Abstract
Automatic segmentation of the prostate in magnetic resonance imaging (MRI) has many applications in prostate cancer diagnosis and therapy. We propose a deep fully convolutional neural network (CNN) to segment the prostate automatically. Our deep CNN model is trained end-to-end in a single learning stage based on prostate MR images and the corresponding ground truths, and learns to make inference for pixel-wise segmentation. Experiments were performed on our in-house data set, which contains prostate MR images of 20 patients. The proposed CNN model obtained a mean Dice similarity coefficient of 85.3%±3.2% as compared to the manual segmentation. Experimental results show that our deep CNN model could yield satisfactory segmentation of the prostate.
Keywords: Magnetic resonance imaging (MRI), prostate segmentation, deep learning, convolutional neural network
1. PURPOSE
It is estimated that there are 180,890 new cases of prostate cancer and 26,120 deaths from prostate cancer in the USA in 2016 [1]. MRI becomes a routine modality for prostate examination [2–5]. Accurate segmentation of the prostate and lesions from MRI has many applications in prostate cancer diagnosis and treatment. However, manual segmentation can be time consuming and subject to inter- and intra-reader variations. In this study, a deep learning method is proposed to automatically segment the prostate on T2-weighted (T2W) MRI.
Recently, deep learning has dramatically changed the landscape of computer vision. The initial work was for object classification [6] using a big data set of natural images called ImageNet [7]. Currently, most deep learning algorithms are for image-level classification [6, 8]. To obtain pixel/voxel-level classification, some researchers [9–15] propose a patch-wise segmentation method, which extracts small patches (e.g., 32×32) from images and then train a convolutional neural network model. In the training stage, each patch is assigned a label, which can be directly fed into the image-level classification framework. In the testing stage, the inferred label of each patch is considered as a label of the center pixel of the patch. The performance of patch-wise methods can be affected by the patch size. A large patch size reduces the localization accuracy and a small patch size can see a small context. In addition, when the number of patches is large (each voxel assigned a patch), there is a high redundant computation that needs to be performed for neighboring patches. To solve these problems, Long et al. [16] proposed an end-to-end pixel-wise, natural image segmentation method based on Caffe [17], a deep learning software. They modified an existing classification CNN to a fully convolutional network (FCN) for object segmentation. A coarse label map can be obtained from the network by classifying every local region, and then perform a simple deconvolution based on bilinear interpolation for pixel-wise segmentation. This method does not make use of post-processing or post-hoc refinement by random fields [18, 19]. In this paper, we fine tune the FCN for the segmentation of the prostate on MR images.
2. METHOD
2.1. Convolutional neural network
In practice, few people train an entire CNN from scratch, since it is difficult to collect a dataset of sufficient size, especially for medical images. In contrast to learn from scratch, it is common to use a pre-trained CNN on a large data set and then retrain an own classifier on top of the CNN for the new data set, named as fine tuning. Tajbakhsh [9] showed that knowledge transfer from natural images to medical image is possible based on CNN. Therefore, we fine tune Long’s FCN model [16] trained on natural images and retrain it based on our medical image dataset for the prostate segmentation, named as PSFCN. Fig. 1 shows the proposed deep learning method.
Fig. 1.
The framework of the proposed deep convolutional neural network (CNN). There are 7 hidden layers in the CNN. The number shows the feature or channel dimension of each hidden layer.
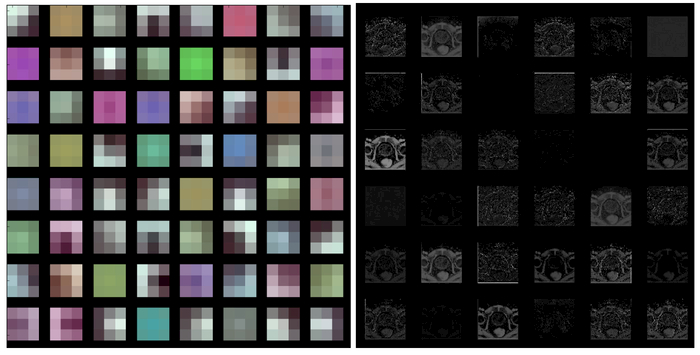
The early layers of a CNN learn low-level generic features that are applicable to most tasks. The late layers learn high-level specific features that are applicable to the application at hand [20, 21], Therefore, we only fine tune the last three layers of the FCN in this work. Fig. 2 shows the filters and the outputs of the first hidden layer. The first layer learns simple features, such as edge, junction and comer. Fig. 3 shows the outputs of late hidden layers. The figure shows that the late layers learn high-level features, which yield highly abstracted output images.
Fig. 2.
Filters and output of the first hidden layer of the CNN. Left: Filters of the first hidden layer (3×3 filters). Right: Outputs of the first hidden layer (first 36 only).
Fig. 3.
Output of the fourth layer (left) and the fifth layer (right).
The proposed PSFCN predicts the probability of each voxel belonging to the prostate or background. For each prostate MR image, the prostate only has a small region compared with the background, which means that the number of the foreground voxels is less than that of the background voxels. This unbalance between the prostate and background regions will cause the learning algorithm to get trapped in local minima with the use of the softmax loss function in Caffe [17], Therefore, the prostate is often missed and the prediction tends to classify prostate voxels as the background. In this work, we use a weighted cross entropy loss function [22], The loss function is formulated as follows.
where Pi represents the ground truth or golden standard, denotes the probability of the voxel i belonging to the prostate. is the weight, which is set as 1/|pixels of class xi = k|. Based on the weighted cross entropy loss function, the class unbalancing problem can be alleviated.
Data augmentation has been proven to improve the performance of deep learning [6, 23, 24], To obtain robustness and increased precision on the test data set, we augment the original training data set by using image translations and horizontal reflections.
2.2. Evaluation metrics
The proposed method was evaluated using our in-house data set and manual segmentation. Four quantitative metrics are used for segmentation evaluation, which are Dice similarity coefficient (DSC), relative volume difference (RVD), Hausdorff distance (HD), and average surface distance (ASD) [25–29],
3. RESULTS
3.1. Data set
The proposed method was evaluated on our in-house prostate MR data set, which has 20 T2-weighted MRI volumes. The voxel sizes of the volumes are from 0.625 mm to 1 mm, while the slice thickness is 3 mm.
3.2. Implementation details
Our algorithm was implemented by using a custom version of the Caffe framework. Training and inference were implemented in Python language. All the experiments ran on an Ubuntu workstation equipped with 512 GB memory, an Intel Xeon E5–2667 v3 CPU, and an Nvidia Quadro M5000 graph card with 8 GB video memory. The training time of the CNN model is 20 hours. Learning rate was set as le-9, while the iteration was 80,000. Each prostate MR image was segmented in about 4 seconds.
3.3. Qualitative evaluation results
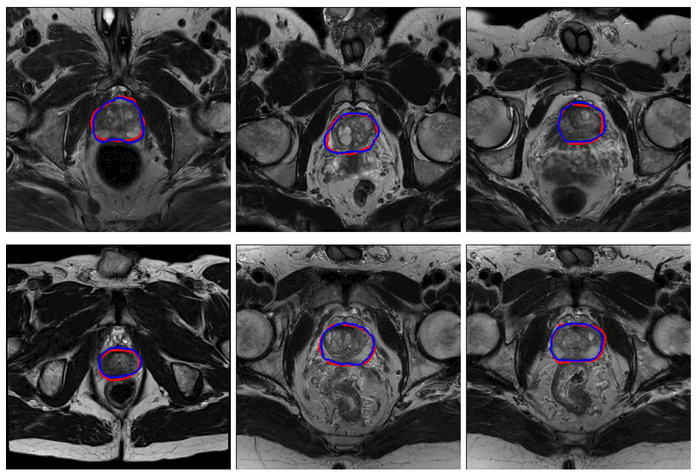
The performance of the proposed deep learning method was evaluated qualitatively by visual comparison with the manually segmented contours. Fig. 3 shows the qualitative results.
3.4. Quantitative evaluation results
The quantitative results are shown in Table 1. Our proposed method yields a mean DSC of 85.3%±3.2% and mean HD of 9.3±2.5 mm.
Table 1.
Quantitative evaluation of the proposed prostate segmentation method.
| DSC(%) | RVD(%) | HD(mm) | ASD(mm) | |
|---|---|---|---|---|
| Avg. | 85.3 | 2.4 | 9.3 | 3.3 |
| Std. | 3.2 | 12.6 | 2.5 | 0.7 |
| Max | 91.5 | 23.0 | 12.9 | 4.7 |
| Min | 77.1 | −14.7 | 4.2 | 2.0 |
4. CONCLUSIONS
We proposed an automatic deep learning method to segment the prostate on MR images. An end-to-end deep CNN model was trained on our in-house prostate MR data set and achieved good performance for prostate MR image segmentation. To the best of our knowledge, this is the first study to fine-tune a fully convolutional network (FCN) that has been pre-trained using a large set of labeled natural images for segmenting the prostate in MR images. We found that the use of pre-trained FCN with fine-tuning could yield satisfactory segmentation results. Our future work will focus on segmenting prostate in ultrasound and CT images.
Fig. 4.
The qualitative results of the proposed method. The red curves represent the prostate contours obtained by the proposed method, while the blue curves represent the contours obtained from manual segmentation by an experienced radiologist.
ACKNOWLEDGMENT
This research is supported in part by NIH grants (CA176684, R01CA156775 and CA204254).
REFERENCES
- 1.Siegel RL, Miller KD, and Jemal A, “Cancer statistics,”. CA: a cancer journal for clinicians, 66(1): p. 7–30, (2016). [DOI] [PubMed] [Google Scholar]
- 2.Fei B, Kemper C, and Wilson DL, “A comparative study of warping and rigid body registration for the prostate and pelvic MR volumes.” Comput Med Imaging Graph, 27(4): p. 267–81, (2003). [DOI] [PubMed] [Google Scholar]
- 3.Fei B, et al. , “Slice-to-volume registration and its potential application to interventional MRI-guided radio-frequency thermal ablation of prostate cancer.” IEEE Trans Med Imaging, 22(4): p. 515–525, (2003). [DOI] [PubMed] [Google Scholar]
- 4.Wu Qiu JY, Ukwatta Eranga, Sun Yue, Rajchl Martin, and Fenster Aaron, “Prostate Segmentation: An Efficient Convex Optimization Approach With Axial Symmetry Using 3-D TRUS and MR Images.” IEEE Transactions on Medical Imaging, 33(4): p. 947–960, (2014). [DOI] [PubMed] [Google Scholar]
- 5.Litjens G, et al. , “Computer-aided detection of prostate cancer in MRI.” IEEE transactions on Medical Imaging, 33(5): p. 1083–1092, (2014). [DOI] [PubMed] [Google Scholar]
- 6.Krizhevsky A, Sutskever I, and Hinton GE. “Imagenet classification with deep convolutional neural networks.” in Advances in neural information processing systems. p. 1907–1105, (2012). [Google Scholar]
- 7.Deng J, et al. “Imagenet: A large-scale hierarchical image database.” in IEEE Conference on Computer Vision and Pattern Recognition p. 248–255, (2009). [Google Scholar]
- 8.He K, et al. , “Deep residual learning for image recognition.” arXiv preprint arXiv:1512.03385, (2015). [Google Scholar]
- 9.Tajbakhsh N, et al. , “Convolutional Neural Networks for Medical Image Analysis: Full Training or Fine Tuning?” IEEE transactions on medical imaging, 35(5): p. 1299–1312, (2016). [DOI] [PubMed] [Google Scholar]
- 10.Zhang W, et al. , “Deep convolutional neural networks for multi-modality isointense infant brain image segmentation.” NeuroImage, 108: p. 214–224, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo Y, Gao Y, and Shen D, “Deformable MR Prostate Segmentation via Deep Feature Learning and Sparse Patch Matching.” IEEE Trans Med Imaging, 35(4): p. 1077–89, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ciresan D, et al. “Deep neural networks segment neuronal membranes in electron microscopy images.” in Advances in neural information processing systems. p. 2843–2851, (2012). [Google Scholar]
- 13.Pereira S, et al. , “Brain Tumor Segmentation using Convolutional Neural Networks in MRI Images.”, IEEE Trans Med Imaging, 35(5), p. 1240–1251, (2016). [DOI] [PubMed] [Google Scholar]
- 14.Kooi T, et al. , “Large Scale Deep Learning for Computer Aided Detection of Mammographic Lesions.” Medical Image Analysis, 35, p. 303–312, (2016). [DOI] [PubMed] [Google Scholar]
- 15.Milletari F, et al. , “Hough-CNN: Deep Learning for Segmentation of Deep Brain Regions in MRI and Ultrasound.” arXiv preprint arXiv:1601.07014, (2016). [Google Scholar]
- 16.Long J, Shelhamer E, and Darrell T. “Fully convolutional networks for semantic segmentation.” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, p. 3431–3440, (2015). [DOI] [PubMed] [Google Scholar]
- 17.Jia Y, et al. “Caffe: Convolutional architecture for fast feature embedding.” in Proceedings of the 22nd ACM international conference on Multimedia p. 675–678, (2014). [Google Scholar]
- 18.Farabet C, et al. , “Learning hierarchical features for scene labeling.” IEEE transactions on pattern analysis and machine intelligence, 35(8): p. 1915–1929, (2013). [DOI] [PubMed] [Google Scholar]
- 19.Hariharan B, et al. “Simultaneous detection and segmentation.”, in European Conference on Computer Vision p. 297–312, (2014). [Google Scholar]
- 20.Yosinski J, et al. “How transferable are features in deep neural networks?” in Advances in neural information processing systems p. 3320–3328, (2014). [Google Scholar]
- 21.Zeiler MD and Fergus R. “Visualizing and understanding convolutional networks” in European Conference on Computer Vision. Springer, p. 818–833, (2014). [Google Scholar]
- 22.Christ Patrick Ferdinand, M.E.E., Ettlinger Florian, et al. , “Automatic liver and lesions segmentation using Cascaded Fully Convolutional Neural Networks and 3D Conditional Random Fields.” MICCAI, p. PS4–18, (2016). [Google Scholar]
- 23.Ciregan D, Meier U, and Schmidhuber J. “Multi-column deep neural networks for image classification.” in IEEE Conference on Computer Vision and Pattern Recognition (CVPR), p. 3642–3649, (2012). [Google Scholar]
- 24.Milletari F, Navab N, and Ahmadi S-A, “V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation.” arXiv preprint arXiv:1606.04797, (2016). [Google Scholar]
- 25.Tian Z, et al. , “Superpixel-Based Segmentation for 3D Prostate MR Images.” IEEE Trans Med Imaging, 35(3): p. 791–801, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Litjens G, et al. , “Evaluation of prostate segmentation algorithms for MRI: the PROMISE12 challenge.” Medical image analysis, 18(2): p. 359–373, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Akbari H, Fei B. “3D ultrasound image segmentation using wavelet support vector machines.” Med Phys. 39(6):2972–84, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang X, Wu S, Sechopoulos I, Fei B. “Cupping artifact correction and automated classification for high-resolution dedicated breast CT images.” Med Phys. 39(10):6397–406, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang H, Fei B. “An MR image-guided, voxel-based partial volume correction method for PET images.” Med Phys. 39(1):179–95, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]