Abstract
Acquisition of the trace element copper (Cu) is critical to drive essential eukaryotic processes such as oxidative phosphorylation, iron mobilization, peptide hormone biogenesis, and connective tissue maturation. The Ctr1/Ctr3 family of Cu importers, first discovered in fungi and conserved in mammals, are critical for Cu+ movement across the plasma membrane or mobilization from endosomal compartments. Whereas ablation of Ctr1 in mammals is embryonic lethal, and Ctr1 is critical for dietary Cu absorption, cardiac function, and systemic iron distribution, little is known about the intrinsic contribution of Ctr1 for Cu+ permeation through membranes or its mechanism of action. Here, we identify three members of a Cu+ importer family from the thermophilic fungus Chaetomium thermophilum: Ctr3a and Ctr3b, which function on the plasma membrane, and Ctr2, which likely functions in endosomal Cu mobilization. All three proteins drive Cu and isoelectronic silver (Ag) uptake in cells devoid of Cu+ importers. Transport activity depends on signature amino acid motifs that are conserved and essential for all Ctr1/3 transporters. Ctr3a is stable and amenable to purification and was incorporated into liposomes to reconstitute an in vitro Ag+ transport assay characterized by stopped-flow spectroscopy. Ctr3a has intrinsic high-affinity metal ion transport activity that closely reflects values determined in vivo, with slow turnover kinetics. Given structural models for mammalian Ctr1, Ctr3a likely functions as a low-efficiency Cu+ ion channel. The Ctr1/Ctr3 family may be tuned to import essential yet potentially toxic Cu+ ions at a slow rate to meet cellular needs, while minimizing labile intracellular Cu+ pools.
Keywords: copper, copper transport, metal homeostasis, liposome, thermophile, Ctr1
Introduction
Copper (Cu) is an essential metal that drives catalysis for a variety of critical biological processes such as neuropeptide and hormone biogenesis, mitochondrial oxidative phosphorylation, pigmentation, neuropeptide maturation, neutrophil development, superoxide disproportionation, and connective tissue maturation (1, 2). Consequently, Cu deficiency in mammals causes defects in iron mobilization and thermoregulation, peripheral neuropathy, skin laxity, hypertrophic cardiomyopathy, and irreversible cognitive impairment (3–7). However, the redox active nature of Cu (Cu+, Cu2+) generates reactive oxygen species that damage proteins, lipids, and nucleic acids, and the electrochemical properties of Cu drive metal–ligand complex formation, causing inappropriate displacement of other metals, such as zinc and iron, from physiological ligands (8–10). Consequently, the influx of Cu must be carefully orchestrated in response to needs, available storage capacity, and the implementation of rheostatically regulated detoxification mechanisms.
Cu acquisition in eukaryotic cells occurs through multiple mechanisms, such as delivery by the multi-Cu oxidase ceruloplasmin in mammals (11), low-affinity transporters identified in fungi (12), and via a family of high-affinity plasma membrane transporters pioneered by the functionally redundant Ctr1 (Cu transporter 1) and Ctr3 (Cu transporter 3) proteins in the baker's yeast, Saccharomyces cerevisiae, that are broadly conserved in mammals and other metazoans (13–15). Ctr1/3 proteins are characterized by a variable length N-terminal extracellular domain (ectodomain) that is often Met- and His-rich, with three transmembrane domains flanking an intracellular loop and a short cytosolic tail (16–18) (Fig. 1A). Genetic, biochemical, and structural studies demonstrate that Ctr1/3 proteins function as a homotrimeric complex in Cu+ acquisition (16, 19–23). Whereas loss or mutagenesis of the Met or His Cu ligands in the ectodomain reduces Cu import but does not ablate activity, mutagenesis of a MXXXM (X is any amino acid) motif in the second transmembrane domain completely abolishes Ctr1 activity (16, 24). Also essential for Ctr1/3 function is a conserved glycine zipper motif in the third transmembrane domain rich in Gly, Ala, and Ser residues that facilitates helical packing in transmembrane proteins (25). Mutation of this motif results in protein instability and proteasomal degradation (22). The substrate for Ctr1/Ctr3-mediated Cu transport is Cu+ rather than Cu2+, demanding the function of cell-surface metallo-reductases that have been identified in fungi and mammals or an extracellular ligand to provide reduced Cu+ to the site of Ctr1/Ctr3-mediated import (26–28). Notably, the only observed difference between Ctr1 and Ctr3 Cu+ import proteins is the utilization of Cys residues, in addition to Met and His, in the ectodomain of Ctr3 (29).
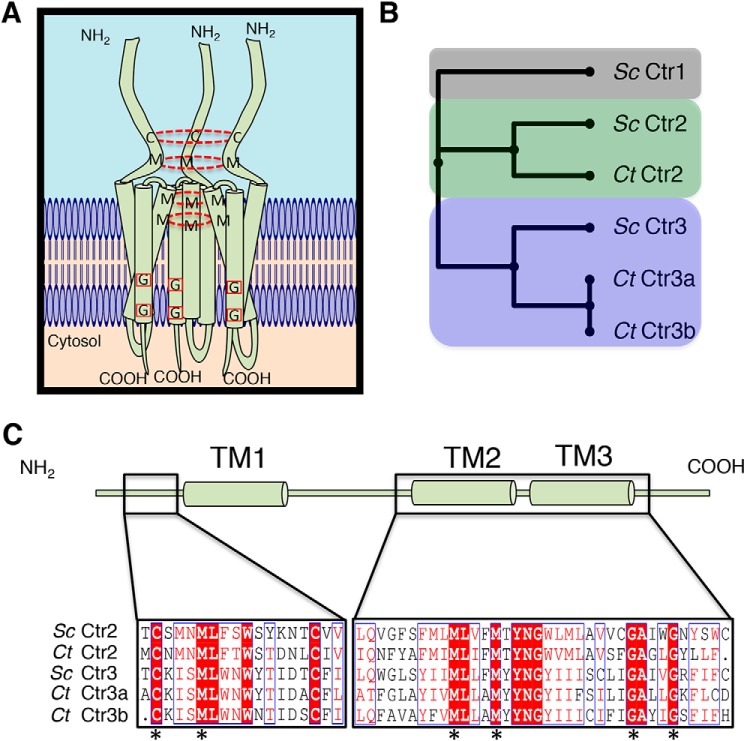
Figure 1.
Bioinformatic analysis identifies C. thermophilum copper transporters. A, model showing a homotrimeric Ctr1/Ctr3 family protein complex with key Cys and Met residues involved in Cu+ transport connected with a dashed red circle and key Gly residues involved in trimer stability via transmembrane helical packing highlighted with a red box. B, putative C. thermophilum Ctr proteins were aligned against characterized S. cerevisiae Ctr proteins with MUSCLE and compiled into a phylogenetic tree. C, C. thermophilum Ctr2 and Ctr3 proteins possess critical functional residues that are hallmarks of Ctr family members. Residues in white type surrounded by red are strictly conserved, residues in red type surrounded by a blue line possess similar biochemical properties, and residues in black type are not conserved. Asterisks indicate strictly conserved residues that are critical for transport activity.
Genetic studies demonstrate the importance of the Ctr1/Ctr3 family of Cu importers in diverse eukaryotes. Loss of both Ctr1/Ctr3 Cu importers in yeast results in Cu deficiency in the cytosol, mitochondria, and secretory compartment (16). Structurally related Cu transporters, Ctr2 in S. cerevisiae and Ctr6 in Schizosaccharomyces pombe, localize to the vacuolar membrane and serve to mobilize vacuolar Cu stores under conditions of extracellular Cu scarcity (30–32). Systemic inactivation of the sole identified Cu transporter in mammals, Ctr1, results in embryonic lethality (33, 34), whereas intestine-specific loss of Ctr1 results in peripheral Cu deficiency and hypertrophic cardiomyopathy (35), which is recapitulated by a cardiac-specific Ctr1 knockout (36). Interestingly, inactivation of Ctr1 in the liver reduces hepatic Cu accumulation, while enhancing urinary Cu excretion, suggesting compensatory homeostatic responses to Ctr1 status (37). In turn, hepatic or cardiac deletion of mitochondrial cytochrome oxidase Cu assembly factor Sco1 results in enhanced Ctr1 turnover or changes in Ctr1 trafficking, respectively (38, 39). Together, these reports demonstrate the critical function and regulation of Ctr1/Ctr3 Cu+ importers in cellular and organismal physiology and Cu homeostasis.
Although we know much about the physiological function of Ctr1/Ctr3 Cu importers, little is known about their mechanism for Cu+ import and the structural features that drive Cu+ acquisition. For instance, eukaryotes possess two cytosolic Cu+ chaperones, Atx1 and CCS, both of which interact with and potentially modulate Ctr1 activity (40–42). Additionally, an increase in the Cu-binding tripeptide GSH, present at millimolar concentrations (43), increases Ctr1-dependent Cu+ uptake in cultured cells (44). Accordingly, depletion of cellular GSH reduces Ctr1-dependent Cu import, suggesting a role for this intracellular Cu-buffering molecule in Ctr1 function. Furthermore, increasing concentrations of both potassium (K+) and protons (H+) increases cellular Ctr1-dependent Cu import, raising the possibility that counterions may be required for Ctr1-mediated transport (18). Although structural changes in Ctr1/3 proteins have been observed in the presence of Cu+ ions (45), and the MXXXM motif in the second transmembrane domain is essential for Cu+ import (16, 18, 29), there is currently no information demonstrating that Ctr1/Ctr3 proteins have intrinsic Cu+ transport activity.
Initially Ctr1/Ctr3 proteins were presumed to function as transporters, defined by the possession of two ion gates on opposite sides of the membrane that open in alternating fashion (46). Indeed, other metal transporters, such as the ZIP and Znt families of zinc transporters (47–50) and the DMT1 family of iron transporters, function in this manner (51, 52). The large conformational changes associated with the precisely timed opening of two alternating gates results in substrate transport on the order of ∼101 to 103 ions/s for bona fide transporters. In vivo kinetic studies of mammalian Ctr1 revealed a concentration-dependent and saturable rate of transport of ∼101 ions/s, consistent with classical transporter proteins (18, 24, 53). This is in contrast to ion channels, which are defined by a single ion gate that allows for substrate ions to permeate down a central pore (46). Due to significantly less structural rearrangement required for channels compared with transporters, channels are capable of substrate transport up to ∼108 ions/s, near the rate of diffusion.
However, the role of Ctr1 as a transporter was challenged by an ∼7-Å structural model of human Ctr1 that revealed a porelike structure reminiscent of classical ion channels, suggesting that Cu+ permeates down the central channel (20, 21). Additional complexity is introduced by heterogeneity of Ctr1 protein cleavage and localization. Ctr1 exists in cells and mammalian tissues as two separate species: a full-length protein and a proteolytically cleaved protein in which the majority of the metal-binding ectodomain has been removed (54). As cells that express the ectodomain truncated form of Ctr1 exhibit decreased Cu+ accumulation (24), it is difficult to interpret in vivo Cu uptake kinetics. Moreover, Cu induces clathrin-mediated Ctr1 endocytosis, further complicating the interpretation of in vivo kinetic studies (55–57). In the absence of a purified in vitro system, the intrinsic nature by which Ctr1/Ctr3 proteins facilitate Cu+ transport across membranes has not been elucidated. One hurdle preventing this level of understanding lies in the difficulties in obtaining pure, stable protein that is amenable to biochemical characterization and the lack of a reconstituted in vitro metal ion transport assay.
To gain mechanistic insights into the mode of action of the Ctr1/Ctr3 family in Cu+ import, we mined the genome of a thermophilic fungus, Chaetomium thermophilum, for Ctr1/Ctr3 family members. Of three homologues identified, two that localize to the plasma membrane functionally substitute for loss of Ctr1 and Ctr3 in baker's yeast. The C. thermophilum proteins are amenable to the addition of affinity tags and biochemical purification and, in contrast to other Ctr1/Ctr3 family members, demonstrate stability after purification. An in vitro reconstituted Ctr1/Ctr3 vesicular metal ion transport assay was developed that validated the intrinsic metal ion transport activity of Ctr1/Ctr3 proteins and the requirement for the MXXXM motif in this process. Moreover, calculation of half-maximal transport (K½) and ion turnover rate for C. thermophilum Ctr3a demonstrates that this Ctr1/Ctr3 family transports with a high affinity but a surprisingly slow intrinsic rate, potentially to mitigate the toxicity associated with rapid Cu+ influx.
Results
The thermophilic fungus C. thermophilum possesses three putative Ctr1/Ctr3 homologues
Functional studies of the Ctr1/Ctr3 family of Cu+ importers have largely focused on the in vivo Cu accumulation, trafficking, and physiological defects in fungal, mammalian cell culture, or mouse genetics experiments. However, the in vitro characterization of Cu+ transport mechanisms has not been explored, in part due to a lack of stable, purified Ctr1/Ctr3 protein preparations that are amenable to biochemical analysis. To address this, the genome of a thermophilic fungus, C. thermophilum, was inspected for Ctr1/Ctr3 family members, following on the observation that proteins obtained from thermophiles tend to be more stable under purification conditions and therefore amenable to biochemical characterization (58). BLAST (59) analysis of the C. thermophilum genome revealed three putative Ctr1/Ctr3 homologues. Multiple Sequence Comparison by Log-Expectation (MUSCLE)3 (60) alignment suggests that two of the proteins are members of the Ctr3 subfamily (herein designated Ct Ctr3a and Ct Ctr3b), with the additional protein classified as a member of the vesicular localized Ctr2/Ctr6 subfamily (herein denoted Ct Ctr2) (Fig. 1B).
It is interesting to note that, unlike in S. cerevisiae, there are no homologues in C. thermophilum that can be assigned to the Ctr1-like subfamily, bearing a methionine-rich ectodomain. However, among the potential metal-binding residues located in the ectodomain and essential for Cu+ transport activity in S. cerevisiae Ctr3 (29), there is conservation of two critical Cys residues and one Met residue in the C. thermophilum Ctr2 and Ctr3 homologues (Fig. 1C). Additionally, the obligatory MXXXM motif in TM2 and the glycine zipper in TM3 required for helical packing are both present in the Ctr2 and Ctr3 homologues. These observations suggest that the three open reading frames identified in C. thermophilum encode members of the Ctr2 and Ctr3 family of Cu+ transporters.
C. thermophilum Ctr3 homologues rescue S. cerevisiae Cu-dependent growth
To test whether the C. thermophilum Ctr2/Ctr3 family members are functional Cu+ transporters in vivo, cDNA sequences for each were cloned into an S. cerevisiae expression vector and transformed into strain MPY17 (61), lacking both high-affinity Ctr1 and Ctr3 plasma membrane–localized Cu+ transporters. Due to loss of both CTR1 and CTR3, this strain exhibits a deficit in Cu acquisition, resulting in the inability to grow on nonfermentable carbon sources, such as ethanol and glycerol (YPD, ethanol (2%), and glycerol (3%) (YPEG) medium), due to a lack of Cu to support mitochondrial cytochrome oxidase activity, with normal growth on a dextrose carbon source (YPD). In S. cerevisiae, this YPEG growth defect is suppressed by exogenous Cu, which is imported via low-affinity cell-surface Cu transporters, such as Fet4 (12, 32).
The human Ctr1 cDNA, fused in-frame to enhanced GFP (hCtr1-EGFP) served as a positive control for the restoration of Cu-dependent growth of the S. cerevisiae ctr1Δ ctr3Δ strain on YPEG (Fig. 2A). Although MPY17 cells expressing Ct Ctr2 are not capable of growth on YPEG, Ct Ctr3a and Ct Ctr3b robustly restore growth to this strain. Interestingly, when the same cells are plated on synthetic complete (SC) ethanol glycerol (SCEG; SC, ethanol (2%), and glycerol (3%)) medium, which has higher levels of bioavailable Cu, Ct Ctr2 modestly rescues growth (Fig. 2B). This phenotype is reminiscent of the S. cerevisiae vacuolar Cu exporter, Ctr2, which is unable to rescue growth on YPEG without exogenous Cu (30) and further supports the hypothesis, based on sequence alignment (Fig. 1B), that Ct Ctr2 may function as a Ctr2-like vacuolar Cu transporter protein. The observation that the addition of 50 μm Cu to YPEG results in reduced growth for cells expressing Ct Ctr3a, but not the other strains, may be due to high levels of Cu import by Ct Ctr3a that lead to cellular toxicity (Fig. 2B). Cells grown in a quantitative liquid assay display similar results, with Ct Ctr2 only able to moderately rescue growth in SCEG media, whereas both Ct Ctr3a and Ct Ctr3b robustly restore Cu-dependent growth (Fig. 2C).
Figure 2.
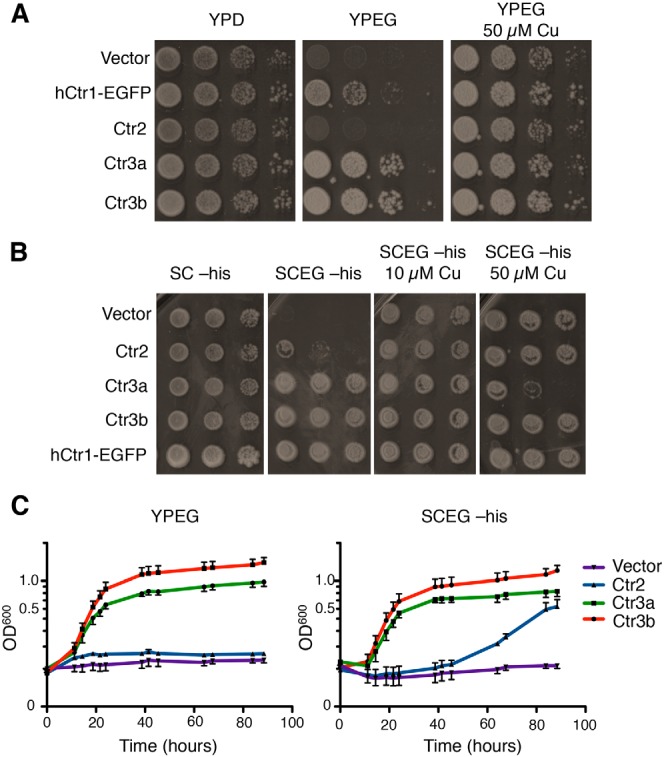
C. thermophilum Ctr2 and Ctr3 homologues rescue growth on nonfermentable medium. A, S. cerevisiae ctr1Δctr3Δ cells (strain MPY17) were transformed with the indicated plasmids and plated on rich agar medium containing either dextrose (YPD), ethanol and glycerol (YPEG), or YPEG 50 μm CuSO4 and photographed after 2 days (YPD) or 5 days (YPEG). B, the same cells as in A were plated on minimal agar medium containing either dextrose (SC), ethanol and glycerol (SCEG), or SCEG plus the indicated final CuSO4 concentrations and photographed after 3 days (SC-His) or 7 days (SCEG-His). C, the same cells as in A and B were inoculated into liquid YPEG or SCEG-His medium, and growth was monitored by optical absorbance.
Differential requirements for N-terminal residues for Cu transport
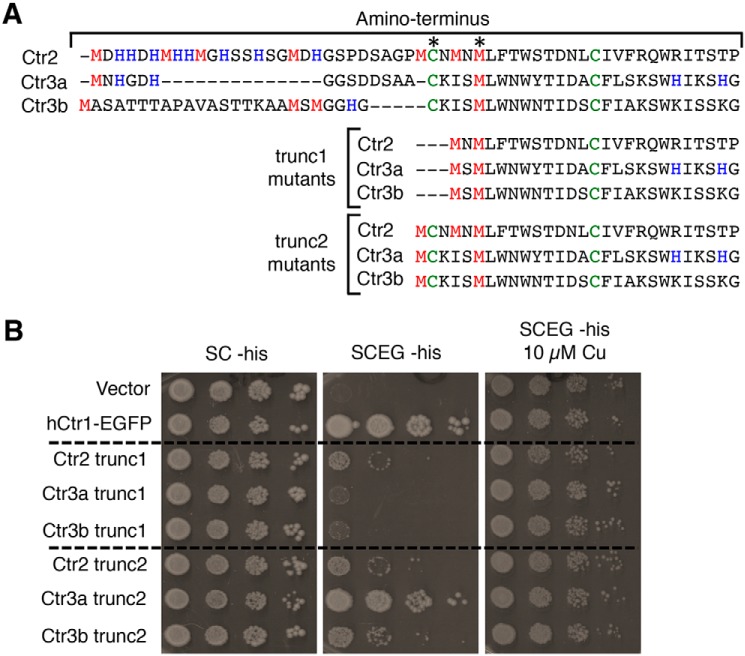
Previous studies from fungi to humans have demonstrated the contribution of the metal-binding ectodomain in Ctr1/Ctr3-mediated Cu+ transport, with a strict requirement for either two methionine residues or a methionine and cysteine residue located proximal to the first transmembrane domain (16, 24, 28) (Fig. 1A). Although less well-conserved and not strictly essential for Cu transport, the ectodomain also typically possesses more distal regions enriched in histidine and methionine residues thought to facilitate accumulation of Cu near the entrance of the transmembrane pore. Interestingly, Ctr1/3 family members with large ectodomains possessing many histidine and methionine regions tend to lack cysteine residues, and members containing cysteine residues tend to have shorter ectodomains with fewer histidine and methionine regions (16, 29). As cysteine shows a higher affinity for Cu+ than methionine and histidine (62–64), the evolutionary or physiological rationale for the use of cysteine as opposed to histidine and methionine may be suited to the particular extracellular environment encountered.
The three C. thermophilum proteins identified in this study possess hybrid ectodomain characteristics, containing both distal regions of histidine and methionine residues while also harboring proximal cysteine residues, and it is not immediately apparent which are required for Cu transport (Fig. 3A). The importance of these ectodomain residues in Cu+ transport was ascertained by first deleting the distal portion of the ectodomain and thus removing the histidine and methionine regions as well as the distal cysteine (trunc1 mutants). In a similar fashion to the trunc1 mutants, a second group of truncation mutants was created that retain only three additional residues compared with the trunc1 mutants (trunc2 mutants). Despite lacking 14 Met, His, and Cys residues, the Ct Ctr2 trunc1 mutant modestly rescues growth on SCEG (Fig. 3B), in a similar fashion to full-length Ct Ctr2 (Fig. 1B). This suggests that the N terminus of Ctr2 is dispensable for Cu transport activity. In contrast, the Ctr3a and Ctr3b trunc1 mutants are unable to rescue growth. All trunc2 mutants rescued growth on SCEG, to varying extents, suggesting that the additional Cys residue present in the trunc2 mutants is essential for Cu transport in Ctr3a and Ctr3b but dispensable for Ctr2. These results reveal key features of Cu transport that differ between the three C. thermophilum Cu+ transporter family members.
Figure 3.
Dependence on metal binding ectodomain for Cu transport function. A, MUSCLE alignment of the Ctr2, Ctr3a, and Ctr3b ectodomains from WT, trunc1, and trunc2 proteins with the predicted metal-binding residues Met, His, and Cys shown in red, blue, and green, respectively. Asterisks, residues predicted to be necessary for Cu transport based on prior work referenced herein. B, S. cerevisiae ctr1Δctr3Δ cells (strain MPY17) were transformed with plasmids expressing the indicated proteins, plated on the indicated media, and photographed after 3 days (SC-His) or 7 days (SCEG-His).
Ctr2 displays characteristics of Ctr2-like family proteins
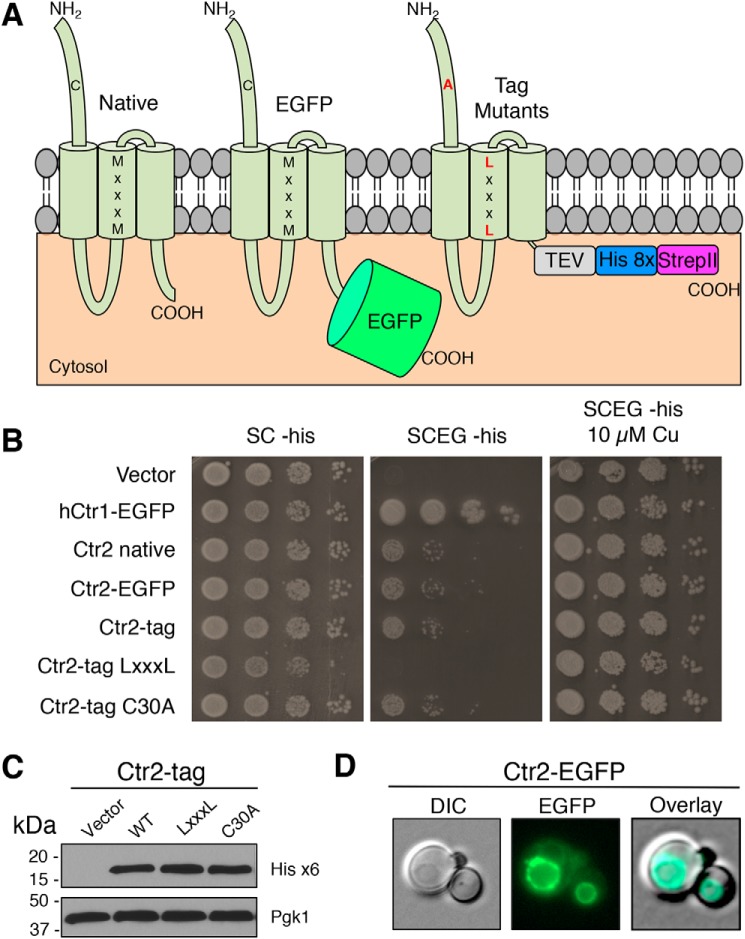
To gain insights into the localization and function of C. thermophilum Ctr2, Ctr3a, and Ctr3b, EGFP was fused to the C terminus of each reading frame to allow for localization. Moreover, to facilitate the generation of protein derivatives amenable to purification and in vitro reconstitution, the intrinsically disordered C terminus was removed and replaced with a tandem His8, StrepII affinity purification tag that is cleavable via TEV protease to allow for protein isolation. In a final set of mutants, the distal cysteine residue of the N terminus was mutated to alanine, and the critical MXXXM motif was mutated to LXXXL. (Fig. 4A). Both the addition of EGFP and the affinity tag do not alter the ability of Ctr2 to modestly rescue growth on YPEG (Fig. 4B). However, mutation of the Ctr2 MXXXM motif to LXXXL in the second transmembrane domain did not support growth on SCEG. As is characteristic for Ctr2 in S. cerevisiae, and in agreement with N-terminal deletion experiments (Fig. 3B), the C30A mutation showed no reduction in growth, further supporting the notion that this protein depends on similar residues as does the S. cerevisiae vacuolar Ctr2 Cu+ exporter (30, 32, 65). Immunoblot analysis reveals that the Ct Ctr2 tag variants are expressed at similar levels (Fig. 4C), and fluorescence microscopy revealed that Ct Ctr2 localizes to the vacuole (Fig. 4D). These results confirm that Ct Ctr2 is a Ctr2-like protein that likely functions in vacuolar Cu+ export.
Figure 4.
C. thermophilum Ctr2 is a vacuolar Cu transporter. A, model depicting protein variants generated in this study. Letters in black indicate WT amino acid residues, and letters in red indicate mutated residues. TEV indicates a TEV protease cleavage site, and His 8x and StrepII indicate the respective affinity purification tags. B, S. cerevisiae ctr1Δctr3Δ cells (strain MPY17) were transformed with plasmids expressing the indicated proteins and plated on the indicated media. C, immunoblot from Triton X-100–solubilized protein extracts from selected transformants in B probed with anti-His6 antibody or anti-PGK antibody. D, cells from B were visualized by fluorescence microscopy and photographed. DIC, differential interference contrast.
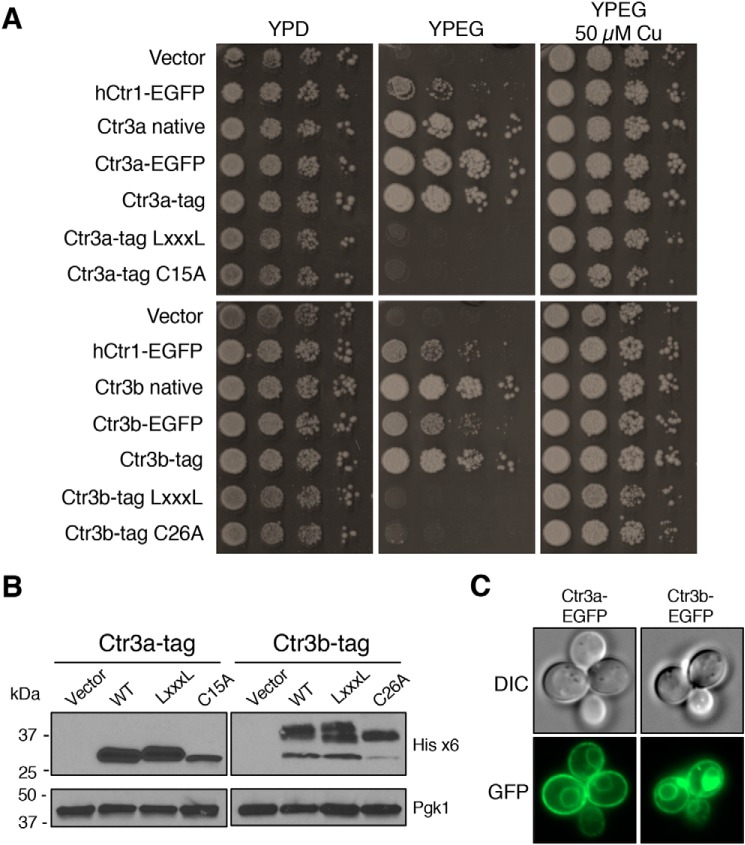
Protein sequence alignment (Fig. 1B) and functional analysis of ectodomain truncation mutants (Fig. 3B) suggest that C. thermophilum Ctr3a and Ctr3b are in the Ctr1/3 family of plasma membrane Cu+ transporters. As expected, mutation of either the MXXXM motif to LXXXL or the distal ectodomain Cys to Ala resulted in loss of function in supporting growth on YPEG (Fig. 5A). Immunoblot analysis demonstrates that the proteins are expressed, with that expressed from the Cys-to-Ala mutant alleles showing decreased protein levels for both Ctr3a and Ctr3b (Fig. 5B), similar to what is observed for the S. cerevisiae Ctr3 Cys mutant (29). Electrophoretic fractionation of Ctr3b resolves multiple species, with the lower band corresponding to the predicted molecular weight of the primary translation product, whereas the others may correspond to glycosylated species that have been observed for Ctr1/3 family members in other fungi (66–68). Additionally, a C-terminal His8/StrepII tag with a cleavable TEV protease site, or fusion to EGFP, allows for rescued growth of S. cerevisiae on YPEG and reveals that the proteins localize to both the plasma membrane and vacuolar membrane, a common feature for membrane proteins overexpressed in S. cerevisiae (69) (Fig. 5C).
Figure 5.
C. thermophilum Ctr3a and Ctr3b are plasma membrane–localized transporters. A, S. cerevisiae ctr1Δctr3Δ cells (strain MPY17) were transformed with plasmids expressing the indicated proteins and plated on media to assay growth. B, immunoblot from Triton X-100–solubilized protein extracts from selected transformants in A probed with anti-His6 antibody or anti-PGK antibody. C, cells from A were visualized by fluorescence microscopy and photographed. DIC, differential interference contrast.
C. thermophilum Ctr3a and Ctr3b support uptake of Cu and Ag in vivo
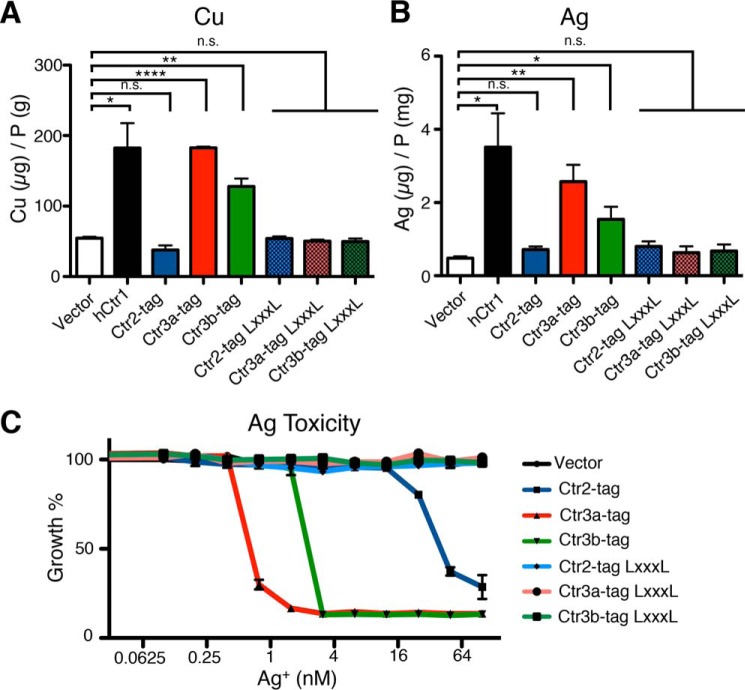
To determine whether expression of the C. thermophilum Ctr2, Ctr3a, and Ctr3b alter Cu levels in S. cerevisiae MPY17, ICP-MS analysis was used to measure total cell-associated Cu levels (Fig. 6A). Expression of human Ctr1 strongly increased cellular Cu levels compared with cells harboring the empty vector. Expression of C. thermophilus Ctr3a and Ctr3b, but not Ctr2, led to increased cell-associated Cu levels, paralleling their ability to rescue growth on nonfermentable carbon sources. In addition, the same trend was observed for Ag accumulation in growing cells that were pulsed with AgNO3 (Fig. 6B). Previous studies have shown that Ctr1/3 family members mediate transport of Ag+, as this ion is isoelectronic to Cu+ (70). Furthermore, growth analysis of cells exposed to increasing concentrations of AgNO3 reveals that both Ctr3a and Ctr3b mediate AgNO3 toxicity at levels under 10 nm, with Ctr2 driving AgNO3 toxicity at higher AgNO3 concentrations (Fig. 6C). Mutation of the MXXXM motif to LXXXL abolished AgNO3 toxicity, indicating that a transport-competent Ctr2 or Ctr3 protein is required for this observation.
Figure 6.
C. thermophilum Ctr3a and Ctr3b mediate cellular Cu and Ag accumulation and Ag toxicity. A, S. cerevisiae ctr1Δctr3Δ cells (strain MPY17) were transformed with plasmids expressing the indicated proteins and grown in liquid YPD medium to mid-log phase before harvest and inductively coupled plasma MS analysis. B, mid-log phase cells from A were grown in the presence of 1 μm AgNO3 in YPD for 60 min before harvest and ICP-MS analysis. C, cells from A were transferred into a sterile 96-well plate containing the indicated final concentration of AgNO3 and allowed to grow for 48 h before A600 measurements. Error bars, S.E. for biological triplicates as analyzed by paired t tests. *, p ≤ 0.05; **, p ≤ 0.01; ****, p ≤ 0.0001. n.s., not significant.
Ctr3a variants are stable and amenable to biochemical characterization
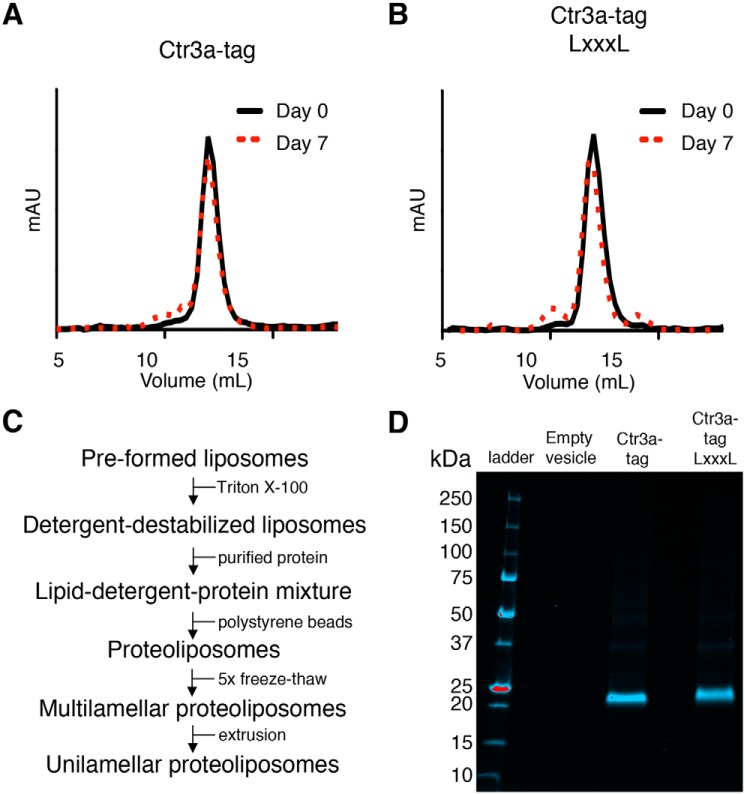
As Ctr3a in MPY17 shows no signs of posttranslational modification and drives the highest level of Cu and Ag accumulation, we sought to purify Ctr3a for in vitro characterization of metal transport activity. The coding region for Ctr3a tag and the inactive Ctr3a tag LXXXL mutant were cloned into a galactose-inducible, glucose-repressible plasmid and transformed into S. cerevisiae strain RSY620. After induction of expression, membrane preparations were isolated, integral membrane proteins were detergent-solubilized with 2% (w/v) dodecyl-β-d-maltopyranoside, and Ctr3a was isolated and purified by Ni-NTA chromatography followed by size-exclusion chromatography. Analytical chromatography revealed a monodisperse protein population that elutes at the predicted size for both trimeric Ctr3a and Ctr3a LXXXL proteins (Fig. 7 (A and B), black traces). To assess their stability, these preparations were stored at 4 ºC for 7 days and re-evaluated by size-exclusion chromatography (Fig. 7 (A and B), red traces). Although small deviations in the distribution profile appeared over time, both proteins remained largely monodisperse. Whereas many cell-based assays and knock out experiments in fungi and metazoans demonstrate a requirement for Ctr1/3 family members in Cu or Ag accumulation, no reports demonstrate protein-intrinsic metal transport activity. Based on the ease of purification of Ctr3a and Ctr3a LXXXL and their observed stability, we sought to use these proteins to create a proteoliposome-based reconstituted in vitro metal transport assay. Initially, a number of lipid variations were evaluated for proteoliposome reconstitution, including the defined synthetic lipids dioleoyl-phosphatidylethanolamine, dioleoyl-phosphatidylglycerol, and dioleoyl-phosphatidylcholine as well as complex polar lipids isolated from Escherichia coli, S. cerevisiae, and chicken egg yolks. Multiple combinations of lipids were evaluated by formation of liposomes followed by introduction of Ctr3a tag and Ctr3a tag LXXXL along with the fluorescent reporter PhenGreen SK (PGSK) (Fig. 7C). PGSK is composed of a phenanthroline-derived moiety, which has broad metal-chelating properties, linked to a fluorescein derivative in a manner that results in fluorescence quenching at 530 nm upon stable binding to metal (71–75). Multiple rounds of freeze-thaw followed by extrusion encapsulated PGSK and induced a scrambled transporter orientation (76). Although many different synthetic lipids as well as complex polar lipids have been utilized in in vitro assays for other metal transporters (77–79), we were not able to obtain desirable results with these lipids despite numerous tested combinations (data not shown). Rather, polar lipids isolated from chicken egg yolks, which have been used to successfully establish an in vitro transport assay for the mitochondrial Cu importer SLC25A3 (80), displayed desirable characteristics. SDS-PAGE analysis of the final unilamellar proteoliposomes shows protein purity and successful insertion into the lipid bilayer (Fig. 7D).
Figure 7.
C. thermophilum Ctr3a purification, stability, and incorporation into proteoliposomes. A, Ctr3a tag was expressed in S. cerevisiae, extracted from solubilized membrane preparations, and purified via Ni-NTA affinity chromatography followed by size-exclusion chromatography (day 0). Peak fractions were pooled. After 7 days at 4 °C, the pooled sample was re-analyzed by size exclusion chromatography (day 7). B, the Ctr3a tag LXXXL protein was purified and analyzed as in A. C, workflow depicting steps for incorporation of purified Ctr3a proteins into unilamellar proteoliposomes. D, stain-free SDS-polyacrylamide gel of proteoliposomes without protein (Empty vesicle), with Ctr3a tag and with Ctr3a tag LXXXL. Molecular weight markers are indicated with mass in kDa.
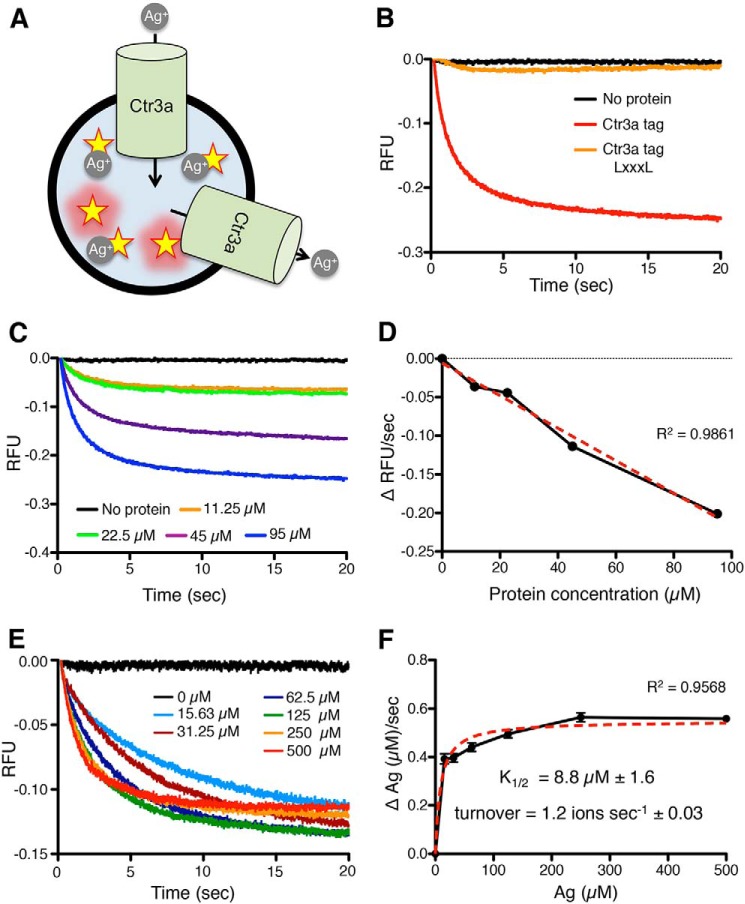
Because the scrambled orientation of Ctr3a is expected to generate both inward-facing and outward-facing proteins, this will allow for an equilibrium of metal ions to be reached between the inside and outside of the vesicle (Fig. 8A). The increase in lumenal metal concentration will result in quenching of PGSK and a decrease in fluorescent signal. Due to the redox nature of Cu and previous reports demonstrating that Ctr1/3 family proteins can transport Ag+, this assay was developed using the redox-stable, Cu+ mimetic Ag+ (70).
Figure 8.
In vitro Ag+ transport assays in reconstituted proteoliposomes. A, model depicting the in vitro reconstituted Ctr3a-dependent Ag+ transport assay. Stars, fluorescent reporter molecules of PGSK that are quenched in the presence of Ag+. Ctr3a is incorporated into liposomes in both inward-facing and outward-facing orientations. B, traces from stopped-flow measurement of PGSK fluorescence (relative fluorescence units (RFU)) over time (in seconds) after the addition of Ag+, with no protein (black), Ctr3a tag (red), or Ctr3a tag LXXXL (orange). C, traces from stopped-flow measurements of PGSK fluorescence from liposomes containing no protein (black) or increasing amounts of Ctr3a tag (11.25 μm (orange), 22.5 μm (green), 45 μm (purple), and 95 μm (blue)). D, initial rate of PGSK fluorescence quenching at a fixed concentration of Ag+ (500 μm) as a function of Ctr3a tag protein concentration incorporated into proteoliposomes. Red dotted line, best-fit linear regression. E, traces from stopped-flow measurements of PGSK fluorescence due to increasing Ag+ concentrations added to proteoliposomes containing a fixed concentration of Ctr3a tag (45 μm). F, Michaelis–Menten plot of initial Ag+ transport velocity versus Ag+ concentration, with K½ and turnover calculations. Red dotted line, best-fit nonlinear regression.
Utilizing a stopped-flow spectrometer for kinetic analysis, proteoliposomes encapsulating PGSK and incorporating either no protein, Ctr3a tag, or Ctr3a tag LXXXL were mixed 1:1 with a 1 mm solution of pH 7.4 AgNO3, and fluorescence was monitored at 532 nm (Fig. 8B). Proteoliposomes containing Ctr3a tag displayed rapid fluorescence quenching, indicating Ag transport across the lipid bilayer, whereas liposomes lacking Ctr3a, as well as proteoliposomes reconstituted with Ctr3a tag LXXXL, displayed little time-dependent fluorescence quenching. Notably, Ag transport was achieved without the addition of a counterbalancing ion, ATP, or additional accessory factors (18). Moreover, the incorporation of multiple concentrations of Ctr3a tag into liposomes demonstrated a protein dose dependence for Ag transport (Fig. 8C). Increasing the amount of Ctr3a protein both increased the rate of transport and altered the steady-state fluorescence levels, indicating that at these concentrations of Ctr3a, there exist a population of vesicles without any transporter. Nonetheless, the initial velocity of transport remained linear over the Ctr3a concentrations used (Fig. 8D).
Having established conditions for Ctr3a-mediated Ag transport in vitro, the rate of Ag transport was determined in the reconstituted system. Proteoliposomes were mixed 1:1 with increasing concentrations of AgNO3, and fluorescence was monitored (Fig. 8E). An AgNO3 dose–dependent fluorescence quenching was observed, and equilibrium between Ag+ transport into and out of proteoliposomes was rapidly achieved. However, at lower concentrations of Ag+ (e.g. 15 μm), transport equilibrium was not reached over the time course of the experiment. Examination of the kinetic parameters of transport indicates an apparent transport affinity of 8.8 μm Ag+, similar to previous reports for both fungal and mammalian Ctr1/3 family members in cultured cells (14, 15, 18, 24, 53, 67, 81) (Fig. 8F). Furthermore, the observed turnover rate for Ag+ was calculated at 1.2 ions/s, in agreement with in vivo studies for human Ctr1 (53), suggesting a conserved rate of transport for Ctr1/Ctr3 family members from fungi to human origin.
Discussion
The Ctr1/3 family of high-affinity Cu+ transporters were discovered nearly 25 years ago via yeast genetics studies (14, 15). Work in fungi and mice has demonstrated the importance of this family for Cu acquisition, because loss of Ctr1/Ctr3 function results in Cu and iron deficiency; defective activation of Cu,Zn-superoxide dismutase, cytochrome oxidase, and other Cu-dependent enzymes such as laccase or tyrosinase; and intestinal Cu absorption and tissue-specific defects in mice (1, 2, 82).
Although the importance of Cu acquisition and utilization for normal physiology is clear, there is currently a poor understanding of the molecular mechanisms by which these proteins facilitate the transduction of Cu+ through the plasma membrane or across endosomal membranes. Mutagenesis experiments have revealed key Ctr1/Ctr3 amino acid residues and regions that are required for Cu+ movement or that support this process, and structural studies provide an overall understanding of the organization of the homotrimeric subunits and a potential channel through which Cu+ ions would permeate (16, 19–22, 24). Our goal here was to identify, characterize in vivo, and purify Ctr1/Ctr3 family members and generate a reconstituted metal ion transport system in vitro using purified components. Using a thermophilic fungus, C. thermophilum, we identified and functionally validated two Ctr3 family members that function in Cu+ import and a Ctr2 family member that likely functions in vacuolar Cu+ mobilization, similar to the Ctr3 and Ctr2 family members found in other fungi (29–31). The newly classified Ctr3a and Ctr3b proteins display characteristics of Ctr1/3-like plasma membrane Cu+ transporters from other fungi in that they require a tract of cysteine/methionine residues located in the ectodomain and second transmembrane domain, with mutations of these residues leading to an inactive or severely dampened transporter (16, 24). The presence of multiple cysteine residues in the ectodomain raises the possibility that oxidation or disulfide bond formation may influence Cu transport. However, because some members of the Ctr1/3 family completely lack cysteine residues in the ectodomain, it is unlikely that their involvement in regulation of Cu transport is widely conserved. C. thermophilum Ctr2 displays characteristics of Ctr2 and Ctr6-like vacuolar transport proteins found in S. cerevisiae and S. pombe, respectively, and does not require the distal cysteine that is required for Ctr3a, Ctr3b, and S. cerevisiae Ctr3 (30, 31). Mutagenesis and functional analysis in S. cerevisiae Cu+ transport–defective mutants support these functional roles and their dependence on regions and residues that are structurally and functionally conserved in high-affinity Cu+ transporters from fungi to mammals. Thus, observations made with the C. thermophilum Cu+ transporters are likely to be mechanistically relevant to those found across a broad swath of evolutionary divergence.
The intrinsic stability of the C. thermophilum Cu+ transporters, and specifically Ctr3a, allowed the facile purification of this protein for incorporation into liposomes. This reconstituted system exploited the metal binding–dependent quenching of PGSK autofluorescence, coupled with the use of Ag+ to mimic Cu+ ions, thereby obviating the need for the inclusion of Cu2+ reductases that stimulate the activity of fungal Cu+ transporters in vivo (26, 65, 83). The use of Ag+ in these assays is further validated by the ability of Ctr3a and Ctr3b, and to a lesser extent Ctr2, to enhance both Cu and Ag accumulation and Ag-induced toxicity, when expressed in S. cerevisiae ctr1Δ ctr3Δ cells. Whereas there may be differences in the transport of Ag+ compared with Cu+ in these assays, the generation of an in vitro Ctr3a-dependent transport assay could make mechanistic predications that could be validated by the use of Cu2+ and a source of reducing power.
The Ctr3a-containing proteoliposomes demonstrated a time–, protein–, and Ag+ concentration–dependent transport of Ag+ into the lumen, as measured by PGSK fluorescence quenching. Due to the bidirectional equilibrium of Ctr3a, an equilibrium of luminal Ag+ ions with that found in the extravesicular environment was achieved over time. Of importance are the observations that Ctr3a and, by inference, other Ctr1/Ctr3 family members have intrinsic metal ion transport activity independent of Cu-binding chaperones or counterions and do not serve as protein co-factors that are essential for the activity of as yet unidentified high-affinity Cu+ transporters. Moreover, consistent with previous reports demonstrating that inhibition of ATP synthesis does not alter Cu uptake dynamics in cell culture (18), no dependence on nucleotide hydrolysis was observed for Ctr3a-mediated Ag+ transport.
It is interesting to note that whereas fungi, including S. cerevisiae, S. pombe, Cryptococcus neoformans, and C. thermophilum, encode two functionally redundant Ctr1/Ctr3 proteins for plasma membrane Cu+ transport and one Ctr2-like protein for vacuolar Cu+ efflux (84), mammals express a single, essential high-affinity Cu+ transporter, Ctr1, that is found both at the plasma membrane and on endosomal compartments (54). Previous work suggests that the metal-binding ectodomain of mouse and human Ctr1 assists with the import of Cu+ across the plasma membrane but is inhibitory for Cu+ mobilization from the interior of an endosomal compartment (16, 18, 24). Furthermore, an endosomal cysteine protease, cathepsin L, works in conjunction with a Ctr1-related protein, Ctr2, to cleave the Ctr1 ectodomain. This creates a truncated form that supports endosomal Cu export in a similar fashion to the dedicated fungal vacuole transporters (54). There are also notable differences between S. cerevisiae and C. thermophilum Ctr2-like proteins and Ctr1/3 proteins in the residues necessary for Cu transport in the metal-binding ectodomain. As the acidic interior of an intracellular vesicle is markedly different from the more neutral pH of the extracellular space, differences in Ctr1/Ctr3 ectodomain composition may be used to properly control Cu+ transport in distinct environments. The tools developed here provide the experimental basis for dissection of the mechanism of action and the role of chaperones and other ligands as well as physiological conditions on Cu+ transport mediated by plasma membrane versus endosomal fungal Cu+ transporters and by full-length versus truncated mammalian Ctr1.
Several additional questions remain regarding the nature of Ctr1/Ctr3-mediated Cu+ transport. The observed rate of transport in vitro and in vivo is on the order of ∼101 ions/s, which is within the range of rates observed for transporters controlled by gates on both sides of the membrane and would support the original classification that this family of proteins functions as a transporter. However, the ∼7-Å cryo-EM structural model for human Ctr1 (20) depicts a central pore though which Cu+ ions would transit, reminiscent of single-gate ion channels that display transport rates of 104 to 108 ions/s. Previous studies suggest that the metal-binding ectodomain assists with transport in vivo, as mutants lacking this ectodomain display significantly slower rates of Cu accumulation (16, 24). Thus, the N terminus of the transporter does not function as a rate-limiting gate and cannot be responsible for the observed slow transport. On the other hand, deletion of the Ctr1 cytosolic C-terminal tail increases the observed rate of Cu transport in mammalian cells (53). Thus, perhaps the C terminus of Ctr1/Ctr3 proteins functions in a gate-like fashion to inhibit the rapid inflow of Cu into cells, which would have the potential for toxicity. Further structural information, ideally in the form of high-resolution models comprising the apo and Cu+-bound states, will be necessary to determine whether Ctr1/3 proteins function as transporters with an ion channel-like central pore or as ion channels with uncharacteristically slow kinetics. Additionally, the assay developed here could be used to assess the relationship between rate of transport and temperature. Because transporters require large protein domain reorganizations to exert their function, they show a much higher dependence on temperature than do channels (85).
Also, now amenable for study is the influence of intra- and extracellular factors on the rate of Ctr1/3-mediated transport. Previous studies have shown that changes in pH can alter Ctr1/3-mediated cellular Cu import (18). By modulating the intra- and extravesicular pH, the in vitro transport assay developed here will enable direct assessment of role that pH plays in Ctr1/3 transport. Furthermore, extracellular Cu donors, such as albumin and ceruloplasmin, have been proposed as the source for cellular Cu import (11, 86). Comparisons in the rate of transport between donor-loaded metal complex versus free metal would enable further exploration of the biological source for Ctr1/3 Cu import. Additionally, the intracellular copper chaperones Atx1 and CCS, as well as GSH, have been suggested to directly interact with the C terminus of Ctr1/3 proteins (40–42, 44). By encapsulating an intracellular Cu acceptor inside Ctr3a proteoliposomes, followed by the addition of PGSK–metal complex and monitoring the increase in fluorescence resulting from Ctr1/3-mediated transport, the effect of intracellular Cu acceptors on the rate of Ctr1/3 transport can be interrogated. Furthermore, cell culture experiments have documented a mutant in human Ctr1, H139R, that significantly increases Cu transport. Because this mutation also decreases Ctr1 endocytosis (53), it is difficult to understand whether the mechanism for increased Cu transport is due to a higher turnover rate or a longer residence time at the plasma membrane. The in vitro metal transport assays developed here can be used to directly address questions about the roles played by specific Ctr1/Ctr3 residues and the influence of intracellular Cu chaperones, GSH, or other ligands on the transport process and to decipher detailed mechanisms for the permeation of Cu+ ions across a variety of cellular compartments.
Experimental procedures
Yeast strains and plasmids
S. cerevisiae strains MPY17 and RSY620 used in this study have been described (14, 61) (Table S1). Cells were routinely grown in selective media with agitation at 30 °C. All yeast plasmids were created by subcloning into the p413GPD or p423Gal backbone at the SpeI and XhoI restriction enzyme sites (87) (Data S1). For the creation of plasmids containing C. thermophilum Ctr2 or Ctr3 cDNA variants, codon-optimized gBlocks (Integrated DNA Technologies) were synthesized. Point mutations were created using the QuikChange II site-directed mutagenesis kit (Agilent Technologies).
Phylogenetic analysis
Ctr2 and Ctr3 protein sequences from the indicated species were identified and retrieved from the NCBI Protein database by iterative BLAST searches (59). A multiple-sequence alignment was created using MUSCLE (60).
Functional complementation assays
S. cerevisiae ctr1Δctr3Δ cells (strain MPY17) were transformed with the indicated plasmids and grown to exponential phase in selective media with agitation at 30 °C. For solid medium assays, 10-fold serial dilutions were spotted on YPEG, YPEG containing 50 μm copper, SCEG, or SCEG containing 10 or 50 μm copper and containing 1.5% agar; incubated for 3–7 days at 30 °C; and photographed. For liquid medium assays, cells were diluted to a starting dilution of 0.002 A600 in either YPEG or SCEG and incubated at 30 °C with shaking. Optical absorbance was measured at the indicated times. Experiments were performed in triplicate.
Fluorescence microscopy
S. cerevisiae ctr1Δctr3Δ cells (strain MPY17) were transformed with the indicated plasmids and grown in selective media to exponential phase at 30 °C, washed at room temperature, and resuspended in PBS before dispensing onto a microscope slide containing an agar pad. The slide was covered with a No. 2 coverslip, sealed with Vaseline, and imaged on a Zeiss Axio Imager microscope.
Protein extraction and immunoblotting
For the preparation of protein extracts, yeast cell pellets were resuspended in ice-cold radioimmune precipitation assay buffer (Cell Signaling Technology) and supplemented with protease inhibitors (Halt Protease Inhibitor Mixture, Thermo Scientific). Cell suspensions were lysed via three cycles of 1 min on, 1 min off bead beating with 0.1-mm glass beads (Sigma-Aldrich) at 4 °C in a Mini-Beadbeater-16 (BioSpec Products) and centrifuged at 20,000 × g at 4 °C for 10 min, and supernatants were collected. Protein concentrations were measured with the BCA protein assay kit (Thermo Scientific). SDS-PAGE and immunoblotting were carried out by standard protocols. Antibodies used were anti-His6 (Sigma-Aldrich) and anti-phosphoglycerate kinase (PGK; Invitrogen). Horseradish peroxidase–conjugated anti-mouse IgG (GE Healthcare) was used as secondary antibody for immunoblotting.
Metal analysis
Total cell-associated copper or silver was measured by ICP-MS (model 7500cs, Agilent, Santa Clara, CA). Briefly, log-phase yeast cells were grown in SC-His medium (MP Biomedicals) normalized to cell number, washed, and harvested into acid-washed 1.5-ml microcentrifuge tubes. For measurement of silver, cells were co-cultured with 1 μm AgNO3 for 60 min before harvest. Yeast pellets were dissolved 1:10 (w/v) with trace analysis grade nitric acid (Sigma). All samples were then heated at 85–95 °C for ∼1 h before analysis.
Statistical analysis
All results are presented as mean with error bars indicating the S.E. Significance was assessed by paired Student's t test and is indicated as follows: *, p < 0.05; **, p < 0.01; ****, p < 0.0001.
Ctr3 expression and purification
Ctr3 proteins were expressed from a p423Gal plasmid with a C-terminal His6/StrepII tag. Plasmids were transformed into S. cerevisiae pep4Δ cells (strain RSY620), as this genetic background contains significantly reduced vacuolar protease activity, and single colonies were selected for overnight growth in 3 ml of SC-His medium (88). 1 ml of culture was inoculated into 25 ml of SC-His medium containing dextrose (repressing conditions). After overnight growth, 25 ml of culture was inoculated into 750 ml of SC-His medium in which the carbon source was raffinose rather than dextrose (nonrepressing, noninducing conditions). When absorbance reached A600 of 0.8, 250 ml of 4× YPGal (galactose, promoter-inducing conditions) was added to induce Ctr3 protein expression. After ∼24 h of growth, cells were harvested and lysed in 50 mm NaPO4 (pH 7.4), 300 ml of NaCl, 10% glycerol, and 5 mm tris(2-carboxyethyl) phosphine via 30 cycles of 1 min on, 1 min off bead beating with 0.1-mm glass beads (Sigma-Aldrich) on ice in a 350-ml BeadBeater (BioSpec Products). Cell debris was pelleted by centrifugation at 6,000 × g for 15 min at 4 °C. Membranes were isolated from the supernatant by ultracentrifugation at 180,000 × g for 1 h at 4 °C and solubilized with lysis buffer containing 2% (w/v) n-dodecyl-β-d-maltopyranoside overnight. Samples were ultracentrifuged at 180,000 × g for 1 h at 4 °C to remove insoluble material, and Ni-NTA chromatography was performed to isolate His6-tagged proteins using lysis buffer with 0.05% (w/v) n-dodecyl-β-d-maltopyranoside. Elutions were collected and fractionated over an S200 size-exclusion column on an ÄKTA Pure FPLC (GE Healthcare). Proteins were concentrated to 1 mg/ml via Amicon Ultra centrifugal filters (EMD Millipore).
Lipid isolation and purification
Two dozen Grade A chicken eggs were purchased from a local grocer, and polar lipids were isolated via a modified Bligh/Dyer extraction (89). Specifically, eggs were cracked, and yolks were mechanically separated into a glass beaker. A 2:1 mixture of chloroform/methanol was added to egg yolks until the final volume was 20 times the initial volume of egg yolks, and the mixture was stirred at room temperature for 20 min. Insoluble material was pelleted by centrifugation at 4,000 × g for 10 min at room temperature. Liquid was decanted into a separatory funnel containing 0.2 volumes of 3 m NaCl and vigorously shaken. After phase separation, the chloroform layer was collected, and this step was repeated until no white fluffy material was present at the phase interface. After sufficient washing, chloroform was evaporated via rotovap to yield a viscous yellow oil. To precipitate polar lipids, a 10× volume of acetone was then added and mixed vigorously. Centrifugation at 4,000 × g for 10 min pelleted the insoluble polar lipids, and the colored liquid was discarded. This step was repeated until no color was present in the acetone wash. Remaining acetone was evaporated under a nitrogen stream, and lipids were weighed.
Proteoliposome formation
Polar egg yolk lipids were then suspended in 10 mm HEPES, pH 7.4, at a concentration of 20 mg/ml via sonication to create small unilamellar vesicles. Vesicles were then subjected to three freeze thaw cycles to create large multilamellar vesicles. Vesicles were then extruded through a 400-μm polycarbonate filter to create large unilamellar vesicles and diluted 5× with 10 mm HEPES, pH 7.4, plus 25% glycerol. Triton X-100 was then added to a final concertation of 2% (v/v) to destabilize vesicles. Ctr proteins were then added to the desired concentration, and the vesicle/protein/detergent mixture was incubated with gentle rocking at room temperature for 30 min. Liposomes were made parallel to proteoliposomes as a protein-free control. To remove detergent, 200 mg of Bio-Beads SM-2 (Bio-Rad) per 5 ml of vesicle/protein/detergent mixture was added. Samples were incubated with gentle rocking for 30 min at 4 °C. An additional 200 mg of Bio-Beads SM-2 per 5 ml was added and incubated with gentle rocking for 60 min at 4 °C. An additional 200 mg of Bio-Beads SM-2 per 5 ml was then added and incubated with gentle rocking overnight at 4 °C. A final addition of 200 mg of Bio-Beads SM-2 per 5 ml was added, and samples were incubated with gentle rocking for 2 h at 4 °C. Bio-Beads were then removed by filtration over disposable Polyprep columns and diluted with 10 mm HEPES, pH 7.4, to a final glycerol concentration of 2%. Proteoliposomes were harvested by ultracentrifugation at 180,000 × g for 1 h, resuspended to a concentration of 20 mg of lipid/ml in 10 mm HEPES, pH 7.4, and PGSK (Thermo Fisher Scientific) at a final concentration of 200 μm. Ctr3 orientation was scrambled, and PGSK was incorporated into proteoliposomes by five freeze thaw cycles and extrusion through a 400-μm polycarbonate filter. Unincorporated PGSK was removed via Zeba Spin desalting columns (Thermo Fisher Scientific), and protein incorporation into the proteoliposomes was validated by SDS-PAGE analysis.
In vitro metal transport assays
Stopped-flow fluorescence kinetic experiments were performed on an Applied Photophysics SX20 instrument at 25 °C. Samples were mixed in an observation cell with a 2-mm path length and excited at 506 nm with a slit width of 1 mm. Emitted light was detected through a 532-nm high-transmitting band pass filter (Andover Corp.). Experiments were performed by mixing proteoliposomes or liposomes with AgNO3 solutions at a ratio of 1:1. Kinetic traces were collected over 20 s. All traces were the cumulative average of three independent recordings. Baseline fluorescence for each sample was measured by mixing proteoliposomes or liposomes with buffer only. Maximum florescence change was measured for each sample by adding 1 mm AgNO3 to proteoliposomes or liposomes before mixing with 1% (w/v) octyl β-d-glucopyranoside (Anatrace) to permeabilize membranes, and all stopped-flow traces were normalized to a maximum response elicited by detergent solubilization. Background traces collected from liposome samples without PGSK were subtracted, yielding net fluorescence responses. Conversion of relative fluorescence to luminal Ag+ concentration in vesicles was calculated by creating a standard curve of the PGSK fluorescence response after the addition of a known AgNO3 concentration in the same buffer conditions in the absence of vesicles. All experiments were performed in triplicate. K½, Vmax, and turnover number were calculated using nonlinear regression curve fitting through the least-squares method of Michaelis–Menten kinetics (GraphPad).
Author contributions
B. L. L. and D. J. T. conceptualization; B. L. L. and D. J. T. data curation; B. L. L. validation; B. L. L. investigation; B. L. L. visualization; B. L. L. and D. J. T. methodology; B. L. L. and D. J. T. writing-original draft; B. L. L. and D. J. T. writing-review and editing; D. J. T. resources; D. J. T. supervision; D. J. T. funding acquisition; D. J. T. project administration.
Supplementary Material
Acknowledgments
We gratefully acknowledge Professors Paul Cobine for critical advice on lipid purification and on methods for in vitro reconstitution of metal transporters and Terrence Oas for advice on transport kinetics modeling and use of the stopped-flow apparatus. We appreciate critical reading of the manuscript by Paul Cobine, Aaron Smith, and Corinna Probst.
This work was supported by National Institutes of Health Grants R01 GM041840 and DK074192 (to D. J. T.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Table S1 and Data S1.
- MUSCLE
- Multiple Sequence Comparison by Log-Expectation
- hCtr1
- human Ctr1
- YPEG
- YPD, ethanol (2%), and glycerol (3%)
- SCEG
- SC, ethanol (2%), and glycerol (3%)
- TEV
- tobacco etch virus
- Ni-NTA
- nickel-nitrilotriacetic acid
- PGSK
- EGFP, enhanced GFP
- PGK
- phosphoglycerate kinase
- ICP
- inductively coupled plasma
- PGSK
- PhenGreen SK (spiro(isobenzofuran-1(3h),90-(9h)xanthene)-5-carboxamide, 20,70-dichloro-30,60-dihydroxy-3-oxo-n-1,10-phenanthrolin-5-yl-, dipotassium salt.
References
- 1. Kim B. E., Nevitt T., and Thiele D. J. (2008) Mechanisms for copper acquisition, distribution and regulation. Nat. Chem. Biol. 4, 176–185 10.1038/nchembio.72 [DOI] [PubMed] [Google Scholar]
- 2. Nevitt T., Ohrvik H., and Thiele D. J. (2012) Charting the travels of copper in eukaryotes from yeast to mammals. Biochim. Biophys. Acta 1823, 1580–1593 10.1016/j.bbamcr.2012.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Madsen E., and Gitlin J. D. (2007) Copper and iron disorders of the brain. Annu. Rev. Neurosci. 30, 317–337 10.1146/annurev.neuro.30.051606.094232 [DOI] [PubMed] [Google Scholar]
- 4. Madsen E., and Gitlin J. D. (2007) Copper deficiency. Curr. Opin. Gastroenterol. 23, 187–192 10.1097/MOG.0b013e32801421bb [DOI] [PubMed] [Google Scholar]
- 5. Lukasewycz O. A., and Prohaska J. R. (1990) The immune response in copper deficiency. Ann. N.Y. Acad. Sci. 587, 147–159 10.1111/j.1749-6632.1990.tb00142.x [DOI] [PubMed] [Google Scholar]
- 6. Medeiros D. M., Davidson J., and Jenkins J. E. (1993) A unified perspective on copper deficiency and cardiomyopathy. Proc. Soc. Exp. Biol. Med. 203, 262–273 10.3181/00379727-203-43599 [DOI] [PubMed] [Google Scholar]
- 7. Krishnamoorthy L., Cotruvo J. A. Jr., Chan J., Kaluarachchi H., Muchenditsi A., Pendyala V. S., Jia S., Aron A. T., Ackerman C. M., Wal M. N., Guan T., Smaga L. P., Farhi S. L., New E. J., Lutsenko S., and Chang C. J. (2016) Copper regulates cyclic-AMP-dependent lipolysis. Nat. Chem. Biol. 12, 586–592 10.1038/nchembio.2098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Garcia-Santamarina S., Uzarska M. A., Festa R. A., Lill R., and Thiele D. J. (2017) Cryptococcus neoformans iron-sulfur protein biogenesis machinery is a novel layer of protection against Cu stress. MBio 8, e01742–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Foster A. W., Dainty S. J., Patterson C. J., Pohl E., Blackburn H., Wilson C., Hess C. R., Rutherford J. C., Quaranta L., Corran A., and Robinson N. J. (2014) A chemical potentiator of copper-accumulation used to investigate the iron-regulons of Saccharomyces cerevisiae. Mol. Microbiol. 93, 317–330 10.1111/mmi.12661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Macomber L., and Imlay J. A. (2009) The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc. Natl. Acad. Sci. U.S.A. 106, 8344–8349 10.1073/pnas.0812808106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ramos D., Mar D., Ishida M., Vargas R., Gaite M., Montgomery A., and Linder M. C. (2016) Mechanism of copper uptake from blood plasma ceruloplasmin by mammalian cells. PLoS One 11, e0149516 10.1371/journal.pone.0149516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hassett R., Dix D. R., Eide D. J., and Kosman D. J. (2000) The Fe(II) permease Fet4p functions as a low affinity copper transporter and supports normal copper trafficking in Saccharomyces cerevisiae. Biochem. J. 351, 477–484 10.1042/bj3510477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhou B., and Gitschier J. (1997) hCTR1: a human gene for copper uptake identified by complementation in yeast. Proc. Natl. Acad. Sci. U.S.A. 94, 7481–7486 10.1073/pnas.94.14.7481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Knight S. A., Labbé S., Kwon L. F., Kosman D. J., and Thiele D. J. (1996) A widespread transposable element masks expression of a yeast copper transport gene. Genes Dev. 10, 1917–1929 10.1101/gad.10.15.1917 [DOI] [PubMed] [Google Scholar]
- 15. Dancis A., Yuan D. S., Haile D., Askwith C., Eide D., Moehle C., Kaplan J., and Klausner R. D. (1994) Molecular characterization of a copper transport protein in S. cerevisiae: an unexpected role for copper in iron transport. Cell 76, 393–402 10.1016/0092-8674(94)90345-X [DOI] [PubMed] [Google Scholar]
- 16. Puig S., Lee J., Lau M., and Thiele D. J. (2002) Biochemical and genetic analyses of yeast and human high affinity copper transporters suggest a conserved mechanism for copper uptake. J. Biol. Chem. 277, 26021–26030 10.1074/jbc.M202547200 [DOI] [PubMed] [Google Scholar]
- 17. Klomp A. E., Tops B. B., Van Denberg I. E., Berger R., and Klomp L. W. (2002) Biochemical characterization and subcellular localization of human copper transporter 1 (hCTR1). Biochem. J. 364, 497–505 10.1042/bj20011803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee J., Peña M. M., Nose Y., and Thiele D. J. (2002) Biochemical characterization of the human copper transporter Ctr1. J. Biol. Chem. 277, 4380–4387 10.1074/jbc.M104728200 [DOI] [PubMed] [Google Scholar]
- 19. De Feo C. J., Mootien S., and Unger V. M. (2010) Tryptophan scanning analysis of the membrane domain of CTR-copper transporters. J. Membr. Biol. 234, 113–123 10.1007/s00232-010-9239-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. De Feo C. J., Aller S. G., Siluvai G. S., Blackburn N. J., and Unger V. M. (2009) Three-dimensional structure of the human copper transporter hCTR1. Proc. Natl. Acad. Sci. U.S.A. 106, 4237–4242 10.1073/pnas.0810286106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Aller S. G., and Unger V. M. (2006) Projection structure of the human copper transporter CTR1 at 6-Å resolution reveals a compact trimer with a novel channel-like architecture. Proc. Natl. Acad. Sci. U.S.A. 103, 3627–3632 10.1073/pnas.0509929103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Aller S. G., Eng E. T., De Feo C. J., and Unger V. M. (2004) Eukaryotic CTR copper uptake transporters require two faces of the third transmembrane domain for helix packing, oligomerization, and function. J. Biol. Chem. 279, 53435–53441 10.1074/jbc.M409421200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dancis A., Haile D., Yuan D. S., and Klausner R. D. (1994) The Saccharomyces cerevisiae copper transport protein (Ctr1p). Biochemical characterization, regulation by copper, and physiologic role in copper uptake. J. Biol. Chem. 269, 25660–25667 [PubMed] [Google Scholar]
- 24. Eisses J. F., and Kaplan J. H. (2005) The mechanism of copper uptake mediated by human CTR1: a mutational analysis. J. Biol. Chem. 280, 37159–37168 10.1074/jbc.M508822200 [DOI] [PubMed] [Google Scholar]
- 25. Kim S., Jeon T. J., Oberai A., Yang D., Schmidt J. J., and Bowie J. U. (2005) Transmembrane glycine zippers: physiological and pathological roles in membrane proteins. Proc. Natl. Acad. Sci. U.S.A. 102, 14278–14283 10.1073/pnas.0501234102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Georgatsou E., Mavrogiannis L. A., Fragiadakis G. S., and Alexandraki D. (1997) The yeast Fre1p/Fre2p cupric reductases facilitate copper uptake and are regulated by the copper-modulated Mac1p activator. J. Biol. Chem. 272, 13786–13792 10.1074/jbc.272.21.13786 [DOI] [PubMed] [Google Scholar]
- 27. Ohgami R. S., Campagna D. R., McDonald A., and Fleming M. D. (2006) The Steap proteins are metalloreductases. Blood 108, 1388–1394 10.1182/blood-2006-02-003681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rubino J. T., Riggs-Gelasco P., and Franz K. J. (2010) Methionine motifs of copper transport proteins provide general and flexible thioether-only binding sites for Cu(I) and Ag(I). J. Biol. Inorg. Chem. 15, 1033–1049 10.1007/s00775-010-0663-9 [DOI] [PubMed] [Google Scholar]
- 29. Pena M. M., Puig S., and Thiele D. J. (2000) Characterization of the Saccharomyces cerevisiae high affinity copper transporter Ctr3. J. Biol. Chem. 275, 33244–33251 10.1074/jbc.M005392200 [DOI] [PubMed] [Google Scholar]
- 30. Rees E. M., Lee J., and Thiele D. J. (2004) Mobilization of intracellular copper stores by the ctr2 vacuolar copper transporter. J. Biol. Chem. 279, 54221–54229 10.1074/jbc.M411669200 [DOI] [PubMed] [Google Scholar]
- 31. Bellemare D. R., Shaner L., Morano K. A., Beaudoin J., Langlois R., and Labbe S. (2002) Ctr6, a vacuolar membrane copper transporter in Schizosaccharomyces pombe. J. Biol. Chem. 277, 46676–46686 10.1074/jbc.M206444200 [DOI] [PubMed] [Google Scholar]
- 32. Portnoy M. E., Schmidt P. J., Rogers R. S., and Culotta V. C. (2001) Metal transporters that contribute copper to metallochaperones in Saccharomyces cerevisiae. Mol. Genet. Genomics 265, 873–882 10.1007/s004380100482 [DOI] [PubMed] [Google Scholar]
- 33. Kuo Y. M., Zhou B., Cosco D., and Gitschier J. (2001) The copper transporter CTR1 provides an essential function in mammalian embryonic development. Proc. Natl. Acad. Sci. U.S.A. 98, 6836–6841 10.1073/pnas.111057298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee J., Prohaska J. R., and Thiele D. J. (2001) Essential role for mammalian copper transporter Ctr1 in copper homeostasis and embryonic development. Proc. Natl. Acad. Sci. U.S.A. 98, 6842–6847 10.1073/pnas.111058698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nose Y., Kim B. E., and Thiele D. J. (2006) Ctr1 drives intestinal copper absorption and is essential for growth, iron metabolism, and neonatal cardiac function. Cell Metab. 4, 235–244 10.1016/j.cmet.2006.08.009 [DOI] [PubMed] [Google Scholar]
- 36. Kim B. E., Turski M. L., Nose Y., Casad M., Rockman H. A., and Thiele D. J. (2010) Cardiac copper deficiency activates a systemic signaling mechanism that communicates with the copper acquisition and storage organs. Cell Metab. 11, 353–363 10.1016/j.cmet.2010.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kim H., Son H. Y., Bailey S. M., and Lee J. (2009) Deletion of hepatic Ctr1 reveals its function in copper acquisition and compensatory mechanisms for copper homeostasis. Am. J. Physiol. Gastrointest. Liver Physiol. 296, G356–G364 10.1152/ajpgi.90632.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Baker Z. N., Jett K., Boulet A., Hossain A., Cobine P. A., Kim B. E., El Zawily A. M., Lee L., Tibbits G. F., Petris M. J., and Leary S. C. (2017) The mitochondrial metallochaperone SCO1 maintains CTR1 at the plasma membrane to preserve copper homeostasis in the murine heart. Hum. Mol. Genet. 26, 4617–4628 10.1093/hmg/ddx344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hlynialuk C. J., Ling B., Baker Z. N., Cobine P. A., Yu L. D., Boulet A., Wai T., Hossain A., El Zawily A. M., McFie P. J., Stone S. J., Diaz F., Moraes C. T., Viswanathan D., Petris M. J., and Leary S. C. (2015) The mitochondrial metallochaperone SCO1 is required to sustain expression of the high-affinity copper transporter CTR1 and preserve copper homeostasis. Cell Rep. 10, 933–943 10.1016/j.celrep.2015.01.019 [DOI] [PubMed] [Google Scholar]
- 40. Kahra D., Kovermann M., and Wittung-Stafshede P. (2016) The C terminus of human copper importer Ctr1 acts as a binding site and transfers copper to Atox1. Biophys. J. 110, 95–102 10.1016/j.bpj.2015.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Levy A. R., Yarmiayev V., Moskovitz Y., and Ruthstein S. (2014) Probing the structural flexibility of the human copper metallochaperone Atox1 dimer and its interaction with the CTR1 c-terminal domain. J. Phys. Chem. B 118, 5832–5842 10.1021/jp412589b [DOI] [PubMed] [Google Scholar]
- 42. Pope C. R., De Feo C. J., and Unger V. M. (2013) Cellular distribution of copper to superoxide dismutase involves scaffolding by membranes. Proc. Natl. Acad. Sci. U.S.A. 110, 20491–20496 10.1073/pnas.1309820110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hwang C., Sinskey A. J., and Lodish H. F. (1992) Oxidized redox state of glutathione in the endoplasmic reticulum. Science 257, 1496–1502 10.1126/science.1523409 [DOI] [PubMed] [Google Scholar]
- 44. Maryon E. B., Molloy S. A., and Kaplan J. H. (2013) Cellular glutathione plays a key role in copper uptake mediated by human copper transporter 1. Am. J. Physiol. Cell Physiol 304, C768–C779 10.1152/ajpcell.00417.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sinani D., Adle D. J., Kim H., and Lee J. (2007) Distinct mechanisms for Ctr1-mediated copper and cisplatin transport. J. Biol. Chem. 282, 26775–26785 10.1074/jbc.M703973200 [DOI] [PubMed] [Google Scholar]
- 46. Gadsby D. C. (2009) Ion channels versus ion pumps: the principal difference, in principle. Nat. Rev. Mol. Cell Biol. 10, 344–352 10.1038/nrm2668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lu M., Chai J., and Fu D. (2009) Structural basis for autoregulation of the zinc transporter YiiP. Nat. Struct. Mol. Biol. 16, 1063–1067 10.1038/nsmb.1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lu M., and Fu D. (2007) Structure of the zinc transporter YiiP. Science 317, 1746–1748 10.1126/science.1143748 [DOI] [PubMed] [Google Scholar]
- 49. Zhang T., Liu J., Fellner M., Zhang C., Sui D., and Hu J. (2017) Crystal structures of a ZIP zinc transporter reveal a binuclear metal center in the transport pathway. Sci. Adv. 3, e1700344 10.1126/sciadv.1700344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Coudray N., Valvo S., Hu M., Lasala R., Kim C., Vink M., Zhou M., Provasi D., Filizola M., Tao J., Fang J., Penczek P. A., Ubarretxena-Belandia I., and Stokes D. L. (2013) Inward-facing conformation of the zinc transporter YiiP revealed by cryoelectron microscopy. Proc. Natl. Acad. Sci. U.S.A. 110, 2140–2145 10.1073/pnas.1215455110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ehrnstorfer I. A., Manatschal C., Arnold F. M., Laederach J., and Dutzler R. (2017) Structural and mechanistic basis of proton-coupled metal ion transport in the SLC11/NRAMP family. Nat. Commun. 8, 14033 10.1038/ncomms14033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ehrnstorfer I. A., Geertsma E. R., Pardon E., Steyaert J., and Dutzler R. (2014) Crystal structure of a SLC11 (NRAMP) transporter reveals the basis for transition-metal ion transport. Nat. Struct. Mol. Biol. 21, 990–996 10.1038/nsmb.2904 [DOI] [PubMed] [Google Scholar]
- 53. Maryon E. B., Molloy S. A., Ivy K., Yu H., and Kaplan J. H. (2013) Rate and regulation of copper transport by human copper transporter 1 (hCTR1). J. Biol. Chem. 288, 18035–18046 10.1074/jbc.M112.442426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Öhrvik H., Logeman B., Turk B., Reinheckel T., and Thiele D. J. (2016) Cathepsin protease controls copper and cisplatin accumulation via cleavage of the Ctr1 metal-binding ectodomain. J. Biol. Chem. 291, 13905–13916 10.1074/jbc.M116.731281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Guo Y., Smith K., Lee J., Thiele D. J., and Petris M. J. (2004) Identification of methionine-rich clusters that regulate copper-stimulated endocytosis of the human Ctr1 copper transporter. J. Biol. Chem. 279, 17428–17433 10.1074/jbc.M401493200 [DOI] [PubMed] [Google Scholar]
- 56. Petris M. J., Smith K., Lee J., and Thiele D. J. (2003) Copper-stimulated endocytosis and degradation of the human copper transporter, hCtr1. J. Biol. Chem. 278, 9639–9646 10.1074/jbc.M209455200 [DOI] [PubMed] [Google Scholar]
- 57. Clifford R. J., Maryon E. B., and Kaplan J. H. (2016) Dynamic internalization and recycling of a metal ion transporter: Cu homeostasis and CTR1, the human Cu+ uptake system. J. Cell Sci. 129, 1711–1721 10.1242/jcs.173351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Amlacher S., Sarges P., Flemming D., van Noort V., Kunze R., Devos D. P., Arumugam M., Bork P., and Hurt E. (2011) Insight into structure and assembly of the nuclear pore complex by utilizing the genome of a eukaryotic thermophile. Cell 146, 277–289 10.1016/j.cell.2011.06.039 [DOI] [PubMed] [Google Scholar]
- 59. Altschul S. F., Gish W., Miller W., Myers E. W., and Lipman D. J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 60. Edgar R. C. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Peña M. M., Koch K. A., and Thiele D. J. (1998) Dynamic regulation of copper uptake and detoxification genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 18, 2514–2523 10.1128/MCB.18.5.2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rubino J. T., and Franz K. J. (2012) Coordination chemistry of copper proteins: how nature handles a toxic cargo for essential function. J. Inorg. Biochem. 107, 129–143 10.1016/j.jinorgbio.2011.11.024 [DOI] [PubMed] [Google Scholar]
- 63. Haas K. L., Putterman A. B., White D. R., Thiele D. J., and Franz K. J. (2011) Model peptides provide new insights into the role of histidine residues as potential ligands in human cellular copper acquisition via Ctr1. J. Am. Chem. Soc. 133, 4427–4437 10.1021/ja108890c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rubino J. T., Chenkin M. P., Keller M., Riggs-Gelasco P., and Franz K. J. (2011) A comparison of methionine, histidine and cysteine in copper(I)-binding peptides reveals differences relevant to copper uptake by organisms in diverse environments. Metallomics 3, 61–73 10.1039/C0MT00044B [DOI] [PubMed] [Google Scholar]
- 65. Rees E. M., and Thiele D. J. (2007) Identification of a vacuole-associated metalloreductase and its role in Ctr2-mediated intracellular copper mobilization. J. Biol. Chem. 282, 21629–21638 10.1074/jbc.M703397200 [DOI] [PubMed] [Google Scholar]
- 66. Yamaguchi-Iwai Y., Serpe M., Haile D., Yang W., Kosman D. J., Klausner R. D., and Dancis A. (1997) Homeostatic regulation of copper uptake in yeast via direct binding of MAC1 protein to upstream regulatory sequences of FRE1 and CTR1. J. Biol. Chem. 272, 17711–17718 10.1074/jbc.272.28.17711 [DOI] [PubMed] [Google Scholar]
- 67. Zhou H., and Thiele D. J. (2001) Identification of a novel high affinity copper transport complex in the fission yeast Schizosaccharomyces pombe. J. Biol. Chem. 276, 20529–20535 10.1074/jbc.M102004200 [DOI] [PubMed] [Google Scholar]
- 68. Sun T. S., Ju X., Gao H. L., Wang T., Thiele D. J., Li J. Y., Wang Z. Y., and Ding C. (2014) Reciprocal functions of Cryptococcus neoformans copper homeostasis machinery during pulmonary infection and meningoencephalitis. Nat. Commun. 5, 5550 10.1038/ncomms6550 [DOI] [PubMed] [Google Scholar]
- 69. Drew D., Newstead S., Sonoda Y., Kim H., von Heijne G., and Iwata S. (2008) GFP-based optimization scheme for the overexpression and purification of eukaryotic membrane proteins in Saccharomyces cerevisiae. Nat. Protoc. 3, 784–798 10.1038/nprot.2008.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bertinato J., Cheung L., Hoque R., and Plouffe L. J. (2010) Ctr1 transports silver into mammalian cells. J. Trace Elem. Med. Biol. 24, 178–184 10.1016/j.jtemb.2010.01.009 [DOI] [PubMed] [Google Scholar]
- 71. Kim H. S., and Ahner B. A. (2006) Calibration of Phen Green for use as a Cu(I)-selective fluorescent indicator. Anal. Chim. Acta 575, 223–229 10.1016/j.aca.2006.05.089 [DOI] [PubMed] [Google Scholar]
- 72. Illing A. C., Shawki A., Cunningham C. L., and Mackenzie B. (2012) Substrate profile and metal-ion selectivity of human divalent metal-ion transporter-1. J. Biol. Chem. 287, 30485–30496 10.1074/jbc.M112.364208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Shingles R., Wimmers L. E., and McCarty R. E. (2004) Copper transport across pea thylakoid membranes. Plant Physiol. 135, 145–151 10.1104/pp.103.037895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Martins V., Hanana M., Blumwald E., and Gerós H. (2012) Copper transport and compartmentation in grape cells. Plant Cell Physiol. 53, 1866–1880 10.1093/pcp/pcs125 [DOI] [PubMed] [Google Scholar]
- 75. Froschauer E. M., Schweyen R. J., and Wiesenberger G. (2009) The yeast mitochondrial carrier proteins Mrs3p/Mrs4p mediate iron transport across the inner mitochondrial membrane. Biochim. Biophys. Acta 1788, 1044–1050 10.1016/j.bbamem.2009.03.004 [DOI] [PubMed] [Google Scholar]
- 76. Geertsma E. R., Nik Mahmood N. A., Schuurman-Wolters G. K., and Poolman B. (2008) Membrane reconstitution of ABC transporters and assays of translocator function. Nat. Protoc. 3, 256–266 10.1038/nprot.2007.519 [DOI] [PubMed] [Google Scholar]
- 77. Long F., Su C. C., Zimmermann M. T., Boyken S. E., Rajashankar K. R., Jernigan R. L., and Yu E. W. (2010) Crystal structures of the CusA efflux pump suggest methionine-mediated metal transport. Nature 467, 484–488 10.1038/nature09395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wijekoon C. J., Udagedara S. R., Knorr R. L., Dimova R., Wedd A. G., and Xiao Z. (2017) Copper ATPase CopA from Escherichia coli: quantitative correlation between ATPase activity and vectorial copper transport. J. Am. Chem. Soc. 139, 4266–4269 10.1021/jacs.6b12921 [DOI] [PubMed] [Google Scholar]
- 79. Chao Y., and Fu D. (2004) Kinetic study of the antiport mechanism of an Escherichia coli zinc transporter, ZitB. J. Biol. Chem. 279, 12043–12050 10.1074/jbc.M313510200 [DOI] [PubMed] [Google Scholar]
- 80. Boulet A., Vest K. E., Maynard M. K., Gammon M. G., Russell A. C., Mathews A. T., Cole S. E., Zhu X., Phillips C. B., Kwong J. Q., Dodani S. C., Leary S. C., and Cobine P. A. (2018) The mammalian phosphate carrier SLC25A3 is a mitochondrial copper transporter required for cytochrome c oxidase biogenesis. J. Biol. Chem. 293, 1887–1896 10.1074/jbc.RA117.000265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Labbé S., Peña M. M., Fernandes A. R., and Thiele D. J. (1999) A copper-sensing transcription factor regulates iron uptake genes in Schizosaccharomyces pombe. J. Biol. Chem. 274, 36252–36260 10.1074/jbc.274.51.36252 [DOI] [PubMed] [Google Scholar]
- 82. Ohrvik H., and Thiele D. J. (2014) How copper traverses cellular membranes through the mammalian copper transporter 1, Ctr1. Ann. N.Y. Acad. Sci. 1314, 32–41 10.1111/nyas.12371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Hassett R., and Kosman D. J. (1995) Evidence for Cu(II) reduction as a component of copper uptake by Saccharomyces cerevisiae. J. Biol. Chem. 270, 128–134 10.1074/jbc.270.1.128 [DOI] [PubMed] [Google Scholar]
- 84. Smith A. D., Logeman B. L., and Thiele D. J. (2017) Copper acquisition and utilization in fungi. Annu. Rev. Microbiol. 71, 597–623 10.1146/annurev-micro-030117-020444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Lin W., Chai J., Love J., and Fu D. (2010) Selective electrodiffusion of zinc ions in a Zrt-, Irt-like protein, ZIPB. J. Biol. Chem. 285, 39013–39020 10.1074/jbc.M110.180620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Shenberger Y., Shimshi A., and Ruthstein S. (2015) EPR spectroscopy shows that the blood carrier protein, human serum albumin, closely interacts with the N-terminal domain of the copper transporter, Ctr1. J. Phys. Chem. B 119, 4824–4830 10.1021/acs.jpcb.5b00091 [DOI] [PubMed] [Google Scholar]
- 87. Mumberg D., Müller R., and Funk M. (1995) Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156, 119–122 10.1016/0378-1119(95)00037-7 [DOI] [PubMed] [Google Scholar]
- 88. Hays F. A., Roe-Zurz Z., and Stroud R. M. (2010) Overexpression and purification of integral membrane proteins in yeast. Methods Enzymol. 470, 695–707 10.1016/S0076-6879(10)70029-X [DOI] [PubMed] [Google Scholar]
- 89. Bligh E. G., and Dyer W. J. (1959) A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917 10.1139/y59-099 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.