Abstract
As alternative indexes of hepatitis B virus (HBV), covalently closed circular DNA (cccDNA) transcriptional activity, hepatitis B surface antigen (HBsAg), hepatitis B core-related antigen (HBcrAg), and peripheral blood RNA known as pgRNA, have been advocated as novel serum markers for prediction of prognosis and treatment response in chronic hepatitis B (CHB). Since the availability of commercial quantitative assays of HBsAg in 2011, HBsAg has been widely used for predicting treatment response of patients with CHB. Patients who received interferon therapy have shown a sharper reduction of HBsAg level than those who received nucleoside drug (NAs) therapy. Upon peginterferon treatment, sustained responders have presented a larger reduction of HBsAg level than the non-responders. An absence of HBsAg decline, together with < 2log reduction in HBV DNA at week 12, can serve as a stopping rule in HBsAg-negative patients infected with genotype D HBV. A sharp reduction of HBsAg titer in the NAs therapy is a predictor of HBsAg clearance in long-term treatment. HBcrAg, which consists of three species of related proteins sharing an identical 149 amino acid sequence, including HbcAg, hepatitis B e antigen (HBeAg), and a truncated 22-kDa precore protein, is still detectable in situations where serum HBV DNA levels become undetectable or HBsAg loss is achieved. Therefore, HBcrAg remains a measurable serum marker to correlate with cccDNA in this situation. The decline in HBcrAg has been observed with NAs therapy and the pattern of decline might provide prognostic information on the risk of HBV post-treatment reactivation. Peripheral blood RNA, which is known as pgRNA, directly derives from cccDNA and reflects intrahepatic cccDNA level. Quantitative pgRNA has been suggested to be helpful in CHB management. However, commercial quantitative assays are lacking. Additionally, the use of simultaneous and continuous clearance of HBV RNA and HBV DNA in serum has been suggested to be a safe stopping rule of NAs therapy for patients with CHB. However, clinical studies of large sample sizes are needed to prove the feasibility and significance of using serum HBV RNA as the assessment standard of antiviral therapy in CHB and the safety of the stopping rule in clinics.
Keywords: Hepatitis B core-related antigen, Response prediction, Chronic hepatitis B, Interferon, Nucleos(t)ide analogs, Hepatitis B surface antigen, Progenome RNA
Core tip: As the surrogate biomarkers of intrahepatic viral replicative activity, hepatitis B surface antigen (HBsAg), hepatitis B core-related antigen (HbcrAg), and serum hepatitis B virus (HBV) RNA levels have been advocated as novel serum markers for treatment response in chronic hepatitis B. Currently, quantitative HBsAg has been widely used for predicting treatment response of chronic hepatitis B. HBcrAg can predict the risk of post-treatment reactivation of HBV as it is detectable in patients whose HBV DNA levels are undetectable or HBsAg loss is achieved. Serum HBV RNA may be useful in monitoring drug withdrawal, but clinical studies with large sample sizes remain necessary to further determine this capability.
INTRODUCTION
Chronic hepatitis B virus (HBV) infection is one of the threats to human health and has become a global health issue. Approximately 0.35 billion of the worldwide population is infected with HBV, and 75% of this number is in the Asia-Pacific region. Yearly, approximately 650 000 people die of hepatic failure, liver cirrhosis, and liver cancer related to HBV infection[1]. Antiviral drugs, which are currently approved for chronic hepatitis B (CHB) patients, include pegylated interferons (Peg-IFN-α)-2a and the following five oral polymerase inhibitors: Three nucleoside [lamivudine (LAM), telbivudine (LDT), and entecavir (ETV)] and two nucleotide [adefovir dipivoxil (ADV) and tenofovir (TDF)] drugs. Oral HBV polymerase inhibitors not only reduce the occurrence rate of corresponding complications by inhibiting HBV duplication for a long time but also increase the survival rates and living quality of patients with chronic HBV. However, these inhibitors cannot completely eliminate covalently closed circular DNA (cccDNA) molecules in hepatic cells, resulting in uncertain treatment periods. Furthermore, most patients may have to take medicines all their lives[2,3].
Long-term virus inhibition, which is manifested by hepatitis B envelope antigen (HBeAg) seroconversion, can induce virus immunity control of some patients. Moreover, hepatitis B surface antigen (HBsAg) clearance or seroconversion may occur in some patients. Therefore, seroconversion of HBeAg and HBsAg is widely accepted as the endpoint of therapy[2,3]. Spontaneous or therapy-induced seroconversion of HBeAg is viewed as the premise of HBsAg clearance or seroconversion, which implies the stable infection of HBV. Such seroconversion is presently acknowledged as the stopping rule[4,5]. However, indexes for predicting seroconversion of HBeAg in antiviral therapy are not completely certain. Indexes of disease activity, namely, tissue inflammation score and alanine aminotransferase (ALT) level, and viral indexes, such as HBV DNA and HBsAg, can be used to predict seroconversion of HBeAg[6-10]. Despite established markers, including serum HBV DNA levels and HBsAg titers, hepatitis B core-related antigen (HBcrAg) and HBV RNA are also considered serum markers of HBV infection. HBV RNA carries virus gene information, and its quantitative assay is not highly influenced by viral antigen and antibody immune compounds. Therefore, HBV RNA is important in the clinical diagnosis and response prediction. In particular, quantitative assay of serum HBV RNA level is superior to HBV DNA in terms of response prediction to HBV polymerase inhibitor based on therapy[11-13]. Serum HBV RNA level has a key role in the prediction of the stopping rule[14] and drug resistance[6]. HBcrAg, which contains three related proteins that share an identical 149 amino acid sequence [HBcAg, HBeAg, and a truncated 22-kDa precore protein (p22Cr)], is detectable in many patients with undetectable HBV DNA and HBsAg seroclearance. Along with the establishment of a fully automated detection method, HBcrAg may be extensively used in monitoring chronic HBV antiviral therapy in the future, particularly in situations where serum HBV DNA becomes undetectable. The major findings and potential clinical applications of HBcrAg in chronic HBV infection have been comprehensively described by Mak et al[15]. Therefore, this study mainly focuses on introducing the role of HBsAg titers and HBV RNA level in the antiviral therapy efficacy prediction of patients with CHB.
SIGNIFICANCE OF HBsAG LEVEL
HBsAg has been viewed as an important diagnosis index of HBV infection since its discovery by Blumberg et al[16] in 1965 and report in 1968. Recently, clinical significance of the HBsAg level has again attracted considerable attention since a corresponding quantitative assay was established and the correlation between HBsAg and cccDNA was confirmed[17,18].
Peg-IFN-α2a treatment
HBeAg-positive chronic hepatitis B: In HBeAg-positive chronic hepatitis B serum, HBsAg level is closely related with intrahepatic cccDNA level and can reflect intrahepatic cccDNA contents. Reduction of HBsAg level implies a decrease in intrahepatic cccDNA[19-21]. Responses of HBeAg-positive patients to Peg-IFN-α2a therapy can be predicted according to HBsAg reduction. In an early study by Janssen et al[22], HBeAg-positive patients with sustained virological response (SVR) to Peg-IFN-α2a therapy showed a dramatic reduction of serum HBsAg. In addition, the HBsAg level in patients with SVR was lower than in non-responders at the end of therapy[22-24].
Seroconversion of HBeAg and SVR can be predicted from the baseline serum HBsAg level and its dynamic changes during Peg-IFN treatment. Early serological responses, which are defined as a low HBsAg level or a dramatic reduction of HBsAg during treatment, implies high seroconversion of HBeAg and HBV DNA suppression six months after treatment[24-26]. Specifically, patients who presented HBsAg < 300 IU/mL and HBeAg positive at week 24 during Peg-IFN-α2a treatment achieved an SVR of 62%, but the SVR of the remaining patients was only 11%[25]. Patients who had HBsAg reduction > 1log10 IU/mL and the absolute HBsAg < 300 IU/mL at week 24 of the therapy achieved an SVR of 75% six months after treatment. However, SVR of those patients without this combined response was only 15% (P < 0.001). This combined HBsAg response generated positive predictive values (PPVs) of 75% and negative predictive values (NPVs) of 85% for achieving SVR in HBeAg-positive patients.
HBsAg level decline at weeks 12 and 24 during treatment is an alternative index to predict SVR in HBeAg-positive patients and identify non-responders. In the phase III registration trial on Peg-IFN-α-2a, the PPV of HBsAg < 1500 IU/mL at week 12 and 24 on-treatment for achieving HBeAg seroconversion six months after treatment were 57% and 54% and NPV was 72% and 76%, respectively[26]. Sonneveld et al[27] discovered that SVR of patients without any decline of HBsAg at week 12 on-treatment was only 3%. Therefore, the absence of any decline in HBsAg at week 12 generated an NPV of 97% for response prediction six months after treatment.
HBeAg-negative chronic hepatitis B: Only a few studies have discussed the baseline response predictors for peginterferon-based therapy in HBeAg-negative patients. According to the existing data, low baseline HBsAg level is associated with SVR (defined as HBV DNA < 2000 IU/mL six mo after treatment)[28]. However, this finding has not been proven by other studies[29,30].
Dynamic monitoring of HBsAg levels in HBeAg-negative patients treated with peginterferon may complement the predicting value of HBV DNA alone[31-33]. Existing clinical data showed that SVR of HBeAg-negative patients to Peg-IFN-α2a can be predicted according to HBsAg reduction or the absolute level at week 12 or 24 during treatment. If HBsAg level decreased by 0.5log10 IU/mL at week 12 and 1log10 IU/mL at week 24, then the corresponding PPVs of SVR were 89% and 92% and NPVs were 90% and 97%, respectively[30]. Retrospective analysis of dynamic HBsAg level changes in 120 HBeAg-negative patients who were enrolled into the Peg-IFN-α-2a registration study found that patients with an HBsAg level decline of more than 10% from baseline at week 12 on-treatment achieved higher virus inhibition rates than those with declines of less than 10% (47% vs 16%, P < 0.01)[34]. However, HBsAg clearance occurred in a considerable proportion of patients who did not achieve more than 10% decline in HBsAg level.
Another study on Peg-IFN-α-2a therapy in HBeAg-negative patients predominantly infected with HBV genotype D indicated that dynamic monitoring of HBV DNA and HBsAg are superior to solely either marker in predicting therapeutic effects[29]. In this study, the absence of HBsAg level decline and HBV DNA reduction of less than 2 logs after 12 wk of PEG-IFN antiviral therapy were associated with no response (defined as HBV DNA > 10 000 copies/mL and ALT remains abnormal 6 mo after treatment). This finding is accepted as a stopping rule and is verified by some studies[29]. Nevertheless, such condition is hardly applicable to patients infected with other HBV genotypes. This result might be related with the changing influences of different genotypes on HBsAg during the treatment. Therefore, specific predictive values of different genotypes must be determined.
Additionally, a few studies have discussed the role of HBsAg level at the end of treatment in the prediction of follow-up SVR and subsequent HBsAg clearance[35]. In this study, 52% of 23 patients with HBsAg level < 10 IU/mL at the end of treatment achieved HBsAg clearance three years after treatment, and only 2% of the remaining patients achieved HBsAg clearance. Notably, HBsAg level at the end of treatment is more important than the HBV DNA level in predicting HBsAg clearance[34].
Conclusion: HBsAg reduction in HBeAg-positive patients at weeks 12 and 24 during Peg-IFN-α-2a therapy is conducive to the prediction of post-treatment SVR and effective identification of non-responders. Generally, HBsAg level identifies non-responders at week 12 and predicts SVR at week 24. The combined HBsAg and HBV DNA reduction in HBeAg-negative patients at week 12 can effectively recognize non-responders, especially patients infected with HBV genotype D.
NAs therapy
Variation trend of HBsAg level during NAs therapy: NAs inhibits HBV replication by directly preventing HBV polymerase without affecting the synthesis of HBsAg. Selective virus gene mutation of NAs might result in changes in S open reading frame[36]. Although no direct evidence exists for influences of these genetic changes on serum HBsAg level, these changes can certainly cause retention of intrahepatic HBsAg and increase carcinogenic risks[37]. These factors affect the value of HBsAg quantification in predicting the antiviral efficacy of NAs. Consequently, a few studies on this topic are available, and numerous heterogeneities exist among these studies[38-43]. Overall, serum HBsAg reduction in NAs therapy is slower and less significant compared with that in the interferon therapy[40,41]. HBeAg-positive patients showed a larger reduction of HBsAg level than HBeAg-negative patients[43].
HBeAg-positive chronic hepatitis B: Wursthorn et al[42] conducted a three-year follow-up observation of 162 HBeAg-positive patients on LDT treatment. After two years of treatment, all patients showed HBV DNA ≤ 60 IU/mL. Moreover, nine patients (6%) achieved HBsAg clearance. HBsAg clearance can be predicted from the sharp reduction of HBsAg (> 1log) after one year of treatment. This study confirmed the importance of quantitative HBsAg monitoring in the prediction of HBsAg clearance during NAs treatment. Similar results have been obtained in the follow-up TDF studies[44,45]. One small Chinese study disclosed that HBsAg < 100 IU/mL at the end of treatment was a sign of HBsAg seroconversion for two years after treatment[43].
HBeAg-negative chronic hepatitis B: Among ETV- and TDF-treated patients, HBeAg-negative patients achieved a smaller reduction of HBsAg level compared with HBeAg-positive patients[44,46]. A study in Hong Kong including 53 HBeAg-negative patients who had an average of 19 mo continuous LAM treatment and at least 12 mo post-treatment follow-up demonstrated that the end-of-treatment HBsAg titer was an independent predictor for 12 mo after treatment sustained viral suppression (HBV DNA ≤ 200 IU/mL)[47]. All five patients with HBsAg ≤ 100 IU/mL and reduction > 1log IU/mL (PPV 100%) and four of the eight patients with either HBsAg ≤ 100 IU/mL or HBsAg reduction > 1log (PPV 50%) achieved 12 mo treatment sustained viral suppression. Moreover, other 40 patients with HBsAg reduction ≤ 1log and the absolute level > 100 IU/mL were recognized as non-responders (NPV 100%) at 12 mo after treatment. HBsAg level at the end of treatment can also predict cumulative sustained response and HBsAg clearance at five years after stopping LAM.
Overall, quantitative assay of serum HBsAg in patients with CHB can provide some references regarding prediction of response to IFN and NAs therapies. However, HBsAg level cannot completely reflect intrahepatic cccDNA activity, which is related with HBsAg synthesis process, quantitative assay, and antiviral drug effects. Therefore, other indexes must be combined to predict the efficacy of HBV antiviral therapy.
HBV RNA LEVEL IN PERIPHERAL BLOOD
Quantitative detection of HBV RNA in peripheral blood
The replication cycle of HBV DNA starts from endonuclear cccDNA transcription of pre-genomic RNA (pgRNA). pgRNA is enveloped in the nucleocapsid during the formation of virus, and HBV DNA polymerase transcripts offspring DNA using pgRNA as the template. The offspring DNAs enter into the cell nucleus and facilitate virus circulation. Some offspring DNAs are assembled in the endoplasmic reticulum into complete virions and secreted from cells[48-51]. The replicative cycle of HBV DNA shows that HBV RNA exists in cells and detecting HBV RNA in serum is difficult. In the early 1990s, some studies reported the detection of HBV RNA in peripheral blood mononuclear cells of patients infected with HBV[51-54]. However, it was until 1996 that Köck et al[55] reported the detection of HBV RNA in peripheral blood virions of patients with chronic HBV infection using the reverse transcription PCR method. In 2001, Su et al[11] detected full-length RNA (fRNA) of HBV and truncated RNA (tRNA) in peripheral blood of patients with chronic HBV infection. They also proved that fRNA is correlated with HBeAg and HBV DNA, while trRNA is independent of HBeAg and is weakly related with HBV DNA. Subsequently, they further studied the various modes of DNA and RNA in peripheral blood in patients with CHB after a short-term LAM therapy. HBV RNA-carrying virions only account for 1% of the total virions in peripheral blood of treatment-naive HBV-infected individuals. However, HBV RNA-carrying virions began to take the dominant role in virions after the LAM therapy. Moreover, HBV DNA level decreased more than HBV RNA level during the LAM therapy[56,57]. Rokuhara et al[58] gained similar results from a follow-up of 24 patients with CHB on LAM treatment. They concluded that HBV RNA level in peripheral blood can reflect cccDNA level. Most recently, Huang et al[59] further examined the correlation between serum HBV RNA and intrahepatic cccDNA level. Their results indicated that serum HBV RNA reflects cccDNA activity in HBeAg-positive CHB, and total serum nucleic acids (HBV DNA plus RNA) can better reflect the activity of intrahepatic cccDNA compared with the serum HBV RNA or HBV DNA level.
Quantitative HBV RNA in peripheral blood during antiviral therapy
As an alternative index that directly reflects intrahepatic cccDNA level, HBV RNA level is one of the HBV antiviral therapy efficacy evaluation markers. HBV RNA level can also be used to predict antiviral drug resistance and relapse after drug withdrawal[12-14,59-61].
Peginterferon treatment
HBeAg-positive chronic hepatitis B: Peginterferon can adjust immunity and directly inhibit virus. It can also inhibit replication of virus DNA, RNA, and cccDNA. Dynamic changes of virus RNA can demonstrate the effects of Peginterferon therapy in patients with CHB. Jansen et al[12] performed dynamic monitoring on virus RNA level in peripheral blood of 13 HBeAg-positive patients on 48 wk of interferon combined with ADV therapy and two years of follow-up visits. They discovered that the baseline HBV RNA level is unrelated with response to the therapy. HBV RNA decreases less compared with HBV DNA level at different time points. HBV RNA level in combined responders (defined as HBeAg clearance, HBV DNA ≤ 2000 IU/mL and normal ALT level at weeks 24 and 144 after treatment) is lower than that in non-responders at all-time points. Statistical differences exist between the two groups in terms of HBV RNA levels at all-time points after week 30 (P < 0.001). Therefore, responses of patients with CHB to the antiviral therapy can be predicted according to HBV RNA level. However, this study failed to disclose the threshold of prediction.
HBeAg-negative chronic hepatitis B: For HBeAg-negative patients, the HBV RNA levels of combined responders (HBV DNA ≤ 2000 IU/mL and persistent ALT normalization at 24 and 144 wk of treatment-free follow-up) are lower than those of non-responders before and during the treatment (P < 0.001). Some combined responders showed a considerable reduction of HBV RNA in the early period. During treatment, HBV RNA level of all combined responders at week six is lower than the minimum limit of detection. On the contrary, HBV RNA level of most non-responders is higher than the minimum limit of detection (1.8 ± 0.2 log10 copies/mL vs 3.7 ± 0.7 log10 copies/mL, P = 0.028) at week six. Therefore, baseline HBV RNA level is an independent predictor of responses to the therapy[12].
Nucleos(t)ide analog treatment: HBV RNA can be detected in HBeAg-positive or -negative patients with CHB before the treatment. During the NAs therapy, the reduction rate of HBV RNA is slower than that of HBV DNA level. At the end of the follow-up (week 120), HBV RNA level in HBeAg-negative patients is lower than that in HBeAg-positive patients[12]. HBV RNA levels of most patients are still higher than the minimum limit of detection when the HBV DNA levels are lower than the minimum limit of detection. However, HBV RNA levels in most HBeAg-negative patients are lower than the minimum limit of detection[12].
Prediction of antiviral drug resistance and relapse after drug withdrawal: Given that NAs cannot directly inhibit cccDNA, many patients easily suffer from relapse after drug withdrawal. They have to take medicines for a long time, even their entire lives[2,3]. Some patients may develop gene mutation related to drug resistance during the long-term antiviral therapy, thus resulting in the re-emergence of HBV DNA, hepatitis reflash, and even hepatic failure. Therefore, prediction of antiviral drug resistance and relapse after drug withdrawal is crucial. Existing evidence suggested that HBV RNA level can be a potential biomarker for monitoring gene mutation related to drug resistance and relapse after drug withdrawal[14,60].
Hatakeyama et al[60] detected HBV RNA levels in peripheral blood of 7 ETV-treated patients and 36 LAM-treated patients and found that the median serum HBV RNA levels were considerably higher in patients with YMDD mutations within one year of treatment (n =6, 1.788log copies/mL) than in those with YMDD mutations with more than one year of treatment (n = 12, 0.456log copies/mL, P = 0.0125) or in those without YMDD mutation (n = 18, 0.688log copies/mL, P = 0.039). The results indicated that high HBV RNA level in the early state is related with YMDD mutation.
In 2013, Tsuge et al[61] studied the correlation between HBV RNA and relapse of HBV DNA after drug withdrawal. Based on a 24-wk follow-up of 36 patients with CHB treated by NAs for at least 6 mo, 19 patients had HBV DNA relapse and 12 patients had ALT level rebound at 24 wk after discontinuation of NAs therapy. Serum total nucleic acids after three months of treatment were markedly correlated with HBV DNA rebound [odds ratio (OR) 9.474, 95% confidence interval (CI): 1.069-83.957, P = 0.015]. It is an independent predictor of virological recovery within 24 wk of NAs withdrawal. Wang et al[14] observed the performance of 33 patients at week 24 after NAs treatment for three or more years. All21 HBV RNA-positive patients experienced HBV DNA rebound at the end of treatment at week 24 after drug withdrawal, but only 3 (25%) of 12 end-of-treatment HBV RNA-negative patients had virological relapse at week 24 after drug withdrawal. According to the multivariate analysis, the end-of-treatment HBV RNA level is related to virological relapse at week 24 after drug withdrawal (P = 0.001). Summaries of the progression and status of antiviral monitor in patients with CHB are shown in Figure 1.
Figure 1.
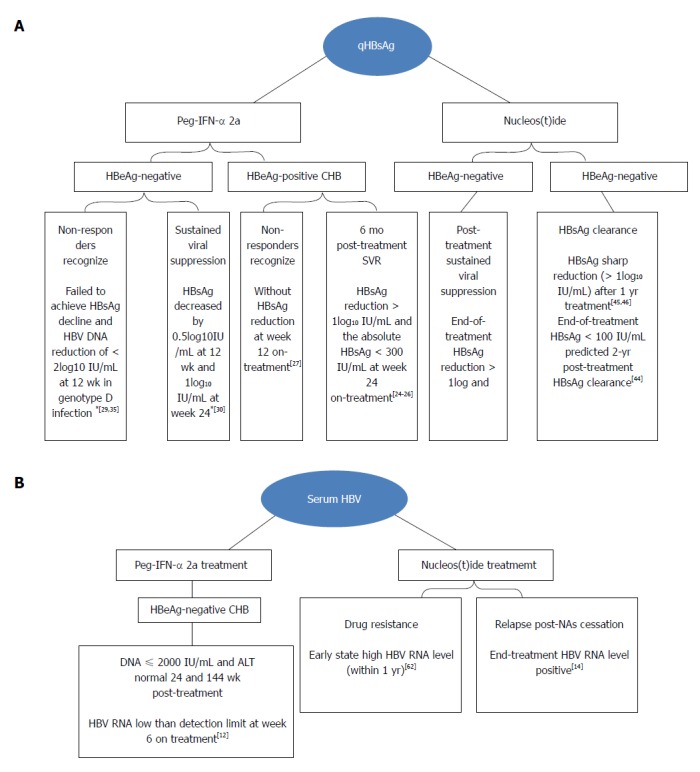
Progression and status of hepatitis B surface antigen (A) and serum hepatitis B virus (B) RNA in antiviral monitor in patients with chronic hepatitis B. HBsAg: Hepatitis B surface antigen; CHB: Chronic hepatitis B; HBV: Hepatitis B virus; Peg-IFN: Pegylated interferons; ALT: Alanine aminotransferase.
CONCLUSION
In summary, HBsAg comes from either cccDNA or integrated gene fragments[62]. HBsAg cannot completely represent the transcription activity of HBV cccDNA. HBV RNA, also known as pgRNA, only comes from cccDNA and can accurately reflect the cccDNA level. With the comprehensive knowledge on HBV RNA, the use of simultaneous continuous clearance of serum HBV DNA and HBV RNA is suggested as the safe stopping rule in patients with CHB on NAs treatment. However, clinical evidence based on a large sample size is needed to prove the feasibility and significance of this stopping rule.
Footnotes
Conflict-of-interest statement: No conflict of interest exists in this review.
Manuscript source: Invited manuscript
Peer-review started: March 20, 2018
First decision: April 11, 2018
Article in press: July 16, 2018
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): D
Grade E (Poor): 0
P- Reviewer: Aizawa Y, Dourakis SP, Otsuka M S- Editor: Ji FF L- Editor: Ma JY E- Editor: Tan WW
Contributor Information
Ya-Yun Liu, Department of Infectious Diseases, Changhai Hospital of Second Military Medical University, Shanghai 200433, China.
Xue-Song Liang, Department of Infectious Diseases, Changhai Hospital of Second Military Medical University, Shanghai 200433, China. liangxuesong2000@163.com.
References
- 1.Maddrey WC. Hepatitis B: an important public health issue. J Med Virol. 2000;61:362–366. doi: 10.1002/1096-9071(200007)61:3<362::aid-jmv14>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 2.European Association for the Study of the Liver. EASL clinical practice guidelines: Management of chronic hepatitis B virus infection. J Hepatol. 2012;57:167–185. doi: 10.1016/j.jhep.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 3.Terrault NA, Bzowej NH, Chang KM, Hwang JP, Jonas MM, Murad MH; American Association for the Study of Liver Diseases. AASLD guidelines for treatment of chronic hepatitis B. Hepatology. 2016;63:261–283. doi: 10.1002/hep.28156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoofnagle JH, Doo E, Liang TJ, Fleischer R, Lok AS. Management of hepatitis B: summary of a clinical research workshop. Hepatology. 2007;45:1056–1075. doi: 10.1002/hep.21627. [DOI] [PubMed] [Google Scholar]
- 5.Buti M, Tsai N, Petersen J, Flisiak R, Gurel S, Krastev Z, Aguilar Schall R, Flaherty JF, Martins EB, Charuworn P, et al. Seven-year efficacy and safety of treatment with tenofovir disoproxil fumarate for chronic hepatitis B virus infection. Dig Dis Sci. 2015;60:1457–1464. doi: 10.1007/s10620-014-3486-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perrillo RP, Lai CL, Liaw YF, Dienstag JL, Schiff ER, Schalm SW, Heathcote EJ, Brown NA, Atkins M, Woessner M, et al. Predictors of HBeAg loss after lamivudine treatment for chronic hepatitis B. Hepatology. 2002;36:186–194. doi: 10.1053/jhep.2002.34294. [DOI] [PubMed] [Google Scholar]
- 7.Marcellin P, Chang TT, Lim SG, Tong MJ, Sievert W, Shiffman ML, Jeffers L, Goodman Z, Wulfsohn MS, Xiong S, et al. Adefovir dipivoxil for the treatment of hepatitis B e antigen-positive chronic hepatitis B. N Engl J Med. 2003;348:808–816. doi: 10.1056/NEJMoa020681. [DOI] [PubMed] [Google Scholar]
- 8.Marcellin P, Heathcote EJ, Buti M, Gane E, de Man RA, Krastev Z, Germanidis G, Lee SS, Flisiak R, Kaita K, et al. Tenofovir disoproxil fumarate versus adefovir dipivoxil for chronic hepatitis B. N Engl J Med. 2008;359:2442–2455. doi: 10.1056/NEJMoa0802878. [DOI] [PubMed] [Google Scholar]
- 9.Wang Y, Thongsawat S, Gane EJ, Liaw YF, Jia J, Hou J, Chan HL, Papatheodoridis G, Wan M, Niu J, et al. Efficacy and safety of continuous 4-year telbivudine treatment in patients with chronic hepatitis B. J Viral Hepat. 2013;20:e37–e46. doi: 10.1111/jvh.12025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lai CL, Gane E, Liaw YF, Hsu CW, Thongsawat S, Wang Y, Chen Y, Heathcote EJ, Rasenack J, Bzowej N, et al. Telbivudine versus lamivudine in patients with chronic hepatitis B. N Engl J Med. 2007;357:2576–2588. doi: 10.1056/NEJMoa066422. [DOI] [PubMed] [Google Scholar]
- 11.Su Q, Wang SF, Chang TE, Breitkreutz R, Hennig H, Takegoshi K, Edler L, Schröder CH. Circulating hepatitis B virus nucleic acids in chronic infection : representation of differently polyadenylated viral transcripts during progression to nonreplicative stages. Clin Cancer Res. 2001;7:2005–2015. [PubMed] [Google Scholar]
- 12.Jansen L, Kootstra NA, van Dort KA, Takkenberg RB, Reesink HW, Zaaijer HL. Hepatitis B Virus Pregenomic RNA Is Present in Virions in Plasma and Is Associated With a Response to Pegylated Interferon Alfa-2a and Nucleos(t)ide Analogues. J Infect Dis. 2016;213:224–232. doi: 10.1093/infdis/jiv397. [DOI] [PubMed] [Google Scholar]
- 13.van Bömmel F, Bartens A, Mysickova A, Hofmann J, Krüger DH, Berg T, Edelmann A. Serum hepatitis B virus RNA levels as an early predictor of hepatitis B envelope antigen seroconversion during treatment with polymerase inhibitors. Hepatology. 2015;61:66–76. doi: 10.1002/hep.27381. [DOI] [PubMed] [Google Scholar]
- 14.Wang J, Shen T, Huang X, Kumar GR, Chen X, Zeng Z, Zhang R, Chen R, Li T, Zhang T, et al. Serum hepatitis B virus RNA is encapsidated pregenome RNA that may be associated with persistence of viral infection and rebound. J Hepatol. 2016;65:700–710. doi: 10.1016/j.jhep.2016.05.029. [DOI] [PubMed] [Google Scholar]
- 15.Mak LY, Wong DK, Cheung KS, Seto WK, Lai CL, Yuen MF. Review article: hepatitis B core-related antigen (HBcrAg): an emerging marker for chronic hepatitis B virus infection. Aliment Pharmacol Ther. 2018;47:43–54. doi: 10.1111/apt.14376. [DOI] [PubMed] [Google Scholar]
- 16.Blumberg BS, Sutnick AI, London WT. Hepatitis and leukemia: their relation to Australia antigen. Bull N Y Acad Med. 1968;44:1566–1586. [PMC free article] [PubMed] [Google Scholar]
- 17.Chan HL, Wong VW, Tse AM, Tse CH, Chim AM, Chan HY, Wong GL, Sung JJ. Serum hepatitis B surface antigen quantitation can reflect hepatitis B virus in the liver and predict treatment response. Clin Gastroenterol Hepatol. 2007;5:1462–1468. doi: 10.1016/j.cgh.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 18.Tseng TC, Kao JH. Clinical utility of quantitative HBsAg in natural history and nucleos(t)ide analogue treatment of chronic hepatitis B: new trick of old dog. J Gastroenterol. 2013;48:13–21. doi: 10.1007/s00535-012-0668-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thompson AJ, Nguyen T, Iser D, Ayres A, Jackson K, Littlejohn M, Slavin J, Bowden S, Gane EJ, Abbott W, et al. Serum hepatitis B surface antigen and hepatitis B e antigen titers: disease phase influences correlation with viral load and intrahepatic hepatitis B virus markers. Hepatology. 2010;51:1933–1944. doi: 10.1002/hep.23571. [DOI] [PubMed] [Google Scholar]
- 20.Chan HL, Wong VW, Wong GL, Tse CH, Chan HY, Sung JJ. A longitudinal study on the natural history of serum hepatitis B surface antigen changes in chronic hepatitis B. Hepatology. 2010;52:1232–1241. doi: 10.1002/hep.23803. [DOI] [PubMed] [Google Scholar]
- 21.Hadziyannis SJ, Papatheodoridis GV. Hepatitis B e antigen-negative chronic hepatitis B: natural history and treatment. Semin Liver Dis. 2006;26:130–141. doi: 10.1055/s-2006-939751. [DOI] [PubMed] [Google Scholar]
- 22.Janssen HL, Kerhof-Los CJ, Heijtink RA, Schalm SW. Measurement of HBsAg to monitor hepatitis B viral replication in patients on alpha-interferon therapy. Antiviral Res. 1994;23:251–257. doi: 10.1016/0166-3542(94)90022-1. [DOI] [PubMed] [Google Scholar]
- 23.Wong VW, Wong GL, Yan KK, Chim AM, Chan HY, Tse CH, Choi PC, Chan AW, Sung JJ, Chan HL. Durability of peginterferon alfa-2b treatment at 5 years in patients with hepatitis B e antigen-positive chronic hepatitis B. Hepatology. 2010;51:1945–1953. doi: 10.1002/hep.23568. [DOI] [PubMed] [Google Scholar]
- 24.Tangkijvanich P, Komolmit P, Mahachai V, Sa-nguanmoo P, Theamboonlers A, Poovorawan Y. Low pretreatment serum HBsAg level and viral mutations as predictors of response to PEG-interferon alpha-2b therapy in chronic hepatitis B. J Clin Virol. 2009;46:117–123. doi: 10.1016/j.jcv.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 25.Chan HL, Wong VW, Chim AM, Chan HY, Wong GL, Sung JJ. Serum HBsAg quantification to predict response to peginterferon therapy of e antigen positive chronic hepatitis B. Aliment Pharmacol Ther. 2010;32:1323–1331. doi: 10.1111/j.1365-2036.2010.04474.x. [DOI] [PubMed] [Google Scholar]
- 26.Martinot-Peignoux M, Asselah T, Marcellin P. HBsAg quantification to optimize treatment monitoring in chronic hepatitis B patients. Liver Int. 2015;35 Suppl 1:82–90. doi: 10.1111/liv.12735. [DOI] [PubMed] [Google Scholar]
- 27.Sonneveld MJ, Rijckborst V, Boucher CA, Hansen BE, Janssen HL. Prediction of sustained response to peginterferon alfa-2b for hepatitis B e antigen-positive chronic hepatitis B using on-treatment hepatitis B surface antigen decline. Hepatology. 2010;52:1251–1257. doi: 10.1002/hep.23844. [DOI] [PubMed] [Google Scholar]
- 28.Takkenberg RB, Jansen L, de Niet A, Zaaijer HL, Weegink CJ, Terpstra V, Dijkgraaf MG, Molenkamp R, Jansen PL, Koot M, et al. Baseline hepatitis B surface antigen (HBsAg) as predictor of sustained HBsAg loss in chronic hepatitis B patients treated with pegylated interferon-α2a and adefovir. Antivir Ther. 2013;18:895–904. doi: 10.3851/IMP2580. [DOI] [PubMed] [Google Scholar]
- 29.Rijckborst V, Hansen BE, Cakaloglu Y, Ferenci P, Tabak F, Akdogan M, Simon K, Akarca US, Flisiak R, Verhey E, et al. Early on-treatment prediction of response to peginterferon alfa-2a for HBeAg-negative chronic hepatitis B using HBsAg and HBV DNA levels. Hepatology. 2010;52:454–461. doi: 10.1002/hep.23722. [DOI] [PubMed] [Google Scholar]
- 30.Moucari R, Mackiewicz V, Lada O, Ripault MP, Castelnau C, Martinot-Peignoux M, Dauvergne A, Asselah T, Boyer N, Bedossa P, et al. Early serum HBsAg drop: a strong predictor of sustained virological response to pegylated interferon alfa-2a in HBeAg-negative patients. Hepatology. 2009;49:1151–1157. doi: 10.1002/hep.22744. [DOI] [PubMed] [Google Scholar]
- 31.Rijckborst V, ter Borg MJ, Cakaloglu Y, Ferenci P, Tabak F, Akdogan M, Simon K, Raptopoulou-Gigi M, Ormeci N, Zondervan PE, et al. A randomized trial of peginterferon alpha-2a with or without ribavirin for HBeAg-negative chronic hepatitis B. Am J Gastroenterol. 2010;105:1762–1769. doi: 10.1038/ajg.2010.186. [DOI] [PubMed] [Google Scholar]
- 32.Marcellin P, Bonino F, Lau GK, Farci P, Yurdaydin C, Piratvisuth T, Jin R, Gurel S, Lu ZM, Wu J, et al. Sustained response of hepatitis B e antigen-negative patients 3 years after treatment with peginterferon alpha-2a. Gastroenterology. 2009;136:2169–2179.e1-4. doi: 10.1053/j.gastro.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 33.Marcellin P, Lau GK, Bonino F, Farci P, Hadziyannis S, Jin R, Lu ZM, Piratvisuth T, Germanidis G, Yurdaydin C, et al. Peginterferon alfa-2a alone, lamivudine alone, and the two in combination in patients with HBeAg-negative chronic hepatitis B. N Engl J Med. 2004;351:1206–1217. doi: 10.1056/NEJMoa040431. [DOI] [PubMed] [Google Scholar]
- 34.Janssen HL, Sonneveld MJ, Brunetto MR. Quantification of serum hepatitis B surface antigen: is it useful for the management of chronic hepatitis B? Gut. 2012;61:641–645. doi: 10.1136/gutjnl-2011-301096. [DOI] [PubMed] [Google Scholar]
- 35.Brunetto MR, Moriconi F, Bonino F, Lau GK, Farci P, Yurdaydin C, Piratvisuth T, Luo K, Wang Y, Hadziyannis S, et al. Hepatitis B virus surface antigen levels: a guide to sustained response to peginterferon alfa-2a in HBeAg-negative chronic hepatitis B. Hepatology. 2009;49:1141–1150. doi: 10.1002/hep.22760. [DOI] [PubMed] [Google Scholar]
- 36.Warner N, Locarnini S. The antiviral drug selected hepatitis B virus rtA181T/sW172* mutant has a dominant negative secretion defect and alters the typical profile of viral rebound. Hepatology. 2008;48:88–98. doi: 10.1002/hep.22295. [DOI] [PubMed] [Google Scholar]
- 37.Warner N, Locarnini S. Can antiviral therapy for chronic hepatitis B enhance the progression to hepatocellular carcinoma? Antivir Ther. 2009;14:139–142. [PubMed] [Google Scholar]
- 38.Kohmoto M, Enomoto M, Tamori A, Habu D, Takeda T, Kawada N, Sakaguchi H, Seki S, Shiomi S, Nishiguchi S. Quantitative detection of hepatitis B surface antigen by chemiluminescent microparticle immunoassay during lamivudine treatment of chronic hepatitis B virus carriers. J Med Virol. 2005;75:235–239. doi: 10.1002/jmv.20262. [DOI] [PubMed] [Google Scholar]
- 39.Manesis EK, Hadziyannis ES, Angelopoulou OP, Hadziyannis SJ. Prediction of treatment-related HBsAg loss in HBeAG-negative chronic hepatitis B: a clue from serum HBsAg levels. Antivir Ther. 2007;12:73–82. [PubMed] [Google Scholar]
- 40.Wiegand J, Wedemeyer H, Finger A, Heidrich B, Rosenau J, Michel G, Bock CT, Manns MP, Tillmann HL. A decline in hepatitis B virus surface antigen (hbsag) predicts clearance, but does not correlate with quantitative hbeag or HBV DNA levels. Antivir Ther. 2008;13:547–554. [PubMed] [Google Scholar]
- 41.Borgniet O, Parvaz P, Bouix C, Chevallier P, Trépo C, André P, Zoulim F. Clearance of serum HBsAg and anti-HBs seroconversion following antiviral therapy for chronic hepatitis B. J Med Virol. 2009;81:1336–1342. doi: 10.1002/jmv.21519. [DOI] [PubMed] [Google Scholar]
- 42.Wursthorn K, Jung M, Riva A, Goodman ZD, Lopez P, Bao W, Manns MP, Wedemeyer H, Naoumov NV. Kinetics of hepatitis B surface antigen decline during 3 years of telbivudine treatment in hepatitis B e antigen-positive patients. Hepatology. 2010;52:1611–1620. doi: 10.1002/hep.23905. [DOI] [PubMed] [Google Scholar]
- 43.Cai W, Xie Q, An B, Wang H, Zhou X, Zhao G, Guo Q, Gu R, Bao S. On-treatment serum HBsAg level is predictive of sustained off-treatment virologic response to telbivudine in HBeAg-positive chronic hepatitis B patients. J Clin Virol. 2010;48:22–26. doi: 10.1016/j.jcv.2010.02.014. [DOI] [PubMed] [Google Scholar]
- 44.Heathcote EJ, Marcellin P, Buti M, Gane E, De Man RA, Krastev Z, Germanidis G, Lee SS, Flisiak R, Kaita K, et al. Three-year efficacy and safety of tenofovir disoproxil fumarate treatment for chronic hepatitis B. Gastroenterology. 2011;140:132–143. doi: 10.1053/j.gastro.2010.10.011. [DOI] [PubMed] [Google Scholar]
- 45.Marcellin P, Buti M, Krastev Z, de Man RA, Zeuzem S, Lou L, Gaggar A, Flaherty JF, Massetto B, Lin L, et al. Kinetics of hepatitis B surface antigen loss in patients with HBeAg-positive chronic hepatitis B treated with tenofovir disoproxil fumarate. J Hepatol. 2014;61:1228–1237. doi: 10.1016/j.jhep.2014.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reijnders JG, Rijckborst V, Sonneveld MJ, Scherbeijn SM, Boucher CA, Hansen BE, Janssen HL. Kinetics of hepatitis B surface antigen differ between treatment with peginterferon and entecavir. J Hepatol. 2011;54:449–454. doi: 10.1016/j.jhep.2010.07.046. [DOI] [PubMed] [Google Scholar]
- 47.Chan HL, Wong GL, Chim AM, Chan HY, Chu SH, Wong VW. Prediction of off-treatment response to lamivudine by serum hepatitis B surface antigen quantification in hepatitis B e antigen-negative patients. Antivir Ther. 2011;16:1249–1257. doi: 10.3851/IMP1921. [DOI] [PubMed] [Google Scholar]
- 48.Ganem D, Prince AM. Hepatitis B virus infection--natural history and clinical consequences. N Engl J Med. 2004;350:1118–1129. doi: 10.1056/NEJMra031087. [DOI] [PubMed] [Google Scholar]
- 49.Pan CQ, Zhang JX. Natural History and Clinical Consequences of Hepatitis B Virus Infection. Int J Med Sci. 2005;2:36–40. doi: 10.7150/ijms.2.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schädler S, Hildt E. HBV life cycle: entry and morphogenesis. Viruses. 2009;1:185–209. doi: 10.3390/v1020185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baginski I, Chemin I, Bouffard P, Hantz O, Trepo C. Detection of polyadenylated RNA in hepatitis B virus-infected peripheral blood mononuclear cells by polymerase chain reaction. J Infect Dis. 1991;163:996–1000. doi: 10.1093/infdis/163.5.996. [DOI] [PubMed] [Google Scholar]
- 52.Lobbiani A, Lalatta F, Lugo F, Colucci G. Hepatitis B virus transcripts and surface antigen in human peripheral blood lymphocytes. J Med Virol. 1990;31:190–194. doi: 10.1002/jmv.1890310304. [DOI] [PubMed] [Google Scholar]
- 53.Leung NW, Tam JS, Lau GT, Leung TW, Lau WY, Li AK. Hepatitis B virus DNA in peripheral blood leukocytes. A comparison between hepatocellular carcinoma and other hepatitis B virus-related chronic liver diseases. Cancer. 1994;73:1143–1148. doi: 10.1002/1097-0142(19940215)73:4<1143::aid-cncr2820730404>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 54.Delfini C, Garbuglia AR, Alfani E, Di Caro A, Sette P, Benedetto A. Heroin addicts infected by HBV and HIV have a low prevalence of HBV DNA in peripheral blood mononuclear cells. J Med Virol. 1993;41:114–119. doi: 10.1002/jmv.1890410206. [DOI] [PubMed] [Google Scholar]
- 55.Köck J, Theilmann L, Galle P, Schlicht HJ. Hepatitis B virus nucleic acids associated with human peripheral blood mononuclear cells do not originate from replicating virus. Hepatology. 1996;23:405–413. doi: 10.1002/hep.510230303. [DOI] [PubMed] [Google Scholar]
- 56.Hacker HJ, Zhang W, Tokus M, Bock T, Schröder CH. Patterns of circulating hepatitis B virus serum nucleic acids during lamivudine therapy. Ann N Y Acad Sci. 2004;1022:271–281. doi: 10.1196/annals.1318.042. [DOI] [PubMed] [Google Scholar]
- 57.Zhang W, Hacker HJ, Tokus M, Bock T, Schröder CH. Patterns of circulating hepatitis B virus serum nucleic acids during lamivudine therapy. J Med Virol. 2003;71:24–30. doi: 10.1002/jmv.10464. [DOI] [PubMed] [Google Scholar]
- 58.Rokuhara A, Matsumoto A, Tanaka E, Umemura T, Yoshizawa K, Kimura T, Maki N, Kiyosawa K. Hepatitis B virus RNA is measurable in serum and can be a new marker for monitoring lamivudine therapy. J Gastroenterol. 2006;41:785–790. doi: 10.1007/s00535-006-1856-4. [DOI] [PubMed] [Google Scholar]
- 59.Huang H, Wang J, Li W, Chen R, Chen X, Zhang F, Xu D, Lu F. Serum HBV DNA plus RNA shows superiority in reflecting the activity of intrahepatic cccDNA in treatment-naïve HBV-infected individuals. J Clin Virol. 2018;99-100:71–78. doi: 10.1016/j.jcv.2017.12.016. [DOI] [PubMed] [Google Scholar]
- 60.Hatakeyama T, Noguchi C, Hiraga N, Mori N, Tsuge M, Imamura M, Takahashi S, Kawakami Y, Fujimoto Y, Ochi H, et al. Serum HBV RNA is a predictor of early emergence of the YMDD mutant in patients treated with lamivudine. Hepatology. 2007;45:1179–1186. doi: 10.1002/hep.21581. [DOI] [PubMed] [Google Scholar]
- 61.Tsuge M, Murakami E, Imamura M, Abe H, Miki D, Hiraga N, Takahashi S, Ochi H, Nelson Hayes C, Ginba H, et al. Serum HBV RNA and HBeAg are useful markers for the safe discontinuation of nucleotide analogue treatments in chronic hepatitis B patients. J Gastroenterol. 2013;48:1188–1204. doi: 10.1007/s00535-012-0737-2. [DOI] [PubMed] [Google Scholar]
- 62.Zucman-Rossi J, Laurent-Puig P. Genetic diversity of hepatocellular carcinomas and its potential impact on targeted therapies. Pharmacogenomics. 2007;8:997–1003. doi: 10.2217/14622416.8.8.997. [DOI] [PubMed] [Google Scholar]


