmarajan V, Lee JH, Patel A, Skalnik DG, and Cosgrove MS (2012). Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases. The Journal of biological chemistry 287, 27275–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee JW, Verdine GL, Allis CD, and Roeder RG (2006). Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nature structural & molecular biology 13, 713–9. [DOI] [PubMed] [Google Scholar]
- Epting D, Slanchev K, Boehlke C, Hoff S, Loges NT, Yasunaga T, Indorf L, Nestel S, Lienkamp SS, Omran H, et al. (2015). The Rac1 regulator ELMO controls basal body migration and docking in multiciliated cells through interaction with Ezrin. Development 142, 174–84. [DOI] [PubMed] [Google Scholar]
- Garrod AS, Zahid M, Tian X, Francis RJ, Khalifa O, Devine W, Gabriel GC, Leatherbury L, and Lo CW (2014). Airway ciliary dysfunction and sinopulmonary symptoms in patients with congenital heart disease. Annals of the American Thoracic Society 11, 1426–32. [DOI] [PubMed] [Google Scholar]
- Gori F, Friedman LG, and Demay MB (2006). Wdr5, a WD-40 protein, regulates osteoblast differentiation during embryonic bone development. Developmental biology 295, 498–506. [DOI] [PubMed] [Google Scholar]
- Houtmeyers E, Gosselink R, Gayan-Ramirez G, and Decramer M (1999). Regulation of mucociliary clearance in health and disease. Eur Respir J 13, 1177–88. [DOI] [PubMed] [Google Scholar]
- Huang T, You Y, Spoor MS, Richer EJ, Kudva VV, Paige RC, Seiler MP, Liebler JM, Zabner J, Plopper CG, et al. (2003). Foxj1 is required for apical localization of ezrin in airway epithelial cells. Journal of cell science 116, 4935–45. [DOI] [PubMed] [Google Scholar]
- Ioannou A, Santama N, and Skourides PA (2013). Xenopus laevis nucleotide binding protein 1 (xNubp1) is important for convergent extension movements and controls ciliogenesis via regulation of the actin cytoskeleton. Developmental biology 380, 243–58. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, and Marshall WF (2011). Ciliogenesis: building the cell’s antenna. Nature reviews. Molecular cell biology 12, 222–34. [DOI] [PubMed] [Google Scholar]
- Kennedy MP, Omran H, Leigh MW, Dell S, Morgan L, Molina PL, Robinson BV, Minnix SL, Olbrich H, Severin T, et al. (2007). Congenital heart disease and other heterotaxic defects in a large cohort of patients with primary ciliary dyskinesia. Circulation 115, 2814–21. [DOI] [PubMed] [Google Scholar]
- Khokha MK, Chung C, Bustamante EL, Gaw LW, Trott KA, Yeh J, Lim N, Lin JC, Taverner N, Amaya E, et al. (2002). Techniques and probes for the study of Xenopus tropicalis development. Developmental dynamics : an official publication of the American Association of Anatomists 225, 499–510. [DOI] [PubMed] [Google Scholar]
- Klos Dehring DA, Vladar EK, Werner ME, Mitchell JW, Hwang P, and Mitchell BJ (2013). Deuterosome-mediated centriole biogenesis. Developmental cell 27, 103–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunimoto K, Yamazaki Y, Nishida T, Shinohara K, Ishikawa H, Hasegawa T, Okanoue T, Hamada H, Noda T, Tamura A, et al. (2012). Coordinated ciliary beating requires Odf2-mediated polarization of basal bodies via basal feet. Cell 148, 189–200. [DOI] [PubMed] [Google Scholar]
- Kulkarni SS, and Khokha MK (2018). WDR5 regulates left-right patterning via chromatin dependent and independent functions. Development (In Press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Klena NT, Gabriel GC, Liu X, Kim AJ, Lemke K, Chen Y, Chatterjee B, Devine W, Damerla RR, et al. (2015). Global genetic analysis in mice unveils central role for cilia in congenital heart disease. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merveille AC, Davis EE, Becker-Heck A, Legendre M, Amirav I, Bataille G, Belmont J, Beydon N, Billen F, Clement A, et al. (2011). CCDC39 is required for assembly of inner dynein arms and the dynein regulatory complex and for normal ciliary motility in humans and dogs. Nature genetics 43, 72–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier A, and Azimzadeh J (2016). Multiciliated Cells in Animals. Cold Spring Harb Perspect Biol 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. (2007). Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448, 553–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell B, Jacobs R, Li J, Chien S, and Kintner C (2007). A positive feedback mechanism governs the polarity and motion of motile cilia. Nature 447, 97–101. [DOI] [PubMed] [Google Scholar]
- Mitchell B, Stubbs JL, Huisman F, Taborek P, Yu C, and Kintner C (2009). The PCP pathway instructs the planar orientation of ciliated cells in the Xenopus larval skin. Current biology : CB 19, 924–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakhleh N, Francis R, Giese RA, Tian X, Li Y, Zariwala MA, Yagi H, Khalifa O, Kureshi S, Chatterjee B, et al. (2012). High prevalence of respiratory ciliary dysfunction in congenital heart disease patients with heterotaxy. Circulation 125, 223242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwkoop PDF,J (1994). Normal Table of Xenopus Laevis (Daudin) Odho, Z., Southall, S.M., and Wilson, J.R. (2010). Characterization of a novel WDR5binding site that recruits RbBP5 through a conserved motif to enhance methylation of histone H3 lysine 4 by mixed lineage leukemia protein-1. The Journal of biological chemistry 285, 32967–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh EC, and Katsanis N (2012). Cilia in vertebrate development and disease. Development 139, 443–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens NDL, Blitz IL, Lane MA, Patrushev I, Overton JD, Gilchrist MJ, Cho KWY, and Khokha MK (2016). Measuring Absolute RNA Copy Numbers at High Temporal Resolution Reveals Transcriptome Kinetics in Development. Cell reports 14, 632–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park TJ, Mitchell BJ, Abitua PB, Kintner C, and Wallingford JB (2008). Dishevelled controls apical docking and planar polarization of basal bodies in ciliated epithelial cells. Nature genetics 40, 871–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A, Dharmarajan V, and Cosgrove MS (2008a). Structure of WDR5 bound to mixed lineage leukemia protein-1 peptide. The Journal of biological chemistry 283, 32158–61. [DOI] [PubMed] [Google Scholar]
- Patel A, Vought VE, Dharmarajan V, and Cosgrove MS (2008b). A conserved arginine-containing motif crucial for the assembly and enzymatic activity of the mixed lineage leukemia protein-1 core complex. The Journal of biological chemistry 283, 32162–75. [DOI] [PubMed] [Google Scholar]
- Pedersen LB, and Rosenbaum JL (2008). Intraflagellar transport (IFT) role in ciliary assembly, resorption and signalling. Curr Top Dev Biol 85, 23–61. [DOI] [PubMed] [Google Scholar]
- Pollard TD (1986). Mechanism of actin filament self-assembly and regulation of the process by actin-binding proteins. Biophys J 49, 149–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD (2016). Actin and Actin-Binding Proteins. Cold Spring Harb Perspect Biol 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuetz A, Allali-Hassani A, Martin F, Loppnau P, Vedadi M, Bochkarev A, Plotnikov AN, Arrowsmith CH, and Min J (2006). Structural basis for molecular recognition and presentation of histone H3 by WDR5. The EMBO journal 25, 4245–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedzinski J, Hannezo E, Tu F, Biro M, and Wallingford JB (2016). Emergence of an Apical Epithelial Cell Surface In Vivo. Developmental cell 36, 24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedzinski J, Hannezo E, Tu F, Biro M, and Wallingford JB (2017). RhoA regulates actin network dynamics during apical surface emergence in multiciliated epithelial cells. Journal of cell science 130, 420–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma N, Berbari NF, and Yoder BK (2008). Ciliary dysfunction in developmental abnormalities and diseases. Curr Top Dev Biol 85, 371–427. [DOI] [PubMed] [Google Scholar]
- Stubbs JL, Davidson L, Keller R, and Kintner C (2006). Radial intercalation of ciliated cells during Xenopus skin development. Development 133, 2507–15. [DOI] [PubMed] [Google Scholar]
- Stubbs JL, Oishi I, Izpisua Belmonte JC, and Kintner C (2008). The forkhead protein Foxj1 specifies node-like cilia in Xenopus and zebrafish embryos. Nature genetics 40, 1454–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland MJ, and Ware SM (2009). Disorders of Left-Right Asymmetry: Heterotaxy and Situs Inversus. Am J Med Genet C 151C, 307–317. [DOI] [PubMed] [Google Scholar]
- Swisher M, Jonas R, Tian X, Lee ES, Lo CW, and Leatherbury L (2011). Increased postoperative and respiratory complications in patients with congenital heart disease associated with heterotaxy. The Journal of thoracic and cardiovascular surgery 141, 637–44, 644 e1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan SY, Rosenthal J, Zhao XQ, Francis RJ, Chatterjee B, Sabol SL, Linask KL, Bracero L, Connelly PS, Daniels MP, et al. (2007). Heterotaxy and complex structural heart defects in a mutant mouse model of primary ciliary dyskinesia. J Clin Invest 117, 3742–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang TK (2013). Centriole biogenesis in multiciliated cells. Nature cell biology 15, 1400–2. [DOI] [PubMed] [Google Scholar]
- Trievel RC, and Shilatifard A (2009). WDR5, a complexed protein. Nature structural & molecular biology 16, 678–80. [DOI] [PubMed] [Google Scholar]
- Vladar EK, and Axelrod JD (2008). Dishevelled links basal body docking and orientation in ciliated epithelial cells. Trends in cell biology 18, 517–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walck-Shannon E, and Hardin J (2014). Cell intercalation from top to bottom. Nature reviews. Molecular cell biology 15, 34–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YY, Liu LJ, Zhong B, Liu TT, Li Y, Yang Y, Ran Y, Li S, Tien P, and Shu HB (2010). WDR5 is essential for assembly of the VISA-associated signaling complex and virus-triggered IRF3 and NF-kappaB activation. Proceedings of the National Academy of Sciences of the United States of America 107, 815–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner ME, Hwang P, Huisman F, Taborek P, Yu CC, and Mitchell BJ (2011). Actin and microtubules drive differential aspects of planar cell polarity in multiciliated cells. The Journal of cell biology 195, 19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner ME, and Mitchell BJ (2013). Using Xenopus skin to study cilia development and function. Methods in enzymology 525, 191–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner ME, Mitchell JW, Putzbach W, Bacon E, Kim SK, and Mitchell BJ (2014). Radial intercalation is regulated by the Par complex and the microtubulestabilizing protein CLAMP/Spef1. The Journal of cell biology 206, 367–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, Roeder RG, Brivanlou AH, and Allis CD (2005). WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell 121, 859–72. [DOI] [PubMed] [Google Scholar]
- Yarmola EG, Somasundaram T, Boring TA, Spector I, and Bubb MR (2000). Actin-latrunculin A structure and function. Differential modulation of actin-binding protein function by latrunculin A275, 28120–7. [DOI] [PubMed] [Google Scholar]
- Yu X, Ng CP, Habacher H, and Roy S (2008). Foxj1 transcription factors are master regulators of the motile ciliogenic program. Nature genetics 40, 1445–53. [DOI] [PubMed] [Google Scholar]
- Zaidi S, Choi M, Wakimoto H, Ma LJ, Jiang JM, Overton JD, RomanoAdesman A, Bjornson RD, Breitbart RE, Brown KK, et al. (2013). De novo mutations in histone-modifying genes in congenital heart disease. Nature 498, 220–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, and Mitchell BJ (2015). Centriole biogenesis and function in multiciliated cells. Methods Cell Biol 129, 103–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Zhu L, Zhu Y, Cao J, Li S, Huang Q, Xu T, Huang X, Yan X, and Zhu X (2013). The Cep63 paralogue Deup1 enables massive de novo centriole biogenesis for vertebrate multiciliogenesis. Nature cell biology 15, 1434–44. [DOI] [PubMed] [Google Scholar]
- Zhu JY, Fu Y, Nettleton M, Richman A, and Han Z (2017). High throughput in vivo functional validation of candidate congenital heart disease genes in Drosophila. eLife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1: Cilia driven epidermal flow visualized using red microbeads over the period of 9 seconds in X. tropicalis embryos. Related to Figure 1.
Embryos were either uninjected (controls) or injected with wdr5 MO, wdr5 MO + WT human WDR5 RNA and wdr5 MO + human WDR5 (S91K) RNA.
Video S2: WDR5 migrates and docks apically as MCC expands. Related to Figure 6.
Video S3: WDR5 and F-actin interact dynamically as WDR5 docks to apical membrane in MCCs. Related to Figure 6.
Video S4: F-actin appears to organize around WDR5 at the apical surface. Related to Figure 6.
The video shows the region marked with a white square in Figure 6.