Abstract
Acute myeloid leukemia (AML) is one of the most common hematological malignancies all around the world. MicroRNAs have been determined to contribute various cancers initiation and progression, including AML. Although microRNA-204 (miR-204) exerts anti-tumor effects in several kinds of cancers, its function in AML remains unknown. In the present study, we assessed miR-204 expression in AML blood samples and cell lines. We also investigated the effects of miR-204 on cellular function of AML cells and the underlying mechanisms of the action of miR-204. Our results showed that miR-204 expression was significantly downregulated in AML tissues and cell lines. In addition, overexpression of miR-204 induced growth inhibition and apoptosis in AML cells, including AML5, HL-60, Kasumi-1 and U937 cells. Cell cycle analysis further confirmed an augmentation in theapoptotic subG1 population by miR-204 overexpression. Mechanistically, baculoviral inhibition of apoptosis protein repeat containing 6 (BIRC6) was identified as a direct target of miR-204. Enforcing miR-204 expression increased the luciferase activity and expression of BIRC6, as well as p53 and Bax expression. Moreover, restoration of BIRC6 reversed the pro-apoptotic effects of miR-204 overexpression in AML cells. Taken together, this study demonstrates that miR-204 causes AML cell apoptosis by targeting BIRC6, suggesting miR-204 may play an anti-carcinogenic role in AML and function as a novel biomarker and therapeutic target for the treatment of this disease.
Keywords: Acute myeloid leukemia, Apoptosis, BIRC6, Cell cycle, microRNA-204
INTRODUCTION
Evasion of apoptosis is a hallmark of acute myeloid leukemia (AML) and other cancer cells. A group of proteins known as the inhibitor of apoptosis proteins (IAPs) play a relevant role in apoptosis resistance in a wide range of cancer cells (1, 2). Increased IAP expression has been observed in a variety of human cancers, including AML (1). Baculoviral inhibition of apoptosis protein (IAP) repeat containing 6 (BIRC6), is the largest member of IAPs family with a unique ubiquitin-conjugating domain, which differs from other members, suggesting a particular role of this protein in the IAP family (3). In childhood AML, higher BIRC6 expression was associated with unfavorable response to therapy (4). Similar results were also obtained by Carter et al. demonstrating that BIRC6 overexpression negatively influenced overall 3-year survival in childhood AML (5). These findings indicate that downregulation of BIRC6 may contribute to anti-carcinogenic effects in AML.
Another critical regulator of cell apoptosis, p53, has been well documented to be involved in carcinogenesis (6). It is worthy to note that p53 is an important downstream effector of BIRC6 (1). In addition to MDM2, BIRC6 is also the most crucial E3 ubiquitin ligase for p53. BIRC6 binds to p53 and ubiquitinates p53 for proteasomal degradation (6). Previous study has been reported the carcinogenic effect of BIRC6 through attenuating p53 function in hepatocellular carcinoma (7). Therefore, the balance between BIRC6 and p53 is significant for p53 expression and function to exert its effect in tumor suppression.
Accumulating evidences have implicated that MicroRNAs (miRNAs) regulation is critical in AML pathogenesis. MiR-204 is reported to play a relevant role in the development of gastric cancer (8), prostate cancer (9, 10), ovarian cancer (11), colorectal cancer (12), cervical cancer (13), and melanoma (14). For example, miR-204 suppressed cell proliferation in gastric cancer by targeting CSK1B, GPRC5A and CXCL1 (8). Moreover, miR-204 was downregulated in cervical cancer tissues which plays an important role in regulating cervical cancer cell proliferation, migration and invasion (13). Interleukin-6 (IL-6) repressed miR-204 via a STAT3-binding sits and in turn increased cisplatin resistance in ovarian cancer cells (11). A recent study observed that miR-204 expression was markedly decreased in hepatocellular carcinoma patients, which suggests a promising significance of the association of miR-204 expression with hepatocellular carcinoma pathological features (15). Similarly, Song et al. found that miR-204 was one of the downregulated miRNAs in prostate cancer tissues that were suitable for predicting the different stage of prostate cancer (10). However, the functional role of miR-204 in AML is still poorly understood and needs for investigation. In the current study, we provide the first evidence demonstrating that miR-204 induces human AML cell apoptosis by directly targeting BIRC6. Our data indicate that miR-204 may be a potential novel therapeutic target for the treatment of AML.
RESULTS
MiR-204 is decreased in clinical AML samples
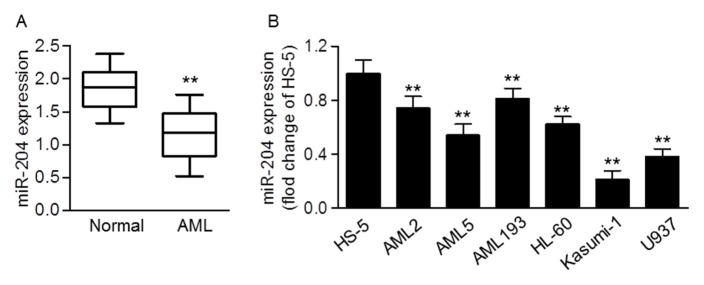
To unveil the role of miR-204 in AML, the expression level of miR-204 was first examined in blood samples of patient with AML. Quantitative real-time PCR results showed that miR-204 expression was significantly decreased in AML samples compared with normal samples (Fig. 1A). In addition, we found lower expression of miR-204 in human AML cell lines such as AML2, AML5, AML193, HL-60, Kasumi-1 and U937 than in normal cell line HS-5 (Fig. 1B). The data indicate that miR-204 may play an anti-tumor role in the progression of AML.
Fig. 1.
MiR-204 expression in clinical AML samples. (A) Quantitative real-time PCR was used to analysis the expression level of miR-204 in AML blood samples (n = 60) compared with normal samples (n = 42). **P < 0.01 vs. control. (B) miR-204 expression in HS-5 normal cells and human AML cell lines, AML2, AML5, AML193, HL-60, Kasumi-1 and U937 was determined by quantitative real-time PCR. **P < 0.01 vs. HS-5 cells, n = 10.
MiR-204 overexpression induces apoptosis in human AML cells
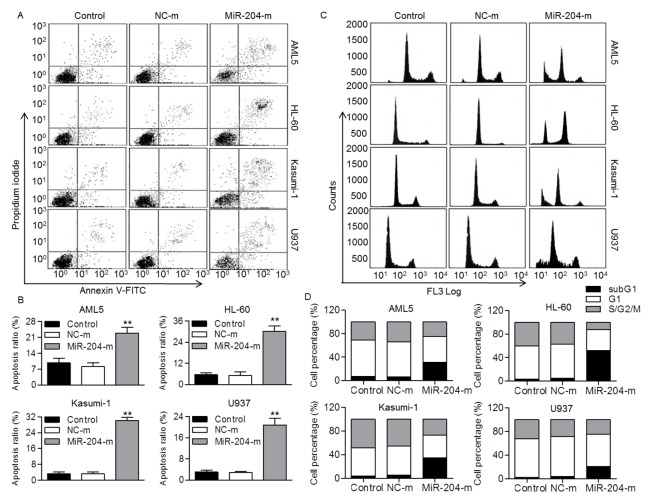
The above findings demonstrated that miR-204 expression in AML5, HL-60, Kasumi-1 and U937 was relative lower than other AML cell lines. Thus, these four cell lines were chosen to investigate the effect of miR-204 on cellular function. The results from CCK-8 assay showed that overexpression of miR-204 significantly suppressed, whereas miR-204 inhibition increased the viability of AML5, HL-60, Kasumi-1 and U937 cells (Fig. S1A and B). We next determine whether miR-204 was able to induce AML cell apoptosis. Our results showed that overexpression of miR-204 markedly induced apoptosis in AML5, HL-60, Kasumi-1 and U937 cells compared with control group, respectively (Fig. 2A and B). To clarify whether miR-204 inhibits AML cell viability through alteration of cell cycle distribution, population in different phases of cell cycle was determined by flow cytometry. The results revealed that the subG1 population representing the apoptotic cells was dramatically increased after miR-204 overexpression (Fig. 2C and D). This was further supported by western blotting results showing that miR-204 overexpression markedly increased cleaved Caspase-3 expression (Fig. S1C). The data indicate miR-204 inhibits AML cell viability via induction of apoptosis rather than inhibition of cell proliferation.
Fig. 2.
MiR-204 upregulation induced human AML cell apoptosis. (A) Human AML cell lines, AML5, HL-60, Kasumi-1 and U937, were transfected with miR-204 mimics (miR-204-m, 20 nM) or mimics negative control (NC-m) for 48 h. Cell apoptosis was examined by Annexin V/PI staining using flow cytometry. (B) The percentage of apoptosis ratio was quantified. **P < 0.01 vs. control, n = 8. Cell cycle was quantified by flow cytometry. **P < 0.01 vs. control, n = 6. (C) Representative flow cytometry histograms displaying the effect of miR-204 overexpression on AML5, HL-60, Kasumi-1 or U937 cell cycle distribution. (D) Graphs correspond to the distribution of cell population in different phases.
MiR-204 upregulaiton activates p53-dependent apoptosis pathway by targeting BIRC6
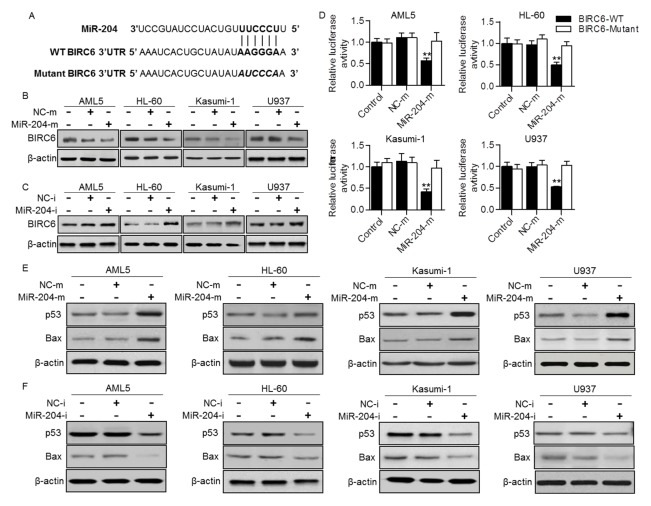
To understand the mechanism by which miR-204 induces AML cell apoptosis, we used computational mRNA target analysis online software (www.Targetscan.org) to predict the potential target gene of miR-204. The results showed that miR-204 may directly target BIRC6, a gene known to be involved in the regulation of cell apoptosis (Fig. 3A). Results from western blotting analysis showed that the protein expression of BIRC6 in AML5, HL-60, Kasumi-1 and U937 was significantly decreased after miR-204 upregulation (Fig. 3B). However, miR-204 inhibition increased BIRC6 protein expression in these AML cell lines (Fig. 3C). To investigate whether miR-204 decreased BIRC6 expression through direct 3′UTR interaction, BIRC6 3′UTR luciferase reporter was constructed and luciferase assay was performed. The results showed that upregulation of miR-204 significantly decreased the luciferase activity in the above four cell lines transfected with wild-type BIRC6 3′UTR, but not in cells transfected with the mutant vector (Fig. 3D).
Fig. 3.
MiR-204 negatively regulated BIRC6 expression and activated p53-dependent apoptosis pathway. (A) BIRC6 was predicted as the target gene of miR-204. The complementary sequences were shown. (B and C) Western blotting analysis of BIRC6 expression in AML cells transfected with miR-204 mimics (miR-204-m) (B) or miR-204 inhibitor (miR-204-i) (C). (D) AML cells were cotransfected with luciferase reporter containing wild-type (WT) BIRC6 3′ UTR or the mutant one, and miR-204 mimics or mimics negative control. Dual luciferase activity assay was performed to examine the luciferase activity of BIRC6 3′ UTR. **P < 0.01 vs. control, n = 6. (E and F) The protein expression of p53 and Bax in AML cells transfected with miR-204 mimics (E) or miR-204 inhibitor (F) was determined.
Given that P53, an important factor in regulating tumor growth and apoptosis, can be ubiquitinated and degraded by BIRC6 (13), we next examined the expression of p53 in these AML cell lines. Western blotting showed that protein expression of p53 in AML5, HL-60, Kasumi-1 and U937 cells was significantly increased after miR-204 upregulation, which exhibited a reciprocal pattern of BIRC6 expression. Moreover, the protein expression of Bax, a p53 target gene was also markedly increased after miR-204 overexpression (Fig. 3E). On the contrary, opposite results were obtained in cells treated with miR-204 inhibitor (Fig. 3F). These results suggest that miR-204 increases p53 expression and transcriptional activity, which subsequently activates its dependent apoptotic pathway.
Restoration of BIRC6 reverses the enhanced effect of miR-204 on AML cell apoptosis
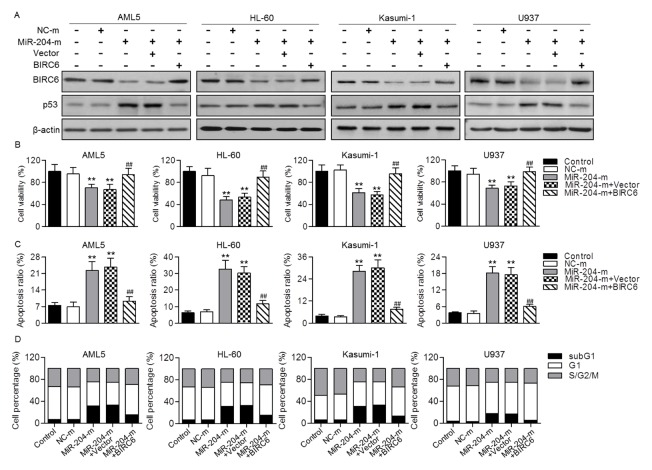
To confirm that whether downregulation of BIRC6 is a major mechanism that responsible for the induction of AML cell apoptosis by miR-204, we constructed BIRC6 plasmid, and then measured its effect on cell viability and apoptosis upon miR-204 overexpression. Expectedly, upregulation of BIRC6 obviously attenuated miR-204 overexpression-induced the decrease of BIRC6 expression and the increase of p53 expression (Fig. 4A). CCK-8 assay showed that the inhibitory effect of miR-204 on AML cell viability was significantly attenuated by transfection of BIRC6 plasmid (Fig. 4B). Moreover, overexpression of BIRC6 inhibited miR-204 mimics-induced the increase of apoptotic ratio in AML5, HL-60, Kasumi-1 and U937 cells (Fig. 4C). Similarly, following restoration of BIRC6 expression, the increased percentage of subG1 phase in miR-204 mimics-treated cells was also reduced (Fig. 4D). These results further support the critical role of BIRC6 in miR-204-induced AML cell apoptosis.
Fig. 4.
The inhibitory effect of miR-204 on AML cell growth was abolished by restoration of BIRC6. (A) AML5, HL-60, Kasumi-1 or U937 cells were cotransfected with miR-204 mimics (miR-204-m) and BIRC6 plasmid for 48 h. The protein expression of BIRC6 and p53 were determined, respectively. (B) Cell viability was determined by CCK-8 assay. (C) Flow cytometry was used to examine cell apoptosis by Annexin V/PI staining. (D) Cell cycle was also quantified by flow cytometry. Graphs correspond to the distribution of cell population in different phases. **P < 0.01 vs. control; ##P < 0.01 vs. miR-204-m, n = 6.
DISCUSSION
Induction of apoptotic cell death has been considered as a conventional therapy for cancer chemoprevention (16). Nevertheless, evasion of apoptotic response in cancer cells promotes tumor initiation, development and drug resistance (1). A lot of researches have indicated that miR-204 plays a role as an anti-carcinogenic gene in many type of cancer studied (9, 11–13, 17). Notably, despite miR-204 is highly expressed in leukemia, its function has not yet reported in AML (17–19). It has been known that most apoptosis signalings are activated by intrinsic mitochondrial apoptosis pathways that regulated by pro-apoptotic or anti-apoptotic protein in Bcl-2 family (9, 20). Among these members, Bcl-2 functions as an anti-apoptotic protein and has been observed to be the target of miR-204 in gastric cancer, colon cancer, neuroblastoma and intrahepatic cholangiocarcinoma (17, 20). Thus, induction of apoptosis seems to be the major mechanism underlying the anti-carcinogenic effect of miR-204. Here, we used four different leukemic cell lines to investigate the function of miR-204 in AML. In addition to AML5 cells, HL-60 is an acute promyelocytic leukemia cell line, Kasumi-1 is an acute myeloblastic leukemia cell line and U937 is one of lymphoma cell lines. The results showed that overexpression of miR-204 decreased viability and induced apoptosis in AML cells. Moreover, miR-204 upregulation in AML cells was associated with cell cycle arrest in the subG1 phase containing subdiploid or apoptotic cells, further confirming that miR-204 suppresses AML cells growth through induction of apoptosis rather than inhibition of cell proliferation.
To clarify the mechanisms by which miR-204 induces AML cell apoptosis, we used custom prediction of TargetScan to predict the target of miR-204. The potential target of miR-204 that could be involved in apoptosis led us to focus on BIRC6, a member of the IAPs family, which protects cells from apoptosis (3). Silencing BIRC6 resulted in growth inhibition of several cancer cells and increased the sensitivity of tumor cells to chemotherapy (6, 21). Several miRNAs have been evidenced to control BIRC6 through binding to specific target sequences in the 3′UTR of the gene, as shown for miR-BART15, miR-342 and miR-446h (22–24). In this study, we found that overexpression of miR-204 was associated with suppression of BIRC expression and luciferase activity, suggesting that miR-204 directly targets to the 3′UTR of BIRC6. Correspondingly, our study showed that miR-204 overexpression increased p53 and its target gene Bax expression. This indicates that miR-204 inhibits BIRC6, which may in turn increase p53 expression and activate p53-dependent apoptotic pathway. Although the anti-carcinogenic role of miR-204 may be partially due to its ability to suppress the expression of Bcl-2 (17), the mechanism of directly targeting BIRC6 provides a new insight into the anti-apoptotic effect of miR-204. Moreover, considering that BIRC6 is the ubiquitin ligase for p53, which causes p53 degradation and inhibits its down-stream mitochondria-dependent apoptosis pathway (2, 3), our findings showing that miR-204 negatively regulates BIRC6 expression seem to be more significance. Similar to previous observations that BIRC6 upregulation was resistant to various anticancer drugs in different cancer cells (2, 4, 25), the effect of miR-204 on apoptosis through BIRC6 was further confirm in our study using BIRC6 plasmid in AML cells. Notably, in addition to ubiquitin degradation function, BIRC6 carries a baculovirus IAP repeat (BIR) domain characteristic for the binding sites of Caspase and confers cell apoptosis (26). Moreover, Caspase in turn mediates BIRC6 degradation, which further decreases chemoresistance in human cancer (27). These findings suggest that BIRC6 also regulates cell apoptosis by a p53-independent pathway. Further study to clarify the different mechanisms of BIRC6 involved in the regulation of apoptosis in cancer cells is warranted.
In summary, our study demonstrated that miR-204 was frequently downregulated in AML blood sample and cell lines. MiR-204 overexpression induced apoptosis in AML cells. We also showed a direct interaction between miR-204 and BIRC6. These results reveal a novel pro-apoptotic role of miR-204 in AML cells, suggesting that miR-204 may be a promising therapeutic target for AML treatment.
MATERIALS AND METHODS
AML tissues
The research protocols were approved by the Ethics Committee of Xi’an University and performed in accordance with the Declaration of Helsinki and Good Clinical Practice. Informed consent was obtained from each patient and healthy volunteer. Human blood samples were collected from the First Affiliated Hospital of Xi’an University between 2013 and 2015 in the Department of Hematology. The patients (n = 60) with AML were diagnosed according to the pathological and clinic features. The normal samples (n = 42) were obtained from healthy volunteers without any diseases.
Cell culture
HS-5 normal cells and human AML cell lines including AML2, AML5, AML193, HL-60, Kasumi-1 and U937 were obtained from American Type Culture Collection (ATCC, VA, USA) and grown at 37°C, 5% CO2, and 95% in RPMI-1640 medium that supplemented with 10% fetal bovine serum (FBS) and 50 μg/ml Gentamicin (all from Gibco, CA, USA).
Quantitative real-time PCR
Whole blood samples or AML cells was isolated using the mirVana™ miRNA Isolation Kit (Ambio, Austin, TX, USA). Total RNA was reversed transcribed using the miRNA Reverse Transcription Kit (Applied Biosystems, CA, USA). cDNA was mixed with TaqMan® Micro Assay Kit and Fast SYBR® Green Master Mix Kit (Applied Biosystems) and amplified using 7500 Fast Real-Time PCR Systems (Applied Biosystems). The primer of detection of miR-204 and U6 were obtained from Rio Biotechnology (Guangzhou, China). The mRNA level of miR-204 was normalized by U6, and calculated using the 2−ΔΔCT method.
Cell transfection
AML cells were transfected with miR-204 mimics, miR-204 inhibitor, mimics negative control or inhibitor negative control at a final concentration of 20 nM using Lipofectamine 3000 (Invitrogen, CA, USA) according to the manufacturer’s instruction. After 48 h transfection, cells were harvested for flow cytometry analysis, western blotting, luciferase assay and cell viability assay.
Cell viability assay
The viability of AML cells was detected using the Cell Counting Kit-8 (CCK-8, Yiyuan Biotechnology, Guangzhou) according to the manufacturer’s instructions. After indicated treatments, the cells were incubated with CCK-8 reagent for 4 h at 37°C, 5% CO2. The absorbance was measured at a wavelength of 490 nm using a microplate reader (Bio-Tek, VY, USA).
Apoptosis detection assay
Cell apoptosis was evaluated using the FITC-Annexin V Apoptosis Detection Kit (Beyotime Institute of Biotechnology, Shanghai, China) by FACS Caliber flow cytometry (Becton Dickinson, CA, USA) according to the manufacturer’s instruction. AML cells were washed with PBS three times and stained with Annexin V-FITC and propidium iodide (PI) for 20 min at room temperature. The apoptotic cells were counted with flow cytometry and analyzed using WinMDI software (The Scripps Research Institute, CA, USA).
Cell cycle analysis
Distribution of AML cells in cell cycle was assessed by flow cytometry using propidium iodide (PI)-stained cells. In brief, cells were washed with PBS and fixed with 70% ethanol for 30 min on ice. After centrifugation, pelleted cells were incubated with 50 μg/ml propidium iodide (PI) dissolved in PBS for 30 min at 37°C. The PI-stained cells were analyzed by using a Becton Dickinson FACScan flow cytometer. The percentages of subG1, G1 and S/G2/M population were quantified with the ModFit software program (Verity Software House, Topsham, VT, USA).
Western blotting analysis
Protein were electrophoresed on 8% SDS polyacrylamide gel and transferred to nitrocellulose membranes (Millipore, MA, USA). Blots were blocked with 5% skim milk in PBS and probed with the indicated primary antibodies as follows: BIRC6, p53, Bax and β-actin (1:1000, Santa Cruz Biotechnology, CA, USA), and cleaved Caspase-3 (Cell Signaling Technology, MA, USA). Afterwards, the membranes were washed with PBS three times and incubated with secondary antibodies conjugated with peroxidase (Beyotime Institute of Biotechnology) for 1 h. Blots were exposed to enhanced chemiluminescence kit according the manufacturer’s instructions (Beyotime Institute of Biotechnology) and quantified by ImageJ software (ImageJ, Version 1.41, NIH, MD, USA).
Luciferase assay
The 3′UTR of BIRC6, which contains the predicted binding site for miR-204, was cloned into the pmirGLO dual luciferase miRNA target expression vector (Promega, WI, USA), referred to as wild-type BIRC6 3′UTR. The mutant 3′UTR of BIRC6 was constructed by substitution of 4 bp from seed region of miR-204. AML cells (1 × 105 cells/well) were plated in 96-well plates. After 24 h, the pmirGLO vector containing the wild-type BIRC6 3′UTR or mutant BIRC6 3′UTR was cotransfected with miR-204 mimics or mimics negative control for 48 h. Luciferase activity were quantified using a dual luciferase reporter system (Promega) according to the manufacturer’s protocols.
Plasmid construction and transfection
Full-length cDNA for human BIRC6 was obtained from OpenBioSystems (AL, USA), and amplified and cloned into pSMCV expression vector (OpenBioSystems). This plasmid was confirmed by sequencing. For overexpressing BIRC6 in AML cells, BIRC6 plasmid or empty vector (pSMCV) was transiently transfected into AML cells using Lipofectamine 3000 for 48 h.
Statistical analysis
Data were expressed as mean value ± standard error of mean (SEM) and compared by two-tailed Student t test or one-way ANOVA, followed by the Bonferroni multiple comparison test. Statistical analysis was performed by SPSS 18.0 software (SPSS Inc., IL, USA). P < 0.05 was considered statistically significant.
ACKNOWLEDGEMENTS
This study was financially supported by the grants from the Research Project of Education Office of Heilongjiang Province (NO. 12541299).
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
REFERENCES
- 1.Fulda S. Inhibitor of Apoptosis (IAP) proteins in hematological malignancies: molecular mechanisms and therapeutic opportunities. Leukemia. 2014;28:1414–1422. doi: 10.1038/leu.2014.56. [DOI] [PubMed] [Google Scholar]
- 2.Chen Z, Naito M, Hori S, Mashima T, Yamori T, Tsuruo T. A human IAP-family gene, apollon, expressed in human brain cancer cells. Biochem Biophys Res Commun. 1999;264:847–854. doi: 10.1006/bbrc.1999.1585. [DOI] [PubMed] [Google Scholar]
- 3.Martin SJ. An Apollon vista of death and destruction. Nat Cell Biol. 2004;6:804–806. doi: 10.1038/ncb0904-804. [DOI] [PubMed] [Google Scholar]
- 4.Ismail EA, Mahmoud HM, Tawfik LM, et al. BIRC6/Apollon gene expression in childhood acute leukemia: impact on therapeutic response and prognosis. Eur J Haematol. 2012;88:118–127. doi: 10.1111/j.1600-0609.2011.01734.x. [DOI] [PubMed] [Google Scholar]
- 5.Sung KW, Choi J, Hwang YK, et al. Overexpression of Apollon, an antiapoptotic protein, is associated with poor prognosis in childhood de novo acute myeloid leukemia. Clin Cancer Res. 2007;13:5109–5114. doi: 10.1158/1078-0432.CCR-07-0693. [DOI] [PubMed] [Google Scholar]
- 6.Lopergolo A, Pennati M, Gandellini P, et al. Apollon gene silencing induces apoptosis in breast cancer cells through p53 stabilisation and caspase-3 activation. Br J Cancer. 2009;100:739–746. doi: 10.1038/sj.bjc.6604927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang W, Xue R, Weng S, et al. BIRC6 promotes hepatocellular carcinogenesis: interaction of BIRC6 with p53 facilitating p53 degradation. Int J Cancer. 2015;136:E475–487. doi: 10.1002/ijc.29194. [DOI] [PubMed] [Google Scholar]
- 8.Shrestha S, Yang CD, Hong HC, et al. Integrated MicroRNA-mRNA Analysis Reveals miR-204 Inhibits Cell Proliferation in Gastric Cancer by Targeting CKS1B, CXCL1 and GPRC5A. Int J Mol Sci. 2017;19 doi: 10.3390/ijms19010087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shu Y, Ren L, Xie B, Liang Z, Chen J. MiR-204 enhances mitochondrial apoptosis in doxorubicin-treated prostate cancer cells by targeting SIRT1/p53 pathway. Oncotarget. 2017;8:97313–97322. doi: 10.18632/oncotarget.21960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song CJ, Chen H, Chen LZ, Ru GM, Guo JJ, Ding QN. The potential of microRNAs as human prostate cancer biomarkers: A meta-analysis of related studies. J Cell Biochem. 2017;119:2763–2786. doi: 10.1002/jcb.26445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu X, Shen H, Yin X, et al. IL-6R/STAT3/miR-204 feedback loop contributes to cisplatin resistance of epithelial ovarian cancer cells. Oncotarget. 2017;8:39154–39166. doi: 10.18632/oncotarget.16610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yin Y, Zhang B, Wang W, et al. miR-204-5p inhibits proliferation and invasion and enhances chemotherapeutic sensitivity of colorectal cancer cells by downregulating RAB22A. Clin Cancer Res. 2014;20:6187–6199. doi: 10.1158/1078-0432.CCR-14-1030. [DOI] [PubMed] [Google Scholar]
- 13.Duan S, Wu A, Chen Z, Yang Y, Liu L, Shu Q. MiR-204 Regulates Cell Proliferation and Invasion by Targeting EphB2 in Human Cervical Cancer. Oncol Res. 2017;26:713–723. doi: 10.3727/096504017X15016337254641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diaz-Martinez M, Benito-Jardon L, Alonso L, Koetz-Ploch L, Hernando E, Teixido J. miR-204-5p and miR-211-5p contribute to BRAF inhibitor resistance in melanoma. Cancer Res. 2017;78:1017–1030. doi: 10.1158/0008-5472.CAN-17-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo YH, Tang W, Zhang X, et al. Promising significance of the association of miR-204-5p expression with clinicopathological features of hepatocellular carcinoma. Medicine (Baltimore) 2017;96:e7545. doi: 10.1097/MD.0000000000007545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reubold TF, Eschenburg S. A molecular view on signal transduction by the apoptosome. Cell Signal. 2012;24:1420–1425. doi: 10.1016/j.cellsig.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 17.Li T, Pan H, Li R. The dual regulatory role of miR-204 in cancer. Tumour Biol. 2016;37:11667–11677. doi: 10.1007/s13277-016-5144-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roldo C, Missiaglia E, Hagan JP, et al. MicroRNA expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. J Clin Oncol. 2006;24:4677–4684. doi: 10.1200/JCO.2005.05.5194. [DOI] [PubMed] [Google Scholar]
- 19.Zanette DL, Rivadavia F, Molfetta GA, et al. miRNA expression profiles in chronic lymphocytic and acute lymphocytic leukemia. Braz J Med Biol Res. 2007;40:1435–1440. doi: 10.1590/S0100-879X2007001100003. [DOI] [PubMed] [Google Scholar]
- 20.Jin Z, El-Deiry WS. Overview of cell death signaling pathways. Cancer Biol Ther. 2005;4:139–163. doi: 10.4161/cbt.4.2.1508. [DOI] [PubMed] [Google Scholar]
- 21.Chu L, Gu J, Sun L, Qian Q, Qian C, Liu X. Oncolytic adenovirus-mediated shRNA against Apollon inhibits tumor cell growth and enhances antitumor effect of 5-fluorouracil. Gene Ther. 2008;15:484–494. doi: 10.1038/gt.2008.6. [DOI] [PubMed] [Google Scholar]
- 22.Choi H, Lee H, Kim SR, Gho YS, Lee SK. Epstein-Barr virus-encoded microRNA BART15-3p promotes cell apoptosis partially by targeting BRUCE. J Virol. 2013;87:8135–8144. doi: 10.1128/JVI.03159-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crippa E, Folini M, Pennati M, Zaffaroni N, Pierotti MA, Gariboldi M. miR-342 overexpression results in a synthetic lethal phenotype in BRCA1-mutant HCC1937 breast cancer cells. Oncotarget. 2016;7:18594–18604. doi: 10.18632/oncotarget.7617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Druz A, Chu C, Majors B, Santuary R, Betenbaugh M, Shiloach J. A novel microRNA mmu-miR-466h affects apoptosis regulation in mammalian cells. Biotechnol Bioeng. 2011;108:1651–1661. doi: 10.1002/bit.23092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Houdt WJ, Emmink BL, Pham TV, et al. Comparative proteomics of colon cancer stem cells and differentiated tumor cells identifies BIRC6 as a potential therapeutic target. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M111.011353. M111 011353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bartke T, Pohl C, Pyrowolakis G, Jentsch S. Dual role of BRUCE as an antiapoptotic IAP and a chimeric E2/E3 ubiquitin ligase. Mol Cell. 2004;14:801–811. doi: 10.1016/j.molcel.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 27.Okumu DO, East MP, Levine M, et al. BIRC6 mediates imatinib resistance independently of Mcl-1. PLoS One. 2017;12:e0177871. doi: 10.1371/journal.pone.0177871. [DOI] [PMC free article] [PubMed] [Google Scholar]