Abstract
Type 2 taste receptors (T2Rs, TAS2Rs) mediate bitterness perception and are involved in diverse defence mechanisms in extraoral tissues. The thyrocyte-expressed T2Rs control thyroid hormone production, and this regulatory role may be associated with susceptibility to thyroid diseases. This study examined whether the variations in TAS2Rs modify the risk of papillary thyroid carcinoma (PTC) and whether such T2R-related PTC risk is associated with genetically modified thyroid function. We conducted a case-control study with 763 Korean females, including 250 PTC cases. Seventy-three single-nucleotide polymorphisms in 13 TAS2R genes and the pre-diagnosis levels of 4 thyroid-related functional markers [total triiodothyronine (TT3), free thyroxine, thyroid-stimulating hormone and thyroglobulin] were analysed. Individuals with TAS2R3/4 CC haplotype (rs2270009 and rs2234001) were at a lower risk for PTC than those with the remaining haplotypes (odds ratio = 0.59, 95% confidence interval: 0.36–0.97). Furthermore, TT3 levels were significantly reduced for TAS2R3/4 CC haplotype carriers compared with other haplotype carriers (p = 0.005). No other genetic variants exhibited critical associations with the PTC phenotype and biomarkers. In summary, genetic variations in T2R3/4 bitterness receptors may modify the PTC risk, and the genetically modified thyroid hormone level by those variations may be linked with the PTC-T2Rs association.
Introduction
Thyroid centres in the endocrine system regulate human development, homeostasis and metabolism. Therefore, functional perturbation of the thyroid may be associated with subsequent abnormalities in various tissues, including thyroid diseases1–3.
Thyroid cancer is one of the most prevalent malignancies in Koreans4. The incidence rate of thyroid cancer has been rapidly increasing by approximately 25% over the last decade, making it one of the highest rates worldwide5. Although over-diagnosis resulting from advanced detection techniques cannot be dismissed5, some factors are reported to be associated with such thyroid health issues. Alterations in environmental factors, including iodine and calcium intake and radiation exposure6–8, genetic factors, such as single-nucleotide polymorphisms (SNPs) in the regions of FOXE1, NRG1 and NKX2-1, and their combined effects are all thought to contribute to a high risk of thyroid cancer9.
Type 2 taste receptors (T2Rs, TAS2Rs) are a class of G protein-coupled receptors (GPCRs) involved in signal transduction on the cellular membrane, especially in response to bitter-tasting compounds. In humans, TAS2R genes are located on chromosomes 5, 7 and 12 and encode approximately 25 different T2R isoforms. These receptors are expressed in oral tissues, where the bitterness sensing of food- or water-borne compounds begins, and in other extraoral tissues, including those in the respiratory, genitourinary, gastrointestinal and nervous systems10. Universally expressed T2Rs differentiate beneficial or noxious exogenous and endogenous molecules and activate subsequent processes to utilize or eliminate those stimuli11, therefore possibly regulating subsequent metabolism and disease risk12.
Growing evidence suggests that T2Rs play a role in disease aetiology, with genetic variants serving as modifying factors. The association between TAS2R38 diplotype and gastrointestinal cancer has been reported in multiple ethnic groups; however, the precise underlying mechanisms are not fully understood to date13–15. A pathogenic role of the genetic variations in bitterness receptors was also evident in the exterior of alimentary systems. Altered bitterness receptor functions are associated with susceptibility to cavities, asthma, respiratory infection and nephropathy16–18. Studies have also expanded to other isoforms of T2Rs beyond T2R38. The differential expression of T2R4 was observed between cancerous and non-cancerous breast cell line models19. The biochemical and pharmacological consequences resulting from coding variations of TAS2Rs have been major research interests20. Additionally, many agonists responding to T2Rs, including T2R8, T2R10 and T2R14, have been revealed and have shown anticancer effects or associations with anticancer stemness and anti-invasive activity21–23.
Recently, the expression of T2Rs and their mechanisms of action were identified in the thyroid24. Thyrocyte-expressed T2Rs, especially T2R4, T2R10 and T2R43, regulated the production of hormones via thyroid-stimulating hormone (TSH)-dependent intracellular calcium and iodine efflux. One common variant allele of TAS2R42 has also been associated with serum concentrations of free triiodothyronine (T3) and thyroxine (FT4)24. Given these findings, it is possible to conjecture that genetic variations in TAS2Rs modify the risk for thyroid malignancy. Furthermore, the alterations in thyroid function due to such TAS2R genetic variants may be associated with its underlying mechanism of action. However, additional epidemiological evidence is required to verify this hypothesis.
In this study, we examined whether TAS2R genetic variations may modify the susceptibility to thyroid cancer in Koreans. Additionally, we evaluated whether genetically modified thyroid function by TAS2R variations may be associated with such modified risks for thyroid cancer. As biomarkers indicative of thyroid-related function, the following four types of indices were selected and examined: total T3 (TT3); FT4; TSH; and thyroglobulin (Tg). Gender disparities have been observed in thyroid cancer25. Furthermore, each subtype of thyroid cancer shows numerous biologically distinctive features26,27. Therefore, for a more unbiased approach and better interpretation of the findings, the study was conducted in pathologically homogeneous subjects, including female controls and females with papillary thyroid carcinoma (PTC), the most common type of thyroid cancer in Korea. In addition, biomarkers were evaluated using samples obtained prior to diagnosis.
Results
General characteristics of the study subjects
Table 1 presents the descriptive data of subjects analysed in the current study. The cases and controls did not exhibit differences in their analysed anthropometric, life style, socio-economical or reproductive characteristics, with the exception of family history of thyroid cancer. PTC cases were more likely to have a family member with thyroid cancer than controls. Therefore, family history of thyroid cancer was considered a covariate in the subsequent analyses.
Table 1.
General characteristics of the study subjects.
| All subjects (n = 763) | Controls (n = 513) | Cases (n = 250) | Pa | |
|---|---|---|---|---|
| Age (years, mean ± SD) | 48.9 ± 8.48 | 48.9 ± 8.51 | 48.9 ± 8.43 | 0.955 |
| Body mass index (kg/m2) | 0.346 | |||
| <23 | 402 (52.7)b | 278 (54.2) | 124 (49.6) | |
| 23 to <25 | 177 (23.2) | 117 (22.8) | 60 (24.0) | |
| ≥25 | 179 (23.5) | 113 (22.0) | 66 (26.4) | |
| Missing | 5 (0.66) | 5 (0.97) | . | |
| Education level | 0.646 | |||
| Elementary school or less | 60 (7.86) | 39 (7.60) | 21 (8.40) | |
| Middle school | 61 (7.99) | 39 (7.60) | 22 (8.80) | |
| High school | 319 (41.8) | 216 (42.1) | 103 (41.2) | |
| College or more | 267 (34.9) | 189 (36.8) | 78 (31.2) | |
| Missing | 56 (7.34) | 30 (5.85) | 26 (10.4) | |
| Monthly household income (10,000 won) | 0.255 | |||
| <200 | 120 (15.7) | 81 (15.7) | 39 (15.6) | |
| 200 to <400 | 208 (27.3) | 135 (26.3) | 73 (29.2) | |
| 400 to <700 | 217 (28.4) | 160 (31.2) | 57 (22.8) | |
| ≥700 | 81 (10.6) | 55 (10.7) | 26 (10.4) | |
| Missing | 137 (17.9) | 82 (15.9) | 55 (22.0) | |
| Marital status | 0.445 | |||
| Married | 644 (84.4) | 439 (85.6) | 205 (82.0) | |
| Unmarried | 21 (2.75) | 15 (2.92) | 6 (2.40) | |
| Divorced/Widowed | 72 (9.44) | 44 (8.58) | 28 (11.2) | |
| Missing | 26 (3.41) | 15 (2.92) | 11 (4.40) | |
| Alcohol drinking | 0.096 | |||
| Never drinkers | 420 (55.1) | 271 (52.8) | 149 (59.6) | |
| Ever drinkers | 317 (41.6) | 223 (43.5) | 94 (37.6) | |
| Missing | 26 (3.41) | 19 (3.70) | 7 (2.80) | |
| Smoking | 0.789 | |||
| Never smokers | 675 (88.5) | 449 (87.5) | 226 (90.4) | |
| Ever smokers | 69 (9.04) | 47 (9.16) | 22 (8.80) | |
| Missing | 19 (2.49) | 17 (3.31) | 2 (0.80) | |
| Family history of thyroid cancer (first-degree relative) | 0.005 | |||
| No | 719 (94.2) | 491 (95.7) | 228 (91.2) | |
| Yes | 28 (3.67) | 12 (2.34) | 16 (6.40) | |
| Missing | 16 (2.10) | 10 (1.95) | 6 (2.40) | |
| Age at menarche (years) | 0.849 | |||
| ≤13 | 170 (22.3) | 114 (22.2) | 56 (22.4) | |
| 14–15 | 294 (38.5) | 202 (39.4) | 92 (36.8) | |
| ≥16 | 281 (36.8) | 187 (36.5) | 94 (37.6) | |
| Missing | 18 (2.36) | 10 (1.95) | 8 (3.20) | |
| Postmenopausal: Yes | 358 (46.9) | 234 (45.6) | 124 (49.6) | 0.229 |
| Age at menopause (years) | 0.608 | |||
| <46 | 80 (22.4) | 53 (22.7) | 27 (21.8) | |
| 46 to <50 | 122 (34.1) | 83 (35.5) | 39 (31.5) | |
| 50 to <52 | 70 (19.6) | 41 (17.5) | 29 (23.4) | |
| ≥52 | 70 (19.6) | 46 (19.7) | 24 (19.4) | |
| Missing | 16 (4.47) | 11 (4.70) | 5 (4.03) | |
| Postmenopausal hormone use (ever) | 0.995 | |||
| Yes | 116 (32.4) | 76 (32.5) | 40 (32.3) | |
| No | 209 (58.4) | 137 (58.6) | 72 (58.1) | |
| Missing | 33 (9.22) | 21 (8.97) | 12 (9.68) | |
| Parity | 0.534 | |||
| Yes | 676 (88.6) | 455 (88.7) | 221 (88.4) | |
| No | 46 (6.03) | 33 (6.43) | 13 (5.20) | |
| Missing | 41 (5.37) | 25 (4.87) | 16 (6.40) |
ap-values for age from Student’s t-test, otherwise from chi-squared tests. bNumbers in brackets represent percentages. SD, standard deviation.
Genotype/diplotype distribution and the association with PTC
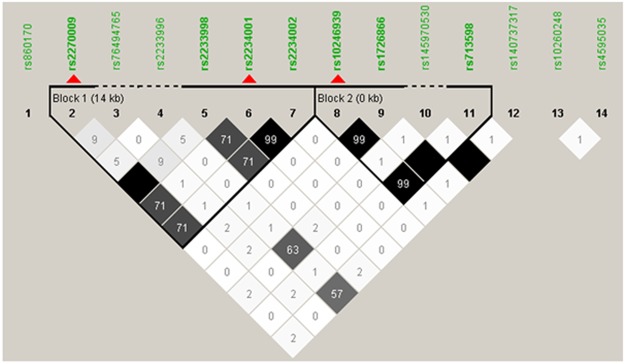
A total of 73 SNPs in 13 TAS2R genes on chromosomes 7 and 12 were obtained as a result of genotyping and imputation (see Supplementary Table S1 for the full list of SNPs). Haplotype-based analysis could provide more comprehensive information for the individual’s genetic background than multiple individual-locus tests. Therefore, we assessed linkage disequilibrium (LD) patterns and tagging loci among the genetic variations for the subsequent analyses. According to Haploview28, two LD blocks were evident across 14 SNPs in 6 TAS2Rs on chromosome 7 (Fig. 1). Two tagging SNPs in TAS2R3 (rs2270009) and TAS2R4 (rs2234001) in the first LD block and 1 tagging locus in TAS2R38 (rs10246939) in the second LD block were identified. In chromosome 12, a single LD block existed, with 2 taggers (rs10772397 and rs1868769) in 2 TAS2R genes (TAS2R50 and TAS2R48) (Supplementary Fig. S1). These tagging SNPs in each LD block were applied to compute the diplotype for the subsequent analyses, with the exception of the TAS2R38 variants. However, the TAS2R38 diplotype consists of three loci (rs10246939, rs1726866 and rs713598) that have been known to well-describe the functional protein changes, such as differential bitterness intensity29. Therefore, the diplotype for TAS2R38 was computed using those three SNPs. Finally, 2 loci of LD block 1 (TAS2R3/4) and 3 loci of LD block 2 (TAS2R38) in chromosome 7 and 2 loci of LD block in chromosome 12 (TAS2R50/48) were applied for diplotype computation (Table 2).
Figure 1.
Linkage disequilibrium patterns and tagging single-nucleotide polymorphisms of TAS2Rs in chromosome 7. Red triangles denote the tagging loci. The numbers in the squares are r2 values (×100) between the loci, with darker shades indicating greater r2 values.
Table 2.
Descriptive data for selected/tagging single-nucleotide polymorphisms analysed in the current study.
| Chr | Associated/ nearest gene | SNP | Obs HET | Pred HET | HW p-val | MAF | Allelesa | SNP type | LD block | Tagging SNP |
|---|---|---|---|---|---|---|---|---|---|---|
| 7 | TAS2R3 | rs2270009 | 0.43 | 0.44 | 0.824 | 0.32 | T:C | synonymous | 1 | Ch7B1_1 |
| TAS2R4 | rs2234001 | 0.36 | 0.38 | 0.273 | 0.26 | C:G | Val96Leu | 1 | Ch7B1_2 | |
| TAS2R38 | rs10246939 | 0.47 | 0.49 | 0.418 | 0.42 | C:T | Ile296Val | Ch7B2_1 | ||
| rs1726866 | 0.47 | 0.49 | 0.455 | 0.43 | G:A | Val262Ala | 2 | . | ||
| rs713598 | 0.47 | 0.49 | 0.455 | 0.43 | G:C | Ala49Pro | . | |||
| 12 | TAS2R50 | rs10772397 | 0.34 | 0.34 | 0.818 | 0.22 | T:C | synonymous | 1 | Ch12_1 |
| TAS2R48 | rs1868769 | 0.15 | 0.15 | 0.913 | 0.08 | A:G | synonymous | 1 | Ch12_2 |
Chr, chromosome; SNP, single-nucleotide polymorphism; Obs HET, observed heterozygosity; Pred HET, predicted heterozygosity; HW p-val, Hardy-Weinberg equilibrium test p-value; MAF, minor allele frequency; LD, linkage disequilibrium. aMajor; minor allele.
As a result of diplotype analyses, 6 and 4 types of diplotype were computed to exist in TAS2R3/4 and TAS2R38 LD blocks, respectively. Nine differential diplotypes were also present in the LD block of chromosome 12. The logistic regression models were only established with diplotypes with 3% or more of subjects to prevent false-positive findings due to a small number of subjects (Table 3). The findings suggested that subjects with the CC/TC TAS2R3/4 diplotype were less likely to have PTC than those with TC/TC, the most frequent diplotype [odds ratio (OR) = 0.43, 95% confidence interval (95% CI): 0.23–0.79]. Furthermore, when the subjects were stratified based on the presence of CC haplotype, the CC haplotype reduced the risk for PTC compared with that of those lacking the CC haplotype (OR = 0.59, 95% CI: 0.36–0.97). However, other TAS2R diplotypes were not significantly associated with PTC susceptibility. Finally, the prognostic ability of TAS2R genetic variation in disease recurrence was analysed using the PTC stage and AGES (age, grade, extent of disease, size), MACIS (distant metastasis, age, complete surgical resection, invasion, size) and AMES (age, metastasis, extent of disease, size) criteria. However, those genetic variations did not influence the severity of PTC (Supplementary Tables S2–4).
Table 3.
Distribution of TAS2R genetic variants and their association with risk for papillary thyroid carcinoma.
| Controls (%) | Case (%) | Odds ratio (95% CI)a | P | |
|---|---|---|---|---|
| TAS2R3/4 diplotype | ||||
| TC/TC | 229 (44.6) | 121 (48.4) | 1.00 (Reference) | |
| TC/CG | 170 (33.1) | 84 (33.6) | 0.89 (0.64–1.27) | 0.549 |
| CC/TC | 63 (12.3) | 14 (5.60) | 0.43 (0.23–0.79) | 0.007 |
| CG/CG | 35 (6.82) | 21 (8.40) | 1.07 (0.59–1.94) | 0.822 |
| CC/CG | 14 (2.73) | 10 (4.00) | — | |
| CC/CC | 2 (0.39) | — | — | |
| */* | 434 (84.6) | 226 (90.4) | 1.00 (Reference) | |
| CC/* | 49 (15.4) | 24 (9.60) | 0.59 (0.36–0.97) | 0.036 |
| TAS2R38 diplotype | ||||
| PAV/PAV | 171 (33.3) | 87 (34.8) | 1.00 (Reference) | |
| PAV/AVI | 252 (49.1) | 109 (43.6) | 0.82 (0.58–1.17) | 0.273 |
| AVI/AVI | 89 (17.4) | 54 (21.6) | 1.26 (0.82–1.95) | 0.289 |
| AVI/AAV | 1 (0.19) | — | ||
| TAS2R diplotype in chromosome 12 b | ||||
| TA/TA | 264 (51.7) | 128 (52.0) | 1.00 (Reference) | |
| CA/TA | 148 (28.9) | 69 (28.1) | 0.97 (0.68–1.41) | 0.938 |
| TA/TG | 48 (9.39) | 25 (10.2) | 1.09 (0.64–1.85) | 0.761 |
| CA/TG | 26 (5.09) | 10 (4.07) | 0.87 (0.40–1.89) | 0.733 |
| CA/CA | 21 (4.11) | 10 (4.07) | 0.88 (0.39–1.98) | 0.751 |
| CA/CG | 3 (0.59) | 1 (0.41) | — | — |
| CG/TG | 1 (0.20) | 1 (0.41) | — | — |
| CG/CG | — | 1 (0.41) | — | — |
| TG/TG | — | 1 (0.41) | — | — |
95% CI, 95% confidence interval. Subjects with diplotypes with a frequency below 3% were excluded from the logistic regression tests based on rarity. aThe odds ratio was adjusted for the family history of thyroid cancer. bSix individuals were excluded due to missing genotype data.
Associations between TAS2R diplotype and biomarkers of thyroid function
No significant differences in the concentrations of TT3, FT4, TSH and Tg were noted between PTC phenotypes (Supplementary Table S5). However, when those biomarkers were analysed with TAS2R genetic characteristics, the TAS2R3/4 diplotype exhibited a distinctive difference in TT3 concentration (Table 4). In the entire study population, the TT3 level for individuals with the CC/TC diplotype was lower than that for individuals with other diplotypes (p = 0.05). This trend towards TT3 and genotype was more clearly evident when the subjects were re-grouped, taking account of the presence of the CC haplotype (CC/*): CC haplotype carriers exhibited significantly lower levels of TT3 than non-carriers (1.04 ± 0.03 and 1.16 ± 0.01, p = 0.005). Further analyses taking account of the presence of the PTC phenotype also supported the association of the CC haplotype and TT3. In both the control and the PTC groups, the level of TT3 for CC haplotype carriers was lower compared with that of CC haplotype non-carriers (1.02 ± 0.04 and 1.14 ± 0.02, p = 0.022), although the difference in PTC cases was not statistically significant (p = 0.111). None of the genetic groups for the TAS2R38 and TAS2R50/48 diplotype exhibited meaningful associations with biomarkers related to thyroid function in all subjects as well as in cases and controls, respectively (Tables 5 and 6).
Table 4.
Levels of biomarkers of thyroid function, taking TAS2R3/4 diplotype and papillary thyroid carcinoma phenotype into account.
| TT3 (ng/mL) | FT4 (ng/dL) | TSH (μIU/mL) | Tg (ng/mL) | |||||
|---|---|---|---|---|---|---|---|---|
| N | Mean (SE) | N | Mean (SE) | N | Mean (SE) | N | Mean (SE) | |
| All subjects | ||||||||
| TC/TC | 95 | 1.13 (0.02) | 95 | 1.31 (0.02) | 95 | 2.13 (0.13) | 55 | 24.98 (7.36) |
| TC/CG | 77 | 1.17 (0.02) | 77 | 1.30 (0.02) | 77 | 2.81 (0.28) | 47 | 14.92 (2.97) |
| CC/TC | 17 | 1.04 (0.03) | 17 | 1.29 (0.05) | 17 | 2.30 (0.29) | 7 | 20.50 (3.75) |
| CG/CG | 15 | 1.19 (0.05) | 15 | 1.26 (0.04) | 15 | 2.69 (0.63) | 8 | 17.50 (3.66) |
| CC/CG | 6 | 1.14 (0.05) | 6 | 1.26 (0.09) | 6 | 2.02 (0.68) | 1 | 4.75 |
| CC/CC | 1 | 0.70 | 1 | 1.47 | 1 | 1.14 | — | — |
| Pa | 0.05 | 0.613 | 0.652 | 0.24 | ||||
| */* | 187 | 1.16 (0.01) | 187 | 1.30 (0.01) | 187 | 2.45 (0.14) | 110 | 20.14 (3.91) |
| CC/** | 23 | 1.04 (0.03) | 24 | 1.29 (0.04) | 24 | 2.18 (0.26) | 8 | 18.53 (3.80) |
| Pb | 0.005 | 0.803 | 0.536 | 0.364 | ||||
| Controls | ||||||||
| TC/TC | 50 | 1.12 (0.02) | 50 | 1.28 (0.03) | 50 | 2.40 (0.20) | 25 | 19.01 (4.39) |
| TC/CG | 40 | 1.17 (0.03) | 40 | 1.29 (0.02) | 40 | 2.59 (0.33) | 25 | 12.19 (2.04) |
| CC/TC | 8 | 1.03 (0.04) | 8 | 1.28 (0.06) | 8 | 2.88 (0.44) | 4 | 25.47 (3.84) |
| CG/CG | 8 | 1.15 (0.08) | 8 | 1.18 (0.07) | 8 | 3.65 (1.16) | 4 | 23.33 (3.66) |
| CC/CG | 3 | 1.10 (0.06) | 3 | 1.21 (0.10) | 3 | 1.80 (0.79) | 1 | 4.75 |
| CC/CC | 1 | 0.70 | 1 | 1.47 | 1 | 1.14 | — | — |
| Pa | 0.159 | 0.327 | 0.496 | 0.084 | ||||
| */* | 98 | 1.14 (0.02) | 98 | 1.27 (0.02) | 98 | 2.57 (0.19) | 55 | 16.30 (2.26) |
| CC/** | 12 | 1.02 (0.04) | 12 | 1.28 (0.05) | 12 | 2.47 (0.38) | 5 | 21.32 (5.10) |
| Pb | 0.022 | 0.996 | 0.974 | 0.317 | ||||
| Cases | ||||||||
| TC/TC | 45 | 1.15 (0.03) | 45 | 1.34 (0.03) | 45 | 1.84 (0.15) | 30 | 29.95 (13.02) |
| TC/CG | 37 | 1.18 (0.03) | 37 | 1.31 (0.03) | 37 | 3.04 (0.45) | 22 | 18.04 (5.93) |
| CC/TC | 9 | 1.04 (0.06) | 9 | 1.30 (0.07) | 9 | 1.78 (0.32) | 3 | 13.87 (5.51) |
| CG/CG | 7 | 1.24 (0.07) | 7 | 1.31 (0.05) | 7 | 1.73 (0.21) | 3 | 7.77 (2.13) |
| CC/CG | 3 | 1.20 (0.10) | 3 | 1.34 (0.20) | 3 | 2.24 (1.28) | — | — |
| Pa | 0.159 | 0.651 | 0.179 | 0.408 | ||||
| */* | 89 | 1.17 (0.02) | 89 | 1.32 (0.02) | 89 | 2.33 (0.21) | 55 | 23.98 (7.49) |
| CC/** | 11 | 1.07 (0.25) | 12 | 1.30 (0.06) | 12 | 1.90 (0.36) | 3 | 13.87 (5.51) |
| Pb | 0.111 | 0.739 | 0.415 | 0.933 | ||||
TT3, total triiodothyronine; FT4, free thyroxine; TSH, thyroid-stimulating hormone; Tg, thyroglobulin, analysed only for anti-thyroglobulin antibody-negative subjects; N, number of subjects; SE, standard error. aP-values come from the generalized linear models adjusted for family history of thyroid cancer (diplotype groups with fewer than 5 subjects were excluded from the comparison because of the rarity). bP-values come from Student’s t-tests between diplotype groups (diplotypes with the CC haplotype versus all other diplotypes).
Table 5.
Levels of biomarkers of thyroid function, taking TAS2R38 diplotype and papillary thyroid carcinoma phenotype into account.
| TT3 (ng/mL) | FT4 (ng/dL) | TSH (μIU/mL) | Tg (ng/mL) | |||||
|---|---|---|---|---|---|---|---|---|
| N | Mean (SE) | N | Mean (SE) | N | Mean (SE) | N | Mean (SE) | |
| All subjects | ||||||||
| PAV/PAV | 78 | 1.14 (0.02) | 78 | 1.29 (0.02) | 78 | 2.56 (0.23) | 52 | 16.13 (2.71) |
| PAV/AVI | 102 | 1.16 (0.02) | 102 | 1.29 (0.02) | 102 | 2.37 (0.18) | 48 | 27.30 (8.36) |
| AVI/AVI | 31 | 1.10 (0.03) | 31 | 1.32 (0.04) | 31 | 2.22 (0.30) | 18 | 11.90 (2.74) |
| Pa | 0.441 | 0.795 | 0.658 | 0.181 | ||||
| Controls | ||||||||
| PAV/PAV | 36 | 1.12 (0.03) | 36 | 1.27 (0.02) | 36 | 2.52 (0.23) | 23 | 17.32 (4.75) |
| PAV/AVI | 59 | 1.14 (0.02) | 59 | 1.27 (0.02) | 59 | 2.67 (0.27) | 25 | 17.70 (1.94) |
| AVI/AVI | 15 | 1.09 (0.04) | 15 | 1.31 (0.05) | 15 | 2.17 (0.46) | 12 | 13.52 (3.88) |
| Pa | 0.682 | 0.593 | 0.816 | 0.263 | ||||
| Cases | ||||||||
| PAV/PAV | 42 | 1.16 (0.03) | 42 | 1.31 (0.03) | 42 | 2.60 (0.39) | 29 | 15.18 (3.14) |
| PAV/AVI | 43 | 1.18 (0.03) | 43 | 1.33 (0.03) | 43 | 1.97 (0.19) | 23 | 37.74 (17.26) |
| AVI/AVI | 16 | 1.12 (0.05) | 16 | 1.32 (0.05) | 16 | 2.27 (0.40) | 6 | 8.67 (2.70) |
| Pa | 0.548 | 0.744 | 0.356 | 0.452 | ||||
TT3, total triiodothyronine; FT4, free thyroxine; TSH, thyroid-stimulating hormone; Tg, thyroglobulin, analysed only for anti-thyroglobulin antibody-negative subjects; N, number of subjects; SE, standard error. aP-values come from the generalized linear models adjusted for family history of thyroid cancer.
Table 6.
Levels of biomarkers of thyroid function, taking TAS2R50/48 diplotype on chromosome 12 and papillary thyroid carcinoma phenotype into account.
| TT3 (ng/mL) | FT4 (ng/dL) | TSH (μIU/mL) | Tg (ng/mL) | |||||
|---|---|---|---|---|---|---|---|---|
| N | Mean (SE) | N | Mean (SE) | N | Mean (SE) | N | Mean (SE) | |
| All subjects | ||||||||
| TA/TA | 106 | 1.13 (0.02) | 106 | 1.30 (0.02) | 106 | 2.38 (0.15) | 59 | 21.97 (6.85) |
| CA/TA | 60 | 1.14 (0.03) | 60 | 1.31 (0.02) | 60 | 2.55 (0.32) | 36 | 20.21 (3.94) |
| TA/TG | 27 | 1.19 (0.04) | 27 | 1.29 (0.03) | 27 | 1.96 (0.26) | 11 | 12.91 (4.07) |
| CA/TG | 11 | 1.12 (0.04) | 11 | 1.29 (0.04) | 11 | 2.52 (0.61) | 8 | 17.25 (3.11) |
| CA/CA | 5 | 1.12 (0.06) | 5 | 1.25 (0.04) | 5 | 2.94 (0.64) | 4 | 14.93 (6.01) |
| CA/CG | 1 | 1.10 | 1 | 1.08 | 1 | 6.59 | — | — |
| CG/TG | 1 | 1.10 | 1 | 1.21 | 1 | 4.45 | — | — |
| Pa | 0.723 | 0.977 | 0.577 | 0.477 | ||||
| Controls | ||||||||
| TA/TA | 52 | 1.12 (0.02) | 52 | 1.26 (0.02) | 52 | 2.49 (0.23) | 29 | 15.22 (1.72) |
| CA/TA | 32 | 1.14 (0.03) | 32 | 1.29 (0.03) | 32 | 2.65 (0.36) | 18 | 20.40 (6.09) |
| TA/TG | 15 | 1.13 (0.05) | 15 | 1.30 (0.04) | 15 | 1.97 (0.41) | 5 | 15.75 (8.25) |
| CA/TG | 8 | 1.14 (0.04) | 8 | 1.26 (0.05) | 8 | 3.21 (0.69) | 6 | 14.89 (3.22) |
| CA/CA | 2 | 1.15 (0.15) | 2 | 1.29 (0.01) | 2 | 2.53 (1.44) | 2 | 13.25 (5.12) |
| CA/CG | 1 | 1.10 | 1 | 1.08 | 1 | 6.59 | — | — |
| Pa | 0.899 | 0.751 | 0.179 | 0.809 | ||||
| Cases | ||||||||
| TA/TA | 54 | 1.15 (0.02) | 54 | 1.33 (0.02) | 54 | 2.27 (0.19) | 30 | 28.50 (13.4) |
| CA/TA | 28 | 1.15 (0.04) | 28 | 1.34 (0.04) | 28 | 2.42 (0.56) | 18 | 20.01 (5.18) |
| TA/TG | 12 | 1.27 (0.06) | 12 | 1.27 (0.05) | 12 | 1.95 (0.31) | 6 | 10.55 (3.61) |
| CA/TG | 3 | 1.07 (0.07) | 3 | 1.37 (0.06) | 3 | 0.70 (0.23) | 2 | 24.34 (7.00) |
| CA/CA | 3 | 1.10 (0.06) | 3 | 1.21 (0.06) | 3 | 3.22 (0.76) | 2 | 16.60 (13.6) |
| CG/TG | 1 | 1.10 | 1 | 1.21 | 1 | 4.45 | — | — |
| Pa | 0.133 | 0.629 | 0.599 | 0.758 | ||||
TT3, total triiodothyronine; FT4, free thyroxine; TSH, thyroid-stimulating hormone; Tg, thyroglobulin, analysed only for anti-thyroglobulin antibody-negative subjects; N, number of subjects; SE, standard error. aP-values come from the generalized linear models adjusted for family history of thyroid cancer (diplotype groups with fewer than 5 subjects were excluded from the comparison because of rarity).
Discussion
This study hypothesized that T2R bitterness receptors may modify susceptibility to PTC and that the T2R-PTC association may be associated with genetically mediated thyroid function. Here, the current findings supported this idea: variations in the TAS2R3 and TAS2R4 genes reduced the risk of PTC, and the differential concentration of TT3 modified by those TAS2R genetic variations may be associated with the potential underlying mechanism of PTC development and progression.
Earlier studies of T2Rs mainly focused on their role in bitterness sensing. However, the presence of extra-orally expressed T2R has been reported and suggests that T2Rs play differential roles beyond bitterness perception that are tailored to various locations30,31. To date, growing evidence suggests that T2R taste receptors possess regulatory effects in human metabolism and disease. The physiological roles of those taste receptors, including innate immunity, secretion, contraction and relaxation of smooth muscle cells, have been conjectured to explain such pathological roles of T2R in disease aetiology31,32; however, most of the mechanisms were only presumptive. A study by Clark et al. confirmed the expression of T2R on thyrocytes and their mechanism of action24. The study verified that T2R agonists inhibited the intracellular levels of calcium and iodine ions, which regulate the production of thyroid hormones, only in the presence of TSH. This finding could provide evidence regarding how T2Rs and their genetic variations influence energy metabolism and body composition. Furthermore, these findings may also imply a defence mechanism employed by the thyroid against noxious molecules24. Many phytotoxins and harmful compounds possess bitterness characteristics23,33. The chemosensing of pernicious bitterness ex-/endogenous molecules in the thyroid may trigger the protective mechanism via functional alterations of thyrocytes, such as the inhibition of hormone production24. Consistent with this finding, our results also suggest that the variant T2R proteins might lead to differential efficacy in chemosensing and subsequent metabolic alterations, hence reducing the risk for PTC.
The effect-modifying haplotype TAS2R3/4 CC consists of two genetic variations: the minor allele C of rs2270009 in TAS2R3 and the major allele C of rs2234001 in TAS2R4. TAS2R3 and TAS2R4 reside closely on chromosome 7. These receptors respond to multiple bitterness substances, including chloroquine, propylthiouracil and denatonium benzoate23. Notably, the C to T change at TAS2R3 rs2270009 is a silent change; this sequence alteration does not lead to an amino acid alteration. However, numerous studies have suggested that synonymous changes lead to subsequent alterations in mRNA splicing and protein folding, ultimately modifying enzyme function34,35. Expression quantitative trait loci database analyses also provide supportive data suggesting that TAS2R3 rs2270009 altered 5 transcription factor binding motifs (E2F, NF-Y, Pax-4, CEBPD, and Pbx-1) and caused decisive changes in the expression of other genes in thyroid tissue (Supplementary Tables S6 and S7)36,37. In contrast, although molecular modelling studies have been revealing the structure-functional influence of genetic variations in TAS2R4, thus far, little is known for V96L, rs223400120. As epidemiological evidence, current findings suggest that the alterations in the secondary structure and stability of T2R3 with the concomitant expression of rs2234001 C–T2R4 may cause altered ligand sensing, mediating the protective effect against PTC and differential TT3 levels. The potential changes in the expression of other genes, including WEE2 and WEE2-AS1, linked with these genetic variations may also contribute to such alteration of the maturation and/or function of thyrocytes and, further, the risk for PTC (Supplementary Table S7)36,38. Additionally, the expression of T2R38 on thyrocytes was evident24, but TAS2R38 taste receptor genetic variation was not associated with any thyroid-related variables examined, the risk for PTC or biomarkers. Although TAS2R38 is the major target of taste receptor sequence variation studies, controversies exist. Studies have reported that TAS2R38 variations showed minimal effects on food intake13,15 or disease susceptibility39–41. Furthermore, other T2R proteins were observed to play tissue-specific physiological roles, and the biochemical and pharmacological characteristics of their variants for responding molecules have been verified20. For better understanding and health/therapeutic applications of bitterness chemosensing protein, more comprehensive approaches are required, including diverse T2Rs and genetic variants; current findings could be referenced in future structure-function studies.
To obtain more precise evidence regarding whether the association between TAS2R genetic variations and reduced PTC risk is associated with thyroid functional changes, we examined the levels of four thyroid function-related markers. The analyses showed that the TAS2R3/4 CC haplotype was associated with TT3 concentration. Overall, CC haplotype carriers, who were at the lower risk for PTC, exhibited lower TT3 concentrations than non-carriers. This trend of TT3 concentration and the haplotype were also retained when PTC phenotype was taken into account. In each phenotype group, CC haplotype carriers showed lower levels of TT3 than non-carriers. Furthermore, the TT3 levels of CC haplotype carriers without PTC were the lowest, while those of non-carrier PTC cases tended to be highest. Although controversial, studies have reported that the T3 level is a prediction marker or risk-modifying factor in disease aetiology. Elevated TT3 levels are positively associated with the risk for breast and renal cancer42–44. In the current study, TT3 levels in PTC cases also tended to be higher than controls, although the difference was not statistically significant (Supplementary Table S5). The precise mechanism for such T3-induced cancer risk has not yet been clearly demonstrated. However, one study suggested a new molecular mechanism45: T3 enhanced TRb1/Oct-1-mediated cyclin D1 transcription, which further promoted PTC cell proliferation. Given these findings, here, we carefully describe a potential association with variant T2R proteins, thyroid function and PTC susceptibility. The structural alteration of the TAS2R3/4 CC haplotype could lead to a change in agonist sensing of T2R3 and T2R4 receptors. The altered efficacy of molecular sensing of variant receptors may regulate the function of thyrocytes and their reduced activity and/or T3 thyroid hormone levels, which may act protectively against potential PTC carcinogenic mechanisms. This hypothesis regarding the association between TAS2R3/4 variation and TT3 levels may explain the findings of differential risk for PTC based on TAS2R3/4 genotype, at least in part.
T3 and T4 are both produced in thyrocytes. T3 is biologically active, but the direct production of T3 in the thyroid is only responsible for 20% of T3; the remaining 80% is produced by conversion from T446,47. In the study of Clark et al. in an Amish population24, TAS2R42 rs5020531 regulated free T3 and T4 levels, and FT4 exhibited a greater association with that genetic factor. However, in these Korean females, the TAS2R diplotype, including TAS2R42, did not exhibit a distinctive influence on examined variables (the analyses with rs5020531 did not reveal significant effects on TT3 and FT4 levels; data not shown). The TAS2R3/4 diplotype exclusively exhibited an association with TT3 concentration but not FT4. Some hypotheses could be suggested to explain the discrepancies between these studies and gene-hormone associations. First, the Amish are a Caucasian and rural-living population that possess significant differences in genetic and milieu factors from Korean females48. Such differential ethnic characteristics could contribute to the differential distribution of LD patterns and tagging SNPs and, ultimately, the genetic variation-hormone association. Second, the differential study design (case-control versus cohort study) may have resulted in discordance between findings. Finally, unlike T2R42, T2R3/4 proteins may regulate only the T3 produced directly in thyrocytes. The thyrocyte-expressed T2R3/4 proteins may selectively affect the production and/or transportation of T3 across the follicular lumen, cell and endothelium and its release into the blood. Additionally, T2R3/4 proteins may also be involved in the conversion of T4 into T3 in each target tissue. T4 bound to plasma protein is transported to the target organ and converted into T3 by deiodinase enzymes. Tissue-specific T2R3/4 bitterness receptors may be related with such a deiodination process, hence leading to differences in TT3 concentrations. However, the TT3 concentration could be modified due to various reasons. The TT3 level is also essentially regulated by the negative feedback mechanism. Lower T3 and T4 levels initiate the production and release of TSH, therefore maintaining the T3 and T4 concentrations. However, in the current study, TAS2R3/4 genetic variations did not modify the level of TSH, only influencing the TT3 concentration. We could not dismiss the potential that TAS2R3/4 genetic variation may be associated with the activity or function of T3 rather than its expression level, but the evidence is limited. The present analyses of hormones and TAS2R variants were only performed in relatively small numbers of subjects, and the findings and hypotheses should be confirmed in a large cohort study and will require mechanistic evidence from experimental models.
The present study provides preliminary epidemiological evidence that genetic variations in TAS2Rs are associated with PTC risk and thyroid function. The subjects with homogeneous characteristics and the analyses of pre-diagnostic levels of biomarkers are the strengths of the study; yet, the work may harbour limitations. First, the study was performed in a relatively small population. The findings have not yet been confirmed in secondary or experimental studies, and the potential effects of multiple statistical tests were not adjusted. Second, the LD patterns, tagging SNPs and study findings could vary depending on the study population. Analyses with additional genetic data and/or in other ethnicities may lead to differential results compared with the current findings. Third, due to limited resources, the measurements of thyroid function and autoimmunity markers were performed only in a selected group of 110 cases and 101 controls. These subjects were only chosen based on the availability of pre-diagnostic serum samples, and unknown bias may exist. Lastly, study subjects were recruited from the Cancer Screening Cohort Study (CSCS), a major cancer study cohort in Korea that has a relatively short history. The mean follow-up time for those PTC cases with pre-diagnostic samples was 3.15 ± 2.79 years; therefore, the biomarkers may lack power as prognostic indicators. For these reasons, the study’s findings should be interpreted with caution.
In conclusion, genetic variations in bitterness receptor TA2Rs may modify susceptibility to PTC in Korean females, and genetic variation-mediated thyroid function may be associated with PTC carcinogenesis. Given the metabolic and therapeutic importance of thyroid and T2R GPCR, the current findings add knowledge regarding chemosensing mechanisms and disease treatment.
Methods
Study population and data collection
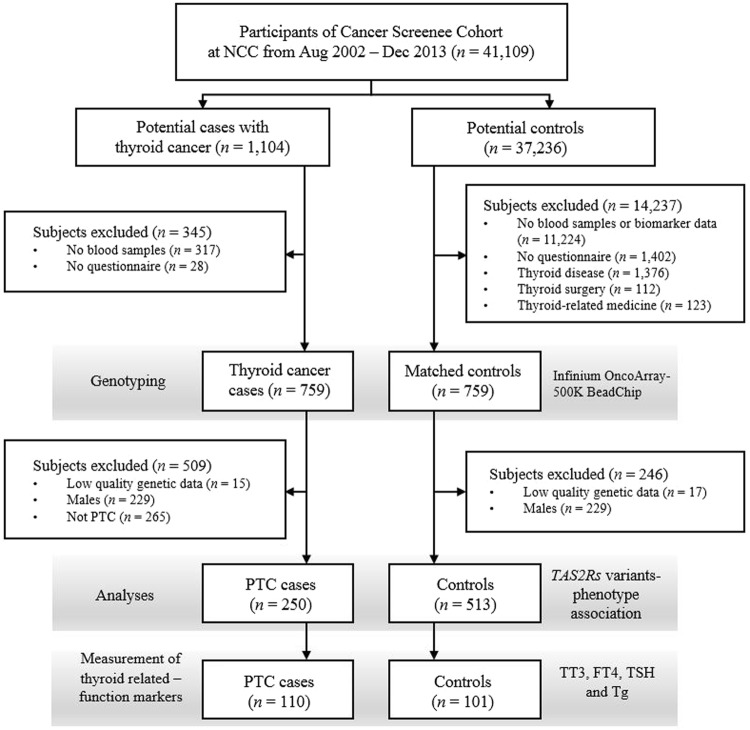
The subjects of this case-control study were recruited from among individuals who visited the National Cancer Center, Korea, between August 2002 and December 2013. The details of the study population have been described previously (Fig. 2)49,50. Briefly, a total of 41,109 volunteers older than 30 years of age who underwent health-screening examinations (a benefit programme of the National Health Insurance) were participants in an ongoing CSCS. At the baseline evaluation, those individuals were requested to complete a self-administered questionnaire with information on socio-demographic characteristics, personal and family medical histories and reproductive and lifestyle factors. Blood samples were also collected at this point. Using the linkage with the Korea Central Cancer Registry database (ICD10 code C73), 1,104 patients from the CSCS were identified as having thyroid cancer, and 37,236 were defined as potential controls. Among these potential cases and controls, 759 thyroid cases and 759 age- (CSCS entry) and sex-matched controls were genotyped. Then, 250 female PTC cases defined by endocrinologists and 513 female controls were subjected to analyses of the association between TAS2R variations and phenotype. The prognostic effect of genetic variations was also estimated using the carcinoma stage and AGES, MACIS and AMES classification systems. The PTC stage and risk prediction score/group were defined following the American Thyroid Association classification system (7th edition)51. Among those subjects, the levels of four thyroid-related functional markers were determined in 110 PTC cases and 101 controls from whom we were able to obtain pre-diagnostic serum samples.
Figure 2.
A simplified flowchart of the current study. NCC, National Cancer Center; PTC, papillary thyroid carcinoma; TT3, total triiodothyronine; FT4, thyroxine; TSH, thyroid-stimulating hormone; Tg, thyroglobulin.
Measurement of biomarkers
The serum samples collected at the baseline evaluation were stored at −196 °C until analyses. Electrochemiluminescence immunoassays (ELCLIA; Molecular Analytics E170, Roche kit, Roche, Mannheim, Germany) were applied for the measurements. The techniques were detailed previously1 and reference range for the biomarkers are presented in Supplementary Table S5.
Genetic data production and analyses
Genomic DNA was extracted using the following commercial kit and instrument: MagAttract DNA Blood M48 Kit (Qiagen, Hilden, Germany) and BioRobot M48 automatic extraction equipment (Qiagen, Hilden, Germany). Genomic DNA samples were stored at −80 °C until the analyses. TAS2R genotypes were determined using 500 ng of genomic DNA and an Infinium OncoArray-500K BeadChip (Illumina Inc., CA, USA). For quality control (QC) of the produced genotypes, loci exhibiting call rates <95%, minor allele frequencies <0.01 and deviation from Hardy-Weinberg equilibrium (p < 1 × 10−6) were eliminated. Strand alignment and phasing were performed using PLINK v1.0752 and SHAPEIT253. Thereafter, imputation was performed with IMPUTE2 software54, referencing the 1000 Genomes EAS Phase III reference panel (integrated variant set release in NCBI build 37, hg19). For QC of the imputed results, only the variants with info scores >0.6 were included in subsequent evaluations. The haplotype and tagging SNPs were evaluated using Haploview28, and the diplotype was computed with FAMHAP55. The functional alterations of T2R variant proteins were estimated using HaploReg v4.137.
Statistical analyses
To compare the general characteristics of the study subjects depending on PTC phenotype, chi-squared and Student’s t-tests were used. ORs and 95% CIs were estimated to predict the association between TAS2R genetic variation and PTC risk using logistic regression models. Analysis of variance (ANOVA) was performed to assess the differences in biomarkers between PTC phenotypes or TAS2R diplotypes. The chi-squared tests and ANOVA were also applied to test the association between TAS2R genetic variation and PTC stage as well as various risk prediction criteria and groups. Those logistic regression and ANOVA models were established in the presence of covariates. For post hoc analyses, Tukey’s test was used. All continuous variables were applied in the statistical model after log-transformation. All statistical analyses were conducted using SAS 9.3 software (SAS Institute Inc., Cary, NC, USA), and a two-sided p-value < 0.05 was considered significant.
Ethics statement
Ethical approval for this study was obtained from the Institutional Review Board of the NCC (#NCC2016-0088). All actual study procedures were conducted following the approved protocol, and informed consent was obtained from participants prior to study commencement.
Electronic supplementary material
Acknowledgements
This study was supported by the Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Science, ICT and Future Planning (NRF-2015R1C1A1A02036717, NRF-2015R1D1A1A09058684 and NRF-2018R1A1A1A05019155) and the National Cancer Center (NCC1510040).
Author Contributions
Conception and design: Jeong-Hwa Choi. Data acquisition: Eun Kyung Lee, Yul Hwangbo and Jeongseon Kim. Data analysis and interpretation: Jeong-Hwa Choi and Sarah Yang. Project administration and constructing databases: Jeonghee Lee. Drafting of the article: Jeong-Hwa Choi. Critical revision, overall content and guarantor: Jeongseon Kim.
Statement of Data Availability
All datasets generated during the present study are available from the corresponding author on reasonable request.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-33338-6.
References
- 1.Cho YA, et al. Biomarkers of thyroid function and autoimmunity for predicting high-risk groups of thyroid cancer: a nested case-control study. BMC Cancer. 2014;14:873. doi: 10.1186/1471-2407-14-873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moeller LC, Fuhrer D. Thyroid hormone, thyroid hormone receptors, and cancer: a clinical perspective. Endocr. Relat. Cancer. 2013;20:R19–29. doi: 10.1530/ERC-12-0219. [DOI] [PubMed] [Google Scholar]
- 3.Haymart MR, et al. Higher serum thyroid stimulating hormone level in thyroid nodule patients is associated with greater risks of differentiated thyroid cancer and advanced tumor stage. J. Clin. Endocrinol. Metab. 2008;93:809–814. doi: 10.1210/jc.2007-2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jung KW, Won YJ, Kong HJ, Lee ES. Cancer statistics in Korea: incidence, mortality, survival, and prevalence in 2015. Cancer Res. Treat. 2018;50:303–316. doi: 10.4143/crt.2018.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee JH, Shin SW. Overdiagnosis and screening for thyroid cancer in Korea. Lancet. 2014;384:1848. doi: 10.1016/S0140-6736(14)62242-X. [DOI] [PubMed] [Google Scholar]
- 6.Cho YA, Lee J, Kim J. Association between nutrient intake and thyroid cancer risk in Korean women. Nutr. Res. Pract. 2016;10:336–341. doi: 10.4162/nrp.2016.10.3.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho YA, Kim J. Dietary factors affecting thyroid cancer risk: a meta-analysis. Nutr. Cancer. 2015;67:811–817. doi: 10.1080/01635581.2015.1040517. [DOI] [PubMed] [Google Scholar]
- 8.Veiga LH, et al. Thyroid cancer after childhood exposure to external radiation: an updated pooled analysis of 12 studies. Radiat. Res. 2016;185:473–484. doi: 10.1667/RR14213.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ren Y, et al. FOXE1 polymorphism interacts with dietary iodine intake in differentiated thyroid cancer risk in the Cuban population. Thyroid. 2016;26:1752–1760. doi: 10.1089/thy.2015.0594. [DOI] [PubMed] [Google Scholar]
- 10.Jaggupilli A, et al. Analysis of the expression of human bitter taste receptors in extraoral tissues. Mol Cell. Biochem. 2017;426:137–147. doi: 10.1007/s11010-016-2902-z. [DOI] [PubMed] [Google Scholar]
- 11.Sternini C, Anselmi L, Rozengurt E. Enteroendocrine cells: a site of ‘taste’ in gastrointestinal chemosensing. Curr. Opin. Endocrinol. Diabetes Obes. 2008;15:73–78. doi: 10.1097/MED.0b013e3282f43a73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaik FA, et al. Bitter taste receptors: extraoral roles in pathophysiology. Int. J. Biochem. Cell Biol. 2016;77:197–204. doi: 10.1016/j.biocel.2016.03.011. [DOI] [PubMed] [Google Scholar]
- 13.Choi JH, et al. Genetic variation in the TAS2R38 bitter taste receptor and gastric cancer risk in Koreans. Sci. Rep. 2016;6:26904. doi: 10.1038/srep26904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carrai M, et al. Association between TAS2R38 gene polymorphisms and colorectal cancer risk: a case-control study in two independent populations of Caucasian origin. PLoS One. 2011;6:e20464. doi: 10.1371/journal.pone.0020464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi JH, et al. Variations in the bitterness perception-related genes TAS2R38 and CA6 modify the risk for colorectal cancer in Koreans. Oncotarget. 2017;8:21253–21265. doi: 10.18632/oncotarget.15512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee RJ, et al. T2R38 taste receptor polymorphisms underlie susceptibility to upper respiratory infection. J. Clin. Invest. 2012;122:4145–4159. doi: 10.1172/JCI64240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wooding SP, et al. Association of a bitter taste receptor mutation with Balkan endemic nephropathy (BEN) BMC Med. Genet. 2012;13:96. doi: 10.1186/1471-2350-13-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gil S, et al. Genotype-specific regulation of oral innate immunity by T2R38 taste receptor. Mol. Immunol. 2015;68:663–670. doi: 10.1016/j.molimm.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh N, Chakraborty R, Bhullar RP, Chelikani P. Differential expression of bitter taste receptors in non-cancerous breast epithelial and breast cancer cells. Biochem. Biophys. Res. Commun. 2014;446:499–503. doi: 10.1016/j.bbrc.2014.02.140. [DOI] [PubMed] [Google Scholar]
- 20.Jaggupilli A, Howard R, Upadhyaya JD, Bhullar RP, Chelikani P. Bitter taste receptors: novel insights into the biochemistry and pharmacology. Int. J. Biochem. Cell Biol. 2016;77:184–196. doi: 10.1016/j.biocel.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 21.Seo Y, Kim YS, Lee KE, Park TH, Kim Y. Anti-cancer stemness and anti-invasive activity of bitter taste receptors, TAS2R8 and TAS2R10, in human neuroblastoma cells. PLoS One. 2017;12:e0176851. doi: 10.1371/journal.pone.0176851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sugantha Priya E, et al. Anti-cancer activity of quercetin in neuroblastoma: an in vitro approach. Neurol. Sci. 2014;35:163–170. doi: 10.1007/s10072-013-1462-1. [DOI] [PubMed] [Google Scholar]
- 23.Meyerhof W, et al. The molecular receptive ranges of human TAS2R bitter taste receptors. Chem. Senses. 2010;35:157–170. doi: 10.1093/chemse/bjp092. [DOI] [PubMed] [Google Scholar]
- 24.Clark AA, et al. TAS2R bitter taste receptors regulate thyroid function. FASEB J. 2015;29:164–172. doi: 10.1096/fj.14-262246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rahbari R, Zhang L, Kebebew E. Thyroid cancer gender disparity. Future Oncol. 2010;6:1771–1779. doi: 10.2217/fon.10.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aboelnaga EM, Ahmed RA. Difference between papillary and follicular thyroid carcinoma outcomes: an experience from Egyptian institution. Cancer Biol. Med. 2015;12:53–59. doi: 10.7497/j.issn.2095-3941.2015.0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lloyd RV, Buehler D, Khanafshar E. Papillary thyroid carcinoma variants. Head Neck Pathol. 2011;5:51–56. doi: 10.1007/s12105-010-0236-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 29.Kim UK, et al. Positional cloning of the human quantitative trait locus underlying taste sensitivity to phenylthiocarbamide. Science. 2003;299:1221–1225. doi: 10.1126/science.1080190. [DOI] [PubMed] [Google Scholar]
- 30.Ji M, et al. Identification of novel compounds for human bitter taste receptors. Chem. Bio.l Drug. Des. 2014;84:63–74. doi: 10.1111/cbdd.12293. [DOI] [PubMed] [Google Scholar]
- 31.Lu P, Zhang CH, Lifshitz LM, ZhuGe R. Extraoral bitter taste receptors in health and disease. J. Gen. Physiol. 2017;149:181–197. doi: 10.1085/jgp.201611637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee RJ, Cohen NA. Taste receptors in innate immunity. Cell Mol. Life Sci. 2015;72:217–236. doi: 10.1007/s00018-014-1736-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ames BN, Profet M, Gold LS. Nature’s chemicals and synthetic chemicals: comparative toxicology. Proc. Natl. Acad. Sci. USA. 1990;87:7782–7786. doi: 10.1073/pnas.87.19.7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shabalina SA, Spiridonov NA, Kashina A. Sounds of silence: synonymous nucleotides as a key to biological regulation and complexity. Nucleic Acids Res. 2013;41:2073–2094. doi: 10.1093/nar/gks1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Supek F, Minana B, Valcarcel J, Gabaldon T, Lehner B. Synonymous mutations frequently act as driver mutations in human cancers. Cell. 2014;156:1324–1335. doi: 10.1016/j.cell.2014.01.051. [DOI] [PubMed] [Google Scholar]
- 36.GTEx Consortium. Human genomics. The genotype-tissue expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science348, 648–660 (2015). [DOI] [PMC free article] [PubMed]
- 37.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gentiluomo M, et al. Taste receptor polymorphisms and male infertility. Hum. Reprod. 2017;32:2324–2331. doi: 10.1093/humrep/dex305. [DOI] [PubMed] [Google Scholar]
- 39.Schembre SM, Cheng I, Wilkens LR, Albright CL, Marchand le L. Variations in bitter-taste receptor genes, dietary intake, and colorectal adenoma risk. Nutr. Cancer. 2013;65:982–990. doi: 10.1080/01635581.2013.807934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Timpson NJ, et al. TAS2R38 (phenylthiocarbamide) haplotypes, coronary heart disease traits, and eating behavior in the British Women’s Heart and Health Study. Am. J. Clin. Nutr. 2005;81:1005–1011. doi: 10.1093/ajcn/81.5.1005. [DOI] [PubMed] [Google Scholar]
- 41.Gallo S, et al. TAS2R38 taste receptor gene and chronic rhinosinusitis: new data from an Italian population. BMC Med. Genet. 2016;17:54. doi: 10.1186/s12881-016-0321-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Sibio MT, et al. Triiodothyronine and breast cancer. World J. Clin. Oncol. 2014;5:503–508. doi: 10.5306/wjco.v5.i3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tosovic A, Bondeson AG, Bondeson L, Ericsson UB, Manjer J. T3 levels in relation to prognostic factors in breast cancer: a population-based prospective cohort study. BMC Cancer. 2014;14:536. doi: 10.1186/1471-2407-14-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Czarnecka AM, et al. Triiodothyronine regulates cell growth and survival in renal cell cancer. Int. J. Oncol. 2016;49:1666–1678. doi: 10.3892/ijo.2016.3668. [DOI] [PubMed] [Google Scholar]
- 45.Perri A, et al. T3 enhances thyroid cancer cell proliferation through TRbeta1/Oct-1-mediated cyclin D1 activation. Mol. Cell. Endocrinol. 2014;382:205–217. doi: 10.1016/j.mce.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 46.Rousset, B., Dupuy, C., Miot, F. & Dumont, J. In Endotext (ed. DeGroot, L. J.) (MDText.com, Inc., 2015).
- 47.Sapin R, Schlienger JL. Thyroxine (T4) and tri-iodothyronine (T3) determinations: techniques and value in the assessment of thyroid function. Ann. Biol. Clin. (Paris). 2003;61:411–420. [PubMed] [Google Scholar]
- 48.Hsueh WC, et al. Diabetes in the Old Order Amish: characterization and heritability analysis of the Amish family diabetes study. Diabetes Care. 2000;23:595–601. doi: 10.2337/diacare.23.5.595. [DOI] [PubMed] [Google Scholar]
- 49.Kim J. Cancer screenee cohort study of the National Cancer Center in South Korea. Epidemiol. Health. 2014;36:e2014013. doi: 10.4178/epih/e2014013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Myung SK, Lee CW, Lee J, Kim J, Kim HS. Risk factors for thyroid cancer: a hospital-based case-control study in Korean adults. Cancer Res. Treat. 2017;49:70–78. doi: 10.4143/crt.2015.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.American Thyroid Association, Thyroid Cancer Staging Calculator (AJCC 7th Edition). https://www. thyroid.org/professionals/calculators/thyroid-cancer-staging-calculator-old/ (2018).
- 52.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Delaneau O, Marchini J. Integrating sequence and array data to create an improved 1000 Genomes Project haplotype reference panel. Nat. Commun. 2014;5:3934. doi: 10.1038/ncomms4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 55.Herold C, Becker T. Genetic association analysis with FAMHAP: a major program update. Bioinformatics. 2009;25:134–136. doi: 10.1093/bioinformatics/btn581. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets generated during the present study are available from the corresponding author on reasonable request.