Abstract
Treatment of hepatitis C virus (HCV) infection has evolved greatly through the recent decade. The availability of direct-acting antiviral agents (DAAs) targeting the functional proteins of HCV has resulted in the introduction of DAA-based combination therapies, providing an optimal rate of treatment success. Among the DAAs, NS5A inhibitors are used in most of the introduced and approved HCV antiviral regimens. Resistance-associated substitutions (RASs) are amino acid substitutions in HCV protein sequences that result in decreased antiviral efficacy of the HCV DAAs. Among the HCV RASs, the NS5A RASs were found to effectively modify and decrease treatment response to NS5A inhibitor-containing regimens. As a baseline predictor of treatment response, NS5A RAS draws attention for pretreatment testing in targeted patient groups. Given NS5A RASs are either naturally-occurring or DAA-selected, the application of NS5A RAS testing can be considered in two settings of NS5A inhibitor-naïve patients and NS5A inhibitor-experienced patients. Less than 5% of NS5A inhibitor-naïve patients harbor naturally-occurring NS5A RAS with high resistance level (> 100X resistance fold-change). In NS5A inhibitor-naïve patients, NS5A RAS testing accompanied by treatment optimization cannot increase treatment response more than 2%-3%, while in NS5A inhibitor-experienced patients, > 75% are found to have NS5A RASs > 100X and NS5A RAS testing in this group of patients seems to be reasonable. This editorial will address the debate on the application of NS5A RAS testing and will discuss if the NS5A RAS testing has any role in clinical management of hepatitis C.
Keywords: Direct-acting antiviral agent, Hepatitis C, NS5A, Resistance, Treatment
Core tip: Hepatitis C virus resistance to NS5A inhibitors is one of the main problems of treatment with NS5A inhibitors. While the treatment of NS5A inhibitor-naïve patients is feasible and efficient, retreatment of NS5A inhibitor-experienced patients is challenging. In the context of failure following treatment with NS5A inhibitor-containing regimens, NS5A resistance-associated substitution testing can help in clinical decision-making, while the usefulness of baseline NS5A resistance-associated substitution testing in NS5A inhibitor-naïve patients is in question.
INTRODUCTION
Antiviral therapy of hepatitis C virus (HCV) infection has been the mainstay of hepatitis C management since application of conventional interferon (IFN) was introduced as the first HCV antiviral agent in 1990s[1,2]. HCV antiviral therapy has transformed greatly over time, from use of immunomodulatory agents, such as IFN, to use of direct-acting antiviral agents (DAAs), such as NS3 protease inhibitors, NS5A inhibitors and NS5B polymerase inhibitors[1]. While the IFN-based regimens result in suboptimal (30%-70%) virologic response, the DAA-based all-oral regimens have efficacy of > 95% in most patient groups[3-5]. Moreover, the treatment response to IFN-based regimens was modified by a large number of different host and virus parameters, such as age, sex, host genetics, liver fibrosis, HCV ribonucleic acid (RNA) level, HCV genotype, variations in HCV genes, etc[3,6,7].
On the other hand, with an increase of response rate to the DAA-based regimens, the modifiers of treatment response have been limited to few parameters, including cirrhosis, history of previous treatment (especially failure with DAA-based regimens), and finally naturally-occurring and DAA-selected resistance-associated substitutions (RASs)[8,9]. Currently, in most of the approved regimens for treatment of HCV infection, NS5A inhibitors are one of the components of the combination therapy with DAAs. The currently available and approved NS5A inhibitors are ledipasvir (LDV), daclatasvir (DCV), ombitasvir, elbasvir, velpatasvir (VEL) and pibrentasvir (PIB). In the context of treatment with NS5A inhibitor-containing regimens, the NS5A RAS is one of the baseline predictors of treatment response and, also, the major finding after treatment failure with these regimens[8,10].
NS5A RASs
Among the nonstructural proteins of HCV, the roles of NS5A protein are not fully known yet; however, it has been found that NS5A binds the RNA-dependent RNA polymerase NS5B and modulates RNA-dependent RNA polymerase activity[11]. The inhibitory activity of NS5A inhibitors on replication of HCV shows the pivotal role of NS5A protein in replication of HCV. The substitutions in the amino acid sequence of NS5A protein causing decreased binding of NS5A inhibitor molecules to NS5A protein can be selected under pressure of the treatment with NS5A inhibitors and subsequently decrease the inhibitory activity of NS5A inhibitors[12].
Generally, the NS5A amino acid substitutions causing > 2.5 resistance fold-change are considered as NS5A RASs. A subset of these NS5A RASs cause > 100 resistance fold-change, and are considered NS5A RASs > 100X[13]. It is worth noting that the resistance fold-change should be defined with consideration to the HCV genotype and subtype (i.e., 1a, 1b, 3a, etc.), NS5A inhibitor type (i.e., LDV, DCV, VEL, etc.) and also the specific amino acid substitution (i.e., Y93H, Y93F, Y93L, etc.). The NS5A RASs are observed as naturally-occurring substitutions in a proportion (< 10% to > 50%) of NS5A inhibitor-naïve patients[13-16]. The rate of detection of NS5A RASs is determined mainly by HCV genotypes and subtypes, and by the method for detection of NS5A RASs (i.e., deep sequencing vs Sanger sequencing)[17,18]. Among patients with NS5A RASs, only a small number (< 5% of NS5A inhibitor-naïve patients) of patients harbor isolates with naturally-occurring NS5A RASs > 100X.
The main finding regarding the prevalence of RASs in NS5A inhibitor-naïve patients with HCV genotype-1 (HCV-1) is the higher prevalence of NS5A RASs in patients with HCV-1b than in those with HCV-1a[19,20]. In the study by Dietz et al[19], the prevalence of NS5A RASs was 7.1% in patients with HCV-1a infection and 17.6% in those with HCV-1b infection. These NS5A RASs could be found at different levels of a mixed population of the mutated and wild-type viruses[21,22]. In NS5A inhibitor-experienced patients, especially those who have undergone a complete course of 12-wk or > 12-wk treatment, DAA-selected NS5A RASs are observed in > 75% of patients following treatment failure[10,23-25]. Most of these patients with failure to NS5A inhibitor-containing regimens harbor isolates with NS5A RASs > 100X[24,26]. Unfortunately, these DAA-selected NS5A RASs are persistent and they are observed even in the follow-up, 96 wk after treatment failure[27].
The impact of naturally-occurring baseline NS5A RASs has been assessed in many studies. These studies showed the impact of NS5A RASs on treatment response to NS5A inhibitor-containing regimens such as LDV/sofosbuvir[23,28], grazoprevir/elbasvir[29-31], ombitasvir/paritaprevir/ritonavir[21] and DCV/asunaprevir[22]; however, treatment with regimens containing the second-generation NS5A inhibitors with medium resistance barrier, including sofosbuvir/VEL[26,32] and glecaprevir/PIB[33], was either not impacted or only minimally modified by the NS5A RASs. In terms of HCV genotypes, it seems that the treatment of patients with HCV-3 infection using first-generation NS5A inhibitors is more influenced by NS5A RASs than that of patients with HCV-1 infection using same NS5A inhibitor[13,26]. This finding can be a result of different resistance fold-change of NS5A RASs by HCV genotypes with higher resistance fold-change for NS5A RASs in HCV-3 isolates than that for NS5A RASs in HCV-1 isolates[34]. Finally, it can be concluded that the higher response rate observed with the optimized treatment using a second-generation NS5A inhibitor with higher resistance barrier (i.e., VEL, PIB, etc.) effectively inhibiting NS5A protein of all HCV genotypes (pan-genotypic regimen) results in a lower chance for observation of treatment response modification by NS5A RASs.
With the knowledge that most of the patients with failure following treatment with NS5A inhibitor-containing regimens harbor NS5A RASs, especially those which confer a high level of resistance, the impact of these DAA-selected NS5A RASs on the efficacy of retreatment were evaluated in a small number of studies; the results showed that NS5A RASs are prominent treatment response predictors, especially when the regimen of the retreatment was not intensified or optimized for eradication of the virus[35,36].
TESTING RESISTANCE TO NS5A INHIBITORS: WHEN AND HOW?
There is a great debate on the application and usefulness of NS5A RAS testing prior to treatment. Generally, NS5A RAS testing can be considered under two different conditions: (1) prior to treatment with NS5A inhibitor-containing regimens in NS5A inhibitor-naïve patients; and (2) prior to retreatment with NS5A inhibitor-containing regimens in patients with a history of treatment failure using NS5A inhibitor-containing regimens. In Figure 1, the strategies with and without pretreatment NS5A RAS testing for treatment of HCV infection using NS5A inhibitor-containing regimens in NS5A inhibitor-naïve patients are presented. Since the overall response rate to the currently available and standard NS5A inhibitor-containing regimens is high (> 95%) and also the prevalence of NS5A RASs conferring high resistance level is low (< 5%), the strategy of pretreatment NS5A RAS testing and optimization of treatment in terms of treatment prolongation, adding ribavirin (RBV) and targeting all three HCV protease, NS5A and polymerase proteins can result in 2%-3% increase of the overall treatment response rate. Considering the costly ($100-$500) and time-consuming (7-30 d) process of NS5A RASs testing, we do not recommend NS5A RAS testing in NS5A inhibitor-naïve patients prior to treatment with NS5A inhibitor-containing regimens.
Figure 1.
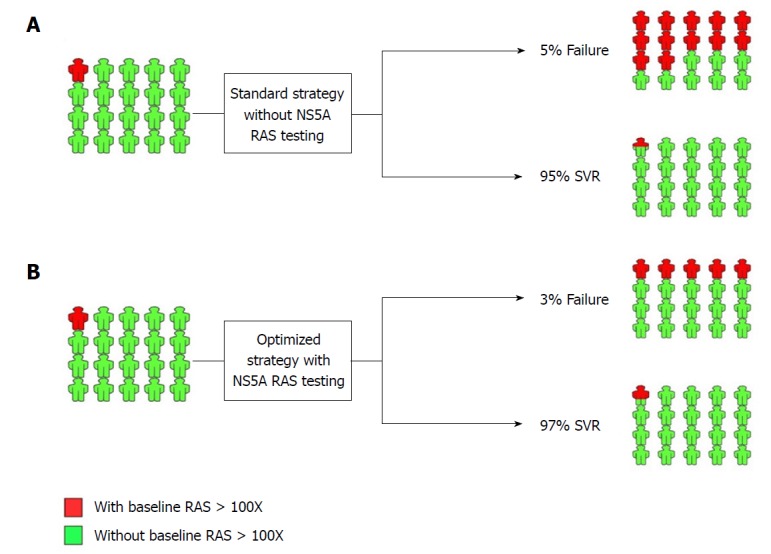
NS5A resistance-associated substitution testing in NS5A inhibitor-naïve patients prior to treatment with NS5A inhibitor-containing regimen. A: Standard treatment strategy and outcome without NS5A resistance-associated substitution (RAS) testing; B: Optimized treatment strategy and outcome with NS5A RAS testing. The implementation of NS5A RAS testing is developed by decision-making based on the presence of RAS > 100X. SVR: Sustained virologic response.
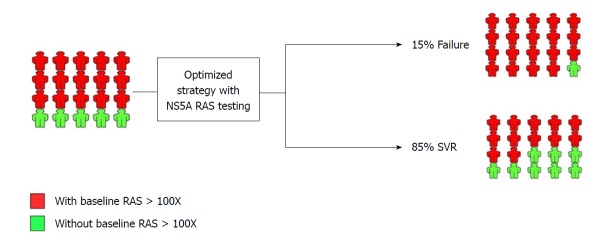
In patients with previous history of treatment with NS5A inhibitor-containing regimens, the scenario is much different with the fact that > 75% of them harbor NS5A RASs. As shown in Figure 2, the NS5A RAS testing can be implemented in the management of patients with previous history of treatment with NS5A inhibitor-containing regimens. If the patient harbors the NS5A RAS, they should be treated with the most intensified and optimized available treatment regimen (adding RBV, prolongation of treatment to 24 wk, and targeting all three protease, NS5A and polymerase proteins). However, the small portion of patients with previous history of treatment with NS5A inhibitor-containing regimens and without detectable NS5A RAS can be treated by treatment prolongation and/or adding RBV without targeting additional HCV protein.
Figure 2.
NS5A resistance-associated substitution testing in NS5A inhibitor-experienced patients prior to retreatment with NS5A inhibitor-containing regimen. Optimized treatment strategy with NS5A resistance-associated substitutions (RAS) testing and the outcome is shown. The implementation of NS5A RAS testing is developed by decision-making based on the presence of RAS > 100X. SVR: Sustained virologic response.
Currently, there are services available for NS5A RAS testing in diagnostic laboratory centers. These services are based on Sanger direct population sequencing or deep sequencing using next-generation sequencing platforms. The main difference between these two available technologies is the limit of detection of the RAS (mutated nucleotide) among the population of the virus without the RAS (wild-type nucleotide). Sanger sequencing can detect the NS5A RASs comprising 20%-30% of the whole population of the virus, while deep sequencing can even detect NS5A RASs comprising < 1% of the whole HCV population. It figures that deep sequencing is much more advanced for detection of NS5A RASs than Sanger sequencing; however, it was found that a small (< 10%-20% of the whole genetic pool of virus) population of HCV with RAS in a patient has no or minimal impact on treatment success[21,22,37]. With this finding, it seems that in the clinical management of hepatitis C, the NS5A RAS testing using Sanger sequencing is reasonable, and the reports of NS5A RAS testing using deep sequencing methods should be accompanied by the percentage of the NS5A RAS in the whole virus population.
CONCLUSION
DAA-based treatment of HCV is one of the greatest recent achievements in the field of hepatology and liver diseases, making hepatitis C a curable disease; however, the main problem with most of the targeted therapies is resistance. With a combination of NS5A inhibitor with an NS5B polymerase inhibitor and/or an NS3 protease inhibitor, the problem of HCV resistance has been mostly resolved in DAA-naïve patients and only a small (< 5%) population of treated patients relapsing after treatment termination. While pretreatment NS5A RAS testing cannot help in management of DAA-naïve patients, it can help in treatment decision-making for patients with failure to NS5A inhibitor-containing regimens. Currently, with the introduction of second-generation NS5A inhibitors, VEL and PIB, and the upcoming NS5A inhibitors in the pipeline with a high barrier to resistance, the problem of resistance to NS5A inhibitors is going to fade. However, unsolved concerns remain, such as transmission of NS5A RASs in populations with high-risk behaviors, including people who inject drugs, which should be considered by clinicians and researchers carefully.
Footnotes
Conflict-of-interest statement: Both authors declare no potential conflict of interest.
Manuscript source: Invited manuscript
Peer-review started: June 7, 2018
First decision: July 11, 2018
Article in press: August 4, 2018
Specialty type: Gastroenterology and hepatology
Country of origin: Iran
Peer-review report classification
Grade A (Excellent): A, A
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor):0
P- Reviewer: Hann HW, Jarcuska P, Inoue K, Quarleri J S- Editor: Dou Y L- Editor: A E- Editor: Tan WW
Contributor Information
Heidar Sharafi, Baqiyatallah Research Center for Gastroenterology and Liver Diseases, Baqiyatallah University of Medical Sciences, Tehran 1435915371, Iran; Middle East Liver Diseases Center, Tehran 1415513651, Iran. h.sharafi@meldcenter.com.
Seyed Moayed Alavian, Baqiyatallah Research Center for Gastroenterology and Liver Diseases, Baqiyatallah University of Medical Sciences, Tehran 1435915371, Iran; Middle East Liver Diseases Center, Tehran 1415513651, Iran.
References
- 1.Hesamizadeh K, Sharafi H, Rezaee-Zavareh MS, Behnava B, Alavian SM. Next Steps Toward Eradication of Hepatitis C in the Era of Direct Acting Antivirals. Hepat Mon. 2016;16:e37089. doi: 10.5812/hepatmon.37089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Strader DB, Seeff LB. A brief history of the treatment of viral hepatitis C. Clin Liver Dis. 2012;1:6–11. doi: 10.1002/cld.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Behnava B, Sharafi H, Keshvari M, Pouryasin A, Mehrnoush L, Salimi S, Karimi Elizee P, Ghazimoghaddam M, Alavian SM. The Role of Polymorphisms Near the IL28B Gene on Response to Peg-Interferon and Ribavirin in Thalassemic Patients With Hepatitis C. Hepat Mon. 2016;16:e32703. doi: 10.5812/hepatmon.32703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alavian SM, Hajarizadeh B, Bagheri Lankarani K, Sharafi H, Ebrahimi Daryani N, Merat S, Mohraz M, Mardani M, Fattahi MR, Poustchi H, et al. Recommendations for the Clinical Management of Hepatitis C in Iran: A Consensus-Based National Guideline. Hepat Mon. 2016;16:e40959. doi: 10.5812/hepatmon.guideline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharafi H, Nikbin M, Alavian SH, Behnava B, Alavian SM. Efficacy and Safety of Generic Sofosbuvir/Ledipasvir Fixed-Dose Combination in Iranian Patients with Chronic Hepatitis C Virus Infection. Hepat Mon. 2017;17:e12216. [Google Scholar]
- 6.Haj-Sheykholeslami A, Keshvari M, Sharafi H, Pouryasin A, Hemmati K, Mohammadzadehparjikolaei F. Interferon-λ polymorphisms and response to pegylated interferon in Iranian hepatitis C patients. World J Gastroenterol. 2015;21:8935–8942. doi: 10.3748/wjg.v21.i29.8935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miri SM, Sharafi H, Pouryasin A, Behnava B, Keshvari M, Salimi S, Karimi Elizee P, Alavian SM. The Role of Polymorphisms Near IFNL3 Gene as Predictors of Residual HCV RNA in Buffy Coat after Successful Antiviral Therapy. Hepat Mon. 2017;17:e12301. [Google Scholar]
- 8.Rezaee-Zavareh MS, Hesamizadeh K, Behnava B, Alavian SM, Gholami-Fesharaki M, Sharafi H. Combination of Ledipasvir and Sofosbuvir for Treatment of Hepatitis C Virus Genotype 1 Infection: Systematic Review and Meta-Analysis. Ann Hepatol. 2017;16:188–197. doi: 10.5604/16652681.1231562. [DOI] [PubMed] [Google Scholar]
- 9.Alavian SM, Sharafi H. Update on Recommendations for the Clinical Management of Hepatitis C in Iran 2017. Hepat Mon. 2017;17:e63956. doi: 10.5812/hepatmon.guideline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wyles D, Dvory-Sobol H, Svarovskaia ES, Doehle BP, Martin R, Afdhal NH, Kowdley KV, Lawitz E, Brainard DM, Miller MD, et al. Post-treatment resistance analysis of hepatitis C virus from phase II and III clinical trials of ledipasvir/sofosbuvir. J Hepatol. 2017;66:703–710. doi: 10.1016/j.jhep.2016.11.022. [DOI] [PubMed] [Google Scholar]
- 11.Shirota Y, Luo H, Qin W, Kaneko S, Yamashita T, Kobayashi K, Murakami S. Hepatitis C virus (HCV) NS5A binds RNA-dependent RNA polymerase (RdRP) NS5B and modulates RNA-dependent RNA polymerase activity. J Biol Chem. 2002;277:11149–11155. doi: 10.1074/jbc.M111392200. [DOI] [PubMed] [Google Scholar]
- 12.Kwon HJ, Xing W, Chan K, Niedziela-Majka A, Brendza KM, Kirschberg T, Kato D, Link JO, Cheng G, Liu X, et al. Direct binding of ledipasvir to HCV NS5A: mechanism of resistance to an HCV antiviral agent. PLoS One. 2015;10:e0122844. doi: 10.1371/journal.pone.0122844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeuzem S, Mizokami M, Pianko S, Mangia A, Han KH, Martin R, Svarovskaia E, Dvory-Sobol H, Doehle B, Hedskog C, et al. NS5A resistance-associated substitutions in patients with genotype 1 hepatitis C virus: Prevalence and effect on treatment outcome. J Hepatol. 2017;66:910–918. doi: 10.1016/j.jhep.2017.01.007. [DOI] [PubMed] [Google Scholar]
- 14.Sarrazin C, Dvory-Sobol H, Svarovskaia ES, Doehle BP, Pang PS, Chuang SM, Ma J, Ding X, Afdhal NH, Kowdley KV, et al. Prevalence of Resistance-Associated Substitutions in HCV NS5A, NS5B, or NS3 and Outcomes of Treatment With Ledipasvir and Sofosbuvir. Gastroenterology. 2016;151:501–512.e1. doi: 10.1053/j.gastro.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 15.Walker A, Siemann H, Groten S, Ross RS, Scherbaum N, Timm J. Natural prevalence of resistance-associated variants in hepatitis C virus NS5A in genotype 3a-infected people who inject drugs in Germany. J Clin Virol. 2015;70:43–45. doi: 10.1016/j.jcv.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 16.Bartolini B, Giombini E, Taibi C, Lionetti R, Montalbano M, Visco-Comandini U, D’Offizi G, Capobianchi MR, McPhee F, Garbuglia AR. Characterization of Naturally Occurring NS5A and NS5B Polymorphisms in Patients Infected with HCV Genotype 3a Treated with Direct-Acting Antiviral Agents. Viruses. 2017;9:pii: E212. doi: 10.3390/v9080212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bagaglio S, Andolina A, Merli M, Uberti-Foppa C, Morsica G. Frequency of Natural Resistance within NS5a Replication Complex Domain in Hepatitis C Genotypes 1a, 1b: Possible Implication of Subtype-Specific Resistance Selection in Multiple Direct Acting Antivirals Drugs Combination Treatment. Viruses. 2016;8:91. doi: 10.3390/v8040091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen ZW, Li H, Ren H, Hu P. Global prevalence of pre-existing HCV variants resistant to direct-acting antiviral agents (DAAs): mining the GenBank HCV genome data. Sci Rep. 2016;6:20310. doi: 10.1038/srep20310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dietz J, Susser S, Berkowski C, Perner D, Zeuzem S, Sarrazin C. Consideration of Viral Resistance for Optimization of Direct Antiviral Therapy of Hepatitis C Virus Genotype 1-Infected Patients. PLoS One. 2015;10:e0134395. doi: 10.1371/journal.pone.0134395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paolucci S, Fiorina L, Mariani B, Gulminetti R, Novati S, Barbarini G, Bruno R, Baldanti F. Naturally occurring resistance mutations to inhibitors of HCV NS5A region and NS5B polymerase in DAA treatment-naïve patients. Virol J. 2013;10:355. doi: 10.1186/1743-422X-10-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krishnan P, Schnell G, Tripathi R, Beyer J, Reisch T, Zhang X, Setze C, Rodrigues L Jr, Burroughs M, Redman R, Chayama K, Kumada H, Collins C, Pilot-Matias T. Analysis of Hepatitis C Virus Genotype 1b Resistance Variants in Japanese Patients Treated with Paritaprevir-Ritonavir and Ombitasvir. Antimicrob Agents Chemother. 2015;60:1106–1113. doi: 10.1128/AAC.02606-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ikeda H, Watanabe T, Okuse C, Matsumoto N, Ishii T, Yamada N, Shigefuku R, Hattori N, Matsunaga K, Nakano H, et al. Impact of resistance-associated variant dominancy on treatment in patients with HCV genotype 1b receiving daclatasvir/asunaprevir. J Med Virol. 2017;89:99–105. doi: 10.1002/jmv.24608. [DOI] [PubMed] [Google Scholar]
- 23.Ogawa E, Furusyo N, Nomura H, Dohmen K, Higashi N, Takahashi K, Kawano A, Azuma K, Satoh T, Nakamuta M, Koyanagi T, Kato M, Shimoda S, Kajiwara E, Hayashi J; Kyushu University Liver Disease Study (KULDS) Group. NS5A resistance-associated variants undermine the effectiveness of ledipasvir and sofosbuvir for cirrhotic patients infected with HCV genotype 1b. J Gastroenterol. 2017;52:845–854. doi: 10.1007/s00535-016-1290-1. [DOI] [PubMed] [Google Scholar]
- 24.Halfon P, Scholtès C, Izopet J, Larrat S, Trimoulet P, Zoulim F, Alric L, Métivier S, Leroy V, Ouzan D, et al. Baseline and post-treatment hepatitis C NS5A resistance in relapsed patients from a multicentric real-life cohort. Antivir Ther. 2017 doi: 10.3851/IMP3184. [DOI] [PubMed] [Google Scholar]
- 25.Starace M, Minichini C, De Pascalis S, Macera M, Occhiello L, Messina V, Sangiovanni V, Adinolfi LE, Claar E, Precone D, et al. Virological patterns of HCV patients with failure to interferon-free regimens. J Med Virol. 2018;90:942–950. doi: 10.1002/jmv.25022. [DOI] [PubMed] [Google Scholar]
- 26.Hezode C, Reau N, Svarovskaia ES, Doehle BP, Shanmugam R, Dvory-Sobol H, Hedskog C, McNally J, Osinusi A, Brainard DM, et al. Resistance analysis in patients with genotype 1-6 HCV infection treated with sofosbuvir/velpatasvir in the phase III studies. J Hepatol. 2018;68:895–903. doi: 10.1016/j.jhep.2017.11.032. [DOI] [PubMed] [Google Scholar]
- 27.Wyles D, Mangia A, Cheng W, Shafran S, Schwabe C, Ouyang W, Hedskog C, McNally J, Brainard DM, Doehle BP, et al. Long-term persistence of HCV NS5A resistance-associated substitutions after treatment with the HCV NS5A inhibitor, ledipasvir, without sofosbuvir. Antivir Ther. 2018;23:229–238. doi: 10.3851/IMP3181. [DOI] [PubMed] [Google Scholar]
- 28.Tsuji K, Kurosaki M, Itakura J, Mori N, Takaki S, Hasebe C, Akahane T, Joko K, Yagisawa H, Takezawa J, et al. Real-world efficacy and safety of ledipasvir and sofosbuvir in patients with hepatitis C virus genotype 1 infection: a nationwide multicenter study by the Japanese Red Cross Liver Study Group. J Gastroenterol. 2018 doi: 10.1007/s00535-018-1455-1. [DOI] [PubMed] [Google Scholar]
- 29.de Lédinghen V, Laforest C, Hézode C, Pol S, Renault A, Alric L, Larrey D, Métivier S, Tran A, Jézéquel C, et al. Retreatment With Sofosbuvir Plus Grazoprevir/Elbasvir Plus Ribavirin of Patients With Hepatitis C Virus Genotype 1 or 4 Who Previously Failed an NS5A- or NS3-Containing Regimen: The ANRS HC34 REVENGE Study. Clin Infect Dis. 2018;66:1013–1018. doi: 10.1093/cid/cix916. [DOI] [PubMed] [Google Scholar]
- 30.Bruchfeld A, Roth D, Martin P, Nelson DR, Pol S, Londoño MC, Monsour H Jr, Silva M, Hwang P, Arduino JM, Robertson M, Nguyen BY, Wahl J, Barr E, Greaves W. Elbasvir plus grazoprevir in patients with hepatitis C virus infection and stage 4-5 chronic kidney disease: clinical, virological, and health-related quality-of-life outcomes from a phase 3, multicentre, randomised, double-blind, placebo-controlled trial. Lancet Gastroenterol Hepatol. 2017;2:585–594. doi: 10.1016/S2468-1253(17)30116-4. [DOI] [PubMed] [Google Scholar]
- 31.Gane E, Nahass R, Luketic V, Asante-Appiah E, Hwang P, Robertson M, Wahl J, Barr E, Haber B. Efficacy of 12 or 18 weeks of elbasvir plus grazoprevir with ribavirin in treatment-naïve, noncirrhotic HCV genotype 3-infected patients. J Viral Hepat. 2017;24:895–899. doi: 10.1111/jvh.12719. [DOI] [PubMed] [Google Scholar]
- 32.von Felden J, Vermehren J, Ingiliz P, Mauss S, Lutz T, Simon KG, Busch HW, Baumgarten A, Schewe K, Hueppe D, et al. High efficacy of sofosbuvir/velpatasvir and impact of baseline resistance-associated substitutions in hepatitis C genotype 3 infection. Aliment Pharmacol Ther. 2018;47:1288–1295. doi: 10.1111/apt.14592. [DOI] [PubMed] [Google Scholar]
- 33.Krishnan P, Schnell G, Tripathi R, Beyer J, Reisch T, Dekhtyar T, Irvin M, Xie W, Fu B, Burroughs M, et al. Integrated Resistance Analysis of CERTAIN-1 and CERTAIN-2 Studies in Hepatitis C Virus-Infected Patients Receiving Glecaprevir and Pibrentasvir in Japan. Antimicrob Agents Chemother. 2018;62:pii: e02217–17. doi: 10.1128/AAC.02217-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gottwein JM, Pham LV, Mikkelsen LS, Ghanem L, Ramirez S, Scheel TKH, Carlsen THR, Bukh J. Efficacy of NS5A Inhibitors Against Hepatitis C Virus Genotypes 1-7 and Escape Variants. Gastroenterology. 2018;154:1435–1448. doi: 10.1053/j.gastro.2017.12.015. [DOI] [PubMed] [Google Scholar]
- 35.Lawitz E, Flamm S, Yang JC, Pang PS, Zhu Y, Svarovskaia E, McHutchison JG, Wyles D, Pockros P. Retreatment of patients who failed 8 or 12 weeks of ledipasvir/sofosbuvir-based regimens with ledipasvir/sofosbuvir for 24 weeks. J Hepatol. 2015;62:S192. doi: 10.1002/hep.27814. [DOI] [PubMed] [Google Scholar]
- 36.Kawakami Y, Ochi H, Hayes CN, Imamura M, Tsuge M, Nakahara T, Katamura Y, Kohno H, Kohno H, Tsuji K, et al. Efficacy and safety of ledipasvir/sofosbuvir with ribavirin in chronic hepatitis C patients who failed daclatasvir/asunaprevir therapy: pilot study. J Gastroenterol. 2018;53:548–556. doi: 10.1007/s00535-017-1380-8. [DOI] [PubMed] [Google Scholar]
- 37.Ide T, Eguchi Y, Harada M, Ishii K, Morita M, Morita Y, Sugiyama G, Fukushima H, Yano Y, Noguchi K, et al. Evaluation of Resistance-Associated Substitutions in NS5A Using Direct Sequence and Cycleave Method and Treatment Outcome with Daclatasvir and Asunaprevir for Chronic Hepatitis C Genotype 1. PLoS One. 2016;11:e0163884. doi: 10.1371/journal.pone.0163884. [DOI] [PMC free article] [PubMed] [Google Scholar]