Abstract
The heterogeneous nuclear ribonucleoproteins (hnRNPs) are a diverse family of RNA binding proteins that function in most stages of RNA metabolism. The prototypical member, hnRNP A1, is composed of three major domains; tandem N-terminal RNA Recognition Motifs (RRMs) and a C-terminal mostly intrinsically disordered region. HnRNP A1 is broadly implicated in basic cellular RNA processing events such as splicing, stability, nuclear export and translation. Due to its ubiquity and abundance, hnRNP A1 is also frequently usurped to control viral gene expression. Deregulation of the RNA metabolism functions of hnRNP A1 in neuronal cells contributes to several neurodegenerative disorders. Because of these roles in human pathologies, the study of hnRNP A1 provides opportunities for the development of novel therapeutics, with disruption of its RNA binding capabilities being the most promising target. The functional diversity of hnRNP A1 is reflected in the complex nature by which it interacts with various RNA targets. Indeed, hnRNP A1 binds both structured and unstructured RNAs with binding affinities that span several magnitudes. Available structures of hnRNP A1-RNA complexes also suggest a degree of plasticity in molecular recognition. Given the reinvigoration in hnRNP A1, the goal of this review is to use the available structural biochemical developments as a framework to interpret its wide-range of RNA functions.
1. Introduction
The life and times of eukaryotic transcripts are determined by dynamic interactions with an ensemble of RNA binding proteins (RBPs). RBPs regulate essential steps in basic RNA metabolism including: 5′ capping, splicing, polyadenylation, stability, subcellular localization and translation [1].
Heterogeneous nuclear ribonucleoproteins (hnRNPs) comprise a diverse family of RBPs that are implicated in most steps in gene regulation [2]. The hnRNPs were first discovered in the isolation of a 40S core RNA-protein complex that contained hnRNP A/B and C [3]. This initial discovery would eventually lead to the identification of at least 20 major types of hnRNPs, named A through U, with molecular weights ranging from 34 kDa to 120 kDa [4]. Each member of the family facilitates the processing of nascent transcripts produced by RNA polymerase II [5].
The hnRNP proteins are composed of modular RNA binding domains (RBD) and intrinsically disordered regions (IDRs) [2]. Each family member differs in the number and orientation of these subunits [5, 6]. The RNA binding domains can be the canonical RNA Recognition Motifs (RRMs), quasi RNA Recognition Motifs (qRRMs) or the more rare KH domain. The IDRs participate in homo and heterotypic protein-protein interactions and also endow certain hnRNP proteins with phase separation properties [7]. Moreover, the modularity of hnRNPs contributes to the combinatorial nature by which these proteins associate with different RNA targets.
The prototypical member, hnRNP A1, is a multipurpose protein involved in normal and pathological RNA processing, including viral gene expression and neurodegenerative disorders [4, 8]. The biological functions of hnRNP A1 are expansive and include both co- and post-transcriptional regulation [2, 9]. Consistent with its multifunctional properties, hnRNP A1 interacts with diverse nucleic acid targets that comprise varying degrees of structural and sequence complexity. Accordingly, one prevailing question is how does hnRNP A1 functionally associate with RNAs that adopt diverse structures and that present multiple binding sites? The overall theme of this review aims to explore this question by using the available structural biochemical developments as a framework. The guiding premise is that hnRNP A1-RNA interactions are idiosyncratic, whereby different modes of molecular recognition contribute to diverse biological outcomes. Several excellent reviews have been written on the cellular functions of hnRNP proteins in general and one specifically on hnRNP A1 [2, 4-6]. Here, we will briefly discuss a subset of the RNA processing activities of hnRNP A1; however, our intention is to summarize the mechanistic information that we believe is necessary to comprehend its molecular function.
2. Structural organization of hnRNP A1
HnRNP A1 exists in the cell as two different isoforms, A1-B (372 aa, 38 kDa) and A1-A (320 aa, 34 kDa), which results from differential processing of the mRNA transcript [10-12]. The predominant isoform, A1-A, has a deletion from residues 253-303 [4]. The domain organization of each isoform consists of N-terminal tandem RRMs and a C-terminal IDR enriched in arginine and glycine residues (Figure 1A). The RRMs form the primary RNA binding surface, whereas the IDR mediates protein-protein interactions. The N-terminal region encompassing the RRMs (residues 1-196) is referred to as unwinding protein 1 (UP1), as it was originally discovered as a duplex-destabilizing protein [13-16]. The RRMs of UP1 are canonical, share a high degree of sequence similarity, and adopt nearly identical three-dimensional structures consisting of the β1α1β2β3α2β4 fold (Figure 1B). The RRM is the most extensively studied RNA binding domain, with ~350 structures in the protein data bank [1, 17]. Generally, RRMs display a wide degree of diversity in their mechanisms of interaction, which leads to further subdivisions (canonical and quasi) that are based on the type of secondary structural elements that form the RNA binding surface [17]. The canonical RRMs of UP1 contain RNP1 and RNP2 submotifs that are located on the central β sheet surface (β1 and β3, respectively) [1, 18]. The RNP submotifs have several conserved aromatic residues that participate in stacking interactions with single strand nucleobases [19, 20].
Figure 1. Structural features of hnRNP A1.
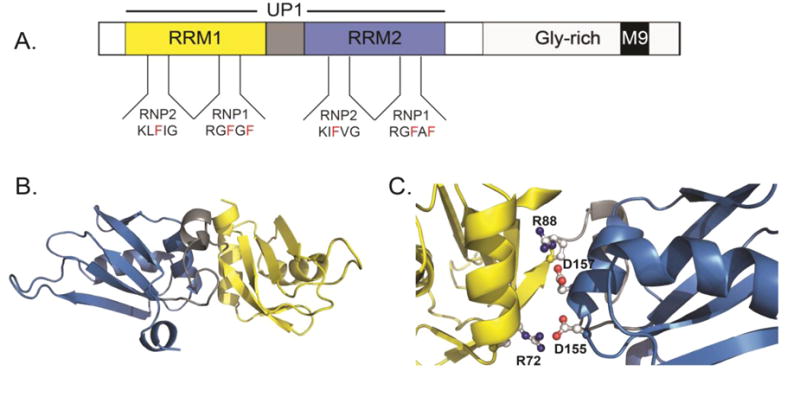
(A) The domain organization of full-length hnRNP A1 showing the N-terminal RNA binding domains (RRM1=yellow, Inter-RRM linker=gray, and RRM2=blue) and the C-terminal domain (light gray) with M9 nuclear localization signal depicted as a black box. The tandem RRMs of hnRNP A1 collectively make up the UP1 protein, residues 1-196. The RNP1 and RNP2 submotifs are also depicted for each RRM. (B) The solution NMR structure (2LYV) of UP1 color-coded as in part A. (C) A zoomed view of the alpha helical side of UP1 showing the conserved salt bridge interactions that stabilize the relative orientation of RRM1 and RRM2.
The C-terminal domain (CTD) of hnRNP A1 is intrinsically disordered and primarily mediates protein-protein interactions [21]. These cooperative protein-protein interactions have been shown to contribute to substrate binding in hnRNP A1 [22]. Although the CTD does not adopt a stable structure, it is glycine-rich and contains several repeats of RGG boxes, interspersed with aromatic residues [23, 24]. The RGG boxes are concentrated in the N-terminal half of the CTD, from residues 190-240 [24, 25]. The RGG box region also interacts with nucleic acid, albeit non-specifically [26, 27]. Post-translational modifications of residues in the CTD modulate the activity of hnRNP A1. Arginine methylation within the RGG box of hnRNP A1 stimulates its activity as an IRES trans acting factor (ITAF) and regulates its ability to associate with stress granules [28, 29]. Phosphorylation of several serine residues (S192, S197, S199, S223, S231) leads to an increase of ordered structure in the CTD [30], while phosphorylation at S199 attenuates its ITAF activity [31]. The CTD also contains a prion-like domain downstream of the RGG box and the combination promotes protein-protein interactions, which are necessary for hnRNP A1 to perform many of its functions [25].
A 38 amino acid M9 nuclear localization signal (NLS) located in the CTD is responsible for shuttling hnRNP A1 between the nucleus and the cytoplasm [32-36]. Nuclear localization of hnRNP A1 is mediated through receptors of the karyopherin-β family, Transportin 1 and Transportin 2, which interact with the nuclear pore complex Nup62. After entering the nucleus, RanGTP disrupts the hnRNP A1:Transportin complex [37]. The interaction of hnRNP A1 with Transportin 1 is regulated through phosphorylation of serine residues in the F peptide, which is the C-terminus of the protein directly downstream from the NLS [38]. O-GlcNAcylation further modulates the interaction with Transportin 1 [39].
In summary, the modular domain architecture along with the differential post-translational modifications (PTMs) contributes to the diversity of biological processes performed by hnRNP A1. In the following section, we will briefly summarize a subset of its RNA functions.
3. RNA Processing
Eukaryotic pre-mRNAs transcribed by RNA polymerase II (RNAP II) undergo multiple modifications that include the addition of a 5′ 7-methylguanosine cap, splicing and 3′ polyadenylation. After processing in the nucleus, mRNA is transported to the cytoplasm where it is utilized for translation [40]. Due to its ability to bind DNA and both nuclear and cytoplasmic RNA, hnRNP A1 fulfills a broad range of biological functions [2, 9]. This section will briefly explore a subset of the roles hnRNP A1 has in the life cycle of an RNA transcript.
3.1 Transcriptional Regulation
The multifunctional properties of hnRNP A1 allow it to play a role in the very beginning of the transcriptional life cycle, the regulation of transcription itself. HnRNP A1 acts as a transcriptional repressor when directly binding DNA promoters. In this process, hnRNP A1 binds specific target sequences on dsDNA before forming higher order protein-protein complexes to suppress transcription [41-44]. Some of the target sequences hnRNP A1 binds were found to be rich with AG dinucleotides, such as the adjacent binding sites found on the human thymidine kinase promoter [43]. The ability of hnRNP A1 to unwind G-quadruplexes also has significance for transcription regulation [45-48]. The formation of G-quadruplexes has been shown to arrest transcription. By unwinding G-quadruplex structures with its UP1 domain, hnRNP A1 acts as a transcriptional activator without the need for forming protein-protein interactions. In contrast to this mechanism, hnRNP A1 can also regulate transcription exclusively through protein-protein interactions, as it has been shown to do with IκBα [49]. Moreover, hnRNP A1 stimulates transcriptional elongation by remodeling the 7SK snRNA to promote the release of the positive transcription elongation factor b (P-TEFb) [50-53]. The 7Sk snRNA is a transcription repressor, which sequesters P-TEFb into an inactive complex through interactions with hexamethylene bisacetamide inducible protein (HEXIM). Binding of hnRNP A1 to 7SK snRNA promotes the release of P-TEFb from the complex, allowing it to stimulate transcriptional elongation by hyper-phosphorylating the C-terminal domain of RNAP II.
3.2 Splicing Control
Proteome diversity in multicellular eukaryotic organisms is largely determined by the extent of alternative splicing [54, 55]. The spliceosome catalyzes the removal of introns and ligation of exons; however, a large number of RBPs modulate the selection of alternative splice sites by binding cis sequences collectively known as splicing regulatory elements (SREs) [56, 57]. The dynamic interactions between RBPs and their cognate SREs across the cellular transcriptome establish a context dependent splicing code [56]. Arguably the major cellular function of hnRNP A1 is to influence the selection of alternative splice sites utilized by the spliceosome [57], although it also facilitates proofreading of constitutive splice sites by forming a stable ternary complex with U2AF and authentic 3′ acceptor sites [58].
The alternative splicing activity of hnRNP A1 was initially identified using model and natural pre-mRNAs, whereby the relative concentrations of hnRNP A1 was shown to modulate alternative 5′ splice site usage [59]. This initial discovery was followed by many other examples, confirming that hnRNP A1 influences alternative splicing in both mammalian and viral systems [60]. The number of alternatively spliced transcripts regulated by hnRNP A1 far exceeds the space allotted here to describe them; however, the Human Immunodeficiency Virus (HIV) has served as a paradigmatic system to understand the mechanisms by which hnRNP A1 regulates splicing [61-63]. Following integration of HIV into the host chromosome, RNAP II synthesizes an ~9 kB polycistronic viral transcript that undergoes several rounds of alternative splicing to generate singly and multiply spliced mRNA isoforms [64]. Each HIV splice site, with the exception of splice donor 1 (SD1), is suboptimal and therefore regulated by SRE-RBP interactions [62, 65, 66]. Human hnRNP A1 represses HIV splicing at several 3′ acceptor sites by binding exonic or intronic splicing silencers (ESS and ISS) [67-70]. The mechanism by which hnRNP A1 affects HIV splicing varies depending on the identity of the 3′ acceptor site and its associated SRE-RBP network [71].
One of the better-characterized HIV alternative splicing events involves processing of the tat mRNA [72]. Cellular levels of the HIV Tat protein are tightly regulated due to its cytotoxic effects [73]. Regulation of Tat expression is achieved in part by controlling the abundance of its multiply spliced mRNA, which involves splicing to downstream acceptor sites A3 and A7. HnRNP A1 regulates the usage of both acceptor sites A3 and A7 through interactions with ESS and ISS elements. At site A3, hnRNP A1 binds directly to the ESS2 element to occlude access of the splicing enhancer protein SRSF2 at its neighboring binding site [72, 74].
Splicing to acceptor site A7 is repressed by the combination of ISS and ESS3a/b elements, both of which bind hnRNP A1 [62, 65, 69, 75]. The prevailing mechanism of action at site A7 posits that hnRNP A1 binds first to the ESS3 element and then spreads cooperatively in a 3′–5′ direction to the ISS element [76-78]. The coating of the RNA region surrounding site A7 has multiple implications for splicing: first hnRNP A1 can effectively displace the splicing enhancer protein SRSF1; second it can destabilize the interaction of U2AF with the upstream polypyrimidine tract; and finally it can block access to the branch point site, thus preventing the formation of the spliceosome E complex [63, 64, 71, 79, 80]. Of note, the RNA regions surrounding sites A3 and A7 adopt conserved secondary structures wherein binding sites for hnRNP A1 map to single strand loops [68, 76, 77]. RNA structure offers a strategy to remove the degeneracy of short consensus sequence motifs (more on this below) recognized by RBPs; however, it’s not clear whether HIV exploits this strategy to specifically recruit hnRNP A1. To this point, hnRNP A1 was shown to effectively unwind RNA hairpins and displace a bound protein, suggesting RNA structure is dispensable [78].
Improper splicing regulation of genes by hnRNP A1 has been found as a cause of several diseases, such as spinal muscular atrophy (SMA) [81-85]. Patients suffering from SMA have an inactive SMN1 gene and depend entirely on the highly homologous SMN2 gene for survival. The SMN2 gene is only able to produce a low level of functional protein due to a single point mutation, C840T, which introduces a binding site for hnRNP A1. Binding at this site causes splicing inhibition and the skipping of exon 7, resulting in a non-functional protein.
The protein-protein interaction properties of hnRNP A1 are essential for its function as a splicing repressor, as shown by recent experiments in which deletion of the CTD in hnRNP proteins led to the loss of splicing repression [86]. After hnRNP A1 binds a high-affinity site on a target RNA, it recruits other proteins through its CTD to block the assembly of the spliceosome [78]. As mentioned above for HIV, hnRNP A1 prevents splicing either by coating the RNA to sterically block the assembly of the spliceosome (or enhancer proteins) or by binding distal sites and looping out the exon from splicing [87].
3.3 MicroRNA Processing
MicroRNAs (miRNAs) are small noncoding transcripts that control gene expression at the level of translation [88, 89]. The biogenesis of miRNAs begins with synthesis by RNAP II as a precursor known as primary miRNA (pri-miRNA), which often contains clusters of stem loops [90]. Pri-miRNAs are initially processed in the nucleus by the RNase III protein Drosha to produce precursor miRNAs that are ~65 nts in length (pre-miRNA) [88, 91-93]. Following nuclear export, pre-miRNAs are further hydrolyzed in the cytoplasm by Dicer to release mature miRNAs, which then associate with Argonaute proteins to form the RNA-induced silencing complex (RISC) [93].
The biogenesis of miRNAs is tightly regulated due to their ability to broadly modulate gene expression during development and tissue homeostasis. The Drosha dependent nuclear processing of pri-miRNAs is one step in the pathway that is specifically regulated by RBPs. Of the RBPs that regulate miRNA biogenesis, hnRNP A1 inhibits the processing of pri-let-7a [94, 95] but stimulates the processing of pri-miR18a [88, 95]. Binding of hnRNP A1 to the apical loop of the pri-let-7a antagonizes the interaction with KSRP, a positive regulatory factor that promotes Drosha cleavage within the adjacent stem region [94]. Conversely, the interaction of hnRNP A1 with the apical loop of pri-miR18a relaxes the secondary structure, creating a more favorable cleavage site for Drosha [88, 96]. In each case, hnRNP A1 executes its regulatory mechanism by binding single strand elements embedded within stable secondary structures, similar to that described above for splicing of the HIV tat pre-mRNA.
3.4 IRES Dependent Translation
Many RNA viruses and some cellular transcripts conditionally bypass cap-dependent translation by using Internal Ribosome Entry Sites (IRES) located within the 5′UTRs of their mRNAs. [97-99]. Viral IRES elements are grouped into four types that depend on the extent of intrinsic long-range RNA secondary structure, the requirements for eukaryotic initiation factors and additional RBPs collectively referred to as IRES trans acting factors (ITAFs) [100]. Enterovirus 71 (EV71) uses a type I IRES structure to facilitate translation of its positive sense mRNA via cellular processes that require hnRNP A1 [101]. While the exact mechanisms by which hnRNP A1 stimulates EV71 translation remain unknown, its site-specific interaction with a conserved bulge of the stem loop II IRES domain appears to contribute [102, 103]. Stem loop II bulge mutations that disrupt the hnRNP A1 interaction in vitro reduces translation efficiency in vivo and suppresses EV71 replication by ~5 log units [102, 103]. HnRNP A1 has also been implicated as an ITAF for several stress-induced cellular IRES elements such as the expression of nuclear factor-interleukin 3 (Nfil3) and myc [29, 104-106]. As with EV71, the mechanisms by which hnRNP A1 stimulates cellular IRES activity are poorly understood.
3.5 Telomere Elongation
Telomeres are tandem repeats of DNA (TTAGGG) that are highly conserved and protect the 3′ ends of linear chromosomes [107-109]. Telomere repeats are replicated via a two-step process that involves synthesis of new repeats followed by capping with the shelterin complex [110, 111]. The shelterin complex is recruited to the DNA by sequence specific interactions between the protein protection of telomeres 1 (POT1) and the telomere repeat sequence, 5′-TTAGGG-3′. The switch between replication and telomere capping requires the displacement of replication protein A (RPA), a process performed by hnRNP A1, but regulated by the telomeric repeat-containing RNA (TERRA), which is known to form a G-quadruplex structure [110-113]. When TERRA associates with the telomere end, it acts as a ‘scaffold’ for sequestering hnRNP A1 and preventing it from removing RPA [112]. The depletion of TERRA in late S phase allows hnRNP A1 to catalyze the switch on the telomere end between RPA and POT1 [110, 111].
3.6 Granule Assembly
Stress granules are cytoplasmic mRNP granules that form from pools of untranslated mRNAs in cells subjected to a variety of stresses [114, 115]. The granules initially assemble from stalled mRNAs, acting to stabilize them [116, 117]. The proteins composing RNP granules often contain both RBP domains as well as IDRs [118]. HnRNP A1 has been found to be a component of stress granules, with both its RNA binding and protein interacting capabilities utilized in granule assembly [115, 119, 120]. The incorporation of hnRNP A1 in cytoplasmic stress granules was originally discovered in vivo, where it was determined RNA binding by the protein was necessary for its localization to stress granules. [115] These findings would later be further supported by in vitro research into the phase separation properties of hnRNP A1 [118-120].
Stress granules are thought to form through liquid-liquid phase separation (LLPS), a process mediated through IDR domains in proteins [121]. Phase separation occurs when electrostatic interactions between the disordered IDR regions gives rise to granule-like droplets [122]. In hnRNP A1, phase separation was mediated by the CTD, with the independent domain able to phase separate on its own [118-120]. The droplets formed by hnRNP A1 proved capable of recruiting other proteins that contained IDRs [119]. Detailed analysis would suggest a contribution by aromatic and electrostatic residues in the molecular interactions necessary for phase separation [120]. As mentioned previously, the CTD contains a region rich in RGG boxes interspersed with aromatic residues, suggesting this area may be necessary for phase separation. The prion-like domain downstream from the RGG boxes was found not to be vital, as a deletion construct for this region still retained the ability to phase separate [119, 120]. While UP1 could not phase separate as an independent domain, experiments with full-length hnRNP A1 revealed RNA binding by UP1 does contribute to phase separation [119, 120]. This data suggests a mechanism by which RNA binding may regulate phase separation, presumably by inducing the assembly of large, cross-linked complexes that are unable to be formed by protein-protein interactions alone [120].
4. Determinants of hnRNP A1-RNA Specificity
As described above, hnRNP A1 participates in numerous RNA processing events. Underlying this functional diversity is the requirement to identify the correct RNA sequence motif embedded within individual transcripts of the cellular transcriptome. These sequence motifs are short, degenerate, can present with variable copy numbers and reside in different structural environments, all of which further complicates the selection process. How then does hnRNP A1 achieve specific recognition of its cognate RNA targets to impact biology? In this section, we summarize what has been learned about the physicochemical basis by which hnRNP A1 binds various RNA elements.
Early efforts to understand hnRNP A1-RNA recognition revealed the protein has a preference for single strand over double strand regions [123, 124], an attribute that is nearly universal for all hnRNPs [125]. These initial studies also determined that the binding sequence is short and that any increase in binding affinity with longer chain lengths results from overlapping sites [123]. The UP1 domain of hnRNP A1 was also shown to accelerate tRNALeu3 renaturation by binding to single strand regions of the molecule, whereby only three exposed nucleotides were sufficient [123, 126]. Results from dynamic fluorescence anisotropy experiments indicated that binding of RNA induces a conformational change in UP1 and that only one of the two RRMs initially interacts with RNA; however, both RRMs are required for high affinity RNA recognition [27, 127, 128].
While these initial studies offered key insights into the properties of hnRNP A1-RNA interactions, its specific RNA sequence motif was determined later by selection/amplication experiments (SELEX), which identified the high affinity 5′-UAGGGA/U-3′ winner sequence [129]. Direct binding analysis demonstrated that hnRNP A1 bound the 5′-UAGGGA/U-3′ sequence with low nanomolar affinity, and that an RNA containing two copies of the motif (20mer and KD = 1 nM) has slightly greater affinity than an RNA with a single copy (18mer and KD = 3 nM). The discovery of the hnRNP A1 winner sequence was very significant in that it was the first demonstration that the protein recognizes a specific motif. Moreover, oligonucleotides harboring the winner sequence were capable of inhibiting splicing of model pre-mRNA constructs, suggesting that hnRNP A1 functions as a general modulator of pre-mRNA processing [129].
Repeat SELEX experiments with the individual RRMs identified slightly different consensus motifs, albeit each contained the 5′-UAG-3′ as a core component [129]. Mutation of the adenosine in the 5′-UAG-3′ core reduced the ability of hnRNP A1 to repress splicing of a β-globin minigene construct, with a cytosine substitution having a larger influence compared to a uracil substitution [78]. Presumably both mutations abolish hnRNP A1 binding to the β-globin minigene [78]. The importance of the adenosine in the 5′-UAG-3′ element for hnRNP A1 recognition of the HIV ESS3 stem loop (SL3ESS3) was also confirmed by calorimetric and NMR titrations [130]. In this system, substitution of the adenosine with cytosine decreased UP1 binding affinity to SL3ESS3 by ~10 fold [130] and the native adenosine showed a large NMR chemical shift perturbation upon complex formation [131]. A similar fold change in UP1 binding affinity for the 5′-AGUAGAUUAGCA-3′ 12mer, which mimics the apical loop of mir-18a, was also observed when either of the two internal AG dinucleotides was mutated to UU [132]. In this system, both RRMs of UP1 interact with the individual 5′-UAG-3′ elements as a 1:1 complex (more on this below). Although not reported, it would be interesting to know the residual binding free energy when both 5′-UAG-3′ sites of the 12mer are mutated.
Interestingly, the ability of the dual copy winner sequence to form a G-quadruplex structure was suggested to also contribute to its affinity for hnRNP A1 [133, 134]. Indeed, a recent photochemical crosslinking study determined that UP1 binds specifically to loops of a telomere RNA G-quadruplex that contains 5′-UUAGGG-3′ repeats reminiscent of the winner sequence [135]. Conversely, intact hnRNP A1 was shown to unwind a stem loop when given a running start by cooperatively spreading from a downstream 5′-UAGGGU-3′ sequence element [135]. NMR studies monitoring UP1 binding directly to stem loops shows the protein only locally perturbs RNA structure [131, 132, 136]. All this data shows the mechanisms by which hnRNP A1 interacts with cognate RNAs are context dependent.
The utilization of cross-linking immunoprecipitation (CLIP) techniques have led to a greater understanding of hnRNP A1-RNA specificity within the cellular environment, and importantly CLIP has confirmed many of the earlier in vitro results. Indeed, CLIP experiments revealed that hnRNP A1 binds pri-miR-18a by making specific contacts with 5′-UAG-3′ core elements [88]. Global application of CLIP across the cellular transcriptome (variations of CLIP-seq) has further validated that hnRNP A1 specifically recognizes RNA targets that contain 5′-UAG-3′ cores, with the AG dinucleotide being the most prevalent signature motif [137][138]. Other high-throughput methods such as RNAcompete, which quantifies a protein’s RNA affinity from a pre-defined pool of molecules, also demonstrated that hnRNP A1 specifically binds RNA sequences containing a 5′-UAGGG-3′ motif [125] [139].
Expanding on these observations, the global affinity distribution of hnRNP A1 for the HIV SL3ESS3 element was determined using high-throughput sequencing analysis of equilibrium (HTS-EQ) binding [136]. HnRNP A1 binds specifically and with low nanomolar affinity (~30 nM) to the 7-nt apical loop of SL3ESS3, which contains the 5′-GAUUAGU-3′ sequence and only one AG dinucleotide throughout the entire molecule [130, 131, 140]. To fully evaluate the determinants by which hnRNP A1 recognizes SL3ESS3, its apical loop was completely randomized yielding 16,384 different variants, and relative binding affinities of UP1 for each variant were determined by HTS-EQ. Despite only having a single 5′-UAG-3′ copy, native SL3ESS3 is optimized to bind hnRNP A1 because it lies on the high affinity side of the distribution. Nevertheless, 722 other variants bind UP1 tighter than native SL3ESS3 of which 72% of those contained at least one AG dinucleotide [136]. Quantitative analysis of the data further confirmed 5′-YAG-3′ (where Y is C or U) as the minimal hnRNP A1 consensus motif, with the AG dinucleotide being the critical specificity determinant. The position of the 5′-YAG-3′ motif within the loop, its copy number, the spacing between multiple 5′-YAG-3′ copies and the formation of new secondary structure each modulates how hnRNP A1 discriminates between multiple competing RNA targets [136]. Interestingly, follow up biophysical titrations revealed the high affinity SL3ESS3 variants bind UP1 with faster second order association rate constants compared to the low affinity variants, thus offering additional mechanistic insights into hnRNP A1-RNA specificity [136].
In sum, hnRNP A1 recognizes a collection of RNA sequences with a range of binding affinities. Despite its apparent promiscuity for RNA, both in vivo and in vitro methods confirm that hnRNP A1 specificity centers on the 5′-YAG-3′ motif but its binding affinity for RNA targets containing this motif can vary several fold, reflecting contextual features that tune hnRNP A1-RNA molecular recognition.
5. Structural Bases of hnRNP A1-RNA Recognition
As described above, biochemical and biophysical studies have revealed valuable insights into the determinants of sequence specific binding by hnRNP A1; however, an understanding of the protein’s mechanism of interaction with RNA requires high-resolution structures or data-inspired models of the complexes. Since the disordered nature of the CTD makes it difficult to obtain structural data, all structural studies have been performed with the UP1 protein. Multiple atomic resolution structures of UP1 in its unbound and bound states have been solved by X-ray crystallography or NMR spectroscopy (Table 1). The high-resolution structures and other biophysical data have provided a framework for the development of several UP1-RNA structural models. This section will summarize what has been learned about the mechanism of RNA binding by UP1 through these various structures.
Table 1.
High-resolution 3D structures reported for hnRNP A1 domains.
| Region | Ligand | Method | PDB code | Reference |
|---|---|---|---|---|
| 1-184 | Free | X-ray Crystallography | 1HA1 | [142] |
| 1-196 | Free | X-ray Crystallography | 1UP1 | [141] |
| 1-196 | Free | X-ray Crystallography | 1L3K | [146] |
| 1-196 | Free | NMR | 2LYV | [144] |
| 1-196 | d(TTAGGG)2 | X-ray Crystallography | 2UP1 | [147] |
| 1-196 | d(TTAGGGTT 7DA GGG) | X-ray Crystallography | 1U1K | [149] |
| 1-196 | d(TTAGGGTT PRN GGG) | X-ray Crystallography | 1U1L | [149] |
| 1-196 | d(TTAGGGTTA 7GU GG) | X-ray Crystallography | 1U1M | [149] |
| 1-196 | d(TTAGGGTTA PRN GG) | X-ray Crystallography | 1U1N | [149] |
| 1-196 | d(TTAGGGTTAG(DI)G) | X-ray Crystallography | 1U1O | [149] |
| 1-196 | d(TTAGGGTTA 2PR GG) | X-ray Crystallography | 1U1P | [149] |
| 1-196 | d(TTAGGGTTA(DI)GG) | X-ray Crystallography | 1U1Q | [149] |
| 1-196 | d(TTAGGGTTAG(2PR)G) | X-ray Crystallography | 1U1R | [149] |
| 1-196 | d(TTAGGGTTAG(6-MI)G) | X-ray Crystallography | 1PGZ | [148] |
| 1-196 | d(TAGG(6MI)TTAGGG) | X-ray Crystallography | 1PO6 | [148] |
| 1-196 | rAGU | X-ray Crystallography | 4YOE | [131] |
| 1-97 (RRM1) | rUUAGGUC | NMR | 5MPG | [150] |
| 95-196(RRM2) | rUCAGUU | NMR | 5MPL | [150] |
5.1 Unbound structures of hnRNP A1
Structural insights into the unbound form of UP1 have been determined by X-ray crystallography and NMR spectroscopy [141-143] [144, 145]. These structures revealed that each RRM domain adopts nearly identical β1α1β2β3α2β4 folds that are spatially oriented in an antiparallel direction (Figure 1B). The inter-RRM linker 89–105 was not observed in any of the crystal structures of the unbound protein, due to its intrinsic flexibility [141, 142, 146]. Heteronuclear15N{1H}-NOE data obtained from solution NMR further confirmed the flexibility of the inter-RRM linker; however, backbone chemical shifts and sparse NOEs indicate the linker partially adopts a helical structure in solution [144]. Similar differences between the crystal and solution structures involving the C-terminal segment (residues 182–187) were also observed [144]. Both the crystal and solution structures show the tandem RRMs are in close contact, albeit with different relative orientations [141, 142, 144]. In the unbound structures, α helix 2 of RRM1 packs against the corresponding helix of RRM2 in an anti-parallel orientation. The packing arrangement is stabilized by the formation of two salt bridge interactions involving R75:D155 and R88:D157 (Figure 1C) and further stabilized by a small hydrophobic cluster composed of Leu13, Ile64, Val90, and Arg88 [141, 142, 144].
The initial crystal structures of unbound UP1 offered early clues as to how hnRNP A1 might use its tandem RRMs to bind RNA. The conserved phenylalanine residues of the RNP submotifs are on the solvent exposed beta sheet surface of the protein, poised to directionally interact with single strand nucleic acids [141, 142]. Interestingly, the crystal structure solved at the highest resolution (1.1 A) showed the phenylalanine side chains adopt alternate conformations in RRM1 and RRM2, suggesting the two domains differentially interact with RNA [146]. Moreover, the loops connecting the elements of secondary structure contain conserved and positively charged amino acids that would allow favorable electrostatic interactions with the phosphodiester backbone of nucleic acids.
5.2 Bound Structures of hnRNP A1
The first structural insights into how hnRNP A1 binds nucleic acids were derived from the crystal structure of its UP1 domain in complex with telomere DNA (5′-TTAGGGTTAGGG-3′) [147]. This was the only structure of a complex of hnRNP A1 with nucleic acid for almost two decades, and consequently, it greatly influenced the mechanistic interpretations of the various RNA functions performed by hnRNP A1. Other crystal structures of UP1 bound to modified variants of the telomere DNA offered additional insights into the determinants of sequence specificity [148, 149].
In the UP1-DNA crystal structures, UP1 forms an extended and open dimer where the tandem RRMs bind two different strands of DNA that run in an anti-parallel direction, giving a complex with a 2:2 stoichiometry (Figure 2A). The primary UP1-DNA contacts occur between the β sheet surface of RRM12 and both copies of the TTAG motif. The 5′ TTAG motif interacts with RRM1 while the 3′ TTAG interacts with RRM2 from the symmetry related monomer. The conserved phenylalanines (Phe17, Phe59, Phe108 and Phe150) of the RNP submotifs interact directly with the central dAG dinucleotides through π−π stacking. Phenylalanines 57 and 148 make additional van der waals contacts with each respective guanosine of the TTAG motifs. Both guanosines adopt the syn conformation in the complex. The crystal structure further revealed a role for nucleic acid recognition by the inter-RRM linker. Serine 95 forms a hydrogen bond with the guanosine of the 5′ dAG dinucleotide while the imidazole ring of His101 stacks above the purine ring of the adenine, which in turn results in the adenine being intercalated between His101 and Phe17. Similar to how the inter-RRM linker folds with DNA, the C-terminal region (residues 183–190) of RRM2 folds to make specific contacts with the 3′ TTAG motif. The side chain of Met186 makes van der Waals interactions with the adenine ring, such that this adenine is also intercalated between Met186 and Phe108. Several other interactions were also observed that revealed the basis for TAG specificity [147].
Figure 2. High-resolution structures of hnRNP A1 complexes with DNA and RNA.
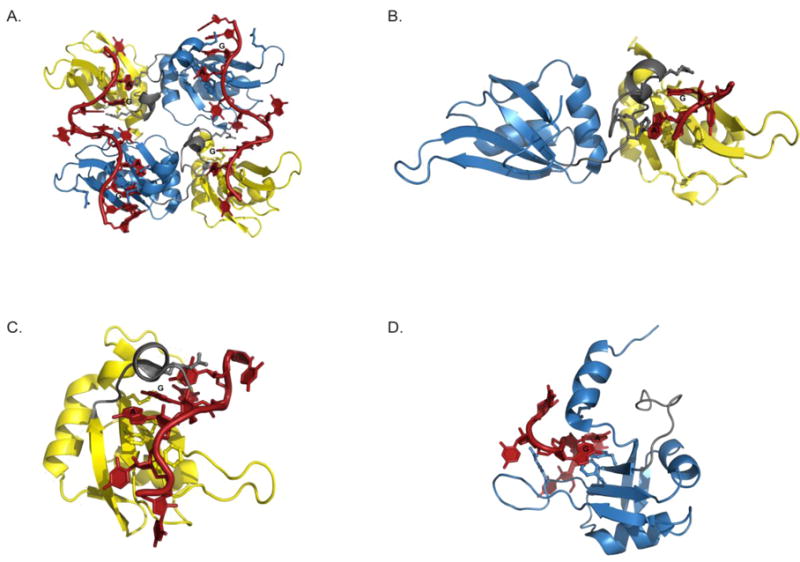
(A) The crystal structure of the UP1-telomeric DNA complex (2UP1). The complex crystalized as a homodimer with a 2:2 stoichiometry. (B) The crystal structure of the UP1-rAGU complex (4YOE). The structure crystalized as a monomer with 1:1 stoichiometry where the 5′-rAGU-3′ trinucleotide was shown to interact only with the RRM1 surface and inter-RRM linker. The NMR solution structures of the (C) RRM1-RNA (5MPG) and (D) RRM2-RNA complexes (5MPL). For each structure, select amino acid side chains that form the binding pockets on the surface of RRM1 and RRM2 of each are shown as ball and sticks. The core rAG dinucleotides that are specifically recognized by hnRNP A1 are also labeled.
The UP1-DNA crystal structures provided a wealth of insight into the determinants of sequence specific recognition; however, the oligomeric state of the protein in the crystals and the 2:2 UP1-DNA stoichiometry do not reflect the solution properties of hnRNP A1 or its mechanisms of interactions with structurally diverse RNA targets. As described above, hnRNP A1 interacts with a range of RNAs that adopt complex structures with variable copies of the 5′-YAG-3′ sequence motif. The first structural insights into hnRNP A1-RNA recognition came from a structure of UP1 bound to an rAGU trinucleotide [131]. A significant difference between the UP1-RNA and DNA complex structures is that UP1 crystallized as a monomer, with the 5′-rAGU-3′ trinucleotide bound through a nucleobase pocket formed by the helix of the inter-RRM linker and RNP residues from the β sheet surface of RRM1; RRM2 does not contact the bound RNA in this structure (Figure 2B). The asymmetric binding mode observed in the UP1-rAGU complex is likely a reflection of the short RNA fragment. Nevertheless, it does suggest that RRM1 and RRM2 have nonequivalent affinities for RNA within the context of UP1. Moreover, the UP1-rAGU structure offered clues as to how hnRNP A1 might bind RNA targets with only a single 5′-YAG-3′ motif, such as the HIV SL3ESS3 element [130, 131, 140] or authentic 3′ splice sites [58].
Besides the differences in oligomeric state and stoichiometry, the key interactions that determine specificity for the central AG dinucleotide are very similar between the RNA and DNA bound structures. As in the UP1-DNA complex, Phe17 and Phe59 stack below the rAG dinucleotide, with residues of the inter-RRM linker (His101 and Arg92) sealing the nucleobase pocket by stacking above (Figure 2C and 2B). However, unlike the DNA bound structure, both rA and rG adopt syn conformations in the UP1-rAGU complex. Additionally, a hydrogen bond interaction between the γ hydroxyl group of Ser95 and the 2′OH of rA is observed, which cannot form in the DNA complex.
The UP1-rAGU structure provides important details as to how hnRNP A1 recognizes RNA as a monomer; it nevertheless lacks information on RRM2-RNA recognition. This information was obtained from the high-resolution NMR structures of isolated RRM1 and RRM2 bound to single strand RNA fragments [150]. The RRM1 co-structure was solved bound to 5′-UUAGGUC-3′, and the overall interactions are very similar to those observed for RRM1 in the UP1-DNA complex [150]. As seen in the UP1-rAGU structure, the core recognition element is the central rAG dinucleotide; however, additional contacts are formed with the longer RNA strand (Figure 2C). Unlike the UP1-rAGU structure, the rA of the dinucleotide adopts an anti conformation and its 2′OH forms an intramolecular hydrogen bond with the neighboring 2-amino group of rG. In the UP1-rAGU complex, the rA 2′OH group hydrogen bonds to the γ hydroxyl group of Ser95.
The RRM2 co-structure was solved in complex with 5′-UCAGUU-3′, and found to have similar protein-RNA interactions as those observed for RRM2 in the UP1-DNA structure (Figure 2D). The central rAG dinucleotide binds through a pocket formed between the β sheet surface and the C-terminus of RRM2. Within the pocket, the rAG dinucleotide stacks with the conserved RNP residues Phe108 and Phe150. The pocket is in turn sealed by Met186, which stacks above rA. Sequence specific hydrogen bonds form between the rAG dinucleotide and the backbone atoms of Lys179 and Leu181. In addition, the phosphate group connecting the rAG dinucleotide forms an electrostatic interaction with the side chain of Arg146. While the rAG dinucleotide anchors the RNA within the binding pocket, the 5′ cytosine interacts in a non-sequence specific manner with the main-chain of Gly111 and the side chain of Glu176. Similar to the RNA induced folding of the inter-RRM linker of UP1, the C-terminal residues (K183-S193) of RRM2 fold into a stable α helix when bound to the 5′-UCAGUU-3′ fragment.
In addition to the structures just described, a crystal structure of UP1 in complex with a single strand 12mer RNA (5′-AGUAGAUUAGCA-3′) that mimics that apical loop of mir-18a was also determined [132]. The UP1-12mer crystalized as a homodimer with 2:2 stoichiometry analogous to the UP1-DNA structure. The contacts observed between UP1 and the 12mer are essentially identical to those already described.
5.3 Structural models of hnRNP A1-RNA complexes
The multi-functionality of hnRNP A1 requires that it interact with RNA targets of varying degrees of structural complexity. At present, there are no high-resolution structures of hnRNP A1 bound to an intact biological RNA element. Instead, several data-inspired structural models of UP1-RNA complexes have been proposed for natural targets (Figure 3). In some cases, the models were determined by integrating multiple data sets along with validation by mutagenesis. While each model offers intriguing perspectives into the mechanisms by which hnRNP A1 binds different RNA elements, they should be interpreted with caution given the nature by which they were determined [131, 132, 135, 150].
Figure 3. Structural models depicting idiosyncratic modes of hnRNP A1-RNA recognition.
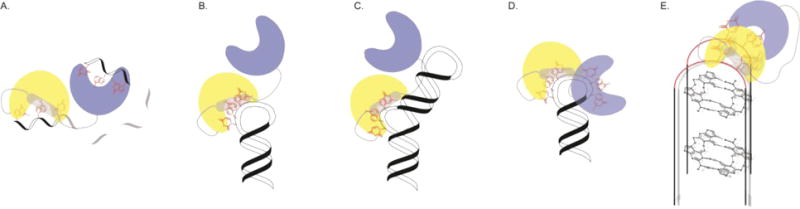
(A) Model of UP1 bound to the single strand ISS-N1 element showing that the RNA directionally loops in a 5′-3′ direction from RRM2 to RRM1. The model was informed by NMR spectroscopy, including long-range PRE restraints and mutagenesis. (B) Model of UP1 bound to the apical loop of the HIV ESS3 stem loop showing the interaction with the RNA occurs through RRM1 and the inter-RRM linker only; RRM is not directly contacting the RNA. This model was informed by X-ray crystallography of the UP1-rAGU structure, NMR spectroscopy, SAXS and mutagenesis. (C) Model of UP1 bound to the bulge loop of the Enterovirus stem loop II IRES domain. This model was informed by X-ray crystallography of the UP1-rAGU structure, NMR spectroscopy, SAXS and mutagenesis. (D) Model of UP1 bound to the apical loop of pri-mir18a showing that both RRM1 and RRM2 contact two UAG elements. This model was informed by X-ray crystallography of the UP1-12mer structure, NMR spectroscopy (including RDCs and PREs), SAXS, SANS and mutagenesis. (E) Model of UP1 bound to an RNA G-quadruplex structure showing that the protein docks onto the connecting loops of a folded G-quadruplex. This model was informed by photochemical crosslinking and mass spec. See manuscripts for additional descriptions and references.
As an example of binding to RNA of low structural complexity, a model of UP1 in complex with the SMN2 intronic splicing silencer (ISS-N1) was determined by incorporating NMR data, including long-range restraints from paramagnetic relaxation enhancements (PREs), and mutagenesis [150]. The ISS-N1 construct (5′-GGACCAGCAUUAUGAAAGGGA-3′) used to build the model contains two AG dinucleotides separated by 9 residues. Since both RRMs of UP1 bind sequences that contain 5′-YAG-3′ elements, the data-inspired model shows that UP1 binds ISS-N1 as a 1:1 complex by directionally looping the RNA in a 5′-3′ direction from RRM2 to RRM1 (Figure 3A). The length of the spacer separating the two AG dinucleotides was shown to be a determinant, with a spacer of at least 4 residues required for looping. Such RNA looping might explain the mechanism by which hnRNP A1 represses splicing of pre-mRNAs [150].
As mentioned above, hnRNP A1 binds the HIV SL3ESS3 element as a 1:1 complex and with low nanomolar affinity. SL3ESS3 folds into a stable stem loop structure that contains only one AG dinucleotide in the entirety of the molecule. A model of UP1 bound to SL3ESS3 was determined by incorporating data from NMR, X-ray crystallography, SAXS and mutagenesis [131]. The key restraints used to build the model were derived from the UP1-rAGU crystal structure (Figure 2B), which showed RNA binding through RRM1 and the inter-RRM linker only. Therefore, structural models of the UP1-SL3ESS3 complex were calculated using this as one condition and then screened against experimental SAXS data. The UP1-SL3ESS3 data-inspired model offers a rationale for how hnRNP A1 interacts with folded RNA elements that contain only one 5′-YAG-3′ motif. Here, the protein docks onto the apical loop of SL3ESS3 using the nucleobase pocket formed by RRM1 and the inter-RRM linker (Figure 3B). This model also suggested that a secondary role for RRM2 is to stabilize the inter-RRM linker in a conformation poised for binding [131]. Indeed, the R75D/R88D UP1 double mutant that destabilizes the RRM1-RRM2 interface, led to an ~18-fold reduction in binding affinity for SL3ESS3. A similar strategy as described for SL3ESS3 was also followed to determine a model of UP1 in complex with the bulge loop of the stem loop II IRES domain from Enterovirus 71 [103]. In this example, UP1 docks onto a central UAG motif within the 5-nt bulge loop (Figure 3C).
The interaction of hnRNP A1 with primary mir-18a represents a more complex binding mode since its apical loop region contains dual 5′-UAG-3′ motifs separated by two spacer residues. A data-inspired model of the UP1-mir-18a complex was determined by integrating NMR (including PRE and RDC restraints), SAXS, mutagenesis and X-ray crystallography [132]. Despite having crystalized as a dimer with a 2:2 stoichiometry, the UP1-12mer structure provided information on the points of contact between the 5′-UAG-3′ motifs and the respective RRMs. The solution properties of the complex were verified to be 1:1 by light scattering and when combined with data from NMR and SAXS, a model of UP1 docked onto the apical loop of mir-18a was determined. The UP1-mir-18a structural model shows that both RRMs interact with the apical loop wherein RRM1 binds the 5′ UAG and RRM2 the 3′ UAG (Figure 3D). The interaction of UP1 with the 3′ UAG requires local relaxing of the mir-18a stem loop structure since this motif is base paired in the unbound form [132].
Lastly, UP1 was shown by photochemical crosslinking and mass spec to bind intact G-quadruplex structures, which mimic the TERRA RNA [135]. In this system, the integrity of the G-quadruplex is maintained because UP1 binds site specifically to the interconnecting loops (Figure 3E). Apparently, hnRNP A1 recognizes various RNA targets using idiosyncratic mechanisms, albeit while preserving sequence specificity. Is it conceivable for these idiosyncrasies to contribute to function? Certainly, idiosyncratic modes of hnRNP A1-RNA recognition would result in complexes with different overall conformations, allowing the assembly of distinct higher-order networks.
6. Diseases Associated with hnRNP A1 and the Potential for Therapeutics
As outlined above, hnRNP A1 functions in multiple aspects of RNA metabolism. Because of its role in these processes, it is involved in several diseases, including those associated with viral infections [37, 64, 71, 79, 101]. Inhibiting hnRNP A1-RNA interactions with novel therapeutics is an emerging area of scientific interest. Indeed, one successful example is the deployment of antisense oligonucleotides to block an hnRNP A1 binding site in the defective SMN2 gene, which is in involved in spinal muscular atrophy (SMA) [138, 151, 152]. In principle, this strategy can be transferable to viruses, where selective inhibition of hnRNP A1 binding to viral transcripts could be employed for the development of novel antivirals [153]. However, the use of antisense therapies may not be effective at blocking hnRNP A1 binding sites, which are embedded in complex RNA secondary structure. As outlined above, hnRNP A1 binds many RNA types where contextual features modulate its affinity for the target and potentially its mechanism of interaction. As such, detailed knowledge of how hnRNP A1 recognizes a specific target will determine the best therapeutic strategy. Small molecule inhibitors, with the potential to disrupt protein-RNA interfaces, represent an alternative approach to antisense therapies [79].
Although not discussed in detail in this review, the CTD of hnRNP A1 has also been shown to play a role in several neurodegenerative diseases [7, 154]. Increased fibrilization of hnRNP A1 is associated with several neural pathologies [25, 155]. Naturally occurring mutations in the CTD at D262 (D262V/D262N) lead to the accumulation of hnRNP A1 fibers, with links to amyotrophic lateral sclerosis (ALS), frontotemporal degeneration (FTD), and inclusion body myositis (IBM) [25, 156]. HnRNP A1 is also involved in the development of multiple sclerosis (MS) [157]. Individuals suffering from MS produce anti-hnRNP A1 antibodies specific to the M9 nuclear localization sequence. These antibodies in turn induce stress granule formation [158]. Thus, the neural diseases caused by aberrant functions of the hnRNP A1 CTD exemplify the need to better understand how the CTD and RBDs cooperate.
7. Conclusions and Future Directions
The goal of this review was to use the available structural biochemical developments on hnRNP A1 as a framework to interpret its wide-range of RNA functions. Historically, the multifunctional nature of hnRNP A1 has led to the general belief that it binds RNA indiscriminately. As discussed above, hnRNP A1 has defined sequence specificity for YAG sequence motifs; however, contextual features of the RNA sequence modulate its mechanisms of interactions. Attempts to understand how hnRNP A1 binds natural RNA targets have been carried out for several decades. An emerging concept from these studies is that hnRNP A1-RNA interactions are idiosyncratic, whereby the sequence composition and structural complexity of the RNA modulates its binding mode (Figure 3). The idea that RNA binding proteins use multiple modes of recognition was recently shown for the Glorund protein from Drosophila. Glorund is a homolog of the hnRNP F/H family, which uses loops of its qRRM domains to bind single strand G-tracts [159]. Structural and biochemical studies revealed that Glorund binds the structured UA-rich nanos RNA using a surface distinct from its G-tract binding interface [159]. Thus, an important future goal is to determine a high-resolution structure of an hnRNP-A1 RNA complex that accurately reflects its functional biochemical state.
Of consideration, the CTD of hnRNP A1 could differentially modulate its multiple modes of RNA recognition. Thus, a complete understanding of the RNA processing functions of hnRNP A1 will not be achieved until the properties of the full-length protein, with the CTD are understood. Because of the disordered nature of the CTD, this has been a challenge; however, the development of new NMR technologies and the integration of complementary techniques are making it possible to study the structural characteristics of intrinsically disordered proteins [160-162]. Characterizing the structural properties of the CTD will provide a more detailed understanding of the mechanisms by which hnRNP A1 interacts with its diverse RNA targets. Along those lines, future studies must take into account the properties of the RNA sequences that promote hnRNP A1 phase separation. It is worth considering that variations in the structural complexity of RNA sequences differentially modulate the ability of hnRNP A1 to phase separate.
Acknowledgments
The authors would like to thank members of the Tolbert group for comments on this manuscript. This work was supported by grants from the National Institutes of Health: R01GM101979, U54GM103297, and R01GM126833.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Afroz T, et al. One, Two, Three, Four! How Multiple RRMs Read the Genome Sequence. Methods Enzymol. 2015;558:235–78. doi: 10.1016/bs.mie.2015.01.015. [DOI] [PubMed] [Google Scholar]
- 2.He Y, Smith R. Nuclear functions of heterogeneous nuclear ribonucleoproteins A/B. Cell Mol Life Sci. 2009;66(7):1239–56. doi: 10.1007/s00018-008-8532-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beyer AL, Christensen ME, Walker BW, LeStourgen WM. Identification and characterization of the packaging proteins of core 40S hnRNP particles. Cell. 1997;11:127–138. doi: 10.1016/0092-8674(77)90323-3. [DOI] [PubMed] [Google Scholar]
- 4.Jean-Philippe J, Paz S, Caputi M. hnRNP A1: the Swiss army knife of gene expression. Int J Mol Sci. 2013;14(9):18999–9024. doi: 10.3390/ijms140918999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han SP, Tang YH, Smith R. Functional diversity of the hnRNPs: past, present and perspectives. Biochem J. 2010;430(3):379–92. doi: 10.1042/BJ20100396. [DOI] [PubMed] [Google Scholar]
- 6.Geuens T, Bouhy D, Timmerman V. The hnRNP family: insights into their role in health and disease. Hum Genet. 2016;135(8):851–67. doi: 10.1007/s00439-016-1683-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harrison AF, Shorter J. RNA-binding proteins with prion-like domains in health and disease. Biochem J. 2017;474(8):1417–1438. doi: 10.1042/BCJ20160499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bekenstein U, Soreq H. Heterogeneous nuclear ribonucleoprotein A1 in health and neurodegenerative disease: from structural insights to post-transcriptional regulatory roles. Mol Cell Neurosci. 2013;56:436–46. doi: 10.1016/j.mcn.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 9.Hamilton BJ, Burns CM, Nichols RC, Rigby WF. Modulation of AUUUA response element binding by heterogenous nuclear ribonucleoprotein A1 in human T lymphocytes. The roles of cytoplasmic location, transcription, and phosphorylation. J Biol Chem. 1997;272(45):28732–41. doi: 10.1074/jbc.272.45.28732. [DOI] [PubMed] [Google Scholar]
- 10.Chabot B, et al. An intron element modulating 5′ splice site selection in the hnRNP A1 pre-mRNA interacts with hnRNP A1. Mol Cell Biol. 1997;17(4):1776–86. doi: 10.1128/mcb.17.4.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blanchette M, Chabot B. A highly stable duplex structure sequesters the 5′ splice site region of hnRNP A1 alternative exon 7B. RNA. 1997;3(4):405–19. [PMC free article] [PubMed] [Google Scholar]
- 12.Hutchison S, et al. Distinct sets of adjacent heterogeneous nuclear ribonucleoprotein (hnRNP) A1/A2 binding sites control 5′ splice site selection in the hnRNP A1 mRNA precursor. J Biol Chem. 2002;277(33):29745–52. doi: 10.1074/jbc.M203633200. [DOI] [PubMed] [Google Scholar]
- 13.Karpel RL, Burchard AC. Physical studies of the interaction of a calf thymus helix-destabilizing protein with the nucleic acids. Biochemistry. 1980;19(20):4674–82. doi: 10.1021/bi00561a021. [DOI] [PubMed] [Google Scholar]
- 14.Herrick G, Delius H, Alberts B. Single-stranded DNA structure and DNA polymerase activity in the presence of nucleic acid helix-unwinding proteins from calf thymus. J Biol Chem. 1976;251(7):2142–6. [PubMed] [Google Scholar]
- 15.Herrick G, Alberts B. Purification and physical characterization of nucleic acid helix-unwinding proteins from calf thymus. J Biol Chem. 1976;251(7):2124–32. [PubMed] [Google Scholar]
- 16.Herrick G, Alberts B. Nucleic acid helix-coil transitions mediated by helix-unwinding proteins from calf thymus. J Biol Chem. 1976;251(7):2133–41. [PubMed] [Google Scholar]
- 17.Clery A, Allain FHT. From structure to function of RNA binding domains. RNA Binding Proteins. 2011 [Google Scholar]
- 18.Clery A, Blatter M, Allain FH. RNA recognition motifs: boring? Not quite. Curr Opin Struct Biol. 2008;18(3):290–8. doi: 10.1016/j.sbi.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 19.Chen Y, Varani G. Engineering RNA-binding proteins for biology. FEBS J. 2013;280(16):3734–54. doi: 10.1111/febs.12375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272(9):2118–31. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 21.Bettany AJE, Eisenstein RS, Munro HN. Mutagenesis of the iron-regulatory element further defines a role for RNA secondary structure in the regulation of ferritin and transferrin receptor expression. Journal of Biological Chemistry. 1992;267(23):16531–16537. [PubMed] [Google Scholar]
- 22.Cobianchi F, Karpel RL, Williams KR, Notario V, Wilson SH. Mammalian heterogenous nuclear ribonucleoprotein complex protein A1: Large-scale overproduction in Escherichia coli and cooperative binding to single-stranded nucleic acids. J Biol Chem. 1988;263(2):1063–71. [PubMed] [Google Scholar]
- 23.Kiledjian M, Dreyfuss G. Primary structure and bindind activity of the hnRNP U protein: binding RNA through RGG box. EMBO J. 1992;11(7):2655–64. doi: 10.1002/j.1460-2075.1992.tb05331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim S, Merrill BM, Rajpurohit R, Kumar A, Stone KL, Papov VV, Schneiders JM, Szer W, Wilson SH, Paik WK, Williams KR. Identification of NG-methylarginine residues in heterogenous RNP protein A1: Phe-Gly-Gly-Gly-Arg-Gly-Gly-Gly/Phe is a preferred recognition motif. Biochemistry. 1997;36:5185–92. doi: 10.1021/bi9625509. [DOI] [PubMed] [Google Scholar]
- 25.Kim HJ, et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495(7442):467–73. doi: 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar A, Casas-Finet JR, Luneau CJ, Karpel RL, Merrill BM, Williams KR, Wilson SH. Mammalian heterogenous nuclear ribonucleoprotein A1: nucleic acid binding properties of the COOH-terminal domain. J Biol Chem. 1990;265(28):17094–100. [PubMed] [Google Scholar]
- 27.Casas-Finet JR, Smith JD, Kumar A, Kim JG, Wilson SH, Karpel RL. Mammalian heterogenous ribonucleoprotein A1 and its constituent domains. Nucelic acid interaction, structural stability and self-association. J Mol Biol. 1993;229(4):873–89. doi: 10.1006/jmbi.1993.1093. [DOI] [PubMed] [Google Scholar]
- 28.Wall ML, Lewis SM. Methylarginines within the RGG-Motif Region of hnRNP A1 Affect Its IRES Trans-Acting Factor Activity and Are Required for hnRNP A1 Stress Granule Localization and Formation. J Mol Biol. 2017;429(2):295–307. doi: 10.1016/j.jmb.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 29.Gao G, Dhar S, Bedford MT. PRMT5 regulates IRES-dependent translation via methylation of hnRNP A1. Nucleic Acids Res. 2017;45(8):4359–4369. doi: 10.1093/nar/gkw1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Idriss H, Kumar A, Casas-Finet JR, Guo H, Damuni Z, Wilson SH. Regulation of in vitro nucleic acid strand annealing activity of heterogenous nuclear ribonucleoprotein A1 by reversible phosphorylation. Biochemistry. 1994;33:11382–90. doi: 10.1021/bi00203a037. [DOI] [PubMed] [Google Scholar]
- 31.Martin J, et al. Phosphomimetic substitution of heterogeneous nuclear ribonucleoprotein A1 at serine 199 abolishes AKT-dependent internal ribosome entry site-transacting factor (ITAF) function via effects on strand annealing and results in mammalian target of rapamycin complex 1 (mTORC1) inhibitor sensitivity. J Biol Chem. 2011;286(18):16402–13. doi: 10.1074/jbc.M110.205096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Izzaurralde E, Jarmolowski A, Beisel C, Mattaj IW, Dreyfuss G, Fisher U. A role for the M9 transport signal of hnRNP A1 in mRNA nuclear export. J Cell Biol. 1997;137(1):27–35. doi: 10.1083/jcb.137.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pollard VW, Michael WM, Nakelny S, Siomi MC, Wang F, Dreyfuss G. A novel receptor-mediated nuclear protein import pathway. Cell. 1996;86(6):985–94. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- 34.Michael WM, Choi M, Dreyfuss G. A nuclear export signal in hnRNP A1: a signal-mediated, temperature-dependent nuclear protein export pathway. Cell. 1995;83(3):415–22. doi: 10.1016/0092-8674(95)90119-1. [DOI] [PubMed] [Google Scholar]
- 35.Siomi H, Dreyfuss G. A nuclear localization domain in the hnRNP A1 protein. J Cell Biol. 1995;129(3):551–60. doi: 10.1083/jcb.129.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weighardt F, Biamonti G, Riva S. Nucleo-cytoplasmic distribution of human hnRNP proteins: a search for the targeting domains in hnRNP A1. J Cell Sci. 1995;108(Pt 2):545–55. doi: 10.1242/jcs.108.2.545. [DOI] [PubMed] [Google Scholar]
- 37.Monette A, et al. Human immunodeficiency virus type 1 (HIV-1) induces the cytoplasmic retention of heterogeneous nuclear ribonucleoprotein A1 by disrupting nuclear import: implications for HIV-1 gene expression. J Biol Chem. 2009;284(45):31350–62. doi: 10.1074/jbc.M109.048736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Allemand E, et al. Regulation of heterogenous nuclear ribonucleoprotein A1 transport by phosphorylation in cells stressed by osmotic shock. Proc Natl Acad Sci U S A. 2005;10210:3605–10. doi: 10.1073/pnas.0409889102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roth S, Khalaila I. The effect of O-GlcNAcylation on hnRNP A1 translocation and interaction with transportin1. Exp Cell Res. 2017;3501:210–217. doi: 10.1016/j.yexcr.2016.11.023. [DOI] [PubMed] [Google Scholar]
- 40.Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;3095740:1514–8. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- 41.Xia H. Regulation of gamma-fibrinogen chain expression by heterogeneous nuclear ribonucleoprotein A1. J Biol Chem. 2005;28013:13171–8. doi: 10.1074/jbc.M414120200. [DOI] [PubMed] [Google Scholar]
- 42.Chen H, Hewison M, Hu B, Adams JS. Heterogenous nuclear ribonucleoprotein (hnRNP) binding to hormone response elements: a cause of vitamin D resistance. Proc Natl Acad Sci U S A. 2003;10010:6109–14. doi: 10.1073/pnas.1031395100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lau JS, Baumeister P, Kim E, Roy B, Hsieh TY, Lai M, Lee AS. Heterogenous nuclear ribonucleotproteins as regulators of gene expression through interactions with the human thymidine kinase promoter. J Cell Biochem. 2000;79:395–406. doi: 10.1002/1097-4644(20001201)79:3<395::aid-jcb50>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 44.Campillos M. Specific interaction of heterogeneous nuclear ribonucleoprotein A1 with the -219T allelic form modulates APOE promoter activity. Nucleic Acids Research. 2003;31(12):3063–3070. doi: 10.1093/nar/gkg435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fukuda H, et al. Unfolding of quadruplex structure in the G-rich strand of the minisatellite repeat by the binding protein UP1. Proc Natl Acad Sci U S A. 2002;99(20):12685–90. doi: 10.1073/pnas.152456899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kan ZY, et al. G-quadruplex formation in human telomeric (TTAGGG)4 sequence with complementary strand in close vicinity under molecularly crowded condition. Nucleic Acids Res. 2007;35(11):3646–53. doi: 10.1093/nar/gkm203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paramasivam M, et al. Protein hnRNP A1 and its derivative Up1 unfold quadruplex DNA in the human KRAS promoter: implications for transcription. Nucleic Acids Res. 2009;37(9):2841–53. doi: 10.1093/nar/gkp138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takimoto M, Tomonaga T, Matunis M, Avigan M, Krutzsch H, Dreyfuss G, Levens D. Specific binding of heterogenous ribonucleoprotein particle protein K to the human c-myc promoter, in vitro. J Biol Chem. 1993;268(24):18249–58. [PubMed] [Google Scholar]
- 49.Hay DC, et al. Interaction between hnRNPA1 and IkappaBalpha is required for maximal activation of NF-kappaB-dependent transcription. Mol Cell Biol. 2001;21(10):3482–90. doi: 10.1128/MCB.21.10.3482-3490.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Diribarne G, Bensaude O. 7SK RNA, a non-coding RNA regulating P-TEFb, a general transcription factor. RNA Biol. 2009;6(2):122–8. doi: 10.4161/rna.6.2.8115. [DOI] [PubMed] [Google Scholar]
- 51.Lemieux B, et al. A Function for the hnRNP A1/A2 Proteins in Transcription Elongation. PLoS One. 2015;10(5):e0126654. doi: 10.1371/journal.pone.0126654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van Herreweghe E, et al. Dynamic remodelling of human 7SK snRNP controls the nuclear level of active P-TEFb. EMBO J. 2007;26(15):3570–80. doi: 10.1038/sj.emboj.7601783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barrandon C, et al. The transcription-dependent dissociation of P-TEFb-HEXIM1-7SK RNA relies upon formation of hnRNP-7SK RNA complexes. Mol Cell Biol. 2007;27(20):6996–7006. doi: 10.1128/MCB.00975-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Warf MB, Berglund JA. Role of RNA structure in regulating pre-mRNA splicing. Trends Biochem Sci. 2010;35(3):169–78. doi: 10.1016/j.tibs.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Park E, et al. The Expanding Landscape of Alternative Splicing Variation in Human Populations. Am J Hum Genet. 2018;102(1):11–26. doi: 10.1016/j.ajhg.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fu XD, Ares M., Jr Context-dependent control of alternative splicing by RNA-binding proteins. Nat Rev Genet. 2014;15(10):689–701. doi: 10.1038/nrg3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136(4):701–18. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 58.Tavanez JP, et al. hnRNP A1 proofreads 3′ splice site recognition by U2AF. Mol Cell. 2012;45(3):314–29. doi: 10.1016/j.molcel.2011.11.033. [DOI] [PubMed] [Google Scholar]
- 59.Mayeda A, Krainer AR. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell. 1992;68(2):365–75. doi: 10.1016/0092-8674(92)90477-t. [DOI] [PubMed] [Google Scholar]
- 60.Fisette JF, et al. hnRNP A1 and hnRNP H can collaborate to modulate 5′ splice site selection. RNA. 2010;16(1):228–38. doi: 10.1261/rna.1890310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Amendt BA, Si ZH, Stoltzfus CM. Presence of exon splicing silencers within human immunodeficiency virus type 1 tat exon 2 and tat-rev exon3: evidence for inhibition mediated by cellular factors. Mol Cell Biol. 1995;15(8):4606–15. doi: 10.1128/mcb.15.8.4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Si ZH, Amendt BA, Stoltzfus CM. Splicing efficiency of human immunovirus type 1 tat RNA is determined by both a suboptimal 3′ splice site and a 10 nucleotide exon splicing silencer element located with tat exon 2. Nucleic Acids Res. 1997;25(4):861–7. doi: 10.1093/nar/25.4.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Domsic JK, et al. Human Immunodeficiency Virus Type 1 hnRNP A/B-Dependent Exonic Splicing Silencer ESSV Antagonizes Binding of U2AF65 to Viral Polypyrimidine Tracts. Molecular and Cellular Biology. 2003;23(23):8762–8772. doi: 10.1128/MCB.23.23.8762-8772.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stoltzfus CM, Madsen JM. Role of Viral Splicing Elements and Cellular RNA Binding Proteins in Regulation of HIV-1 Alternative RNA Splicing. Curr HIV Res. 2006;4(1):53–65. doi: 10.2174/157016206775197655. [DOI] [PubMed] [Google Scholar]
- 65.Staffa A, Cochrane A. The tat/rev intron of human immudeficiency virus type is inefficiently spliced because of suboptimal signals in the 3′ splice site. J Virol. 1994;68(5):3071–9. doi: 10.1128/jvi.68.5.3071-3079.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.O’Reilly MM, McNally MT, Beemon KL. Two strong 5′ splice sites and competing, suboptimal 3′ splice sites involved in alternative splicing of human immunodeficiency virus type 1 RNA. Virology. 1995;213(2):373–85. doi: 10.1006/viro.1995.0010. [DOI] [PubMed] [Google Scholar]
- 67.Bilodeau PS, et al. RNA Splicing at Human Immunodeficiency Virus Type 1 3′ Splice Site A2 Is Regulated by Binding of hnRNP A/B Proteins to an Exonic Splicing Silencer Element. Journal of Virology. 2001;75(18):8487–8497. doi: 10.1128/JVI.75.18.8487-8497.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jacquenet S, et al. A second exon splicing silencer within human immunodeficiency virus type 1 tat exon 2 represses splicing of Tat mRNA and binds protein hnRNP H. J Biol Chem. 2001;276(44):40464–75. doi: 10.1074/jbc.M104070200. [DOI] [PubMed] [Google Scholar]
- 69.Tange TO, et al. The hnRNP A1 protein regulates HIV-1 tat splicing via a novel intron silencer element. EMBO J. 2001;20(20):5748–58. doi: 10.1093/emboj/20.20.5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Caputi M, et al. hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J. 1999;18(14):4060–7. doi: 10.1093/emboj/18.14.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Saliou JM, Bourgeis CF, Ayadi-Ben L, Ropers D, Jacquenet S, Marchand V, Stevenin J, Branlant C. Role of RNA structure and protein factors in the control of HIV-1 splicing. Front Biosci. 2009;14:2714–29. doi: 10.2741/3408. [DOI] [PubMed] [Google Scholar]
- 72.Erkelenz S, et al. Balanced splicing at the Tat-specific HIV-1 3′ss A3 is critical for HIV-1 replication. Retrovirology. 2015;12:29. doi: 10.1186/s12977-015-0154-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fields JA, Dumaop W, Crews L, Adame A, Spencer B, Metcalf J, He J, Rockenstein E, Masliah E. Mechanisms of HIV-1 tat neurotoxicity cia CKK5 translocation and hyper-activation: role in HIV-associated neurocognitive disorders. Curr HIV Res. 2015;13(1):43–54. doi: 10.2174/1570162x13666150311164201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hallay H, et al. Biochemical and NMR study on the competition between proteins SC35, SRp40, and heterogeneous nuclear ribonucleoprotein A1 at the HIV-1 Tat exon 2 splicing site. J Biol Chem. 2006;281(48):37159–74. doi: 10.1074/jbc.M603864200. [DOI] [PubMed] [Google Scholar]
- 75.Zhu J, Mayeda A, Krainer AR. Exon identity established through differential antagonism between exonic splicing silencer-bound hnRNP A1 and enhancer-bound SR proteins. Molecular Cell. 2001;8(6):1351–1361. doi: 10.1016/s1097-2765(01)00409-9. [DOI] [PubMed] [Google Scholar]
- 76.Marchand V, et al. A Janus Splicing Regulatory Element Modulates HIV-1 tat and rev mRNA Production by Coordination of hnRNP A1 Cooperative Binding. Journal of Molecular Biology. 2002;323(4):629–652. doi: 10.1016/s0022-2836(02)00967-1. [DOI] [PubMed] [Google Scholar]
- 77.Damgaard CK, Tange TO, Kjems J. hnRNP A1 controls HIV-1 mRNA splicing through cooperative binding to intron and exon splicing silencers in the context of a conserved secondary structure. Rna-a Publication of the Rna Society. 2002;8(11):1401–1415. doi: 10.1017/s1355838202023075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Okunola HL, Krainer AR. Cooperative-binding and splicing-repressive properties of hnRNP A1. Mol Cell Biol. 2009;29(20):5620–31. doi: 10.1128/MCB.01678-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tazi J, et al. Alternative splicing: regulation of HIV-1 multiplication as a target for therapeutic action. FEBS J. 2010;277(4):867–76. doi: 10.1111/j.1742-4658.2009.07522.x. [DOI] [PubMed] [Google Scholar]
- 80.Chiou NT, Shankarling G, Lynch KW. hnRNP L and hnRNP A1 induce extended U1 snRNA interactions with an exon to repress spliceosome assembly. Mol Cell. 2013;49(5):972–82. doi: 10.1016/j.molcel.2012.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cartegni L, Krainer AR. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat Genet. 2002;30(4):377–84. doi: 10.1038/ng854. [DOI] [PubMed] [Google Scholar]
- 82.Cartegni L, Hastings ML, Calarco JA, de Stanchina E, Krainer AR. Determinants of exon 7 splicing in the spinal muscular atrophy gene, SMN1 and SMN2. Am J Hum Genet. 2006;78(1) doi: 10.1086/498853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kashima T, Manley JL. A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat Genet. 2003;34(4):460–3. doi: 10.1038/ng1207. [DOI] [PubMed] [Google Scholar]
- 84.Kashima T, et al. hnRNP A1 functions with specificity in repression of SMN2 exon 7 splicing. Hum Mol Genet. 2007;16(24):3149–59. doi: 10.1093/hmg/ddm276. [DOI] [PubMed] [Google Scholar]
- 85.Singh NK, et al. Splicing of a critical exon of human Survival Motor Neuron is regulated by a unique silencer element located in the last intron. Mol Cell Biol. 2006;26(4):1333–46. doi: 10.1128/MCB.26.4.1333-1346.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hillebrand F, et al. Differential hnRNP D isoform incorporation may confer plasticity to the ESSV-mediated repressive state across HIV-1 exon 3. Biochim Biophys Acta. 2017;1860(2):205–217. doi: 10.1016/j.bbagrm.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 87.Lee Y, Rio DC. Mechanisms and Regulation of Alternative Pre-mRNA Splicing. Annu Rev Biochem. 2015;84:291–323. doi: 10.1146/annurev-biochem-060614-034316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Guil S, Caceres JF. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat Struct Mol Biol. 2007;14(7):591–6. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- 89.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 90.Lee Y, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23(20):4051–60. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Trabucchi M, et al. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459(7249):1010–4. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Treiber T, et al. A Compendium of RNA-Binding Proteins that Regulate MicroRNA Biogenesis. Mol Cell. 2017;66(2):270–284 e13. doi: 10.1016/j.molcel.2017.03.014. [DOI] [PubMed] [Google Scholar]
- 93.Ha M, V, Kim N. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15(8):509–24. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 94.Michlewski G, Caceres JF. Antagonistic role of hnRNP A1 and KSRP in the regulation of let-7a biogenesis. Nat Struct Mol Biol. 2010;17(8):1011–8. doi: 10.1038/nsmb.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Castilla-Llorente V, Nicastro G, Ramos A. Terminal loop-mediated regulation of miRNA biogenesis: selectivity and mechanisms. Biochem Soc Trans. 2013;41(4):861–5. doi: 10.1042/BST20130058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Michlewski G, et al. Posttranscriptional regulation of miRNAs harboring conserved terminal loops. Mol Cell. 2008;32(3):383–93. doi: 10.1016/j.molcel.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Komar AA, Hatzoglou M. Cellular IRES-mediated translation: the war of ITAFs in pathophysiological states. Cell Cycle. 2011;10(2):229–40. doi: 10.4161/cc.10.2.14472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lopez-Lastra M, et al. Translation initiation of viral mRNAs. Rev Med Virol. 2010;20(3):177–95. doi: 10.1002/rmv.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fitzgerald KD, Semler BL. Bridging IRES elements in mRNAs to the eukaryotic translation apparatus. Biochim Biophys Acta. 2009;1789(9–10):518–28. doi: 10.1016/j.bbagrm.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Niepmann M. Internal translation initiation of picornaviruses and hepatitis C virus. Biochim Biophys Acta. 2009;1789(9–10):529–41. doi: 10.1016/j.bbagrm.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 101.Lin JY, et al. hnRNP A1 interacts with the 5′ untranslated regions of enterovirus 71 and Sindbis virus RNA and is required for viral replication. J Virol. 2009;83(12):6106–14. doi: 10.1128/JVI.02476-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Levengood JD, et al. High-affinity interaction of hnRNP A1 with conserved RNA structural elements is required for translation and replication of enterovirus 71. RNA Biol. 2013;10(7):1136–45. doi: 10.4161/rna.25107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tolbert M, et al. HnRNP A1 Alters the Structure of a Conserved Enterovirus IRES Domain to Stimulate Viral Translation. J Mol Biol. 2017;429(19):2841–2858. doi: 10.1016/j.jmb.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shi Y, et al. Therapeutic potential of targeting IRES-dependent c-myc translation in multiple myeloma cells during ER stress. Oncogene. 2016;35(8):1015–24. doi: 10.1038/onc.2015.156. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 105.Jo OD, et al. Heterogeneous nuclear ribonucleoprotein A1 regulates cyclin D1 and c-myc internal ribosome entry site function through Akt signaling. J Biol Chem. 2008;283(34):23274–87. doi: 10.1074/jbc.M801185200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kim HJ, et al. Heterogeneous nuclear ribonucleoprotein A1 regulates rhythmic synthesis of mouse Nfil3 protein via IRES-mediated translation. Sci Rep. 2017;7:42882. doi: 10.1038/srep42882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Le PN, et al. TERRA, hnRNP A1, and DNA-PKcs Interactions at Human Telomeres. Front Oncol. 2013;3:91. doi: 10.3389/fonc.2013.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Meyne J, Ratliff RL, Moyzis RK. Conservation of the human telomere sequence (TTAGGG)n among vertebrates. Proc Natl Acad Sci U S A. 1989;86:7049–53. doi: 10.1073/pnas.86.18.7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.de Lange T. How telomeres solve the end-protection problem. Science. 2009;326(5955):948–52. doi: 10.1126/science.1170633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Flynn RL, et al. TERRA and hnRNPA1 orchestrate an RPA-to-POT1 switch on telomeric single-stranded DNA. Nature. 2011;471(7339):532–6. doi: 10.1038/nature09772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Flynn RL, Chang S, Zou L. RPA and POT1: friends or foes at telomeres? Cell Cycle. 2012;11(4):652–7. doi: 10.4161/cc.11.4.19061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yamada T, et al. Spatiotemporal analysis with a genetically encoded fluorescent RNA probe reveals TERRA function around telomeres. Sci Rep. 2016;6:38910. doi: 10.1038/srep38910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sui J, et al. DNA-PKcs phosphorylates hnRNP-A1 to facilitate the RPA-to-POT1 switch and telomere capping after replication. Nucleic Acids Res. 2015;43(12):5971–83. doi: 10.1093/nar/gkv539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Protter DS, Parker R. Principles and Properties of Stress Granules. Trends Cell Biol. 2016;26(9):668–79. doi: 10.1016/j.tcb.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Guil S, Long JC, Caceres JF. hnRNP A1 relocalization to the stress granules reflects a role in the stress response. Mol Cell Biol. 2006;26(15):5744–58. doi: 10.1128/MCB.00224-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Buchan JR, Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol Cell. 2009;36(6):932–41. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shah KH, et al. A Hybrid-Body Containing Constituents of Both P-Bodies and Stress Granules Forms in Response to Hypoosmotic Stress in Saccharomyces cerevisiae. PLoS One. 2016;11(6):e0158776. doi: 10.1371/journal.pone.0158776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kato M, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149(4):753–67. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lin Y, et al. Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Mol Cell. 2015;60(2):208–19. doi: 10.1016/j.molcel.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Molliex A, et al. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163(1):123–33. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wheeler JR, et al. Distinct stages in stress granule assembly and disassembly. Elife. 2016;5 doi: 10.7554/eLife.18413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Elbaum-Garfinkle S, Kim Y, Szczepaniak K, Chen CC, Eckmann CR, Myong S, Brangwynne CP. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc Natl Acad Sci U S A. 2015;112(23):7189–94. doi: 10.1073/pnas.1504822112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Karpel RL, Miller NS, Fresco JR. Mechanistic studies of ribonucleic acid renaturation by a helix-destabilizing protein. Biochemistry. 1982;21(9):2102–8. doi: 10.1021/bi00538a019. [DOI] [PubMed] [Google Scholar]
- 124.Nadler SG, Merrill BM, Roberts WJ, Keating KM, Lisbin MJ, Barnett SF, Wilson SF, Williams KR. Interactions of the A1 heterogenous nuclear ribonucleotprotein and its proteolytic derivative, UP1, with RNA and DNA: evidence for multiple RNA binding domains and salt-dependent binding mode transitions. Biochemistry. 1991;30:2968–76. doi: 10.1021/bi00225a034. [DOI] [PubMed] [Google Scholar]
- 125.Dominguez D, et al. Sequence, structure and context preferences of human RNA binding proteins. bioRxiv. 2017 doi: 10.1101/201996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Karpel RL, et al. Acceleration of RNA renaturation by nucleic acid unwinding proteins. Brookhaven. Symp Biol. 1975;26(26):165–74. [PubMed] [Google Scholar]
- 127.Casas-Finet J, et al. Physical studies of tyrosine and tryptophan residues in mammalian A1 heterogeneous nuclear ribonucleoprotein Support for a segmented structure. Journal of Molecular Biology. 1991;221(2):693–709. doi: 10.1016/0022-2836(91)80081-5. [DOI] [PubMed] [Google Scholar]
- 128.Shamoo Y, et al. Both Rna-Binding Domains in Heterogenous Nuclear Ribonucleoprotein A1 Contribute toward Single-Stranded-Rna Binding. Biochemistry. 1994;33(27):8272–8281. doi: 10.1021/bi00193a014. [DOI] [PubMed] [Google Scholar]
- 129.Burd CG, Dreyfuss G. RNA binding specificity of hnRNP A1: significance of hnRNP A1 high-affinity binding sites in pre-mRNA. EMBO J. 1994;13(5):1197–204. doi: 10.1002/j.1460-2075.1994.tb06369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Rollins C, et al. Thermodynamic and Phylogenetic Insights into hnRNP A1 Recognition of the HIV-1 Exon Splicing Silencer 3 Element. Biochemistry. 2014;53(13):2172–84. doi: 10.1021/bi500180p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Morgan CE, et al. The First Crystal Structure of the UP1 Domain of hnRNP A1 Bound to RNA Reveals a New Look for an Old RNA Binding Protein. J Mol Biol. 2015;427(20):3241–57. doi: 10.1016/j.jmb.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kooshapur H, et al. Structural basis for terminal loop recognition and processing of pri-mRNA-18a by hnRNP A1. bioRxiv. 2017 doi: 10.1101/178855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Abdulmanan N, O’Malley SM, Williams KR. Origins of binding specificity of the A1 heterogenous nuclear ribonucleoprotein. Biochemistry. 1996;35:3545–54. doi: 10.1021/bi952298p. [DOI] [PubMed] [Google Scholar]
- 134.AbdulManan N, Williams KR. hnRNP A1 binds promiscuously to oligoribonucleotides: Utilization of random and homo-oligonucleotides to discriminate sequence from base-specific binding. Nucleic Acids Research. 1996;24(20):4063–4070. doi: 10.1093/nar/24.20.4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Liu X, et al. Structure-Dependent Binding of hnRNPA1 to Telomere RNA. J Am Chem Soc. 2017;139(22):7533–7539. doi: 10.1021/jacs.7b01599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Jain N, Lin HC, Morgan CE, Harris ME, Tolbert BS. Rules of RNA specificity of hnRNP A1 revealed by global and quantitative analysis of its affinity distribution. Proc Natl Acad Sci. 2017;114(9):2206–2211. doi: 10.1073/pnas.1616371114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Huelga SC, et al. Integrative genome-wide analysis reveals cooperative regulation of alternative splicing by hnRNP proteins. Cell Rep. 2012;1(2):167–78. doi: 10.1016/j.celrep.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bruun GH, et al. Global identification of hnRNP A1 binding sites for SSO-based splicing modulation. BMC Biol. 2016;14:54. doi: 10.1186/s12915-016-0279-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ray D, et al. A compendium of RNA-binding motifs for decoding gene regulation. Nature. 2013;499(7457):172–7. doi: 10.1038/nature12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Levengood JD, et al. Solution structure of the HIV-1 exon splicing silencer 3. J Mol Biol. 2012;415(4):680–98. doi: 10.1016/j.jmb.2011.11.034. [DOI] [PubMed] [Google Scholar]
- 141.Xu RM, Jokhan L, Cheng X, Mayeda A, Krainer AR. Crystal structure of human UP1, the domain of hnRNP A1 that contains two RNA-recognition motifs. Structure. 1997;5(4):559–70. doi: 10.1016/s0969-2126(97)00211-6. [DOI] [PubMed] [Google Scholar]
- 142.Shamoo Y, Krueger U, Rice LM, Williams KR, Steitz TA. Crystal structure of the two RNA binding domains of human hnRNP A1 at 1.74 A resolution. Nat Struct Biol. 1997;4(3):215–22. doi: 10.1038/nsb0397-215. [DOI] [PubMed] [Google Scholar]
- 143.Kessler H, Mronga S, Gemmecker G. Multi-dimensional NMR experiments using selective pulses. Magnetic Resonance in Chemistry. 1991;29:527–557. [Google Scholar]
- 144.Barraud P, Allain FH. Solution structure of the two RNA recognition motifs of hnRNP A1 using segmental isotope labeling: how the relative orientation between RRMs influences the nucleic acid binding topology. J Biomol NMR. 2013;55(1):119–38. doi: 10.1007/s10858-012-9696-4. [DOI] [PubMed] [Google Scholar]
- 145.Garrett DS, Lodi PJ, Shamoo Y, Williams KR, Clore GM, Gronenborn AM. Determination of the secondary structure and folding topology of an RNA binding domain of mammalian hnRNP A1 protein using three-dimensional heteronuclear magnetic resonance spectroscopy. Biochemistry. 1994;33:2852–58. doi: 10.1021/bi00176a015. [DOI] [PubMed] [Google Scholar]
- 146.Vitali J, Ding J, Jiang J, Zhang Y, Krainer AR, Xu RM. Correlated alternative side chain conformations in the RNA-recognition motif of heterogenous nuclear ribonucleoprotein A1. Nucleic Acids Res. 2002;30(7):1531–38. doi: 10.1093/nar/30.7.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ding J, Hayashi MK, Zhang Y, Manche L, Krainer AR, Xu RM. Crystal structure of the two-RRM domain of hnRNP A1 (UP1) complexed with single-stranded telomeric DNA. Genes Dev. 1999;13(9):1102–15. doi: 10.1101/gad.13.9.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Myers JC, Moore SA, Shamoo Y. Structure-based incorporation of 6-methyl-8-(2-deoxy-beta-ribofuranosyl)isoxanthopteridine into the human telomeric repeat DNA as a probe for UP1 binding and destabilization of G-tetrad structures. J Biol Chem. 2003;278(43):42300–6. doi: 10.1074/jbc.M306147200. [DOI] [PubMed] [Google Scholar]
- 149.Myers JC, Shamoo Y. Human UP1 as a model for understanding purine recognition in the family of proteins containing the RNA recognition motif (RRM) J Mol Biol. 2004;342(3):743–56. doi: 10.1016/j.jmb.2004.07.029. [DOI] [PubMed] [Google Scholar]
- 150.Beusch I, Barraud P, Moursy A, Ciery A, Allain FH. Tandem hnRNP A1 RNA recognition motifs act in concert to repress the splicing of survival motor neuron exon 7. Elife. 2017;25(6):e25736. doi: 10.7554/eLife.25736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Seo J, et al. Spinal muscular atrophy: an update on therapeutic progress. Biochim Biophys Acta. 2013;1832(12):2180–90. doi: 10.1016/j.bbadis.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Singh NN, et al. A short antisense oligonucleotide masking a unique intronic motif prevents skipping of a critical exon in spinal muscular atrophy. RNA Biol. 2009;6(3):341–50. doi: 10.4161/rna.6.3.8723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Dlamini Z, Hull R. Can the HIV-1 splicing machinery be targeted for drug discovery? HIV AIDS (Auckl) 2017;9:63–75. doi: 10.2147/HIV.S120576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Romano M, et al. Evolutionarily conserved heterogeneous nuclear ribonucleoprotein (hnRNP) A/B proteins functionally interact with human and Drosophila TAR DNA-binding protein 43 (TDP-43) J Biol Chem. 2014;289(10):7121–30. doi: 10.1074/jbc.M114.548859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.King OD, Gitler AD, Shorter J. The tip of the iceberg: RNA-binding proteins with prion-like domains in neurodegenerative disease. Brain Res. 2012;1462:61–80. doi: 10.1016/j.brainres.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kapeli K, Martinez FJ, Yeo GW. Genetic mutations in RNA-binding proteins and their roles in ALS. Hum Genet. 2017 doi: 10.1007/s00439-017-1830-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Salapa HE, et al. Contribution of the Degeneration of the Neuro-Axonal Unit to the Pathogenesis of Multiple Sclerosis. Brain Sci. 2017;7(6) doi: 10.3390/brainsci7060069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Douglas JN, et al. Antibodies to the RNA Binding Protein Heterogeneous Nuclear Ribonucleoprotein A1 Colocalize to Stress Granules Resulting in Altered RNA and Protein Levels in a Model of Neurodegeneration in Multiple Sclerosis. J Clin Cell Immunol. 2016;7(2):402. doi: 10.4172/2155-9899.1000402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Tamayo JV, et al. The Drosophila hnRNP F/H Homolog Glorund Uses Two Distinct RNA-Binding Modes to Diversify Target Recognition. Cell Rep. 2017;19(1):150–161. doi: 10.1016/j.celrep.2017.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Sibille N, Bernado P. Structural characterization of intrinsically disordered proteins by the combined use of NMR and SAXS. Biochem Soc Trans. 2012;40(5):955–62. doi: 10.1042/BST20120149. [DOI] [PubMed] [Google Scholar]
- 161.Meier S, Blackledge M, Grzesiek S. Conformational distributions of unfolded polypeptides from novel NMR techniques. J Chem Phys. 2008;128(5):052204. doi: 10.1063/1.2838167. [DOI] [PubMed] [Google Scholar]
- 162.Aznauryan M, et al. Comprehensive structural and dynamical view of an unfolded protein from the combination of single-molecule FRET, NMR, and SAXS. Proc Natl Acad Sci U S A. 2016;113(37):E5389–98. doi: 10.1073/pnas.1607193113. [DOI] [PMC free article] [PubMed] [Google Scholar]


