Abstract
Light signal provides the spatial and temporal information for plants to adapt to the prevailing environmental conditions. Alterations in light quality and quantity can trigger robust changes in global gene expression. In Arabidopsis thaliana, two groups of key factors regulating those changes in gene expression are CONSTITUTIVE PHOTOMORPHOGENESIS/ DEETIOLATED /FUSCA (COP/DET/FUS) and a subset of basic helix-loop-helix transcription factors called PHYTOCHROME-INTERACTING FACTOR (PIFs). Recently, a rapid progress has been made in characterizing the E3 Ubiquitin ligases for the light-induced degradation of PIF1, PIF3 and PIF4. However, the E3 ligase(s) for PIF5 is still unknown. Here, we show that CUL4COP1-SPA complex is necessary for the red light-induced degradation of PIF5. Furthermore, COP1 and SPA proteins stabilize PIF5 in the dark, but promote the ubiquitination and degradation of PIF5 in response to red light through the 26S proteasome pathway. Genetic analysis illustrates that overexpression of PIF5 can partially suppress both cop1–4 and spaQ seedling de-etiolation phenotypes in the dark and red light conditions. In addition, PIF5 protein level cycles under both diurnal and constant light conditions, which is also defective in the cop1–4 and spaQ backgrounds. Both cop1–4 and spaQ show defect in diurnal growth pattern. Overexpression of PIF5 partially restores the growth defects in the cop1–4 and spaQ under diurnal conditions, suggesting that COP1-SPA complex plays an essential role in photoperiodic hypocotyl growth, partly through regulating PIF5 level. Taken together, our data illustrate how CUL4COP1-SPA E3 ligase dynamically controls PIF5 level to regulate plant development.
Keywords: bHLH transcription factor, E3 Ubiquitin ligase, photomorphogenesis, Proteasome degradation, photoperiodic hypocotyl growth
INTRODUCTION
Seedlings experience two contrasting developmental growth programs: skotomorphogenesis (etiolation) in the dark and photomorphogenesis (de-etiolation) under light. Etiolated seedlings grown in the dark display long hypocotyl, closed and yellowish cotyledons. On the other hand, when grown under light, plants develop de-etiolated morphology including short hypocotyl, open, expanded and green cotyledons (Gommers and Monte 2018). The transition from skoto- to photo-morphogenesis is regulated by a diverse group of photoreceptors that perceives and responds to different wavelengths of ambient light (Galvão and Fankhauser 2015). Among those photoreceptors, phytochromes function as pivotal sensors for red/far-red region of the light spectrum by allosterically altering the conformation from an inactive (Pr) to the active (Pfr) form (Bae and Choi 2008). The active Pfr form translocates into the nucleus (Klose et al. 2015) and interacts with multiple proteins including a small family of basic helix-loop-helix (bHLH) transcription factors called PHYTOCHROME INTERACTING FACTORS (PIFs) (Castillon et al. 2007; Huq and Quail 2005; Leivar and Quail 2011). These interactions result in a transcriptional reprogramming to switch to photomorphogenic development (Quail 2007).
Previous genetic studies have shown that each of the four major PIFs (PIF1, PIF3, PIF4 and PIF5) function distinctly as well as redundantly in controlling early seedlings development (Castillon, et al. 2007; Leivar and Monte 2014; Leivar and Quail 2011). In an overlapping manner, PIF1, PIF3, PIF4 and PIF5 inhibit photomorphogenic development in darkness as pifQ mutant display constitutive photomorphogenic phenotypes in the dark (Leivar et al. 2008; Shin et al. 2009) (Paik et al. 2017). Individually, PIF3 functions as a negative regulator of seedling de-etiolation and chlorophyll biosynthesis (Kim et al. 2003; Ogawa et al. 2003; Stephenson et al. 2009). It also positively regulates anthocyanin biosynthesis in response to light (Shin et al. 2007). PIF1 negatively regulates light-induced seed germination and chlorophyll biosynthesis (Huq et al. 2004; Oh et al. 2004). PIF4 acts as a negative regulator of red and far-red light signaling at the seedling stage (Huq and Quail 2002; Lorrain et al. 2009). Along with PIF5, PIF4 also regulates shade avoidance responses (Hersch et al. 2014; Lorrain et al. 2008), leaf senescence (Sakuraba et al. 2014; Song et al. 2014; Zhang et al. 2015), anthocyanin biosynthesis (Liu et al. 2015), and diurnal growth patterns (Kunihiro et al. 2011; Nozue et al. 2007). Recently, a number of studies have shown that PIF4 plays a critical role in the multiple signal integration, including light, temperature and hormonal signaling to regulate plant growth and development (Franklin et al. 2011; Gangappa et al. 2017; Kumar et al. 2012; Paik, et al. 2017). Apart from overlapping functions with PIF4, PIF5 acts as a negative regulator of red light signaling and as a positive regulator of ethylene biosynthesis (Khanna et al. 2007). Thus, PIF4 and PIF5 function as an integrator of multiple signaling pathways to optimize the growth and development.
In addition to PIFs, another group of negative regulators of photomorphogenesis is called CONSTITUTIVE PHOTOMORPHOGENIC/DEETIOLATED/FUSCA (COP/DET/FUS) (Lau and Deng 2012; Xu et al. 2015). These proteins have been shown to function as negative regulators of photomorphogenic development as their loss-of-function mutants develop as light-grown plants even under darkness (Deng et al. 1992; Lau and Deng 2012). COP1-SPA complex acts not only independently but also synergistically with PIFs to destabilize HY5, HFR1 and possibly other positive regulators to prevent photomorphogenesis in darkness (Hoecker 2017; Li et al. 2015; Xu et al. 2017). To promote photomorphogenesis, phytochromes repress these negative regulators through several distinct mechanisms. Phytochromes inhibit COP1-SPA complex by rapidly inducing their reorganization (Lu et al. 2015; Sheerin et al. 2015; Xu, et al. 2015) and/or nuclear exclusion under prolonged light (Subramanian et al. 2004). On the other hand, phytochromes inhibit PIF functions by inducing their degradation through the ubiquitin-proteasome system (UPS)-mediated pathway and also through sequestration of PIFs, eventually preventing them from binding to their target gene promoters (Park et al. 2018; Park et al. 2012; Pham et al. 2018). Thus, phytochromes induce rapid destabilization of the negative regulators (e.g., PIFs) and stabilization of the positive geulators (e.g., HY5, HFR1 and others) to promote photomorphogenesis.
For post-translational regulation of PIF abundance, a number of kinases and E3 Ubiquitin ligases have recently been described (Pham, et al. 2018). Among the E3 ligases, CUL3LRB induces PIF3 degradation in response to light, where the LIGHT-RESPONSE Bric-a-Brack/Tramtrack/Broad (LRB1, LRB2, and LRB3) acts as substrate recognition components (Ni et al. 2014). Furthermore, a recent study showed PIF3 is ubiquitinated and degraded by the SCFEBF1/EBF2 Ubiquitin ligase (Dong et al. 2017), where EIN3-BINDING F BOX PROTEIN 1 (EBF1) and EBF2 act as substrate recognition components. CUL3-based E3 ligase is also involved in the ubiquitination and degradation of PIF4 using the BLADE ON PETIOLE (BOP1/2) as the substrate recognition components (Zhang et al. 2017). CUL4-based E3 Ubiquitin ligase with the substrate recognition components, COP1-SPA promotes the light-induced ubiquitination and degradation of PIF1 in response to light (Zhu et al. 2015). In addition, CUL1CTG10 promotes the degradation of PIF1 to regulate seed germination (Majee et al. 2018). In this case, COLD TEMPERATURE GERMINATING10 (CTG10), an F-box protein acts as a substrate recognition component. However, an E3 Ubiquitin ligase for PIF5 has not been described yet. Here, we show that CUL4COP1-SPA E3 ubiquitin ligase promotes the light-induced ubiquitination and subsequent degradation of PIF5 in red light. Light promotes in vivo interaction between COP1-SPA and PIF5 through the active phyB binding domain (APB). Strikingly, our results also suggest that COP1-SPA complex plays an essential role in controlling the photoperiodic hypocotyl growth, partly through post-translational regulation of the PIF5 abundance. Taken together, we propose a model where dynamic regulation of PIF5 by COP1-SPA complex optimizes not only seedling de-etiolation, but also diurnal regulation of plant growth and development in response to light.
RESULTS
CUL4 COP1-SPA complex promotes the degradation of PIF5 in response to red light
PIF5 is relatively stable in the dark; however, light signal induces rapid phosphorylation and subsequent degradation of PIF5 in a phytochrome-dependent manner (Shen et al. 2007). phyA, phyB and phyD regulate the light-induced degradation of PIF5 in a redundant manner, as phyAB and phyABD mutants show significantly delayed PIF5 degradation under red light. Moreover, the light-induced degradation of PIF5 is reduced by treatment with the proteasome inhibitor, suggesting that PIF5 is degraded through the 26S proteasome pathway (Shen, et al. 2007). However, the E3 Ubiquitin ligase necessary for PIF5 degradation is still unknown. Because there are hundreds of possible combinations of E3 Ubiquitin ligases in plants (Vierstra 2009), we focused on three CULLIN mutants (cul1–6, cul3ab and cul4cs) to narrow down the possible substrate adaptor components required for PIF5 degradation. We performed immunoblots to detect PIF5 abundance in cul4cs (Chen et al. 2010; Zhang et al. 2008), cul1–6 (Moon et al. 2007), and cul3ab (Thomann et al. 2005) mutant seedlings, grown in the dark and dark-grown seedlings exposed to red light. It was observed that the rate of degradation of PIF5 is slower only in the cul4cs background but not in the cul1–6 or cul3ab mutant backgrounds (Figure S1). These data suggest that CUL4-based E3 Ubiquitin ligase promotes the degradation of PIF5 during early light responses.
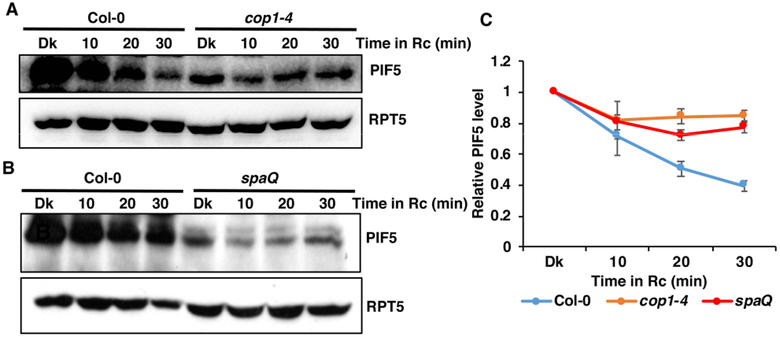
Recently, we have shown that CUL4COP1-SPA complex promotes the ubiquitination and degradation of PIF1 in response to light (Zhu, et al. 2015). To examine whether COP1-SPA complex induces the degradation of PIF5, we performed immunoblots to test the degradation rate of PIF5 in wild-type (Col-0), cop1–4 and spaQ mutants in response to red light. As reported in our earlier studies, PIF5 level is significantly lower in both cop1–4 and spaQ mutants in the dark (Xu, et al. 2017). However, the light-induced degradation is significantly reduced in both cop1–4 and spaQ mutants (Figure 1A, B,B; Figure S2). COP1 appears to play a more prominent role in regulating PIF5 compared to SPA proteins, as PIF5 is more stable in cop1–4 than spaQ background. We also tested PIF5 level under far-red light and shade mimicking (low R/FR ratio) conditions. As consistent with previous studies, PIF5 is stable under far-red and the low R/FR ratio conditions (Figure S3) (Lorrain, et al. 2008). To examine if the red light-induced reduction in PIF5 level is due to a reduction of PIF5 transcript level, we performed qRT-PCR for PIF5 transcript. The results show that PIF5 mRNA level is significantly higher in both cop1–4 and spaQ mutants, compared to wild -type both in the dark and red light, possibly due to the constitutively photomorphogenic phenotypes of these mutants (Figure S4A). However, PIF5 mRNA level is comparable between dark and 30 min of red light treated samples of wild-type and cop1–4 mutant, suggesting that light does not regulate PIF5 expression under these conditions in these backgrounds (Figure S4A). These data clearly illustrate that the light signal triggers the degradation of PIF5 protein through the CUL4COP1-SPA complex.
Figure 1. Red light induced PIF5 degradation is slower in cop1–4 and spaQ mutants.
(A-B) Immunoblots showing the degradation pattern of endogenous PIF5 in response to dark and constant Rc in cop1–4 (A) and spaQ (B) mutants, compared to the wild-type seedlings. Four-day-old dark-grown seedlings were treated with either dark (Dk) or 2 μmolm−2 s−1 constant red light (Rc) for the indicated period of time (min). Total protein was extracted and resolved on 8% SDS-PAGE gel. Proteins were transferred to PVDF membrane and sequentially probed with anti-PIF5 and anti-RPT5.
(C) Line-graph shows the relative rate of degradation of endogenous PIF5 in response to constant Rc over the dark treatment. Band intensities of PIF5 and RPT5 from three replicates were measured using ImageJ tool. For each of the genotype, PIF5 level in the dark (Dk) was set to 1and the relative PIF5 level in response to constant Rc was calculated.
We also tested PIF5 protein level in cop1–4 and spaQ mutants under prolonged red light conditions. Since, PIF5 expression is regulated by circadian clock, we crossed 35S:PIF5-Myc into cop1–4 and spaQ mutants (PIF5-Myc/cop1–4 and PIF5-Myc/spaQ) and PIF5-Myc degradation patterns is examined under prolonged light conditions (Figure S5). The level of PIF5 protein in wild-type is reduced ~50% within 10 min under Rc (2 μmolm−2 s−1) and reached the trough level after 3hrs of continuous red light treatment. Interestingly, starting at 6 hrs, PIF5 level increases to peak at 24 hrs under continuous red light. Re-accumulation of PIF5 at subjective dawn under continuous light suggests that PIF5 protein level might be regulated by the circadian clock or diurnally (Figure S5A). To test whether PIF5 re-accumulates in the cop1–4 and spaQ backgrounds, we performed immunoblots for PIF5-Myc in these backgrounds (Figure S5B, C). The results show that the light-induced degradation of PIF5 is reduced in these backgrounds compared to wild-type. Moreover, the re-accumulation of PIF5 under continuous red light at presumptive dawn is also defective in these mutants. These data suggest that COP1 and SPA proteins are involved in regulating PIF5 level under both short-term and prolonged light conditions.
PIF5 target gene expression is affected in cop1–4 and spaQ
Since PIF5 is a transcription factor that regulates the expression of genes involved in the light response pathways (Kunihiro, et al. 2011; Liu et al. 2013; Sakuraba, et al. 2014), we tested whether PIF5 target gene expression is affected in cop1–4 and spaQ mutants using qRT-PCR assays for three PIF5-induced genes (AT5G02580, ATHB2 and ARF8) and one PIF5-repressed gene (RGF9) (Zhang et al. 2013). The wild type dark level of expression for each gene was set at “1” and then the relative fold-change in expression of these genes in cop1–4 and spaQ backgrounds under darkness and dark-grown seedlings exposed to 30 min of R light were calculated. Consistent with the PIF5 protein level in these mutants, the qRT-PCR data show that the fold-change in PIF5 target gene expression in response to red light is reduced in cop1–4 and spaQ compared to wild-type (Fig. S4B). This is consistent with the reduced PIF5 degradation in cop1–4 and spaQ compared to wild type under red light.
PIF5 is ubiquitinated via 26S proteasome pathway in response to light
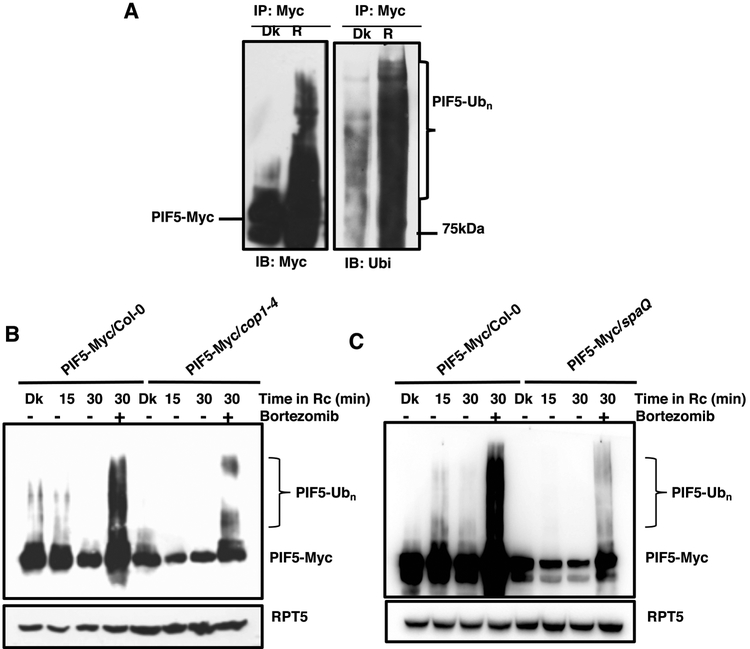
Light–induced degradation of PIFs is mediated through the UPS pathway and involves rapid phosphorylation and ubiquitination of PIFs (Ni, et al. 2014; Shen et al. 2008; Zhu, et al. 2015). To determine whether PIF5 is ubiquitinated in response to light, we immunoprecipitated PIF5-Myc from dark-grown (Dk) seedlings and dark-grown seedlings treated with continuous red light for 30 min (R) and then performed immunoblots using anti-Myc and anti-Ubi antibodies (Figure 2A, right panel). The immunoblots show that PIF5 is strongly ubiquitinated in response to the light.
Figure 2. Red light-induced ubiquitination of PIF5 is reduced in cop1–4 and spaQ mutants.
(A) Immunoblots showing the ubiquitination pattern of PIF5-Myc in response to dark or red light treatment. Four-day-old dark-grown seedlings were treated with either dark (Dk) or 2 μmolm−2 s−1 constant red light (R) for 30 min. Total protein was extracted in native buffer and PIF5-Myc was immunoprecipitated using anti-Myc antibody. Samples were resolved on 6.5% SDS-PAGE and transferred to PVDF membrane. Membranes were blotted with anti-Myc (Left panel) and anti-Ub (Right panel) antibodies.
(B-C) Immunoblots showing the relative ubiquitination status of PIF5-Myc in response to dark and constant Rc in cop1–4 (B) and spaQ (C) mutants, compared to the wild-type. Four-day-old dark-grown seedlings were treated with either dark (Dk) or 2 μmolm−2 s−1 constant red light (Rc) for the indicated period of time (min). One of the samples in each of the genotype was pretreated with 40 μM proteasome inhibitor (Bortezomib) for 3 hrs prior to Rc treatment. Total protein was extracted and samples were resolved on 6.5% SDS-PAGE and transferred to PVDF membrane. Membranes were sequentially blotted with anti-Myc and anti-RPT5 antibodies.
We also examined whether the light-induced ubiquitination of PIF5 is affected in cop1–4 and spaQ mutants in the dark and red light. Four-day-old dark-grown seedlings overexpressing PIF5 in these backgrounds were either kept in the dark or exposed to red light for the indicated period of time. One of the samples was pretreated with a protease inhibitor (40 μM Bortezomib) before exposing to red light for the indicated time. The samples with Bortezomib treatment show strong ubiquitination pattern, which suggests that the protease inhibitor efficiently inhibited the degradation of PIF5 under red light, thereby, increased the ubiquitinated PIF5 level (Figure 2A). Strikingly, the level of PIF5 ubiquitination is reduced in the cop1–4 and spaQ backgrounds compared to wild-type (Figure 2B, C). The phosphorylation of PIF5 is not observable under these conditions for this tagged version. These data suggest that COP1 and SPA proteins promote the ubiquitination of PIF5 to induce the rapid degradation in response to red light. Previous studies showed that PIFs are not directly ubiquitinated by COP1 in vitro (Jang et al. 2010; Xu et al. 2014). However, PIF1 is ubiquitinated by the CUL4COP1-SPA complex in response to light in vivo (Zhu, et al. 2015). Thus, it is possible that the light-induced degradation of PIF5 also requires CUL4COP1-SPA complex similar to PIF1.
PIF5 interacts with COP1 and SPA1 in yeast and in vivo
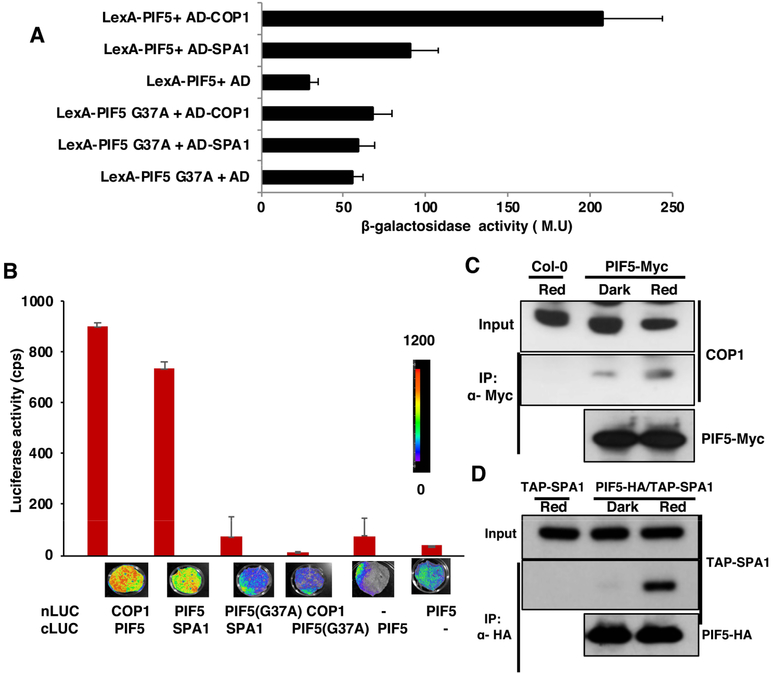
To examine if PIF5 can interact with COP1 and SPA1, we performed yeast two-hybrid (Y2H) assays using full-length PIF5 and COP1-SPA1 proteins. We also created a mutation in the APB motif of PIF5 (PIF5 G37A) and used it in Y2H assays. This residue was shown to be necessary for the physical interaction between PIF5 and phyB (Khanna et al. 2004). Results show that the wild-type PIF5 can interact with both COP1 and SPA1 proteins (Figure 3A). However, the G37A mutation in PIF5 reduced the interaction with both COP1 and SPA1. These results suggest that the APB domain of PIF5 is indeed necessary for the interaction with COP1 and SPA1.
Figure 3. PIF5 interacts with COP1 and SPA1 through the APB domain.
(A) Quantitative yeast two-hybrid assay showing interactions of COP1 and SPA1 with wild-type PIF5 and Active phyB binding domain (APB) mutagenized PIF5 (PIF5G37A). PIF5 and PIF5G37A were cloned into LexA vector containing the DNA binding domain. COP1 and SPA1 were cloned into a vector containing activation domain (AD). Empty AD vector was used to read the background level. Liquid β-galactosidase assays were performed in triplicate and Miller Units (M.U; measure of β-galactosidase activity) was determined.
(B) Split luciferase imaging (LCI) assay (Lower panel) and quantitative assay (Upper panel) showing in vivo interactions of COP1 and SPA1 with PIF5 in N. benthamiana leaves. Full-length PIF5, PIF5-G37A, COP1 and SPA1 were fused to either cLUC or nLUC as indicated in the figure. Empty vectors were used as negative controls.
(C and D) Co-immunoprecipitation (Co-IP) assay showing in vivo interaction of PIF5-Myc with COP1 (C) and PIF5-HA with TAP-SPA1 (D). Five-day-old dark-grown seedlings were pretreated with 40μM bortezomib for 4hrs and then treated with either dark or red light (3000 μmolm−2). Total protein was extracted in native buffer. PIF5-Myc and PIF5-HA were immunoprecipitated with anti-Myc and anti-HA antibodies, respectively. Interacting partners COP1 and SPA1 were detected using anti-COP1 and anti-Myc antibodies, respectively.
In vitro interaction between PIF5 and COP1 has been described earlier (Jang, et al. 2010). To test in planta interaction, we performed transient luciferase complementation imaging assay (LCI) in tobacco (Nicotiana benthamiana) leaves. Similar to Y2H assays, it was observed that the PIF5 but not the PIF5 (G37A) interact with both COP1 and SPA1 (Figure 3B). These data suggest that PIF5 interacts with COP1 and SPA1 in planta.
To promote ubiquitination and degradation of PIF5 in response to light, COP1-SPA1 interaction with PIF5 is expected to be induced by light. To test this possibility, we performed in vivo co-immunoprecipitation (Co-IP) assays using seedlings grown in the dark for four days and four-day-old dark-grown seedlings exposed to red light (3000 μmolm−2). The results show that PIF5 can interact with both COP1 and SPA1, and that the strength of interaction is significantly enhanced by red light treatment (Figure 3C, 3D). Taken together these results clearly indicate that PIF5 does interact with COP1 and SPA1 and these interactions are regulated by light.
PIF5 partially suppresses the cop1 and spaQ phenotypes
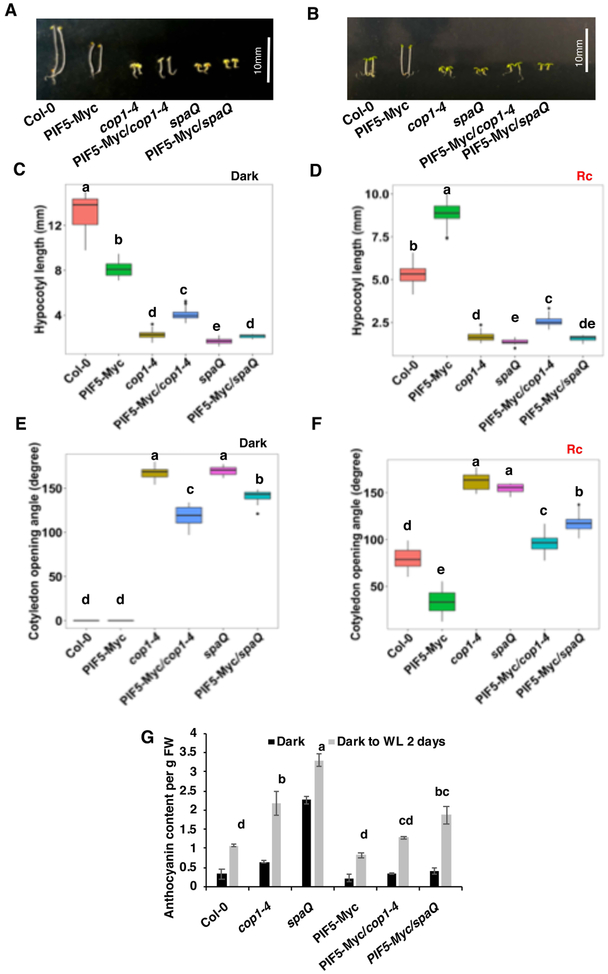
To examine the significance of PIF5 degradation by the COP1-SPA complex, we tested the phenotypes of the PIF5-Myc/cop1–4 and PIF5-Myc/spaQ compared to those of cop1–4 and spaQ phenotypes under both dark and red light conditions. Since PIF5 functions as a negative regulator of photomorphogenesis in the dark, increasing the level of PIF5 in the cop1 and spaQ is expected to suppress photomorphogenesis. In the dark, overexpression of PIF5 mimics triple responses including short hypocotyl and exaggerated apical hooks, due to the induction of ethylene biosynthesis in seedlings (Khanna, et al. 2007). Strikingly, the hypocotyl length of PIF5-Myc/cop1–4 seedlings growing in the dark is significantly longer than cop1–4 (Figure 4A, C). The hypocotyl length of PIF5-Myc/spaQ is slightly longer compared to spaQ in the dark (Fig. 4A, C). In addition, overexpression of PIF5 in the cop1–4 and spaQ partially repressed the cotyledon opening phenotype of both cop1–4 and spaQ in the dark (Figure 4E). Similarly, overexpression of PIF5 in cop1–4 mutant results in slightly longer hypocotyl than cop1–4 mutant under continuous red light (Figure 4B, D). However, the hypocotyl length of PIF5-Myc/spaQ is similar to that of spaQ under continuous red light. By contrast, the cotyledon opening angle of both cop1–4 and spaQ is partly suppressed in PIF5-Myc/cop1–4 and PIF5-Myc/spaQ seedlings (Figure 4F).
Figure 4. COP1 and SPA partially repress photomorphogenesis through PIF5.
(A-F) Phenotypic variations among seedlings of different genotypes, grown under either continuous dark (A) or red light. Box plots represent the mean hypocotyl length of seedlings growing in either dark (C) or continuous red light (D) and mean cotyledon opening angle of seedlings growing in either dark (E) or continuous red light (F). Surface sterilized seeds were plated onto MS medium without sucrose and incubated in either dark or red light for 4-day for seedling growth. Hypocotyl lengths and cotyledon angles were measured from more than 30 seedlings (n>30). Statistical significance among different genotypes was determined using one way ANOVA and Tukey’s HSD tests, and is indicated by different letters. White bar = 10mm.
(G) Bar graph showing the anthocyanin content among seedlings of different genotypes. Two-day-old dark-grown seedlings were incubated in either dark or continuous white light for another two days. Crude samples of anthocyanin were prepared and measured as described in materials and methods. Statistical significance among different genotypes was determined using single factor ANOVA and Tukey’s HSD tests, and is indicated by different letters.
Earlier it was shown that the PIF5 also function as a negative regulator of anthocyanin accumulation (Liu, et al. 2015); thus, light-induced degradation of PIF5 is expected to relieve this inhibition to promote anthocyanin biosynthesis. In the agreement with the previous study, during the dark to light transition, PIF5 overexpressed seedlings display very low level of anthocyanin. However, cop1–4 and spaQ mutants display a high level of anthocyanin. Interestingly, PIF5 overexpression in cop1–4 and spaQ mutants exhibits much less anthocyanin content compared to cop1–4 and spaQ both in the dark and light-exposed seedlings (Figure 4G). Thus, PIF5 partially suppresses the hypocotyl length, cotyledon opening angle, and anthocyanin level phenotypes of the cop1–4 and spaQ mutants.
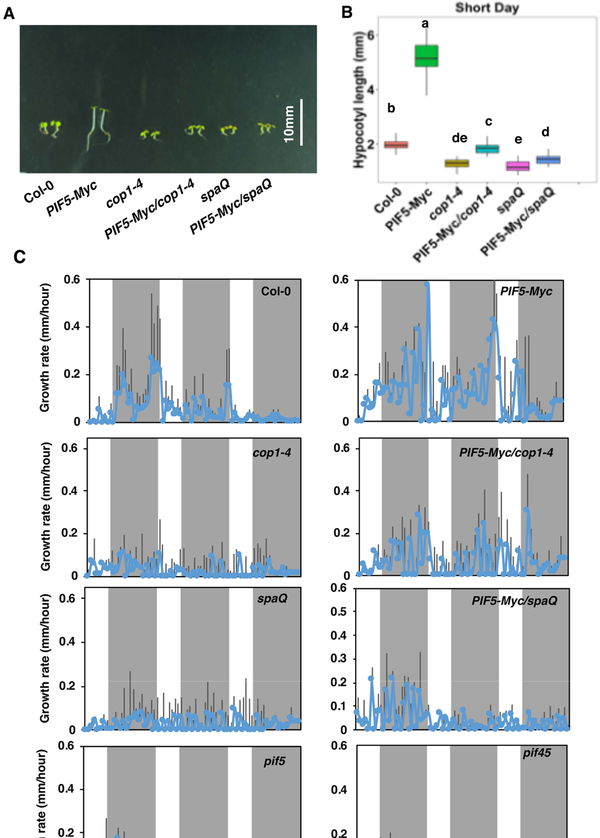
Photoperiodic control of hypocotyl growth by COP1-SPA complex via PIF5
The diurnal regulation of hypocotyl elongation by PIFs have been well documented (Ezer et al. 2017; Nozue, et al. 2007; Nusinow et al. 2011; Soy et al. 2016). However, it is still unknown whether COP1 and SPA proteins regulate diurnal growth patterns in a photoperiodic manner. To test this, we first measured the hypocotyl elongation phenotype under short day (SD) conditions. Overexpression of PIF5-Myc in seedlings displayed much longer hypocotyl, while cop1–4 and spaQ mutants showed much shorter hypocotyl compared to wild-type (Figure 5A, B). However, when PIF5-Myc was introduced into the cop1–4 and spaQ backgrounds, the hypocotyl lengths are partly rescued compared to the cop1–4 and spaQ mutant backgrounds (Figure 5A, B, Figure S6). These data are qualitatively very similar to those under continuous red light conditions. To measure the growth defect in these mutants, we measured growth rate using an infrared imaging system (Nozue, et al. 2007). The hypocotyl growth rate was measured starting from day 2, when the hypocotyl lengths are visible. In Col-0 and PIF5-Myc seedlings, the growth rate reaches the peak during night-time (ZT18-ZT24). In pif5 and pif45 mutants, the growth rate is reduced as previously shown (Nozue, et al. 2007). Strikingly, the growth rate is impaired in the cop1–4 and spaQ mutants during these periods (Figure 5C). However, overexpression of PIF5 in these mutant backgrounds partly rescues the growth rate under these conditions, suggesting that PIF5 acts downstream of these factors to regulate diurnal growth rate.
Figure 5. Photoperiodic control of hypocotyl growth by PIF5 and COP1-SPA complex.
(A) Hypocotyl length of seedlings of different genotypes, growing under short-day (SD) condition.
(B) Box plot showing measurement of hypocotyl length of seedlings shown in (A). Surface sterilized seeds were plated onto MS medium without sucrose and incubated in SD (8h light/16h dark) for 4-day. Statistical significance among different genotypes was determined using one way ANOVA and Tukey’s HSD tests, and is indicated by different letters. White Bar=10mm.
(C) Rate of growth of hypocotyls from seedlings of different genotypes, growing under SD conditions. Surface sterilized seeds were plated onto MS medium without sucrose and incubated in SD (8h light/16h dark) for 4 days. Starting from day 2, infrared images were acquired every 1 hr and images were analyzed using ImageJ tool. Mean and SD were calculated and plotted into graphs.
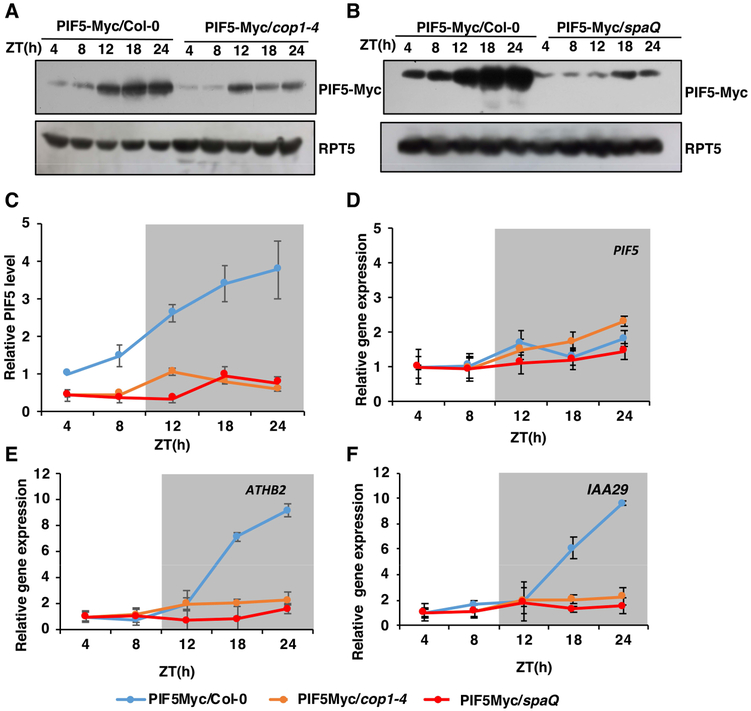
To correlate growth rate with PIF5 abundance, we measured PIF5-Myc protein and mRNA levels in the wild-type, cop1–4 and spaQ mutants under the same conditions (Figure 6A-D). At dawn, light treatment triggers the degradation of PIF5, so that the level of PIF5 dropped at a minimum level at ZT4–8. PIF5 started to re-accumulate at the beginning of the dark period, reaching the maximum level after 16–20h of dark (ZT18-ZT24) (Figure 6A-C). The maximum level of PIF5 protein at the end of the dark period correlates well with the growth rate in PIF5-Myc seedlings supporting the role for PIF5 in diurnal growth regulation (Figure 5C). However, in cop1–4 and spaQ mutants, the level of PIF5 protein was significantly reduced under both prolonged white light and dark period. Moreover, the PIF5 protein level failed to reach the peak at ZT18-ZT24 (Figure 6A-C). Therefore, PIF5 oscillation pattern is dramatically impaired in the cop1–4 and spaQ mutant backgrounds compared to wild-type. Contrary to protein level, qRT-PCR data show that there is little or no significant difference in PIF5 mRNA level in wild-type, cop1–4, and spaQ (Figure 6D). These data suggest that the defect in PIF5 and possibly other PIF levels in the cop1–4 and spaQ backgrounds might correlate with the growth defects in cop1–4 and spaQ under diurnal conditions.
Figure 6. COP1-SPA complex regulates diurnal oscillation of PIF5 protein level.
(A, B) Immunoblots showing the diurnal oscillation pattern of PIF5-Myc protein level in cop1–4 (A) and spaQ (B) mutants, compared to the wild-type seedlings. (C) Line graph showing diurnal oscillation of PIF5-Myc protein abundance in Col-0, cop1–4 and spaQ background. Seedlings were grown under SD conditions (8h light/ 16h dark) for 3 days and samples were collected on 4th day at the specific time ZT (h). Total protein was isolated and separated on 6.5% gel. Proteins were transferred to PVDF membrane and were sequentially blotted with anti-Myc and anti-RPT5 antibodies. For line graph, band intensities of PIF5-Myc and RPT5 from three replicates were measured using ImageJ tool and PIF5-Myc/RPT5 ratio was calculated. PIF5-Myc level in Col-0 at ZT4 was set to 1 and corresponding values were determined.
(D-F) Relative expression level of PIF5 (D), ATHB2 (E) and IAA29 (F) in 4-day-old SD-grown seedlings. Seedlings were grown under SD conditions (8h light/ 16h dark) for 3 days and samples were collected on 4th day at the specific time ZT (h). Total RNA was isolated and cDNA was prepared from it. qRT-PCR was performed using gene specific primers and PP2A was used as an internal control.
PIF5 has been shown to regulate growth-related ATHB2 and IAA29 gene expression during diurnal growth (Kunihiro, et al. 2011). To examine if the expression of these genes correlate with the growth defects in cop1–4 and spaQ mutants, we performed qRT-PCR for these genes in the wild-type, cop1–4 and spaQ backgrounds grown under SD conditions. Surprisingly, the expression level of these two genes is significantly impaired during ZT20–24 in PIF5-Myc/cop1–4 and PIF5Myc/spaQ mutants compared to PIF5-Myc seedlings (Figure 6E-F). Taken together, these data illustrate that COP1-SPA complex is involved in regulating diurnal growth pattern via controlling PIF5 level.
DISCUSSION
CUL4COP1-SPA complex promotes the light–induced degradation of PIF5
The genetic, biochemical and photobiological data presented here strongly suggest that CUL4COP1-SPA complex functions as an E3 Ubiquitin ligase for the light-induced degradation of PIF5. First, the light-induced degradation of PIF5 is reduced in the cul4cs, cop1–4 and spaQ mutants compared to wild-type. Second, both COP1 and SPA1 interact with PIF5 in a light stimulated manner. Third, the light-induced ubiquitination of PIF5 is defective in cop1–4 and spaQ backgrounds. Fourth, overexpression of PIF5 suppresses the photomorphogenic phenotypes of cop1–4 and spaQ mutants. Taken together, these data suggest that COP1 and SPA proteins act as substrate recognition factors to recruit PIF5 in the CUL4COP1-SPA E3 ubiquitin ligase for the light-induced ubiquitination and subsequent degradation.
Different E3 ligase for the degradation of different PIFs
Three main classes of CULLIN (CUL) RING UBIQUITIN LIGASEs (CRLs) have been shown to be involved in the light-induced degradation of PIFs (Pham, et al. 2018). CUL1EBF1/2 and CUL3LRB have been shown to regulate PIF3 abundance (Ni, et al. 2014), while CUL3BOP1/2 promotes the degradation of PIF4 (Zhang, et al. 2017). CUL1CTG10 and CUL4COP1-SPA complexes induce the degradation of PIF1 in response to light to promote seed germination (Majee, et al. 2018; Zhu, et al. 2015). Our study provides evidence that CUL4COP1-SPA Ubiquitin ligase is also involved in the ubiquitination and degradation of PIF5. The light-induced degradation of PIF5 does not appear to be defective in the cul1–6 and cul3ab mutants under these conditions. This does not preclude their involvement in regulating PIF5 under other conditions. In fact, PIF5 is still degraded in the cop1–4 and spaQ mutants under prolonged light conditions, suggesting that other E3 ubiquitin ligases might also be involved at those conditions.
A trend appears when we compare the rate of light-induced degradation of different PIFs and the involvement of different E3 ubiquitin ligases. Among all PIFs, the rate of degradation of PIF1 and PIF5 is the fastest with a half-life of <5 min under red light (Shen, et al. 2008; Shen, et al. 2007). However, the light-induced degradation of PIF3 and PIF4 is relatively slower than that of PIF1 and PIF5 (Al-Sady et al. 2006; Lorrain, et al. 2008). Thus, CUL4COP1-SPA complex might be involved in early and rapid light-induced degradation of PIF1 and PIF5, whereas other E3 Ubiquitin ligases are involved in the degradation of other PIFs, either at a later time and/or under higher irradiation conditions. The facts that both PIF1 and PIF5 bind to COP1 and SPA1 in the dark, and their physical interaction is enhanced by light, suggest that the CUL4COP1-SPA E3 Ubiquitin ligase is poised to initiate the ubiquitination process (Xu, et al. 2014; Zhu, et al. 2015). This is also the case for PIF3-EBF1/2 interaction; however, the SCF formation requires the light-induced phosphorylation, suggesting that SCFEBF1/2 is not functional in the dark (Dong, et al. 2017). These differences might partly explain the faster rate of degradation of PIF1 and PIF5 compared to the other PIFs under these conditions. Regardless of these details, the division of labor for PIF degradation among different E3 Ubiquitin ligases ensures temporal distribution of ubiquitin ligase machineries to optimize the inhibition of skotomorphogenesis and promotion of photomorphogenesis.
PIF5 is not only degraded under light but also in darkness
PIF5 level is much lower in the cop1–4 and spaQ mutants compared to wild-type in the dark, suggesting that COP1-SPA might play a role in stabilizing PIF5 in the dark. Other PIFs including PIF1 and PIF3 are also unstable in the cop1–4 and spa123 backgrounds in the dark (Bauer et al. 2004; Leivar, et al. 2008; Ni, et al. 2014). A recent study showed that COP1-SPA complex stabilizes PIF3 by inhibiting the phosphorylation of PIF3 by BIN2 (Ling et al. 2017). We have also shown that the increased abundance of HFR1 in the cop1–4 mutant induces degradation of PIF1 and PIF5 in the dark in a heterodimerization-dependent manner (Xu, et al. 2017). Collectively, these data suggest that PIF5 is degraded either directly or indirectly in cop1–4 and spaQ background in the dark through the 26S proteasome pathway.
COP1-SPA complex regulates hypocotyl growth under diurnal conditions
PIF5 along with PIF4 functions as an integrator of light and circadian clock, where circadian clock regulates the expression of PIF5 and PIF4, while light signal regulates the PIF5 and PIF4 protein abundance (Nozue, et al. 2007). Circadian clock regulates the diurnal plant growth including hypocotyl elongation, which is more pronounced under short-day (SD) conditions. During early night, the expression of PIF5 and PIF4 is inhibited by the ELF3-ELF4-LUX evening complex (Nusinow, et al. 2011). However, closer to dawn, the clock-mediated repression of PIF5 and PIF4 expression is suppressed. The increased expression of PIF5 and PIF4 induces hypocotyl growth during the end of the night. In consistent with these data, diurnal growth defect in pif4pif5 mutant has been well documented (Ezer, et al. 2017; Nozue, et al. 2007; Nusinow, et al. 2011). PIF3 is also involved in diurnal growth control; however, the mechanism is different. In this case, TOC1 interacts with PIF3 during early evening and inhibits PIF3 function. At dawn, TOC1 expression is reduced, which relieves this inhibition resulting in the promotion of hypocotyl growth by PIF3 (Soy, et al. 2016). In fact, a wave of PSEUDO-RESPONSE REGULATORS (PRR9/7/5) also inhibits PIF activity to repress growth during the day time (Martín et al. 2018). Although, PIFs are at the center stage for diurnal regulation of growth, a growth defect for cop1 and spaQ mutants under diurnal conditions have not been shown yet. Our data show that cop1 and spaQ mutants are not only defective under continuous dark and light conditions, but also under diurnal conditions. Strikingly, this defect is also mediated through regulating PIF5 level post-translationally under these conditions. As previously shown, PIF5 level increases post-translationally toward the end of the dark period under diurnal conditions. However, this increase is defective in the cop1–4 and spaQ mutants, suggesting COP1-SPA complex stabilizes PIF5 under these conditions. Moreover, overexpression of PIF5 in the cop1–4 and spaQ backgrounds partially rescues the diurnal growth defect of cop1–4 and spaQ mutants compared to wild-type. Overall, these data suggest that COP1-SPA complex is involved in regulating diurnal growth pattern through dynamic regulation of PIF5 and possibly other PIF levels.
MATERIALS AND METHODS
Plant growth conditions
Seeds were surface sterilized with 10% commercial bleach and 0.3% SDS for 5 min, washed 5 times with water and then plated on Murashige–Skoog (MS) growth medium containing 0.9% agar without sucrose. Plates were kept in the dark for 3 days at 4°C to stratify. Plates were then exposed to white light for 3hrs at room temperature before being placed in the dark or red light for 4 days. Specific light conditions for each of the assays are described in figure legends.
For high R/FR and low R/FR conditions, seedlings were grown under short-day (8hrs of high R/FR light/16hrs dark) for 5 days. At dawn on day 6th, seedlings were either kept in high R/FR condition (R/FR =4.51) or transferred to low R/FR condition. Low R/FR condition was provided by supplementary FR light (λ=735 nm) (R/FR = 0.1) (Lorrain, et al. 2008).
Measurement of hypocotyl lengths and anthocyanin content
For the measurement of hypocotyl length and cotyledon opening angle under dark and red light (2μmolm−2s−1), images of 90 seedlings (30 seedlings for each experiment with 3 independent biological replicates) were taken and then measured using the publicly available ImageJ software (http://rsb.info.nih.gov/ij/). For the measurement of anthocyanin content, 50 seeds were plated on MS medium containing 2% sucrose and kept in the dark for 3 days at 4°C. Seeds were exposed to 3hrs of white light to induce germination and then kept in the dark for 2 days. One set of seedlings was kept in the dark and another was transferred to continuous white light and grown for another 2 days. Anthocyanin content was measured following the protocol described previously (Neff and Chory 1998; Schmidt and Mohr 1981). Anthocyanin content was calculated by [Abs530 - (0.25 x Abs657)]. Data represents the anthocyanin content per gram fresh weight.
Construction of vectors and generation of transgenic plants
Preparation of PIF5-Myc transgenic line (35S:PIF5-Myc/Col-0) was described previously (Sakuraba, et al. 2014). PIF5-Myc transgenic line was crossed with cop1–4 and spaQ mutants. PIF5-Myc/cop1–4 and PIF5-Myc/spaQ homozygous lines were selected from F3 population. Primer used for cop1–4 mutant sequencing and spaQ genotyping are listed in Table S1.
To prepare double transgenic line PIF5-HA/TAP-SPA1, full length PIF5 CDS without a stop codon was amplified from a cDNA library using primers listed in Table S1, and directionally cloned into pENTR vector (Invitrogen Inc., Carlsbad, CA). Coding sequence was subcloned into pGWB14 binary vector (Karimi et al. 2005) and then transformed into Agrobacterium tumefaciens strain GV3101. This construct was then transformed into p35S:TAP-SPA1 transgenic plants using Agrobacterium-mediated transformation as previously described (Clough and Bent 1998) and double homozygous lines were selected in T3 generation.
Preparation of constructs for LCI assay: pCAMBIA split luciferase vectors were kindly provided by Dr. Jian-Min Zhou (Chen et al. 2008). p35S: nLUC vector was digested with EcoRI/HindIII and ligated into pZP121 vector to create pZP121: nLUC vector. For COP1-PIF5 interaction, full length COP1 CDS was cloned into KpnI/SalI sites of the PZP121-nLUC and full length PIF5/PIF5-G37A CDS was cloned into KpnI/SalI sites of pCAMBIA1300-cLUC vector.
For PIF5-SPA1 interaction, SPA1 coding sequence was cloned into EcoRI/ XhoI sites of pCAMBIA1300-cLUC. PIF5 and PIF5-G37A coding sequencing were cloned into KpnI/SalI sites of the pCAMBIA1300-nLUC vectors. The sequences of pCAMBIA1300-nLUC and pCAMBIA1300-cLUC vectors are available on TAIR website. All the constructs were confirmed by sequencing. The PIF5-G37A mutation was introduced using a site-directed mutagenesis kit (Stratagene, La Jolla, CA). Primers used for vector constructions and also site-directed mutagenesis are listed in Table S1.
Luciferase complementation imaging (LCI)
Luciferase complementation imaging (LCI) for testing the interaction of PIF5 with COP1 and SPA1 in N. benthamiana was performed as previously described with few modifications (Chen, et al. 2008). A. tumefaciens strain GV3101 was used for the injection of different constructs into N. benthamiana leaves. 1mM Luciferin was sprayed into the N. benthamiana leaves and kept in the dark for 5 min. Low-light cooled CCD camera (NightOWL IILB 983 NC 100, BERTHOLD) was used to capture luciferase luminesence. An exposure time of 2 min with 2×2 binning was used. Relative luciferase was equivalent to luminescence intensity / 0.2mm2 leaf area.
Yeast two-hybrid and quantitative β-galatosidase assays
Full-length PIF5 and PIF5-G37A coding sequences were amplified by PCR using the primers listed in Table S1. PIF5 and PIF5-G37A CDS were then cloned into EcoRI/ SalI sites of the LexA vector (pEG202) containing the DNA binding domain. COP1 and SPA1 were cloned into EcoRI/XhoI sites of the activation domain vector (pB42AD). These plasmids were transformed into yeast strain EGY48 and selected on dropout medium without Uracine, Leucine and Tryptophan. The quantitative β-galactosidase assay was performed according to the manufacturer’s instructions (Matchmaker Two-Hybrid System, Clonetech Laboratories Inc.,).
Protein extraction and immunoblotting
To analyze PIF5 abundance in wild-type and different mutant seedlings, total protein was extracted in denaturing buffer as described earlier (Zhu, et al. 2015). Total protein was separated on 8% SDS-PAGE gel and transferred to PVDF membrane. Membranes were blotted with anti-Myc (dilution 1/1000, Catalog number OP10–200UG, EMD Millipore, Billerica, MA) or anti-PIF5 (AS122112, Agrisera, Vannas, Sweden) or anti-RPT5 (dilution 1/3000, Catalog number: BML-PW8245–0100, Enzo Life Sciences, Farmingdale, NY) to detect PIF5-Myc, endogenous PIF5 and RPT5, respectively. In order to provide the true-dark condition, seeds were treated with far-red light for 5 min (6.5 μmolm−2s−1) after 21hrs in the dark and put them back to the dark for an additional 3 days.
For ubiquitination assays 4-day-old dark-grown seedlings were either kept in the dark or exposed to the constant red (Rc; 2 μmolm−2s−1) for 15 and 30 min. One batch of samples from each genotype tested were pretreated with 40 μM Bortezomib (B-1408, LC Laboratories, Woburn, MA) for 4 hrs before being exposed to 30 min Rc. Total protein from 50 seedlings were extracted in 50μl of urea extraction buffer (48% urea (w/v), 0.1M phosphate buffer pH 6.8, 10mM Tris-Cl pH 6.8, 1 mM PMSF and 1× protease inhibitor cocktail). Samples were centrifuged at 16,000g for 10 min and the supernatant was collected. The supernatant was boiled for 10 min at 65°C and were resolved on 6.5 % SDS-PAGE gels. Proteins were transferred to PVDF membranes and blotted with anti-Myc and anti-RPT5.
In vivo Co-immunoprecipitation assays
For detecting the ubiquitination pattern of PIF5-Myc in vivo, four-day-old dark-grown seedlings were kept in the dark or treated with 2 μmolm−2 s−1 constant red light (Rc) for 30 min. Total protein was extracted in native buffer as described previously (Zhu, et al. 2015) and PIF5-Myc was immunoprecipitated using anti-Myc antibody (1μg, 71D10, Cell Signaling Technologies, Danvers, MA). Samples were resolved on 6.5% SDS-PAGE, transferred to PVDF membrane and blotted with anti-Myc (1/1000 dilution, EMD Milliore, Billerica, MA) and anti-Ubi antibodies (1/1000 dilution, sc-8017, Santa Cruz Biotechnology, Santa Cruz, CA).
To perform in vivo Co-IP assays 5-day-old dark-grown seedlings were pretreated with 40μM bortezomib for 4hrs and then treated with either dark or red light (3000 μmolm−2). Total protein was extracted in native buffer and immunoprecipitation was performed as described earlier (Zhu, et al. 2015). For COP1-PIF5 interaction assays, total protein was isolated from PIF5-Myc seedlings. PIF5-Myc was immunoprecipitated using anti-Myc (EMD Milliore, Massachusetts) antibody and interacting partner COP1 was detected using anti-COP1 antibody. For SPA1-PIF5 interaction, total protein was isolated from TAP-SPA1/PIF5-HA double transgenic lines. PIF5-HA was immunoprecipitated using anti-HA (1μg, catalog number ab9110, Abcam, Cambridge, MA) and interacting partner TAP-SPA1 was detected using anti-Myc antibodies (EMD Milliore, Billerica, MA).
RNA isolation and Quantitative RT-PCR assay
Total RNA was extracted from 4 day-old dark-grown seedlings or dark-grown seedlings exposed to 30 min of Rc (2 μmolm−2 s−1) using the Sigma spectrum total RNA kit (Sigma-Aldrich, St. Louis, MO). One μg of total RNA was treated with DNase 1 (NEB, Ipswich, MA) and reversed transcribed using M-MLV reverse transcriptase (ThermoFisher Scientific, Waltham, MA). qRT-PCR was performed using Power SYBR® green (Applied Biosystems, Foster City, CA) and gene specific primers, as listed in Table S1. PP2A was used as the control for normalization the expression of other genes. The cycle threshold (Ct) values were used for calculation of the levels of expression of different genes relative to PP2A.
Hypocotyl and growth rate measurement under SD condition
The infrared images were taken using the Marlin camera (Allied Vision Technologies, Exton, PA) with the lenses fitted with IR long-pass cut filters (NT54–755, Edmund Optics). Seedling samples were back-lit with an IR LED back-light emitting at 880 nm (Advanced Illumination). Seeds were grown under SD and the pictures were acquired at the beginning of day 2. Image acquisition was archived using AVT SmartView software (Allied Vision Technologies, Exton, PA) every 1h for 3 days. Images were analyzed using ImageJ tool. The hypocotyl growth rates were measured using 4 seedlings for each genotype at each time point, and three replicates have been done. The mean and SD shown in the graph indicates the mean and SD for one replicate.
Supplementary Material
Figure S1: Red light induced degradation of PIF5 is slower in cul4cs and but not in cul1–6 and cul3ab mutants.
Figure S2: Prolonged red light induces the degradation of PIF5 in both cop1–4 and spaQ mutants with slower rate compared to wild-type
Figure S3: PIF5 level is stable under far-red (FR) light and shade mimicking (low R/FR) conditions.
Figure S4: COP1-SPA post-translationally regulates PIF5 abundance both in the dark and red light
Figure S5: Fluctuation of the PIF5-Myc level during the prolonged red light treatment
Figure S6: Overexpression of PIF5-Myc partially rescue hypocotyl elongation in cop1–4 and spaQ.
Primer sequences used in experiments described in manuscript.
Significance Statement.
This study illustrates two key findings: 1) CUL4COP1-SPA acts as an E3 Ubiquitin ligase for the light-induced degradation of PIF5, and 2) COP1-SPA complex regulates diurnal growth pattern via dynamic regulation of PIF5. These findings fill an important gap in our knowledge on how PIFs are regulated by light, and how COP1-SPA complex plays crucial roles not only under continuous dark and light conditions, but also under diurnal conditions to regulate plant growth and development.
ACKNOWLEDGEMENTS
We thank members of the Huq laboratory for critical reading of the manuscript, Drs. Giltsu Choi for sharing myc-PIF5 seeds, Xing Wang Deng for cop1–4 seeds, Ute Hoecker for spaQ seeds, Woe Yeon Kim for COP1 antibody and Jian-Min Zhou for pCAMBIA split luciferase vectors. This work was supported by grants from National Institute of Health (NIH) (1R01 GM-114297), National Science Foundation (MCB-1543813) and United States Israel Bilateral Science Foundation (2015316) to E. H.
Footnotes
The Authors declare no conflicts of interest.
REFERENCES
- Al-Sady B, Ni W, Kircher S, Schafer E and Quail PH (2006) Photoactivated Phytochrome Induces Rapid PIF3 Phosphorylation Prior to Proteasome-Mediated Degradation. Mol Cell, 23, 439–446. [DOI] [PubMed] [Google Scholar]
- Bae G and Choi G (2008) Decoding of light signals by plant phytochromes and their interacting proteins Annu Rev Plant Biol, 59, 281–311. [DOI] [PubMed] [Google Scholar]
- Bauer D, Viczian A, Kircher S, Nobis T, Nitschke R, Kunkel T, Panigrahi KC, Adam E, Fejes E, Schafer E and Nagy F (2004) Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell, 16, 1433–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillon A, Shen H and Huq E (2007) Phytochrome Interacting Factors: central players in phytochrome-mediated light signaling networks. Trends Plant Sci, 12, 514–521. [DOI] [PubMed] [Google Scholar]
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X and Zhou JM (2008) Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol, 146, 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HD, Huang X, Gusmaroli G, Terzaghi W, Lau OS, Yanagawa Y, Zhang Y, Li JG, Lee JH, Zhu DM and Deng XW (2010) Arabidopsis CULLIN4-Damaged DNA Binding Protein 1 Interacts with CONSTITUTIVELY PHOTOMORPHOGENIC1-SUPPRESSOR OF PHYA Complexes to Regulate Photomorphogenesis and Flowering Time. Plant Cell, 22, 108–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ and Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J, 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Deng X-W, Matsui M, Wei N, Wagner D, Chu AM, Feldman KA and Quail PH (1992) COP1, an arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a Gβ homologous domain. Cell, 71, 791–801. [DOI] [PubMed] [Google Scholar]
- Dong J, Ni W, Yu R, Deng XW, Chen H and Wei N (2017) Light-Dependent Degradation of PIF3 by SCFEBF1/2 Promotes a Photomorphogenic Response in Arabidopsis. Current Biology, 27, 2420–2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezer D, Jung J-H, Lan H, Biswas S, Gregoire L, Box MS, Charoensawan V, Cortijo S, Lai X, Stöckle D, Zubieta C, Jaeger KE and Wigge PA (2017) The evening complex coordinates environmental and endogenous signals in Arabidopsis. Nature Plants, 3, 17087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Lee SH, Patel D, Kumar SV, Spartz AK, Gu C, Ye S, Yu P, Breen G, Cohen JD, Wigge PA and Gray WM (2011) PHYTOCHROME-INTERACTING FACTOR 4 (PIF4) regulates auxin biosynthesis at high temperature. Proceedings of the National Academy of Sciences, 108, 20231–20235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvão VC and Fankhauser C (2015) Sensing the light environment in plants: photoreceptors and early signaling steps. Current Opinion in Neurobiology, 34, 46–53. [DOI] [PubMed] [Google Scholar]
- Gangappa SN, Berriri S and Kumar SV (2017) PIF4 Coordinates Thermosensory Growth and Immunity in Arabidopsis. Current Biology, 27, 243–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gommers CMM and Monte E (2018) Seedling establishment: a dimmer switch-regulated process between dark and light signaling. Plant Physiol, 176, 1061–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersch M, Lorrain S, de Wit M, Trevisan M, Ljung K, Bergmann S and Fankhauser C (2014) Light intensity modulates the regulatory network of the shade avoidance response in Arabidopsis. Proceedings of the National Academy of Sciences, 111, 6515–6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker U (2017) The activities of the E3 ubiquitin ligase COP1/SPA, a key repressor in light signaling. Current Opinion in Plant Biology, 37, 63–69. [DOI] [PubMed] [Google Scholar]
- Huq E, Al-Sady B, Hudson M, Kim C, Apel K and Quail PH (2004) Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science, 305, 1937–1941. [DOI] [PubMed] [Google Scholar]
- Huq E and Quail PH (2002) PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. Embo J, 21, 2441–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E and Quail PH (2005) Phytochrome signaling In Handbook of Photosensory Receptors (Briggs WR and Spudich JL eds). Weinheim, Germany: Wiley-VCH, pp. 151–170. [Google Scholar]
- Jang I-C, Henriques R, Seo HS, Nagatani A and Chua N-H (2010) Arabidopsis PHYTOCHROME INTERACTING FACTOR Proteins Promote Phytochrome B Polyubiquitination by COP1 E3 Ligase in the Nucleus. Plant Cell, 22, 2370–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Meyer BD and Hilson P (2005) Modular cloning in plant cells. Trends Plant Sci, 10, 103–105. [DOI] [PubMed] [Google Scholar]
- Khanna R, Huq E, Kikis EA, Al-Sady B, Lanzatella C and Quail PH (2004) A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors. Plant Cell, 16, 3033–3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R, Shen Y, Marion CM, Tsuchisaka A, Theologis A, Schäfer E and Quail PH (2007) The Basic Helix-Loop-Helix Transcription Factor PIF5 Acts on Ethylene Biosynthesis and Phytochrome Signaling by Distinct Mechanisms. Plant Cell, 19, 3915–3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Yi H, Choi G, Shin B, Song PS and Choi G (2003) Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction. Plant Cell, 15, 2399–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose C, Viczián A, Kircher S, Schäfer E and Nagy F (2015) Molecular mechanisms for mediating light‐dependent nucleo/cytoplasmic partitioning of phytochrome photoreceptors. New Phytologist, 206, 965–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar SV, Lucyshyn D, Jaeger KE, Alos E, Alvey E, Harberd NP and Wigge PA (2012) Transcription factor PIF4 controls the thermosensory activation of flowering. Nature, 484, 242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunihiro A, Yamashino T, Nakamichi N, Niwa Y, Nakanishi H and Mizuno T (2011) Phytochrome-interacting factor 4 and 5 (PIF4 and PIF5) activate the homeobox ATHB2 and auxin-inducible IAA29 genes in the coincidence mechanism underlying photoperiodic control of plant growth of Arabidopsis thaliana. Plant Cell Physiol, 52, 1315–1329. [DOI] [PubMed] [Google Scholar]
- Lau OS and Deng XW (2012) The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci, 17, 584–593. [DOI] [PubMed] [Google Scholar]
- Leivar P and Monte E (2014) PIFs: Systems Integrators in Plant Development Plant Cell, 26, 56–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E, Oka Y, Liu T, Carle C, Castillon A, Huq E and Quail PH (2008) Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol, 18, 1815–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P and Quail PH (2011) PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci, 16, 19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li FW, Melkonian M, Rothfels CJ, Villarreal JC, Stevenson DW, Graham SW, Wong GK, Pryer KM and Mathews S (2015) Phytochrome diversity in green plants and the origin of canonical plant phytochromes. Nat Commun, 6, 7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling J-J, Li J, Zhu D and Deng XW (2017) Noncanonical role of Arabidopsis COP1/SPA complex in repressing BIN2-mediated PIF3 phosphorylation and degradation in darkness. Proceedings of the National Academy of Sciences, 114, 3539–3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Chen C-Y, Wang K-C, Luo M, Tai R, Yuan L, Zhao M, Yang S, Tian G, Cui Y, Hsieh H-L and Wu K (2013) PHYTOCHROME INTERACTING FACTOR3 Associates with the Histone Deacetylase HDA15 in Repression of Chlorophyll Biosynthesis and Photosynthesis in Etiolated Arabidopsis Seedlings. Plant Cell, 25, 1258–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Zhang Y, Wang J, Li P, Zhao C, Chen Y and Bi Y (2015) Phytochrome-interacting factors PIF4 and PIF5 negatively regulate anthocyanin biosynthesis under red light in Arabidopsis seedlings. Plant Sci, 238, 64–72. [DOI] [PubMed] [Google Scholar]
- Lorrain S, Allen T, Duek PD, Whitelam GC and Fankhauser C (2008) Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J, 53, 312–323. [DOI] [PubMed] [Google Scholar]
- Lorrain S, Trevisan M, Pradervand S and Fankhauser C (2009) Phytochrome interacting factors 4 and 5 redundantly limit seedling de-etiolation in continuous far-red light. Plant Journal, 60, 449–461. [DOI] [PubMed] [Google Scholar]
- Lu X-D, Zhou C-M, Xu P-B, Luo Q, Lian H-L and Yang H-Q (2015) Red-Light-Dependent Interaction of phyB with SPA1 Promotes COP1–SPA1 Dissociation and Photomorphogenic Development in Arabidopsis. Molecular Plant, 8, 467–478. [DOI] [PubMed] [Google Scholar]
- Majee M, Kumar S, Kathare PK, Wu S, Gingerich D, Nayak NR, Salaita L, Dinkins R, Martin K, Goodin M, Dirk LMA, Lloyd TD, Zhu L, Chappell J, Hunt AG, Vierstra R, Huq E and Downie AB (2018) KELCH F-BOX protein positively influences Arabidopsis seed germination by targeting PHYTOCHROME-INTERACTING FACTOR1. Proceedings of the National Academy of Sciences, 115, E4120–E4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín G, Rovira A, Veciana N, Soy J, Toledo-Ortiz G, Gommers CMM, Boix M, Henriques R, Minguet EG, Alabadí D, Halliday KJ, Leivar P and Monte E (2018) Circadian Waves of Transcriptional Repression Shape PIF-Regulated Photoperiod-Responsive Growth in Arabidopsis. Current Biology, 28, 311–318.e315. [DOI] [PubMed] [Google Scholar]
- Moon J, Zhang W, Gray WM, Zhao Y, Huq E and Estelle M (2007) A new CUL1 mutant has altered responses to hormones and light in Arabidopsis thaliana. Plant Physiol, 143, 684–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff MM and Chory J (1998) Genetic Interactions between Phytochrome A, Phytochrome B, and Cryptochrome 1 during Arabidopsis Development. Plant Physiology, 118, 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni W, Xu S-L, Tepperman JM, Stanley DJ, Maltby DA, Gross JD, Burlingame AL, Wang Z-Y and Quail PH (2014) A mutually assured destruction mechanism attenuates light signaling in Arabidopsis. Science, 344, 1160–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozue K, Covington MF, Duek PD, Lorrain AA, Fankhauser C, Harmer SL and Maloof JN (2007) Rhythmic growth explained by coincidence between internal and external cues. Nature, 448, 358–361. [DOI] [PubMed] [Google Scholar]
- Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, Farre EM and Kay SA (2011) The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature, 475, 398–U161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Hanada A, Yamauchi Y, Kuwahara A, Kamiya Y and Yamaguchi S (2003) Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell, 15, 1591–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Kim J, Park E, Kim JI, Kang C and Choi G (2004) PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell, 16, 3045–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik I, Kathare PK, Kim J-I and Huq E (2017) Expanding Roles of PIFs in Signal Integration from Multiple Processes. Mol Plant, 10, 1035–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E, Kim Y and Choi G (2018) Phytochrome B Requires PIF Degradation and Sequestration to Induce Light Responses across a Wide Range of Light Conditions. The Plant Cell, 30, 1277–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E, Park J, Kim J, Nagatani A, Lagarias JC and Choi G (2012) Phytochrome B inhibits binding of phytochrome-interacting factors to their target promoters. Plant J, 72, 537–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham VN, Kathare PK and Huq E (2018) Phytochromes and Phytochrome Interacting Factors. Plant Physiology, 176, 1025–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail PH (2007) Phytochrome-regulated Gene Expression. J. Integr. Plant Biol, 49, 11–20. [Google Scholar]
- Sakuraba Y, Jeong J, Kang M-Y, Kim J, Paek N-C and Choi G (2014) Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nature communications, 5, 4636. [DOI] [PubMed] [Google Scholar]
- Schmidt R and Mohr H (1981) Time-dependent changes in the responsiveness to light of phytochrome-mediated anthocyanin synthesis. Plant, Cell & Environment, 4, 433–437. [Google Scholar]
- Sheerin DJ, Menon C, zur Oven-Krockhaus S, Enderle B, Zhu L, Johnen P, Schleifenbaum F, Stierhof Y-D, Huq E and Hiltbrunner A (2015) Light-activated phytochrome A and B interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex. Plant Cell, 27, 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Ling Z, Castillon A, Majee M, Downie B and Huq E (2008) Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME INTERACTING FACTOR 1 depends upon its direct physical interactions with photoactivated phytochromes. Plant Cell, 20, 1586–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Khanna R, Carle CM and Quail PH (2007) Phytochrome induces rapid PIF5 phosphorylation and degradation in response to red-light activation. Plant Physiol, 145, 1043–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Kim K, Kang H, Zulfugarov IS, Bae G, Lee CH, Lee D and Choi G (2009) Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc. Nat. Acad. Sci. U S A, 106, 7660–7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Park E and Choi G (2007) PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis. Plant J, 49, 981–994. [DOI] [PubMed] [Google Scholar]
- Song Y, Yang C, Gao S, Zhang W, Li L and Kuai B (2014) Age-Triggered and Dark-Induced Leaf Senescence Require the bHLH Transcription Factors PIF3, 4, and 5. Molecular Plant, 7, 1776–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soy J, Leivar P, Gonzalez-Schain N, Martin G, Diaz C, Sentandreu M, Al-Sady B, Quail PH and Monte E (2016) Molecular convergence of clock and photosensory pathways through PIF3-TOC1 interaction and co-occupancy of target promoters. Proc Natl Acad Sci U S A, 113, 4870–4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson PG, Fankhauser C and Terry MJ (2009) PIF3 is a repressor of chloroplast development. Proc Natl Acad Sci U S A, 106, 7654–7659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian C, Kim BH, Lyssenko NN, Xu X, Johnson CH and von Arnim AG (2004) The Arabidopsis repressor of light signaling, COP1, is regulated by nuclear exclusion: mutational analysis by bioluminescence resonance energy transfer. Proc Natl Acad Sci USA, 101, 6798–6802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomann A, Brukhin V, Dieterle M, Gheyeselinck J, Vantard M, Grossniklaus U and Genschik P (2005) Arabidopsis CUL3A and CUL3B genes are essential for normal embryogenesis. Plant J, 43, 437–448. [DOI] [PubMed] [Google Scholar]
- Vierstra RD (2009) The ubiquitin–26S proteasome system at the nexus of plant biology. Nature Rev. Mol. Cell Biol, 10, 385–397. [DOI] [PubMed] [Google Scholar]
- Xu X, Kathare PK, Pham VN, Bu Q, Nguyen A and Huq E (2017) Reciprocal proteasome-mediated degradation of PIFs and HFR1 underlies photomorphogenic development in Arabidopsis. Development, 144, 1831–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Paik I, Zhu L, Bu Q, Huang X, Deng XW and Huq E (2014) PHYTOCHROME INTERACTING FACTOR1 Enhances the E3 Ligase Activity of CONSTITUTIVE PHOTOMORPHOGENIC1 to Synergistically Repress Photomorphogenesis in Arabidopsis. Plant Cell, 26, 1992–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Paik I, Zhu L and Huq E (2015) Illuminating Progress in Phytochrome-Mediated Light Signaling Pathways. Trends in plant science, 20, 641–650. [DOI] [PubMed] [Google Scholar]
- Zhang B, Holmlund M, Lorrain S, Norberg M, Bakó L, Fankhauser C and Nilsson O (2017) BLADE-ON-PETIOLE proteins act in an E3 ubiquitin ligase complex to regulate PHYTOCHROME INTERACTING FACTOR 4 abundance. eLife, 6, e26759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Feng SH, Chen FF, Chen HD, Wang J, McCall C, Xiong Y and Deng XW (2008) Arabidopsis DDB1-CUL4 ASSOCIATED FACTOR1 forms a nuclear E3 ubiquitin ligase with DDB1 and CUL4 that is involved in multiple plant developmental processes. Plant Cell, 20, 1437–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu Z, Chen Y, He JX and Bi Y (2015) PHYTOCHROME-INTERACTING FACTOR 5 (PIF5) positively regulates dark-induced senescence and chlorophyll degradation in Arabidopsis. Plant Sci, 237, 57–68. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Mayba O, Pfeiffer A, Shi H, Tepperman JM, Speed TP and Quail PH (2013) A Quartet of PIF bHLH Factors Provides a Transcriptionally Centered Signaling Hub That Regulates Seedling Morphogenesis through Differential Expression-Patterning of Shared Target Genes in Arabidopsis. Plos Genetics, 9, e1003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Bu Q, Xu X, Paik I, Huang X, Hoecker U, Deng XW and Huq E (2015) CUL4 forms an E3 ligase with COP1 and SPA to promote light-induced degradation of PIF1. Nature commun, 6, 7245. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Red light induced degradation of PIF5 is slower in cul4cs and but not in cul1–6 and cul3ab mutants.
Figure S2: Prolonged red light induces the degradation of PIF5 in both cop1–4 and spaQ mutants with slower rate compared to wild-type
Figure S3: PIF5 level is stable under far-red (FR) light and shade mimicking (low R/FR) conditions.
Figure S4: COP1-SPA post-translationally regulates PIF5 abundance both in the dark and red light
Figure S5: Fluctuation of the PIF5-Myc level during the prolonged red light treatment
Figure S6: Overexpression of PIF5-Myc partially rescue hypocotyl elongation in cop1–4 and spaQ.
Primer sequences used in experiments described in manuscript.