ABSTRACT
The ubiquitous fungus Aspergillus flavus is notorious for contaminating many important crops and food-stuffs with the carcinogenic mycotoxin, aflatoxin. This fungus is also the second most frequent Aspergillus pathogen after A. fumigatus infecting immunosuppressed patients. In many human fungal pathogens including A. fumigatus, the ability to defend from toxic levels of copper (Cu) is essential in pathogenesis. In A. fumigatus, the Cu-fist DNA binding protein, AceA, and the Cu ATPase transporter, CrpA, play critical roles in Cu defense. Here, we show that A. flavus tolerates higher concentrations of Cu than A. fumigatus and other Aspergillus spp. associated with the presence of two homologs of A. fumigatus CrpA termed CrpA and CrpB. Both crpA and crpB are transcriptionally induced by increasing Cu concentrations via AceA activity. Deletion of crpA or crpB alone did not alter high Cu tolerance, suggesting they are redundant. Deletion of both genes resulted in extreme Cu sensitivity that was greater than that following deletion of the regulatory transcription factor aceA. The ΔcrpAΔcrpB and ΔaceA strains were also sensitive to ROI stress. Compared to wild type, these mutants were impaired in the ability to colonize maize seed treated with Cu fungicide but showed no difference in virulence on non-treated seed. A mouse model of invasive aspergillosis showed ΔcrpAΔcrpB and to a lesser degree ΔaceA to be significantly reduced in virulence, following the greater sensitivity of ΔcrpAΔcrpB to Cu than ΔaceA.
KEYWORDS: P1-type ATPase, ROI stress, copper homeostasis, AceA, CrpA, CrpB
Introduction
Copper (Cu) is an essential metal serving as a cofactor for enzymes that function in numerous biological processes in both prokaryotes and eukaryotes [1,2]. The property of Cu to have both reductive (Cu+) and oxidative (Cu2+) states is important in several biological functions [1]. For example, the reducing ability of Cu+ is critical for the activities of Cu-dependent enzymes like Cu/Zn superoxide dismutase (SOD), cytochrome oxidase and laccase [3]. On the other hand, high concentrations of Cu are toxic due to Fenton chemistry, by which Cu can generate harmful reactive oxygen species such as hydroxyl radicals [4,5]. Cu also has a high affinity to displace other metals from their cognate proteins leading to protein inactivation [2,6]. Thus, Cu homeostasis is tightly controlled by cells.
Since Cu leads to toxic oxidative stress, which could cause DNA damage and protein dysfunction, Cu as a sole source or in mixtures such as the Bordeaux mixture has been used as an antimicrobial agent to kill plant pathogens, employed to sterilize wounds or to treat surfaces both external or internal (e.g. implants) susceptible to microbial contamination [7–10]. The anti-microbial properties of Cu are also used by mammalian host cells as defense mechanisms. For example, Cu plays an important role in innate immune defense to microbial infection in mammals [8,11,12]. Previous studies demonstrated that, upon activation by a pathogen, macrophages elevate the expression levels of high-affinity Cu+ importer Ctr1 and P-type Cu ATPase, ATP7A, leading to increased levels of Cu in the phagosome of IFN-γ stimulated macrophages [13,14]. These high Cu levels, together with Fe could react chemically with ROIs and reactive nitrogen intermediates (RNIs), potentially magnifying the Cu mediated toxicity.
On the other hand, Cu homeostasis has been shown to support the virulence of many fungal pathogens, which share many of the elements found in mammalian Cu regulatory systems including Cu-binding transcriptional factors (TFs), Cu-importers/exporters and Cu-metallothioneins (MTs)[2,15]. Fungal Cu-binding TFs tightly control expression levels of genes involved in Cu uptake and export under Cu-deficient conditions or Cu-excess respectively. In Saccharomyces cerevisiae, Cu importers Ctr1 and Ctr3, located in the plasma membrane, as well as the Cu-MTs Fre1 and Fre7 are activated by the Cu-binding TF Mac1 under Cu-insufficient conditions [8,16]. Under excess Cu, the S. cerevisiae Cu-binding TF Ace1 induces the MT-encoding genes CUP1 and CRS5, and the Cu/Zn SOD-encoding gene SOD1 [8,17]. In the human pathogen Cryptococcus neoformans, MTs play a major role of in Cu detoxification [7]. Two MTs, Cmt1 and Cmt2, were shown to be important for fungal virulence in C. neoformans, both of which are regulated by the Cu-binding TF Cuf1 [2,18]. In Candida albicans, a membrane-localized P-type Cu ATPase, Crp1, is responsible for Cu resistance along with the MT Cup1, both of which are transcriptionally activated by Ace1 under high Cu conditions [19–21]. Recent studies also illustrated the regulation of Cu homeostasis in the filamentous fungi Aspergillus fumigatus and A. nidulans, both of which possess the same Cu export machinery [15,22,23]. In A. fumigatus and A. nidulans, the Cu-fist binding TF AceA senses high levels of Cu, and induces the Cu P-type ATPase CrpA as a detoxification mechanism [15,22]. Both AceA and CrpA are virulence factors for the human pathogenic A. fumigatus. Currently there are no studies of the roles of P-type ATPases in plant pathogenic fungi although a CCC-2 type ATPase, BcCCC2, important in melanization in Botrytis cinerea is necessary for fungal development and virulence of that pathogen [24]. The BcCCC2 homolog in A. fumigatus, CtpA, is also required for spore melanin synthesis in that fungus [25].
The ubiquitous fungus Aspergillus flavus is the second most common Aspergillus spp. to infect immunosuppressed patients after A. fumigatus [26] as well as being a frequent cause of trauma-associated keratitis in agricultural workers [27]. Aspergillus flavus is also notorious for its ability to colonize lipid-rich seed resulting in contamination of food and feed crops with the carcinogenic mycotoxin, aflatoxin [28] This dual ability to cause disease in both animals and plants, leading to enormous agricultural economic losses and health problems, exacerbates the importance of A. flavus as an opportunistic pathogen.
As we have recently found that deletion of the Cu export machinery severely impairs A. fumigatus virulence [15], we queried if the same proteins were also important for virulence of A. flavus, both in its role as a plant and mammalian pathogen. Unexpectedly we found that A. flavus possesses two copies of the Cu exporter P1-type ATPase, CrpA and CrpB which results in greater tolerance of Cu in this species than other Aspergilli. We demonstrate that both crpA and crpB are transcriptionally induced by the Cu-fist TF AceA. Only the double deletion of crpA and crpB resulted in a strain sensitive to low concentrations of Cu, indicating a redundant function of these two proteins. Neither the ΔcrpAΔcrpB or ΔaceA mutants showed any defect in colonizing host maize seed but, as expected, both strains failed to colonize maize corn treated with Cu fungicide. On the other hand, the ΔcrpAΔcrpB mutant was significantly reduced in virulence in a murine model of invasive aspergillosis with loss of AceA showing a notable but lesser influence on fungal invasion. This difference in murine virulence between these two mutants reflects the relative sensitivity of these mutants to Cu concentrations.
Results
Aspergillus flavus and A. parasiticus tolerate higher concentrations of Cu
Most Aspergillus species are considered as soil-borne fungi living in environments with fluctuating levels of heavy metals including Cu. This is particularly true for the agricultural plant pathogens A. flavus and A. parasiticus which may be found in fields treated with Cu fungicides [29]. To test whether Aspergillus species exhibit similar growth patterns to Cu extremes, six Aspergillus species (A. flavus NRRL3357, A. parasiticus SU-1, A. fumigatus Af293, A. nidulans FGSC4A, A. niger CBS 113.46 and A. terreus NIH2624) were selected and grown on GMM media supplemented with different concentrations of Cu. In all species, Cu is a cofactor for laccases required for conidia pigmentation and thus, as expected, all species could not produce normal pigment without Cu (Figure 1). The sensitivity of A. fumigatus became apparent at 500 μM Cu on GMM medium after 3 days (Figure 1). Aspergillus nidulans, A. niger and A. terreus exhibited total growth inhibition at 1000 μM Cu, while A. flavus and A. parasiticus remained quite tolerant to this level of Cu (Figure 1).
Figure 1.

Growth phenotype of different Aspergillus strains on different Cu concentrations. 2000 spores of indicated Aspergillus strains grown on solidified GMM under indicated Cu concentrations for 72 h at 37°C.
We were curious to see if this enhanced resistance to copper was a general feature of A. flavus and A. parasiticus. We thus grew four A. oryzae strains (a clade of A. flavus), five more A. flavus strains, four more A. parasiticus strains, five more A. fumigatus strains and then a few other Aspergillus species. Growth phenotypes of all A. oryzae, flavus and parasiticus strains at 1 mM copper was only almost unaltered compared to lower concentrations, while none of the A. fumigatus strains grew at this copper concentration (Figure S1). Of the five other species assessed, two, A. carbonarius and A. brasiliensis also tolerated 1 mM Cu. Intriguingly, when the concentration of Cu was increased to 2 mM, both A. carbonarius and A. brasiliensis could still grow, but slightly less than at lower concentrations, while the tested A. oryzae, flavus and parasiticus strains showed almost no growth impairment (Figure S2).
Aspergillus flavus and A. parasiticus possess a redundant copy of the P type Cu-exporting atpase
The Cu exporter P1-type ATPase is the primary means for Cu detoxification in A. fumigatus [15]. To identify the Cu P1-type ATPase in Aspergillus species, the available protein sequences of the heavy-metal ATPases (HMA) from Aspergillus species available from the NCBI database were retrieved (http://www.aspgd.org). The A. fumigatus CrpA (AfuA_3g12740 [15]) sequence was used to perform a BLASTP [30] analysis. All sequences with an e-value below 1−20 were aligned using MAFFT [31]. The conserved E1-E2 ATPase domains were extracted, re-aligned and a phylogenetic tree was created using fasttree [32]. The analysis shows the three distinct groups including Ccc2-type, CrpA-type and Pca1-type ATPases (Figure 2). Surprisingly, we found that A. flavus and A. parasiticus have two predicted protein sequences (XP_002376116.1/AFL2T_03712/AFLA_020960 and XP_002383472.1/AFL2T_10544/AFLA_053470) in the CrpA clade of Cu-exporting ATPases. As AFL2T_03712 from A. flavus fell into a subgroup with CrpA from A. fumigatus, we named it CrpA. Consequently, AFL2T_10544 was termed CrpB. Notably, other Aspergillus species (A. arachidicola, A. glaucus, and A. cristatus) also harbor two predicted Crp-type proteins. Interestingly, A. sydowii and A. calidoustus seem to harbour even more predicted proteins that fall in this group of Cu-exporting ATPases (Figure 2). Some of the analyzed Aspergillus species (A. sydowii, A. turcosus, and A. calidoustus) also show multiple proteins in the Pca1-type group. The Pca1-type group derived its named from the Saccharomyces cerevisiae cadmium transporting ATPase Pca1p [33]. This is in contrast to the Ccc2-type group (named after the S. cerevisiae Cu transporting ATPase of the trans-Golgi network Ccc2p [34], where only one predicted protein from each analyzed Aspergillus species was found (Figure 2).
Figure 2.

Phylogenetic analysis of heavy metal ATPases (HMA). The phylogenetic tree based on the available HMA sequences from Aspergillus sequences available through NCBI was constructed by MAFFT and Fasttree using the Neighbour-joining method as described in Material and Methods. Bootstrap analysis was performed with 1000 replicates. Aspergillus flavus proteins are shown in red, A. fumigatus in green, A. parasiticus in orange, and A. nidulans in green.
CrpA and CrpB play redundant roles in Cu detoxification in A. flavus
Previous studies had demonstrated that the expression level of the Cu exporter CrpA is induced in high levels of Cu in Aspergillus spp [15,22]. To determine if both crpA and crpB are Cu-inducible in A. flavus, their expression levels were assessed after A. flavus wild-type strain NRRL3357 was treated with 200 μM Cu. As shown in Figure 3A, crpA and crpB were not transcribed in the absence of Cu, but were strongly expressed under excess Cu treatment.
Figure 3.

crpA and crpB were induced by Cu and were redundant in Cu detoxification. (A) Northern blot analysis of crpA and crpB in A. flavus wild-type strain. The wild-type strain was grown in liquid GMM without Cu for 24 h at 37°C and then mycelium mass was divided in half where one-half was grown in medium with no Cu and one-half was grown in 200 μM Cu for 1 h before harvesting. rRNA visualization is loaded as control. (B) Growth assay of A. flavus crp mutants on solidified GMM for 72 h at 37°C under indicated Cu concentrations.
To test whether CrpA or CrpB affected A. flavus Cu tolerance in growth studies, we generated single-deletion mutants of each gene. Diagnostic PCR and Southern analysis were performed to verify correct gene replacement (Figure S3A). Both ΔcrpA and ΔcrpB mutants showed wildtype arginine heterotrophy, indicating that the argB selectable marker was fully functional and unaffected by its chromosomal location (Figure S4). Both mutants exhibited WT sensitivity to high concentrations of Cu (Figure 3(B)). To determine whether ΔcrpA and ΔcrpB are redundant in A. flavus, we generated the double deletion mutant ΔcrpAΔcrpB, verified by Southern blot analysis (Figure S3A). As shown in Figure 3(B), the double deletion mutant ΔcrpAΔcrpB was highly sensitive to Cu (1 μM), indicating that CrpA and CrpB have an overlapping function in Cu detoxification.
The Cu fist transcriptional factor acea regulates crpA and crpB expression during Cu homeostasis
The A. fumigatus Cu fist DNA binding domain protein AceA regulates responses to excess Cu and activates the Cu exporter CrpA for Cu detoxification [15]. BlastP analyses using the protein sequence of A. fumigatus AceA (XP_750669.1) as query allowed the identification of the AceA homolog in A. flavus, which is encoded by AFLA_036460 and contains the conserved Cu fist DNA binding domain at its N-terminal (Figure S5). To test its impact on Cu homeostasis in A. flavus, the deletion mutant of aceA was generated and confirmed by Southern blot analysis (Figure S3A). The ΔaceA mutants did not exhibit marker gene effects and one aceA mutant (TKY5.3) was used for growth assays as well as for generating complementation strains. The ΔaceA mutant exhibited reduced growth on GMM + 5 μM Cu after 3 days (Figure 4A) although it was not as sensitive as the ΔcrpAΔcrpB mutant which could not grow at this Cu concentration (Figure 3(B)). Wild-type Cu resistance was restored in the ΔaceA complemented strain (Figure 4(A)). These results indicate that AceA plays an important role in regulating Cu homeostasis in A. flavus.
Figure 4.

Functional analysis of AceA and its regulation on crpA and crpB. (A) Growth assay of ΔaceA and aceAC mutants on solidified GMM for 72 h at 37°C under indicated Cu concentrations. (B) Northern blot analysis of crpA and crpB in A. flavus wild-type strain. The wild-type strain grown in liquid GMM without Cu for 24 h at 37°C, then to one-half of the cultures, Cu was added to a final concentration of 200 μM for 1 h before harvesting. rRNA visualization is loaded as control. (C) Phenotypic analysis of the strains on solidified GMM under the indicated Cu concentrations for 48 h at 37°C. (D) Total Cu amount from A. flavus mycelia. All strains (5 × 106 conidia/L) were incubated in 50 mL of liquid GMM without Cu in four replicates at 37°C and 200 rpm shaking for 12h. To one-half of the cultures, Cu was added to a final concentration of 50 μM for 12 h before harvesting. Error bars represent standard deviations, asterisk “*, “**” or “***” represent significant differences at p < 0.05, p < 0.01 and p < 0.001, respectively, according to t-test.
To investigate whether the Cu-inducible expression of crpA and crpB depends on AceA, we tested the expression levels of crpA and crpB in the aceA deficient mutant under excess Cu conditions. Our results showed that expression of crpB was not detectable in the ΔaceA mutant with crpA showing marginal expression in the ΔaceA mutant (Figure 4(B)). This demonstrated that AceA is necessary for the activation of crpA and crpB in Cu detoxification. We next constitutively expressed both crpA and crpB in the ΔaceA mutant and wild-type strain, which were verified first by Southern blot analysis (Figure S3C) and then gene expression confirmed by northern analysis (Figure 4(B)). The growth analysis on GMM medium with high Cu showed that constitutive expression of crpA or crpB in the ΔaceA background largely restored Cu tolerance (Figure 4(C)). Notably, overexpression of crpB resulted in a general reduction of fungal growth independent of Cu concentration. Taken together, these data confirm that the A. flavus Cu fist TF AceA is responsible for activating genes related to Cu detoxification under Cu excess.
Next, we determined and compared total Cu levels in the mycelium of all four strains. At the Cu depleted condition, the ΔaceA mutant showed a significant increase in Cu concentration compared to wild-type strain (Figure 4(D)). While treated with 50 µM Cu, both the ΔaceA mutant and the double deletion mutant ΔcrpAΔcrpB displayed an excess accumulation of the intracellular Cu levels (Figure 4(D)), which indicated that the abnormal accumulation of Cu in these mutants make them more sensitive to Cu.
Cu detoxification mediated by AceA increases ROI stress
As noted, excess Cu generates reactive oxygen species such as hydroxyl radicals. To test the role of the Cu detoxification machinery in dealing with ROI stress, all of the mutants were incubated on GMM media with increasing Cu and two ROI stressors: the superoxide generator, menadione or hydrogen peroxide. As shown in Figure 5, when grown on 5 μM menadione and increasing Cu, all strains exhibited growth inhibition, with ΔcrpAΔcrpB in general showing the most severe phenotype. This trend was generally similar in hydrogen peroxide treatment although ΔcrpAΔcrpB showed more similar growth to ΔaceA (Figure S6A). Addition of the reductant l-glutathione (GSH) in the Cu-containing GMM media, mitigated inhibition in all strains (Figure 5), with suggestion that damage to the fungal Cu detoxification pathway enhances ROI toxicity (more noticeable with hydrogen peroxide treatment and 5 μM copper, Figure S6B).
Figure 5.
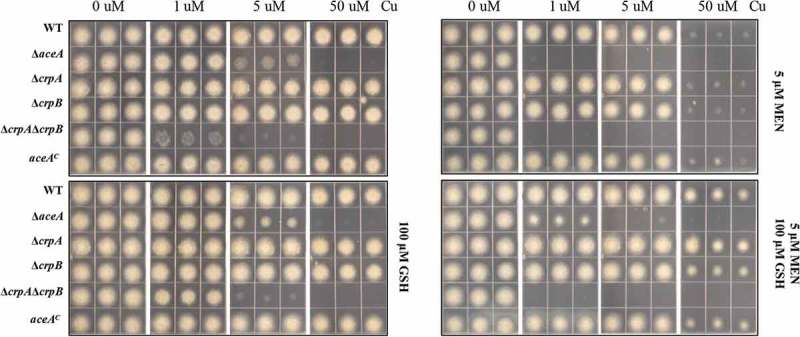
ROS stress increases Cu toxicity in ΔaceA and double ΔcrpAΔcrpB mutants. Growth phenotypes of indicated strains on solidified GMM under indicated concentration of Cu plus supplements as indicated for 48 h at 37°C. MEN = menadione, GSH = l-glutathione.
Cu fungicide reduces pathogenicity of ∆acea and ∆crpA∆crpB mutants
Aflatoxin contamination of many important seed crops causes enormous agricultural economic losses [35]. Several antimicrobial agents including Cu fungicides have been examined for A. flavus growth inhibition as potential post-harvest interventions [36]. Here we examined the A. flavus ΔcrpAΔcrpB and ΔaceA mutants for their abilities to invade both untreated and Cu fungicide-treated maize kernels. As shown in Figure 6 the mutants grew vigorously on maize seeds not treated with fungicide. However, when the maize kernels were treated with the Cu fungicide (Champion wettable powder), the growth of all the A. flavus strains on maize kernels was inhibited (Figure 6(A) lower panel) with statistically less conidia produced than on untreated seed (Figure 6(B,C)). A total growth inhibition was found both in the ΔaceA and ΔcrpAΔcrpB mutants on the Cu fungicide treated maize kernels (Figure 6(B,C)).
Figure 6.
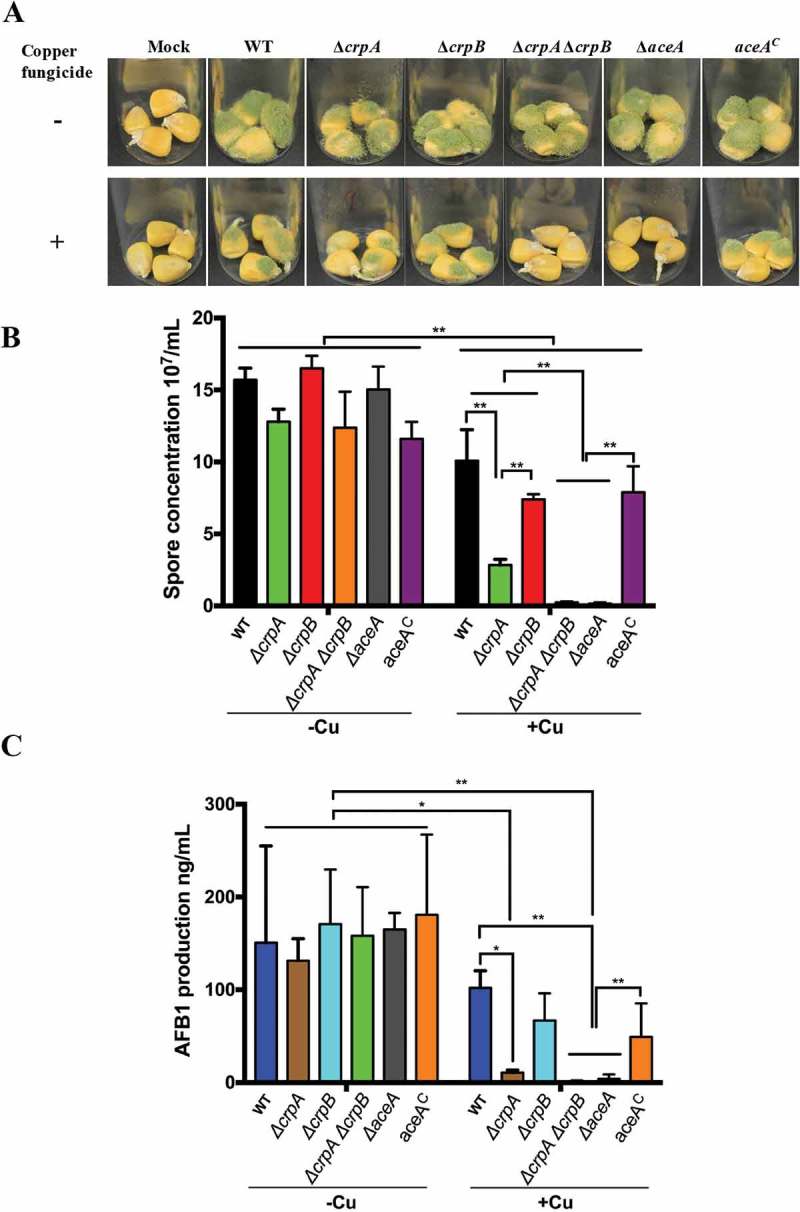
Corn infection with indicated strains. (A) 200 ul of a 106 spore/ml suspension of spores in 0.01% Tween 20 were inoculated on corn and the vials kept in a moist incubator at 29℃ with 12 hours’ light/dark cycling for 5 days. (B) Conidial production assessed from infected maize kernel with and without Cu fungicide treatment. The spores were washed off the seeds with 2.5 mL 100% MeOH and counted. Each sample has four replicates. (C) Aflatoxin extracted from corn and quantified by HPLC. Asterisk “*” or “**” represent significant differences at p < 0.01 and p < 0.001, respectively, according to t-test.
Of interest, the ΔcrpA strain, although not showing defects of growth when grown on high Cu-containing media (Figure 3), grew less well and exhibited a significant drop in conidial production compared to the wild-type strain when grown on the treated maize kernels (Figure 6(A,C)). Taken together, these results suggested that the Cu exporters CrpA and CrpB, activated by the Cu-sensing TF AceA, function as Cu defense machinery in hyperaccumulating Cu environments with possibly CrpA playing a slightly more prominent role in Cu export.
The ΔcrpAΔcrpB mutant is attenuated in virulence in a murine model of invasive aspergillosis
We have previously shown that deletion of AceA or CrpA in A. fumigatus strongly attenuates virulence in murine models of pulmonary aspergillosis [15]. Here, we tested the virulence of the A. flavus WT, ΔaceA, AceAc and ΔcrpAΔcrpB strains for virulence in a steroid model of invasive pulmonary aspergillosis. Survival curves show that, unlike in A. fumigatus, ΔaceA of A. flavus shows similar virulence to the WT and complemented strains as measured by mortality (p = 0.34, Figure 7A). In contrast, deletion of both crpA and crpB (ΔcrpAΔcrpB) resulted in significantly reduced mortality rate compared to the WT and AceAc strains (Figure 7(A)). However, lung fungal burden after 48 h of infection was significantly reduced in both ΔaceA (~ 2-fold p = 0.0027) and ΔcrpAΔcrpB (~ 4-fold, p = 0.0001) (Figure 7(B)) suggesting a need for AceA in colonization although not to the degree as the transporters. Histological staining with GMS (stains fungal elements black) or H&E (stains neutrophils dark purple) revealed large fungal lesions with extensive neutrophil infiltration in A. flavus WT and AceAc, smaller fungal lesions with less extensive neutrophil infiltration in ΔaceA and very few small fungal lesions with weak neutrophil infiltration in ΔcrpAΔcrpB (Figure 7(C)). These results support a major role for the Cu transporters CrpA and CrpB and a lesser role for AceA in virulence. These results also correlate well with the greater in vitro Cu-sensitivity of the ΔcrpAΔcrpB strain compared to ΔaceA (Figure 5).
Figure 7.

Deletion of A. flavus crpA and crpB results in attenuated virulence in lung-infected immunocompromised mice. (A) Survival rates of ICR mice immunocompromised with cortisone acetate and infected intranasally with 5 × 105 A. flavus WT (n = 19 animals), ΔcrpAΔcrpB (n = 10), ΔaceA (n = 19) or aceAc complemented (n = 8) strains, and survival monitored over 18 days. Virulence was only attenuated in the ΔcrpAΔcrpB strain (p = 0.0004) and not in the ΔaceA strain (p = 0.56). (B) Colony forming unit (CFU) of wild type and copper mutants from infected mice lungs. (C) Histopathology of infected mice lungs stained with Grocott’s methenamine silver stain (GMS; fungal staining) and hematoxylin and eosin (H&E; tissue and nuclear staining). Bar = 200 µm.
Discussion
Although a scan of the literature indicates that numerous fungi may crossover to infect both plants and animals, this is a very rare event [37]. However, a prominent fungal exception to this rarity is A. flavus which not only causes toxic seed diseases but also is a common agent of keratitis as well as a frequent pathogen of immunosuppressed patients [26,38]. This fungus is common in agricultural soils and hard to eliminate from that environment due to formation of resistant overwintering structures called sclerotia. The fungus is also hard to treat, both as a seed pathogen and human pathogen due to a limited ability to apply fungicides to edible portions of plants and its emerging resistance to several antifungals [39]. Thus, it is important to identify A. flavus genes and processes important for virulence in both plant and animal disease.
Several recent studies have shown that fungal nutrient and element acquisition pathways may be critical for pathogenicity of both plant and human pathogens [40,41]. For example, all living organisms utilize Cu as cofactor of enzymes functioning in many cellular biochemical processes [3]. However, hyperaccumulation of Cu generates toxic hydroxyl radicals that pose severe cellular damage [42]. Thus, Cu homeostasis, including Cu uptake, utilization and detoxification is tightly controlled by cells. In A. fumigatus, six putative Cu transporters have been identified [15,23], where two are required for Cu import (CtrA2, Afu6g02810 and CtrC, Afu2g03730 [23], one for Cu export (CrpA, Afu43g12740 [15], one for melanin synthesis (CtpA, Afu4g12620 [25] and two others remain characterized (Afu3g13660 and Afu3g08180)[43]. A double deletion of ctrA2 and ctrC showed decreased superoxide dismutase and laccase activity, but did not have an impact on fungal virulence [23]. In contrast, deletion of CrpA resulted in a significant reduction in virulence in murine models of invasive aspergillosis [15]. Recent studies in A. fumigatus and A. nidulans have demonstrated that the Cu binding transcription factor AceA is responsible for sensing excess Cu, and induces expression of crpA as a detoxification mechanism [15,22]. Our study here demonstrates a similar mechanism exists in A. flavus, however with the twist of the presence of an additional AceA regulated Crp protein where the two proteins appear to play a redundant function in protecting the fungus from excess Cu. Previous studies have reported on gene duplications of heavy metal transporters in yeast species [44,45]. While in some cases the gene duplications lead to neofunctionalization [44], in others they can lead to extreme halotolerance [45]. As we find an increased Cu tolerance of A. flavus and A. parasiticus compared to other Aspergillus species that only harbor one Crp-type ATPase protein (Figures 1 and 2), we speculate that in A. flavus and A. parasiticus a gene duplication contributed to increased Cu-export efficiency and not to a neofunctionalization. Nevertheless, this may not be the only factor as two other species, A. carbonarius and A. brasiliensis, also grew on high concentrations of Cu without a Crp-type ATPase gene duplication. Possibly a higher expression level of crp gene or Crp-type ATPase activities might exist in these two fungi.
In Saccharomyces cerevisiae, cells are protected from the toxic effects of excess Cu by protein metallothioneins that contain cysteine rich residues, detoxifying Cu by coordination with multiple Cu ions [46]. While metallothioneins appear to play the major role in Cu resistance in S. cerevisiae, a putative metallothionein, CrdA, identified in A. nidulans did not exhibit a role in Cu detoxification [22]. The homolog in A. fumigatus, CmtA, is not expressed in either Cu deficient or Cu excess conditions, nor is it regulated by AceA, thus suggesting it is also not important in Cu homeostasis in that fungus [15]. Instead of metallothioneins, the P-type Cu ATPase, Crp1 in Candida albicans and CrpA in A. fumigatus/nidulans, was found to be responsible for Cu resistance [15,19,22]. Crp1/CrpA proteins contain GMXCXXC and CXXC consensus motifs that bind free Cu iron or receive chaperone-bound Cu and pump Cu out of the cell through a transmembrane channel [2,19,22]. In this study, we found that most of the analyzed Aspergillus species including A. nidulans and A. fumigatus possess just one CrpA homolog, while one or more redundant copies of the CrpA homolog were identified in A. flavus, A. oryzae and A. parasiticus, likely leading to the high copper tolerance of these three fungi (Figures 1 and 2, Supplementary Figure S1). Other species (A. sydowii, A. glaucus, A. arachidicola, A. calidoustus, and A. cristatus) also harbour additional copies of the CrpA homolog (Figure 2) suggesting that they may also exhibit increased Cu tolerance.
As Cu is an essential trace element for most living organisms, and can also be toxic to cells due to its redox properties, Cu is an effective antimicrobial agent and also used by mammalian host cells as defense mechanisms [12]. In the phagolysosome, where macrophages compartmentalize microbial invaders, reactive oxygen intermediates (ROIs), reactive nitrogen intermediates (RNIs) and low pH conditions are generated to kill pathogens. To attack the invading microbes with excess Cu at the infection site of lung, macrophages elevated the Cu level by inducing expression of both ATP7A and Ctr1 [7,12]. A significant induction of Ctr1 was found in A. fumigatus challenged macrophages, indicating that Cu is mobilized against the fungal invader [15]. The accumulating Cu works with ROIs, generating the toxic hydroxyl radicals and killing the invading pathogens. Thus, loss of either aceA or crpA decreased virulence of A. fumigatus in a murine model of invasive aspergillosis. Here we found that while ΔcrpAΔcrpB showed decreased murine virulence in all measurable parameters (mortality, fungal load and histology), ΔaceA only showed an intermediate attenuated virulence characterized by a reduction in fungal load and slightly less invasive growth as evidenced by histology. This result suggests that some CrpA/CrpB activity remains in the ΔaceA mutant. An additional line to pursue in the future could include assessing virulence of these A. flavus mutants in a chronic granulomatous disease (CGD) murine model. CGD patients, particularly susceptible to infections by Aspergillus, are impaired in ROI generation [47] and possibly at differential risk to strains of Aspergillus unable defend from Cu generated ROI.
Although many studies have shown that the Cu detoxification system plays important roles in virulence of human pathogens [15,48,49], to date there hasn’t been a study of a role for CrpA in a plant pathogenic fungus, nor is there any evidence the plant cells use Cu as a defense mechanism. The only study investigating any role of a Cu transporter in a plant pathogen is where a Cu golgi transporter, BcCCC2, in the pathogen Botrytis cinerea was found important for virulence in that fungus [24]. Here we found no role for AceA or CrpA/B in virulence of A. flavus on host corn seed. Whether this is due to a general lack of a Cu based defense system in plants or a consequence of the somewhat dormant nature of seed responses to pathogens is unknown. However, not surprisingly, we found that the aceA and crpA double mutants failed to colonize on maize kernel seeds treated with the Cu fungicide.
In conclusion, we identified two copies of the Cu exporter P1-type ATPase in A. flavus, leading to a greater tolerance of Cu in this species than other Aspergilli. Both crpA and crpB are transcriptionally regulated by the Cu-fist TF AceA and both need to be deleted to detect sensitivity to Cu or ROI species. Further, our study illustrates that genes/processes important for virulence of this crossover pathogen in the animal Kingdom may not be a factor in infections of the plant Kingdom.
Materials and methods
Strains and culture conditions
All Aspergillus strains used in this study are listed in Table 1. Aspergillus strains were grown on glucose minimal medium (GMM) [50] with 1 μM Cu at 37°C or 30°C with appropriate supplements as indicated. For argB auxotrophs, 1 g/L arginine were added in the growth medium. For pyrG auxotrophs, 5 mM uridine and 5 mM uracil were added in the medium. Liquid GMM with 0.5% yeast extract was used to extract genomic DNA. Conidia were harvested freshly in 0.01% Tween 80, and were counted using a hemocytometer. For growth assay, 2 μL 106 conidia/mL of indicated strains were spotted onto the solidified GMM (Noble Agar, Difco, BD) plates under different Cu concentrations. For Cu tolerance assays, non-EDTA trace elements were used.
Table 1.
Aspergillus strains used in this study.
| Name of strain | Genotype | Source |
|---|---|---|
| NRRL 3357 | A. flavus Wild-type | Keller lab |
| GD4.4 | A. flavus Wild-type | Keller lab |
| SRRC 28 | A. flavus Wild-type | Keller lab |
| TXZ118.2 | A. flavus Wild-type | Keller lab |
| PC70 | A. flavus Wild-type | Keller lab |
| PC42 | A. flavus Wild-type | Keller lab |
| Af293 | A. fumigatus Wild-type | Keller lab |
| SRRC44 | A. fumigatus Wild-type | Keller lab |
| SRRC46 | A. fumigatus Wild-type | Keller lab |
| SRRC43 | A. fumigatus Wild-type | Keller lab |
| SRRC 2006 | A. fumigatus Wild-type | Keller lab |
| CEA10 | A. fumigatus Wild-type | Keller lab |
| FGSC4A | A. nidulans Wild-type | Keller lab |
| CBS 513.65 | A. clavatus Wild-type | Keller lab |
| CBS 506.65 | A. zonatus Wild-type | Keller lab |
| DTO 11-B6 | A. carbonarius Wild-type | Keller lab |
| CBS 101740 | A. brasiliensis Wild-type | Keller lab |
| CBS 172.66 | A. aculeatus Wild-type | Keller lab |
| CBS 113.46 | A. niger Wild-type | Keller lab |
| NIH 2624 | A. terreus Wild-type | Keller lab |
| SRRC 2104 | A. oryzae Wild-type | Keller lab |
| SRRC 2098 | A. oryzae Wild-type | Keller lab |
| SRRC 266 | A. oryzae Wild-type | Keller lab |
| SRRC 2103 | A. oryzae Wild-type | Keller lab |
| SU-1 | A. parasiticus Wild-type | Keller lab |
| ATCC 56774 | A. parasiticus Wild-type | Keller lab |
| SRRC 141 | A. parasiticus Wild-type | Keller lab |
| SRRC 1039 | A. parasiticus Wild-type | Keller lab |
| SRRC 143 | A. parasiticus Wild-type | Keller lab |
| SRRC 164 | A. parasiticus Wild-type | Keller lab |
| TJW 149.27 | pyrG1, Δku70::AfpyrG | [57] |
| TJES20.1 | pyrG1, Δku70, ΔargB::AfpyrG | [58] |
| TXZ 21.3 | pyrG1, Δku70, ΔargB | [58] |
| TJES19.1 | pyrG1, Δku70, | [58] |
| TKY6.1 | pyrG1, Δku70, ΔcrpA::argB | This study |
| TKY6.3 | pyrG1, Δku70, ΔargB::AfpyrG, ΔcrpA::argB | This study |
| TKY7.1 | pyrG1, Δku70, ΔargB::AfpyrG, ΔcrpB::argB | This study |
| TKY11.4 | pyrG1, Δku70, ΔcrpA::argB, ΔcrpB::AfpyrG | This study |
| TKY5.1 | pyrG1, Δku70, ΔaceA::argB | This study |
| TKY5.3 | pyrG1, Δku70, ΔargB::AfpyrG, ΔaceA::argB | This study |
| TKY10.3 | pyrG1, Δku70, ΔaceA::argB, aceA::AfpyrG | This study |
| TKY20.2 | pyrG1, Δku70, gpdA(p)::crpA::AfpyrG | This study |
| TKY21.1 | pyrG1, Δku70, gpdA(p)::crpB::AfpyrG | This study |
| TKY22.4 | pyrG1, Δku70, ΔaceA::argB, gpdA(p)::crpA::AfpyrG | This study |
| TKY23.3 | pyrG1, Δku70, ΔaceA::argB, gpdA(p):: crpB:: AfpyrG | This study |
Strain construction
Aspergillus flavus gene deletion and transformation experiments were conducted following previously described protocols [51]. For the disruption of crpA, crpB and aceA, a homologous recombination strategy was used to replace each gene with A. flavus argB in the parental strain TJES20.1 and TXZ21.3 strain protoplasts. The double-joint fusion PCR was performed to generate the disruption constructs [52]. All primers used for gene deletion are listed in Table S2. The primers P1 and P3 were used to amplify the 5ʹ flanking fragment of the gene in questions, while primers P4 and P6 were used to amplify the 3ʹ flanking fragment of the gene. Aspergillus flavus argB was amplified from genomic DNA using primers argB/F and argB/R. The nested primers P2 and P5 were used to generate entire disruption constructs. The primers crpB/P1 and crpB-pyrG/P3 were used to amplify crpB 5ʹ flanking fragment, while primers crpB-pyrG/P4 and crpB/P6 were used to amplify crpB 3ʹ flanking fragment. The nested primers crpB/P2 and crpB/P5 were used to generate entire crpB disruption construct. To generate a ΔcrpAΔcrpB mutant, the purified fusion PCR constructs were co-transformed into TKY6.1 (pyrG auxotrophs) strain protoplasts.
crpA and crpB were each overexpressed, driven by the A. nidulans gpdA promoter, at their native loci using double-joint PCR constructs. The marker gene A. fumigatus pyrG was used, and was amplified with the primers PyrG/F and PyrG/R from A. fumigatus Af293 genomic DNA. The gpdA promoter amplified from A. nidulans gemonic DNA was inserted directly upstream of the ATG start site. The crpA 5ʹ flank was amplified with primers crpA/P1 and OE-crpA/P3, and the first 1.0 kb fragment of the crpA coding region was amplified with primers OE-crpA/P4 and OE-crpA/P6. The 5ʹ and 3ʹ fragments for the crpB overexpression constructs were amplified with the primers crpB/P1&OE-crpB/P3 and OE-crpB/P4&OE-crpB/P6, respectively. The nested primers crp/P2 and OE-crp/P5 were used to generate entire overexpression constructs. The purified fusion PCR constructs were co-transformed into TJES19.1 and TKY5.1 protoplasts.
To generate an aceAC complemented strain, the A. fumigatus pyrG selective marker amplified from its genomic DNA was used. The aceAC complemented cassette was inserted at the ku70 loci using double-joint PCR constructs. A 3.2-kb PCR product (1.7-kb aceA coding sequence, 1.2-kb upstream sequence and 0.3-kb aceA terminator region) was amplified from A. flavus wild-type genomic DNA using primers aceA-CM/F and aceA-CM/F, and the two 1-kb flanking fragments of ku70 was amplified using primers KU70-CM/P1& KU70-CM/P3 and KU70-CM/P4& KU70-CM/P6, respectively. The nested primers KU70-CM/P2 and KU70-CM/P5 were used to generate the entire complemented construct. The purified fusion PCR constructs then were transformed into ΔaceA 1-1 to create ΔaceA complemented strains TKY10.3. All transformants were confirmed by diagnostic PCR and Southern analysis (Figure S3B). For the Southern analysis, both the 5ʹ and 3ʹ flanking fragment were used as probes.
Phylogenetic analysis
For phylogenetic analysis, BLASTP [30] hits with an e-value below 1−20 against Aspergillus species derived from the NCBI database were aligned using MAFFT (Katho et al, 2002). Phylogenetic analysis was performed using fasttree (Price et al., 2009) using the extracted E1-E2 domain from all identified ATPAses. Bootstrap analysis was performed with 1000 replicates.
Pathogenicity assays on corn
Fungal infection was performed using previously described methods [53,54]. The corn kernels were incubated with 200 μL 106 conidia/mL of indicated strains in a 29°C incubator with a 12-h-light/dark photoperiod for 5 days. Four replicates were conducted for each strain. After incubation, the infected kernels were collected in 2.5 mL methanol to remove the conidia. 100 μL spore suspension was removed for spore amount quantification. Two V of chloroform was added into the vials, and incubated in the dark overnight for aflatoxin extraction. Two mL of the extract was removed, dry down, and resuspended in 1 mL 80:20 water:acetonitrile. HPLC analysis was used for AF analysis according to the published protocol[53].
For application of Cu fungicide, kernels were pretreated with 2.4 g/L Cu powder (wettable powder Champion (Nufarm Americas Inc., IL), and the infection assay was carried out as described above.
RNA extraction and northern analysis
For Northern expression analysis, all strains at 3 × 106 conidia/L were incubated in 30 mL of liquid GMM without Cu in triplicate at 37°C and 200 rpm shaking for 24 h. To one-half of the cultures, Cu was added to a final concentration of 200 μM for 1 h before harvesting. Mycelia were collected and washed with sterile water, frozen in liquid nitrogen and lyophilized 24 h. Total RNA was extracted with QIAzol Lysis Reagent (QIAGEN), according to the manufacturer’s protocol. Northern analysis was performed as previously described[55]. Probes for Northern analysis were constructed using the primers listed in Table S1 and labeled with dCTP α32P.
Copper quantification
To quantify Cu from the A. flavus mycelium, all strains at 5 × 106 conidia/L were incubated in 50 mL of liquid GMM without Cu in four replicates at 37°C and 200 rpm shaking for 12h, then Cu was added to a final concentration of 50 μM for 12 h before harvesting in one half of the treatments. The quantification of copper from the indicated strains was carried out according to our previously publication [15], with the incorporation of sulfur as a mass index, as previously reported, in order to increase accuracy [56].
Animal studies
Six-week-old female ICR mice were immunocompromised by subcutaneous injection with cortisone acetate (300 mg/kg) 3 days prior to infection, on the day of infection, and 3, 7, and 11 days post-infection. The mice were infected intranasally with 5 × 105 dormant spores, which had been suspended in 20 μL of PBS plus 0.2% Tween 20 (10 μL in each nostril). Survival was monitored for up to 21 days. For histopathology, mice were sacrificed two days after infection and their lungs were removed for histological staining with Grocott’s methenamine silver stain (GMS; fungal staining, counter stained with Light Green SM solution) and hematoxylin and eosin (H&E; tissue and nuclear staining). For fungal burden, infected mice were sacrificed on the second day post infection, their lungs were removed and homogenized, and the homogenates were plated on YAG. The plates were incubated at 37°C for 24 h, and the numbers of colony forming unit (CFU) were counted. Experiments were ethically approved by the Ministry of Health (MOH) Animal Welfare Committee, Israel.
Statistical analysis
All data were presented as the means ± standard deviation (SD). Statistical and significance analysis were performed using the GraphPad Prism 5. The statistical differences for mouse survival were calculated using the Log-Rank (Mantel-Cox) test and for organ fungal load using the unpaired t test. One-way ANOVA and the least significant difference (LSD) tests was used to determine significant differences among group means. A p-value less than 0.05 was recognized as statistically significant.
Funding Statement
This work was supported in part by the program of Fujian Agriculture and Forestry University of doctoral students going aboard to be joint trained (No.324-112110069), the Scientific Research Foundation of Graduate School of Fujian Agriculture and Forestry University (YB2014007) and “Excellent initiative” for graduate students of Life Science College to KY, and in part by support by the National Institute of Food and Agriculture, United States Department of Agriculture, Hatch project 232876 and 1006780 to N.P.K. and Food Research Institute support to N.P.K, in part by the Israel Ministry of Health Infect-ERA (Grant 11080) to NO. We thank Agilent Technologies for ICP-MS/MS instrumental support.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Festa RA, Thiele DJ.. Copper at the front line of the host-pathogen battle. PLoS Pathog. 2012. September;8(9):e1002887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Ding C, Ra Festa, Ts Sun, et al. Iron and copper as virulence modulators in human fungal pathogens. Mol Microbiol. 2014. July;93(1):10–23. [DOI] [PubMed] [Google Scholar]
- [3].Vulpe CD, Packman S. Cellular copper transport. Annu Rev Nutr. 1995;15:293–322. [DOI] [PubMed] [Google Scholar]
- [4].Jomova K, Valko M. Advances in metal-induced oxidative stress and human disease. Toxicology. 2011. May 10;283(2–3):65–87. [DOI] [PubMed] [Google Scholar]
- [5].Benov L. How superoxide radical damages the cell. Protoplasma. 2001;217(1–3):33–36. [DOI] [PubMed] [Google Scholar]
- [6].Macomber L, Imlay JA. The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. PNAS. 2009. May 19;106(20):8344–8349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ding C, Ra Festa, Yl Chen, et al. Cryptococcus neoformans copper detoxification machinery is critical for fungal virulence. Cell Host Microbe. 2013. March 13;13(3):265–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Mi Samanovic, Ding C, Dj Thiele, et al. Copper in microbial pathogenesis: meddling with the metal. Cell Host Microbe. 2012. February 16;11(2):106–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Bergemann C, Zaatreh S, Wegner K, et al. Copper as an alternative antimicrobial coating for implants - An in vitro study. World J Transplant. 2017. June 24;7(3):193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Weaver L, Michels HT, Keevil CW. Potential for preventing spread of fungi in air-conditioning systems constructed using copper instead of aluminium. Lett Appl Microbiol. 2010. January;50(1):18–23. [DOI] [PubMed] [Google Scholar]
- [11].Munoz C, Rios E, Olivos J, et al. Iron, copper and immunocompetence. Br J Nutr. 2007. October;98(Suppl 1):S24–S28. [DOI] [PubMed] [Google Scholar]
- [12].Besold AN, Culbertson EM, Culotta VC, et al. Yang of copper during infection. J Biol Inorg Chem. 2016. April;21(2):137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].White C, Lee J, Kambe T, et al. A role for the ATP7A copper-transporting ATPase in macrophage bactericidal activity. J Biol Chem. 2009. December 04;284(49):33949–33956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Palmgren MG, Nissen P. P-type ATPases. Annu Rev Biophys. 2011;40:243–266. [DOI] [PubMed] [Google Scholar]
- [15].Wiemann P, Perevitsky A, Lim FY, et al. Aspergillus fumigatus copper export machinery and reactive oxygen intermediate defense counter host copper-mediated oxidative antimicrobial offense. Cell Rep. 2017. May 02;19(5):1008–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Jungmann J, Reins HA, Lee J, et al. MAC1, a nuclear regulatory protein related to Cu-dependent transcription factors is involved in Cu/Fe utilization and stress resistance in yeast. EMBO J. 1993. December 15;12(13):5051–5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Cyert MS, Philpott CC. Regulation of cation balance in Saccharomyces cerevisiae. Genetics. 2013. March;193(3):677–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Waterman SR, Hacham M, Hu G, et al. Role of a CUF1/CTR4 copper regulatory axis in the virulence of Cryptococcus neoformans. J Clin Invest. 2007. March;117(3):794–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Weissman Z, Berdicevsky I, Cavari BZ, et al. The high copper tolerance of Candida albicans is mediated by a P-type ATPase. PNAS. 2000. March 28;97(7):3520–3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Schwartz JA, Olarte KT, Michalek JL, et al. Regulation of copper toxicity by Candida albicans GPA2. Eukaryot Cell. 2013. July;12(7):954–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Mackie J, Szabo EK, Urgast DS, et al. Host-imposed copper poisoning impacts fungal micronutrient acquisition during systemic Candida albicans infections. PloS One. 2016;11(6):e0158683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Antsotegi-Uskola M, Markina-Inarrairaegui A, Ugalde U. Copper resistance in Aspergillus nidulans relies on the PI-type ATPase CrpA, regulated by the transcription factor AceA. Front Microbiol. 2017;8:912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Park YS, Lian H, Chang M, et al. Identification of high-affinity copper transporters in Aspergillus fumigatus. Fungal Genet Biol. 2014;73:29–38. [DOI] [PubMed] [Google Scholar]
- [24].Saitoh Y, Izumitsu K, Morita A, et al. A copper-transporting ATPase BcCCC2 is necessary for pathogenicity of Botrytis cinerea. Mol Genet Genomics. 2010. July;284(1):33–43. [DOI] [PubMed] [Google Scholar]
- [25].Upadhyay S, Torres G, Lin X. Laccases involved in 1,8-dihydroxynaphthalene melanin biosynthesis in Aspergillus fumigatus are regulated by developmental factors and copper homeostasis. Eukaryot Cell. 2013;Dec;12(12):1641–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Hedayati MT, Pasqualotto AC, Warn PA, et al. Aspergillus flavus: human pathogen, allergen and mycotoxin producer. Microbiology. 2007. June;153(Pt(6)):1677–1692. [DOI] [PubMed] [Google Scholar]
- [27].Chakrabarti A, Singh R. The emerging epidemiology of mould infections in developing countries. Curr Opin Infect Dis. 2011. December;24(6):521–526. [DOI] [PubMed] [Google Scholar]
- [28].Amare MG, Keller NP. Molecular mechanisms of Aspergillus flavus secondary metabolism and development. Fungal Genet Biol. 2014. May;66:11–18. [DOI] [PubMed] [Google Scholar]
- [29].Meite F, Alvarez-Zaldivar P, Crochet A, et al. Impact of rainfall patterns and frequency on the export of pesticides and heavy-metals from agricultural soils. Sci Total Environ. 2017. November 8;616–617:500–509. [DOI] [PubMed] [Google Scholar]
- [30].Altschul SF, Gish W, Miller W, et al. Basic local alignment search tool. J Mol Biol. 1990. October 5;215(3):403–410. [DOI] [PubMed] [Google Scholar]
- [31].Katoh K, Misawa K, Kuma K, et al. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002. July 15;30(14):3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009. July;26(7):1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Shiraishi E, Inouhe M, Joho M, et al. The cadmium-resistant gene, CAD2, which is a mutated putative copper-transporter gene (PCA1), controls the intracellular cadmium-level in the yeast S. cerevisiae. Curr Genet. 2000. February;37(2):79–86. [DOI] [PubMed] [Google Scholar]
- [34].Huffman DL, O’Halloran TV. Energetics of copper trafficking between the Atx1 metallochaperone and the intracellular copper transporter, Ccc2. J Biol Chem. 2000. June 23;275(25):18611–18614. [DOI] [PubMed] [Google Scholar]
- [35].Wu F, Groopman JD, Pestka JJ. Public health impacts of foodborne mycotoxins. Annu Rev Food Sci Technol. 2014;5:351–372. [DOI] [PubMed] [Google Scholar]
- [36].Pettit RE, Taber RA, Schroeder HW, et al. Influence of fungicides and irrigation practice on aflatoxin in peanuts before digging. Appl Microbiol. 1971. October;22(4):629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Gauthier GM, Keller NP. Crossover fungal pathogens: the biology and pathogenesis of fungi capable of crossing kingdoms to infect plants and humans. Fungal Genet Biol. 2013. December;61:146–157. [DOI] [PubMed] [Google Scholar]
- [38].Lalitha P, Sun CQ, Prajna NV, et al. In vitro susceptibility of filamentous fungal isolates from a corneal ulcer clinical trial. Am J Ophthalmol. 2014. February;157(2):318–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Rudramurthy SM, Seyedmousavi S, Dhaliwal M, et al. Pharmacodynamics of voriconazole against wild-type and azole-resistant Aspergillus flavus isolates in a nonneutropenic murine model of disseminated aspergillosis. Antimicrob Agents Chemother. 2017. January; 61(1):e01491-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Jung WH, Kronstad JW. Iron and fungal pathogenesis: a case study with Cryptococcus neoformans. Cell Microbiol. 2008. February;10(2):277–284. [DOI] [PubMed] [Google Scholar]
- [41].Perez N, Johnson R, Sen B, et al. Two parallel pathways for ferric and ferrous iron acquisition support growth and virulence of the intracellular pathogen Francisella tularensis Schu S4. Microbiol Open. 2016. June;5(3):453–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Valko M, Morris H, Cronin MT. Metals, toxicity and oxidative stress. Curr Med Chem. 2005;12(10):1161–1208. [DOI] [PubMed] [Google Scholar]
- [43].Park JI, Grant CM, Dawes IW. The high-affinity cAMP phosphodiesterase of Saccharomyces cerevisiae is the major determinant of cAMP levels in stationary phase: involvement of different branches of the ras-cyclic AMP pathway in stress responses. Biochem Biophys Res Commun. 2005. February 4;327(1):311–319. [DOI] [PubMed] [Google Scholar]
- [44].Logeman BL, Wood LK, Lee J, et al. Gene duplication and neo-functionalization in the evolutionary and functional divergence of the metazoan copper transporters Ctr1 and Ctr2. J Biol Chem. 2017. July 7;292(27):11531–11546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Lenassi M, Gostincar C, Jackman S, et al. Whole genome duplication and enrichment of metal cation transporters revealed by de novo genome sequencing of extremely halotolerant black yeast Hortaea werneckii. PloS One. 2013;8(8):e71328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Palacios O, Atrian S, Capdevila M. Zn- and Cu-thioneins: a functional classification for metallothioneins? J Biol Inorg Chem. 2011. October;16(7):991–1009. [DOI] [PubMed] [Google Scholar]
- [47].Lanternier F, Cypowyj S, Picard C, et al. Primary immunodeficiencies underlying fungal infections. Curr Opin Pediatr. 2013. December;25(6):736–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Parisot D, Dufresne M, Veneault C, et al. clap1, a gene encoding a copper-transporting ATPase involved in the process of infection by the phytopathogenic fungus Colletotrichum lindemuthianum. Mol Genet Genomics. 2002. October;268(2):139–151. [DOI] [PubMed] [Google Scholar]
- [49].Hodgkinson V, Petris MJ. Copper homeostasis at the host-pathogen interface. J Biol Chem. 2012. April 20;287(17):13549–13555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Shimizu K, Keller NP. Genetic involvement of a cAMP-dependent protein kinase in a G protein signaling pathway regulating morphological and chemical transitions in Aspergillus nidulans. Genetics. 2001. February;157(2):591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Yang K, Liang L, Ran F, et al. The DmtA methyltransferase contributes to Aspergillus flavus conidiation, sclerotial production, aflatoxin biosynthesis and virulence. Sci Rep. 2016;6:23259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Lim FY, Sanchez JF, Wang CC, et al. Toward awakening cryptic secondary metabolite gene clusters in filamentous fungi. Methods Enzymol. 2012;517:303–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Christensen S, Borrego E, Shim WB, et al. Quantification of fungal colonization, sporogenesis, and production of mycotoxins using kernel bioassays. J Visualized Exp. 2012;23:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Luo X, Affeldt KJ, Keller NP. Characterization of the Far Transcription Factor Family in Aspergillus flavus. G3 (Bethesda, Md).. 2016. August 17;6(10):3269–3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Green MR, Sambrook J. Molecular cloning: a laboratory manual. New York: Cold Spring Harbour Laboratory Press; 2012. [Google Scholar]
- [56].Figueroa JAL, Stiner CA, Radzyukevich TL, et al. Metal ion transport quantified by ICP-MS in intact cells. Sci Rep. 2016. February;03(6):20551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Khalid S, Baccile JA, Spraker JE, et al. NRPS-derived isoquinolines and lipopetides mediate antagonism between plant pathogenic fungi and bacteria. ACS Chem Biol. 2018. January 19;13(1):171–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Pfannenstiel BT, Zhao X, Wortman J, et al. Revitalization of a forward genetic screen identifies three new regulators of fungal secondary metabolism in the genus Aspergillus. mBio. 2017. September 5; 8(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


