Abstract
Folate derivatives are important cofactors for enzymes in several metabolic processes. Folate-related inhibition and resistance mechanisms in bacteria are potential targets for antimicrobial therapies and therefore a significant focus of current research. Here, we report that the activity of Escherichia coli poly-γ-glutamyl tetrahydrofolate/dihydrofolate synthase (FolC) is regulated by glutamate/glutamine-sensing uridylyltransferase (GlnD), THF-dependent tRNA modification enzyme (MnmE), and UDP-glucose dehydrogenase (Ugd) as shown by direct in vitro protein–protein interactions. Using kinetics analyses, we observed that GlnD, Ugd, and MnmE activate FolC many-fold by decreasing the Khalf of FolC for its substrate l-glutamate. Moreover, FolC inhibited the GTPase activity of MnmE at low GTP concentrations. The growth phenotypes associated with these proteins are discussed. These results, obtained using direct in vitro enzyme assays, reveal unanticipated networks of allosteric regulatory interactions in the folate pathway in E. coli and indicate regulation of polyglutamylated tetrahydrofolate biosynthesis by the availability of nitrogen sources, signaled by the glutamine-sensing GlnD protein.
Keywords: folate, enzyme kinetics, GTPase, gram-negative bacteria, metabolism, allosteric regulation, cytoplasmic GTP, FolC, G-protein MnmE, tetrahydrofolate polyglutamylation, GlnD, Ugd, MnmE, folate metabolism, amino acid sensing
Introduction
Protein–protein interactions regulate many processes in Escherichia coli and probably in all living cells. Carbon source availability is a signal for the recently identified global regulation of glycolysis and energy (ATP) balance by the phosphocarrier protein of the bacterial phosphotransferase system (PTS) HPr (1). Coregulation of carbon and nitrogen metabolism resulting, for example, in activation of glucosamine 6-P deaminase, NagB, by HPr, synergistically with uridylylated PII under limiting nitrogen conditions, is important for nutrient homeostasis (2). We have collaboratively published evidence for the existence of a network of protein–protein interactions (the interactome) in E. coli (3). This work provided the incentive and a guide for the research reported in this and previous works describing allosteric regulatory phenomena (1, 2).
Bifunctional folylpoly-γ-glutamate synthase, or dihydrofolate (DHF)3 synthase (FolC), catalyzes polyglutamylation of DHF; tetrahydrofolate (THF); 5,10-methylene-THF; and 10-formyl-THF using two additional substrates, ATP and l-glutamate. The enzyme also catalyzes glutamylation of 7,8-dihydropteroate, the biosynthetic precursor of folic acid. E. coli is able to synthesize DHF de novo, but eukaryotes must take up extracellular folate using FolT from the reduced folate carrier family (Transporter Classification Database (TC) no. 2.A.48). Bacteria use vitamin uptake transporters of the ECF family (TC no. 2.A.88) (4) or a member of the folate-biopterin transporter (FBT) family present in protists, cyanobacteria, and plants (TC no. 2.A.71) (5–9). Once inside the cell, folate is reduced via DHF reductase (FolA) to DHF and then to THF (Fig. 1), the precursor to many folate derivative cofactors (5). Upon reduction to THF, FolC catalyzes the addition of glutamyl residues, resulting in the formation first of THF-monoglutamate and then of THF-polyglutamate derivatives; the latter exhibit full cofactor activity. This process is tightly regulated in mammals (10, 11). Folate derivatives are important cofactors for enzymes that are involved in several metabolic processes including serine–glycine interconversion, methionine recycling, purine biosynthesis, the thymidylate cycle, and one-carbon metabolism (11, 12). As a result of their involvements in these key processes, folate derivatives are universally required for cell growth (13). However, humans lack the ability to synthesize these compounds de novo and must rely on their diets to acquire adequate levels of folate (10).
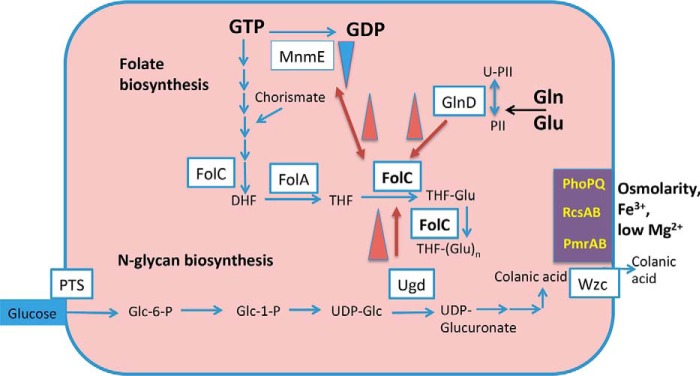
Figure 1.
De novo biosynthesis of folate from GTP, and FolC activation by the GlnD, MnmE, and Ugd proteins of E. coli. Inhibition of the MnmE GTPase activity by FolC and activation of FolC are shown with red arrows. Transcriptional activation of ugd by the PhoPQ, RscAB, and PmrAB two-component systems are shown in violet blocks. The abbreviations are: PS, polysaccharides; Glc, d-glucose; Glu, l-glutamate; Wzc, exporter of colanic acid; FolA, DHF reductase; PTS, phosphotransferase system.
Previous interactome studies have revealed potential physical interactions between FolC and other proteins such as the 1) uridylyltransferase/uridylyl-removing enzyme (GlnD), 2) UDP-glucose 6-dehydrogenase (Ugd), and 3) a tRNA modification enzyme (MnmE).
GlnD senses the cytoplasmic l-glutamine and l-glutamate concentrations, and signal transduction systems deliver output signals, even in complex media (14). Variations in the concentrations of proteins and nutrients stimulate signal transmission for optimal enzymatic activity and growth. GlnD catalyzes uridylylation of the PII protein but has not been known as a regulator of folate biosynthesis. We show that it interacts with and activates FolC in the presence of low concentrations of glutamate (Fig. 1). These observations may explain the fact that an experimental approach showed that GlnD, but not GlnA, GlnB, or GlnE, is an essential protein in E. coli (15, 16), suggesting that it could be important for the design and characterization of inhibitors acting on the FolC/GlnD complex.
Ugd produces UDP-glucuronate, a precursor of colanic acid (17), an enterobacterial exopolysaccharide produced in response to stress conditions. With low cytoplasmic Mg2+ concentrations, E. coli Wzc, a bifunctional two domain tyrosine kinase/colanate exporter responsible for the export of colanic acid (18–23), phosphorylates Ugd. Under these conditions, charge repulsion between lipopolysaccharide molecules increases, preventing aggregation of cells (18, 23). The participation of several combinations of regulatory systems, such as PhoP (in response to low Mg2+ concentration) (21, 24), RcsB, and PmrA, control ugd gene expression in response to a variety of signals including changes in temperature, osmolarity, and overexpression of membrane proteins.
MnmE is a THF-dependent tRNA modification GTPase found in complex with MnmG, another tRNA modification enzyme (25, 26). Its abnormally high GTPase activity depends on the cytoplasmic potassium concentration; however, classical small GTP-binding proteins are usually regulated by activating proteins and guanine nucleotide-exchange factors (25, 27). Typically, GTP-binding proteins show low intrinsic GTPase activity, but MnmE is the exception, being active in the homodimeric state in the presence only of potassium ions.
All four nucleotides in tRNAs can be posttranslationally modified, and modification of the anticodon is particularly important (26). Specifically, position 34, U34, known as the wobble position, is most often modified. The 5-carboxy-methyl-aminomethyl-2-thio–type modification ensures fidelity of regions ending in A or G. Unmodified U34 will interact with all four nucleotides and lead to misincorporation of amino acids into the growing peptide, leading, for example, to poor growth of Salmonella enterica serovar Typhimurium (28). The pathway of U34 modification is complex and involves many proteins. Depending on the MnmEG substrate, an unmodified U34 is converted to aminomethyluridine using ammonium or 5-carboxy-methyl-aminomethyluridine using glycine. MnmE has also been found to play a potential role in virulence interactions of the hosts with S. enterica serovar Typhimurium and Pseudomonas syringae (29).
Interestingly, GadE is a key regulator of the major acid resistance system in E. coli which is composed of the glutamate decarboxylase isoenzymes GadA and GadB and a dedicated glutamic acid/GABA antiporter GadC. During the bacterial response to acid stresses, GadA and GadB catalyze the decarboxylation of glutamic acid, yielding GABA, which is subsequently exported by GadC in exchange for another molecule of glutamic acid. The anaerobic transcriptional activation of the gadE-mdtEF operon in E. coli is largely dependent on the global regulators ArcA and MnmE, as deletion of MnmE causes a significant decrease of the transcription of gadE-mdtEF (30, 31).
Cytoplasmic GTP availability is very important for bacteria, and as shown here, MnmE GTPase activity, associated with tRNA modification, is inhibited by FolC at 75 μm aminopterin (a THF analogue) when the GTP concentration is low (Fig. 1).
Folate pathway–related inhibition and resistance mechanisms have been actively studied in bacteria (32–35). The previously mentioned metabolic processes involving THF operate more slowly with monoglutamylated folyl coenzymes than with polyglutamylated cofactors because folate-dependent enzymes exhibit low affinity for the monoglutamylated cofactors (36). As such, folylpoly-γ-glutamate synthase (FolC) is required for normal cell growth (37). Taken together, this makes FolC an ideal target for antimicrobial therapy, although resistance to FolC inhibitors in Mycobacteria has been documented (38).
We here demonstrate that activation of FolC by Ugd, MnmE, and GlnD occurs via novel regulatory mechanisms, impacting the activation of one-carbon metabolism in response to stress conditions. FolC has the same binding site for THF and dihydropteroate (39). The modeling of the Ugd–GlnD–FolC interactions revealed the probable residues involved in this interaction. We also show that the GTPase activity of the tRNA modification enzyme, MnmE, is inhibited by FolC, thus revealing a novel mode of functional protein–protein interactions in E. coli. The importance of these interactions was confirmed by in vivo growth experiments. For the first time, regulation of polyglutamylated THF biosynthesis by the availability of a nitrogen source, sensed by the glutamine-sensing GlnD protein, is demonstrated for E. coli.
Results
THF synthase (FolC) regulation
FolC polyglutamylates (derivatizes with glutamate) THF, yielding the active cofactor. To study FolC regulation, FolC, MnmE, GlnD, Ugd, TdcB (serine/threonine dehydratase), HybD (hydrogenase 2 maturation protease), and the PII protein were all purified (see “Experimental Procedures”). TdcB and PII were refolded as described in Ref. 1. Interactome data (3) had suggested that FolC interacts with proteins: Ugd, GlnD, MnmE, TdcB, and HybD. They were examined for their effects on FolC activity as shown in Figs. 2 and 3. GlnD activated at least 4-fold, whereas TcdB, HybD, and PII did not activate or inhibit appreciably under the conditions used for the experiment, using 350 μm aminopterin and 2 mm l-glutamate. To investigate the effect of l-glutamate on the activation by GlnD, we measured the kinetics under constant conditions but with varying concentrations of l-glutamate with THF (Fig. 2B). Additionally, experiments were conducted with aminopterin, and the concentration of substrate that produces half-maximal enzyme velocity (Khalf) was measured at 0.2 mm aminopterin (Fig. S1). GlnD activated FolC, reducing the Khalf for glutamate around 10-fold (Table 1). FolC activation by GlnD at low concentrations of l-glutamate and 0.15 mm tetrahydrofolate was possible only in the presence of 7 mm l-glutamine (Fig. 2B). Titration of l-glutamine for FolC activation by GlnD was shown (Fig. S2). The presence of 1.5 mm α-ketoglutarate abolished activation by GlnD in the presence of 7 mm l-glutamine.
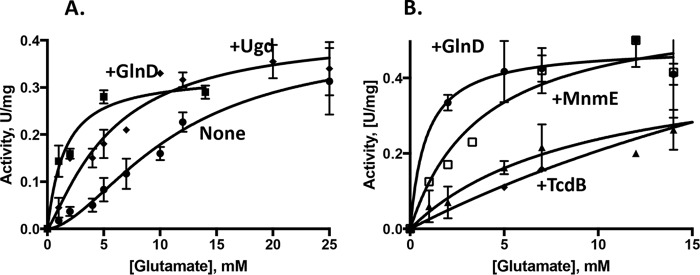
Figure 2.
Allosteric activation of FolC by the GlnD, MnmE, and Ugd proteins. A, the kinetics were measured as a function of the glutamate concentration (0–25 mm) at an aminopterin concentration of 200 μm in the presence of 0.5 μm GlnD (squares) or 0.3 μm Ugd (diamonds) or in their absence (circles). The assay mixture contained Tris-HCl, pH 8.7, 3 mm DTT, 200 mm KCl, 10 mm MgSO4, and 2.2 mm ATP. B, the kinetics were measured as a function of the glutamate concentration (0–15 mm) at a THF concentration of 180 μm in the presence of 0.5 μm GlnD (circles), 0.5 μm MnmE (squares), and TdcB (triangles) or in their absence (diamonds) in the same assay mixture; 10% DMSO was present. The experiments were repeated two times, and error bars indicate standard deviations.
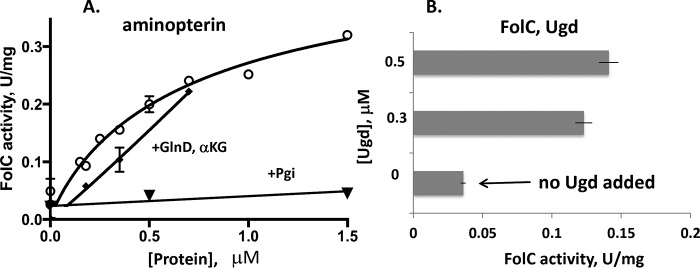
Figure 3.
Allosteric activation of FolC by the GlnD protein. A, activity was measured at 4 mm l-glutamate with 400 μm aminopterin in an assay mixture containing Tris-HCl, pH 8.7, 3 mm DTT, 50 mm KCl, 10 mm MgSO4, and 5 mm ATP in the presence of 0–1.5 μm GlnD (circles) or phosphoglucose isomerase, Pgi (inverted triangles), as a negative control. The α-ketoglutarate (αKG) effect on FolC activity was measured with an l-glutamate concentration of 3 mm and 120 μm aminopterin in the presence of 0–0.7 μm GlnD and αKG (3.5 mm; squares). B, the kinetics were measured as a function of the Ugd (0–0.5 μm) at 1 mm glutamate and an aminopterin concentration of 200 μm. The experiments were repeated two times, and error bars indicate standard deviations.
Table 1.
Kinetic parameters of the FolC-catalyzed reaction with respect to l-glutamate, with aminopterin (Amn) or THF present in the presence or absence of GlnD, MnmE, or Ugd (columns 1–6) and MnmE with respect to GTP (columns 7 and 8). U = μmol/min at 37 °C
| 1. FolC + MnmE | 2. FolC + GlnD | 3. FolC − GlnD − MnmE | 4. FolC − GlnD − Ugd | 5. FolC + Ugd | 6. FolC + GlnD | 7. MnmE − FolC | 8. MnmE + FolC | |
|---|---|---|---|---|---|---|---|---|
| Substrate | Glutamate (THF) | Glutamate (THF) | Glutamate (THF) | Glutamate (Amn) | Glutamate (Amn) | Glutamate (Amn) | GTP | GTP |
| Vmax, (U/mg) | 0.59 ± 0.09 | 0.51 ± 0.04 | 0.29 ± 0.05 | 0.39 ± 0.05 | 0.32 ± 0.03 | 0.33 ± 0.05 | 0.19 ± 0.02 | 0.14 ± 0.01 |
| h, Hill coefficient | 1 | 1 | 1.9 | 1.7 | 1.3 | 1.05 | 3.1 | 2.5 |
| Khalf, mm | 3.8 ± 1.7 | 1 ± 0.4 | 9.2 ± 3.3 | 11 ± 2.9 | 5.6 ± 1.6 | 1.54 ± 0.55 | 0.28 ± 0.03 | 0.46 ± 0.04 |
Titration with GlnD at 1 mm l-glutamate and aminopterin, using the Pgi protein as a negative control, is shown in Fig. 3A. Twelve-fold activation by GlnD (but not by Pgi) using a 0.5 μm protein concentration was found. FolC activation by Ugd at 4 mm l-glutamate and 0.4 mm aminopterin was most efficient at 5 mm ATP (Fig. 3B). We tested separately the effect of l-glutamine (1–30 mm) on activation of FolC by GlnD (0.3 μm) with 0.5 mm l-glutamate, but no inhibitory or activation effect was found using aminopterin as substrate (data not shown). A small inhibitory effect on FolC activity was noticed for α-ketoglutarate at a concentration of 3 mm when using 1 mm l-glutamate and 0.12 mm aminopterin in the presence of GlnD (Fig. 3A). No synergistic or additive effect was observed for Ugd and GlnD (data not shown) suggesting either that both proteins bind to the same FolC allosteric-binding site, or that their binding sites overlap. The kinetics with varying concentrations of aminopterin are shown at Fig. S1.
Activation of FolC by MnmE and reciprocal inhibition of MnmE by FolC
To investigate the effect of l-glutamate on the activation by MnmE, we measured the kinetics under constant conditions but with varying concentrations of l-glutamate with THF or aminopterin (Fig. 2B). Activation by MnmE using a 0.8 μm protein concentration was not observed with aminopterin as substrate, but activation using a 0.5 μm protein concentration with THF was substantial. The kinetics followed the Michaelis-Menten model, and GraphPad Prism 7 was used for calculations to fit the equation. The Michaelis constant Km measured for l-glutamate without a protein-effector was 9.8 mm. MnmE substantially decreased the Km to 3.8 mm in the presence of 10 mm l-glutamine.
MnmE GTPase activity was measured using the assay described in “Experimental Procedures” in the presence and absence of aminopterin or l-glutamate, and also with and without FolC. These were added in different combinations. An increase in the GTPase activity of the methyl-THF–binding tRNA-modification protein MnmE was observed (2-fold) when the THF analogue aminopterin was added (data not shown). The presence of FolC in the assay mixture reduced the activation by aminopterin. Titration with GTP (Fig. 4) confirmed FolC-dependent inhibition of MnmE activity in the presence of the THF analogue aminopterin.
Figure 4.
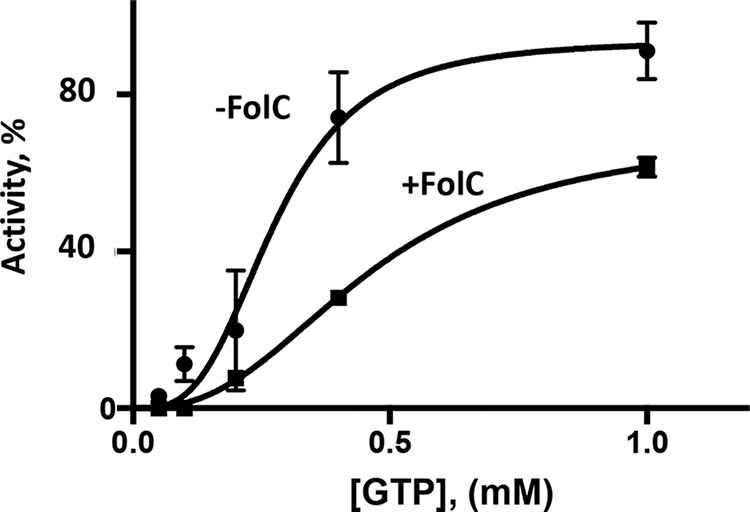
Allosteric inhibition of the MnmE GTPase activity by FolC. The kinetics were measured as a function of the GTP concentration. The assay mixture contained 80 mm Tris-HCl, pH 8.0, 2 mm DTT, 1 mm EDTA, 50 mm KCl, 10% glycerol, 20 nm MnmE, 75 μm aminopterin, and 10 mm MgCl2. The assay was conducted at 37 °C and incubated for 1 h. Pi formation, Pi (μm), was measured (see “Experimental Procedures”). The experiments were repeated two times, and error bars indicate standard deviations.
FolC/MnmE/Ugd docking model
The model for FolC binding showed both Ugd and MnmE binding to the C-terminal (smaller) domain of FolC. Interestingly, although the docking was performed separately for Ugd and MnmE, they showed considerable shape complementarity and possible interfaces with each other (Fig. 5). Also, their binding did not appear to sterically hinder access to the FolC active site in the N-terminal/C-terminal domain cleft. Residues involved in these interactions are listed in Table S1.
Figure 5.
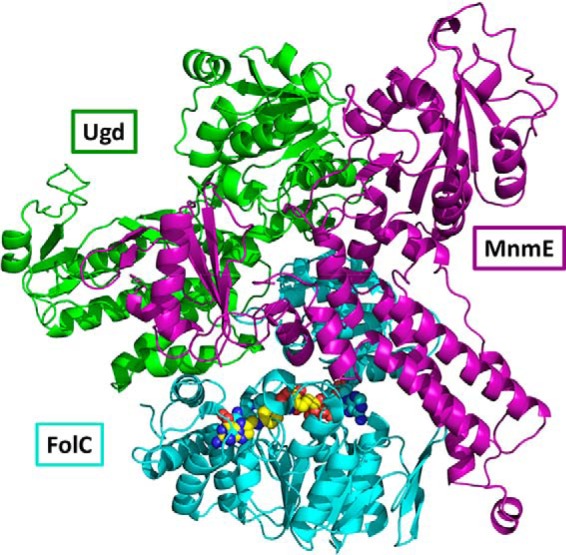
The FolC-binding model shows the potential for simultaneous binding of partners Ugd and MnmE. The C-terminal domain of FolC (teal, upper portion) binding both Ugd (green) and MnmE (purple) is shown. In both cases, protein binding does not block access to the interdomain cleft containing the active site (with attached ADP and dihydropteroate/DHPP shown as spheres). Ugd and MnmE are in close proximity to each other in the model, although with minimal steric overlap. Structures used are PDB ID 1W78:A (FolC) and homology models were based on PDB IDs 3PID:A (Ugd) and 2GJ8:A (MnmE).
folC, glnD, and ugd genetically interact with each other as well as with genes involved in metabolic and folate biosynthetic pathways
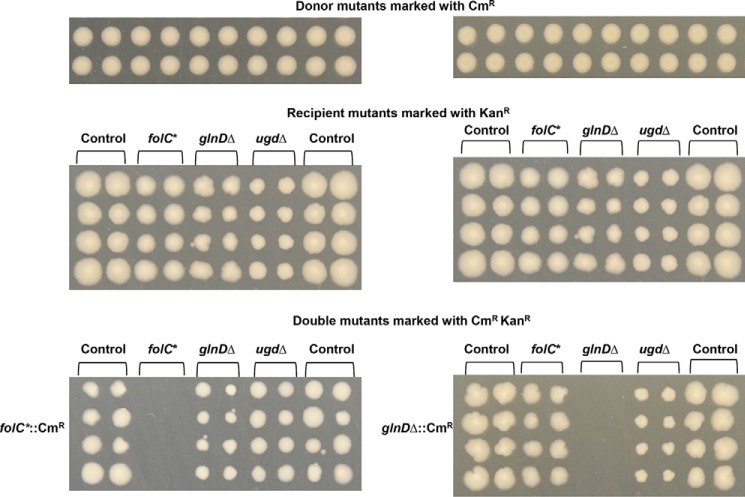
To identify E. coli genes that genetically interact with folC, glnD, and ugd, we used our previously published genetic interaction dataset using the E. coli Synthetic Genetic Array (eSGA) approach (40). Using this approach, double mutants were derived by conjugating the donor HfrC strain (marked with Δ::CmR) with single gene deletions or hypomorphic (i.e. partial loss of gene function) mutant strains of an essential gene with all other recipient nonessential or essential E. coli genes (marked with Δ::KanR) under both prototrophic (rich medium) and auxotrophic (minimal medium) culture conditions. The colony growth and relative fitness of the double mutants that survived the double antibiotic selection were digital imaged and quantified to generate genetic interaction scores. This resulted in cases of synthetic sick or lethal (double mutants grew more slowly than expected) or alleviating (double mutants grew more rapidly than expected) growth phenotypes. In accordance with our protein–protein interaction data and enzyme kinetics (Fig. 2), folC or ugd displayed strong synthetic sick or lethal with the carbon metabolism genes (accBC, frdBCD) and alleviating interactions with the genes involved in folate biosynthesis (folAM, pabC) and glutamate metabolism (purB, glmS) (Table S2). Additionally, we confirmed the recipient glnD and ugd with a modest synthetic sick growth defect when combined with folC or glnD donor mutant strains (Fig. 6). This observation appears to be bona fide as no detectable growth defects were observed when folC or glnD donor mutants were combined with a functionally unrelated gene (40). Taken together, our results suggest that folC, glnD, and ugd function redundantly with each other and with the carbon metabolic genes, and they cooperate with genes involved in glutamate and folate biosynthesis.
Figure 6.
Genetic screening among folC, glnD, and ugd interaction pairs. The Hfr Cavalli donor mutants, folC and glnD, were crossed with the indicated F− recipient mutant strains (*, essential hypomorph), and the resulting double mutants were selected on rich medium plates containing chloramphenicol (CmR) and kanamycin (KanR). The same recipient plate was used for screening of folC and glnD donors.
Discussion
Glutamine metabolism is regulated by the PII protein (GlnB), uridylylated by GlnD (41–43). The cytoplasmic l-glutamate/l-glutamine concentrations determine the rates of GlnD uridylyltransferase activity (14). We measured activation of FolC by GlnD, working as a potential sensor of the glutamate/glutamine concentrations in the cell. The binding of GlnD to an allosteric site in FolC reduced the Khalf of FolC for glutamate maximally about 10-fold (Fig. 2, Table 1).
PII and GlnE (bifunctional glutamine synthetase adenylyltransferase/adenylyl-removing enzyme) are not essential proteins in E. coli, but GlnD is essential (15, 16). We suggest that the mechanism of GlnD essentiality may involve activation of FolC. FolC activity decreased substantially without GlnD, and FolC activity is essential for E. coli growth, thus possibly rendering GlnD essential. Other possible mechanisms explaining GlnD essentiality may involve GlnD interactions with other essential enzymes, such as RnhB (RNase HII), which degrades DNA–RNA hybrids, and/or XerC (tyrosine recombinase) (3). Such potential interactions could also render GlnD protein–protein interactions essential, but these possibilities have not been examined.
The colanic acid transporter Wzc regulates the activity of Ugd, activating it by phosphorylating a tyrosyl residue in the protein (19). Stress conditions, such as low magnesium, have been shown to activate expression of the ugd gene (21), and now we find that similar conditions affect one-carbon metabolism via protein–protein interactions involving Ugd and FolC, activating FolC.
Among the proteins interacting with FolC (3), MnmE activated FolC (Fig. 5), but PII and several other proteins tested had no obvious effect (data not shown). These results clearly substantiate some of the published interactome data (3) and suggest that they are physiologically important. The consequences of the other interactions reported have not yet yielded positive results. Future studies will be required to determine whether these proteins do regulate FolC or if FolC regulates the activities of some of these other proteins.
As noted above, regulatory effects of FolC on MnmE GTPase activity were detected (Fig. 5). This is the first reported protein-dependent regulatory effect so far observed for MnmE. Typically, GTP-binding proteins have very low GTPase activity compared with MnmE (44). They require GTPase-activating proteins and GTP/GDP exchange factors to maximize their GTP hydrolase activities. In contrast, the G-domain of E. coli MnmE has high activity without added factors and has low affinity for GDP. The FolC inhibiting effect on the MnmE GTPase activity may allow growth by preventing GTP depletion. Thus, tetrahydrofolate-dependent activation of MnmE and compensatory inhibition of MnmE by FolC may be essential for the proper functioning of the MnmEG–protein complex. It appears that FolC activation by GlnD, but not by Ugd, is essential for the growth of E. coli strains (Fig. 6).
Experimental procedures
Protein purification
The clones encoding FolC, GlnD, Ugd, MnmE, TdcB, HybD, and GlnB (PII) were used from the ASKA collection (45) following verification by sequencing. For protein overproduction, the cells were grown at 37 °C, induced with isopropyl β-d-1-thiogalactopyranoside for 4 h, and harvested. Then cells were resuspended in 20 mm HEPES buffer, pH 7, containing 100 mm NaCl, 2 mm β-mercaptoethanol, and 0.3% Brij 35 with 2 mm phenylmethylsulfonyl fluoride present. Cells were lysed by incubation with lysozyme (1 mg/ml) for 30 min, followed by a freeze-thaw cycle and sonication as described (1). After centrifugation, the supernatant was loaded onto a nickel-nitrilotriacetic acid agarose minicolumn (0.3 ml) from Qiagen (Valencia, CA). After bound proteins were washed with 2 ml of At-buffer containing 50 mm Tris-HCl buffer (pH 8), 0.5 m NaCl, 5 mm imidazole, and 0.3% Brij 35, they were eluted with 0.4 ml of the same buffer supplemented with 300 mm imidazole. Protein size, expression level, and purity were monitored by SDS-PAGE. All proteins were obtained in sufficient yield (∼0.3–0.5 mg) and purity (80 to 90% pure). Protein concentrations were measured using the Bradford assay kit (Bio-Rad).
FolC enzymatic activity measurements using an improved FolC assay
A novel assay was developed to test the effects of four different proteins and the negative control protein, phosphoglucoisomerase (Pgi), separately and simultaneously on FolC activity. This assay depends on the detection of ADP. At pH 8.7, where FolC activity is low, the interactions with Ugd and GlnD activate FolC substantially (>10-fold) (see “Results”). FolC catalyzes the ATP-dependent addition of l-glutamate to the γ-carboxyl moiety of a glutamyl residue present in THF. FolC converts ATP to ADP while glutamylating THF, and ADP can be used by pyruvate kinase and lactate dehydrogenase to convert phosphoenolpyruvate to pyruvate and further to lactate, following the decrease in absorbance at 340 nm, resulting from the oxidation of NADH in a coupled assay. We added FolC (120–200 nm) to 100 μl of a reaction mixture containing 50 mm KCl, 50 mm Tris-HCl buffer, pH 8.7, 10 mm MgSO4, 0–150 μm aminopterin (the 4-amino derivative of folic acid) or THF, 0–25 mm glutamate, 3 mm DTT, 1.2 mm ATP, 1.2 mm phosphoenolpyruvate, 0.3 mm NADH, 1.2 units of pyruvate kinase and lactate dehydrogenase. Reaction rates were compared with controls in which glutamate or aminopterin was absent. The kinase activity detection kit was used as described (46). When tetrahydrofolate instead of aminopterin was the substrate, DMSO was added to 10%, and PBS, diluted 10-fold, was used in addition to the other conditions described above for the FolC assay.
MnmE activity measurements
For the MnmE GTPase assay, the formation of phosphate was detected using a malachite green assay as described in Ref. 47. This sensitive and reproducible assay was used for the detection of Pi in 96-well format. MnmE activity was measured as a function of GTP concentration, and FolC inhibition was quantitated. The assay mixture contained 80 mm Tris-HCl, pH 8.0, 2 mm DTT, 1 mm EDTA, 50 mm KCl, 10% glycerol, 0–75 μm aminopterin or THF, and 10 mm MgCl2, assayed at 37 °C and incubated for 1 h. The malachite green reagent was added to stop the reaction, and the absorbance at A595 nm was measured. Low FolC GTPase activity was observed relative to MnmE activity, and this was used as the background value.
FolC docking
FolC was docked to Ugd and MnmE. For FolC, PDB ID 1W78 chain A was used, whereas for Ugd a homology model using PDB ID 3PID chain A (83% sequence identity) was used, and for MnmE a homology model using PDB ID 3GEH chain A (38% sequence identity) was used. Both homology models were full length. For GlnD, a homology model could be generated for the central HD domain, but this could not be docked because of nonspecific predicted binding residues.
Binding residues were predicted for FolC, Ugd, and MnmE structures using the CPORT web server (50) with the highest sensitivity setting. Docking was performed employing the HADDOCK web server (51) using the aforementioned structures as well as CPORT predictions as input.
Selection of representative complexes from docking output was performed slightly differently for the two pairs, FolC-Ugd and FolC-MnmE. For FolC-Ugd, the top cluster was the best-scoring across nearly all criteria and was a clear choice. For FolC-MnmE, the top four clusters were all significant, but three of the four were very close spatially (low root mean square deviation) and the fourth was an outlier. The top-scoring cluster was therefore also used for FolC-MnmE. The representative structures for FolC-Ugd and FolC-MnmE were aligned in Pymol by the FolC chains. This aligned pair of complexes is referred to as the “model” for FolC binding.
Genetic crosses
Mini-array screens were performed following the E. coli Synthetic Genetic Array strategy (48) in rich medium conditions using folC and glnD as HfrC donor mutant strains. The folC hypomorphic allele was constructed with the C-terminal SPA (Sequential Peptide Affinity) tag extension engineered by homologous recombination essentially as described previously (40, 49).
Author contributions
I. A. R. and M. B. conceptualization; I. A. R., N. G., A. H., M. B., P. U., and M. H. S. data curation; I. A. R. and M. B. formal analysis; I. A. R. and N. G. validation; I. A. R. and A. H. investigation; I. A. R. and N. G. methodology; I. A. R., N. G., J. D., M. B., and M. H. S. writing-original draft; M. B., P. U., and M. H. S. supervision; M. B., P. U., and M. H. S. funding acquisition; M. B. project administration; M. H. S. writing-review and editing.
Supplementary Material
Acknowledgment
We thank Harry Zhou for help in submission of the manuscript.
This work was supported by National Institutes of Health Grant R01 GM109895 (to M. S. and P. U.) and Natural Sciences and Engineering Research Council of Canada Grant DG-20234 (to M. B.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1 and S2 and Tables S1 and S2.
- DHF
- dihydrofolate
- THF
- tetrahydrofolate.
References
- 1. Rodionova I. A., Zhang Z., Mehla J., Goodacre N., Babu M., Emili A., Uetz P., and Saier M. H. Jr. (2017) The phosphocarrier protein HPr of the bacterial phosphotransferase system globally regulates energy metabolism by directly interacting with multiple enzymes in Escherichia coli. J. Biol. Chem. 292, 14250–14257 10.1074/jbc.M117.795294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rodionova I. A., Goodacre N., Babu M., Emili A., Uetz P., and Saier M. H. Jr. (2017) The nitrogen regulatory PII protein (GlnB) and N-acetylglucosamine 6-phosphate epimerase (NanE) allosterically activate glucosamine 6-phosphate deaminase (NagB) in Escherichia coli. J. Bacteriol. 200, e00691–17 10.1128/JB.00691-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Babu M., Bundalovic-Torma C., Calmettes C., Phanse S., Zhang Q., Jiang Y., Minic Z., Kim S., Mehla J., Gagarinova A., Rodionova I., Kumar A., Guo H., Kagan O., Pogoutse O., et al. (2017) Global landscape of cell envelope protein complexes in Escherichia coli. Nat. Biotechnol. 36, 103–112 10.1038/nbt.4024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rodionov D. A., Hebbeln P., Eudes A., ter Beek J., Rodionova I. A., Erkens G. B., Slotboom D. J., Gelfand M. S., Osterman A. L., Hanson A. D., and Eitinger T. (2009) A novel class of modular transporters for vitamins in prokaryotes. J. Bacteriol. 191, 42–51 10.1128/JB.01208-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. de Crécy-Lagard V. (2014) Variations in metabolic pathways create challenges for automated metabolic reconstructions: Examples from the tetrahydrofolate synthesis pathway. Comput. Struct. Biotechnol. J. 10, 41–50 10.1016/j.csbj.2014.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Saier M. H. Jr., Reddy V. S., Tsu B. V., Ahmed M. S., Li C., and Moreno-Hagelsieb G. (2016) The Transporter Classification Database (TCDB): Recent advances. Nucleic Acids Res. 44, D372–D379 10.1093/nar/gkv1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Saier M. H. Jr., Tran C. V., and Barabote R. D. (2006) TCDB: the Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res. 34, D181–D186 10.1093/nar/gkj001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Austin M. U., Liau W. S., Balamurugan K., Ashokkumar B., Said H. M., and LaMunyon C. W. (2010) Knockout of the folate transporter folt-1 causes germline and somatic defects in C. elegans. BMC Dev. Biol. 10, 46 10.1186/1471-213X-10-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhao Q., Wang C., Wang C., Guo H., Bao Z., Zhang M., and Zhang P. (2015) Structures of FolT in substrate-bound and substrate-released conformations reveal a gating mechanism for ECF transporters. Nat. Commun. 6, 7661 10.1038/ncomms8661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Green J. M., and Matthews R. G. (2007) Folate biosynthesis, reduction, and polyglutamylation and the interconversion of folate derivatives. EcoSal Plus 2, 10.1128/ecosalplus.3.6.3.6 [DOI] [PubMed] [Google Scholar]
- 11. Shane B. (1989) Folylpolyglutamate synthesis and role in the regulation of one-carbon metabolism. Vitam. Horm. 45, 263–335 10.1016/S0083-6729(08)60397-0 [DOI] [PubMed] [Google Scholar]
- 12. Bailey L. B., and Gregory J. F. 3rd (1999) Folate metabolism and requirements. J. Nutr. 129, 779–782 10.1093/jn/129.4.779 [DOI] [PubMed] [Google Scholar]
- 13. Bailey S. W., and Ayling J. E. (2009) The extremely slow and variable activity of dihydrofolate reductase in human liver and its implications for high folic acid intake. Proc. Natl. Acad. Sci. U.S.A. 106, 15424–15429 10.1073/pnas.0902072106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang Y., Pohlmann E. L., Serate J., Conrad M. C., and Roberts G. P. (2010) Mutagenesis and functional characterization of the four domains of GlnD, a bifunctional nitrogen sensor protein. J. Bacteriol. 192, 2711–2721 10.1128/JB.01674-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gerdes S., Edwards R., Kubal M., Fonstein M., Stevens R., and Osterman A. (2006) Essential genes on metabolic maps. Curr. Opin. Biotechnol. 17, 448–456 10.1016/j.copbio.2006.08.006 [DOI] [PubMed] [Google Scholar]
- 16. Gerdes S. Y., Scholle M. D., Campbell J. W., Balázsi G., Ravasz E., Daugherty M. D., Somera A. L., Kyrpides N. C., Anderson I., Gelfand M. S., Bhattacharya A., Kapatral V., D'Souza M., Baev M. V., Grechkin Y., et al. (2003) Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J. Bacteriol. 185, 5673–5684 10.1128/JB.185.19.5673-5684.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sutherland I. W. (1969) Structural studies on colanic acid, the common exopolysaccharide found in the enterobacteriaceae, by partial acid hydrolysis. Oligosaccharides from colanic acid. Biochem. J. 115, 935–945 10.1042/bj1150935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Groisman E. A., Kayser J., and Soncini F. C. (1997) Regulation of polymyxin resistance and adaptation to low-Mg2+ environments. J. Bacteriol. 179, 7040–7045 10.1128/jb.179.22.7040-7045.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lacour S., Bechet E., Cozzone A. J., Mijakovic I., and Grangeasse C. (2008) Tyrosine phosphorylation of the UDP-glucose dehydrogenase of Escherichia coli is at the crossroads of colanic acid synthesis and polymyxin resistance. PLoS One 3, e3053 10.1371/journal.pone.0003053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rubin E. J., Herrera C. M., Crofts A. A., and Trent M. S. (2015) PmrD is required for modifications to Escherichia coli endotoxin that promote antimicrobial resistance. Antimicrob. Agents Chemother. 59, 2051–2061 10.1128/AAC.05052-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mouslim C., Latifi T., and Groisman E. A. (2003) Signal-dependent requirement for the co-activator protein RcsA in transcription of the RcsB-regulated ugd gene. J. Biol. Chem. 278, 50588–50595 10.1074/jbc.M309433200 [DOI] [PubMed] [Google Scholar]
- 22. Wösten M. M., Kox L. F., Chamnongpol S., Soncini F. C., and Groisman E. A. (2000) A signal transduction system that responds to extracellular iron. Cell 103, 113–125 10.1016/S0092-8674(00)00092-1 [DOI] [PubMed] [Google Scholar]
- 23. Soncini F. C., García Véscovi E., Solomon F., and Groisman E. A. (1996) Molecular basis of the magnesium deprivation response in Salmonella typhimurium: Identification of PhoP-regulated genes. J. Bacteriol. 178, 5092–5099 10.1128/jb.178.17.5092-5099.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen H. D., and Groisman E. A. (2013) The biology of the PmrA/PmrB two-component system: The major regulator of lipopolysaccharide modifications. Annu. Rev. Microbiol. 67, 83–112 10.1146/annurev-micro-092412-155751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Armengod M. E., Moukadiri I., Prado S., Ruiz-Partida R., Benitez-Paez A., Villarroya M., Lomas R., Garzon M. J., Martinez-Zamora A., Meseguer S., and Navarro-Gonzalez C. (2012) Enzymology of tRNA modification in the bacterial MnmEG pathway. Biochimie 94, 1510–1520 10.1016/j.biochi.2012.02.019 [DOI] [PubMed] [Google Scholar]
- 26. Fislage M., Wauters L., and Versées W. (2016) Invited review: MnmE, a GTPase that drives a complex tRNA modification reaction. Biopolymers 105, 568–579 10.1002/bip.22813 [DOI] [PubMed] [Google Scholar]
- 27. Lawson C. D., and Ridley A. J. (2018) Rho GTPase signaling complexes in cell migration and invasion. J. Cell Biol. 217, 447–457 10.1083/jcb.201612069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nilsson K., Jäger G., and Björk G. R. (2017) An unmodified wobble uridine in tRNAs specific for glutamine, lysine, and glutamic acid from Salmonella enterica serovar Typhimurium results in nonviability—due to increased missense errors? PLoS One 12, e0175092 10.1371/journal.pone.0175092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shippy D. C., and Fadl A. A. (2014) tRNA modification enzymes GidA and MnmE: Potential role in virulence of bacterial pathogens. Int. J. Mol. Sci. 15, 18267–18280 10.3390/ijms151018267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sayed A. K., and Foster J. W. (2009) A 750 bp sensory integration region directs global control of the Escherichia coli GadE acid resistance regulator. Mol. Microbiol. 71, 1435–1450 10.1111/j.1365-2958.2009.06614.x [DOI] [PubMed] [Google Scholar]
- 31. Deng Z., Shan Y., Pan Q., Gao X., and Yan A. (2013) Anaerobic expression of the gadE-mdtEF multidrug efflux operon is primarily regulated by the two-component system ArcBA through antagonizing the H-NS mediated repression. Front. Microbiol. 4, 194 10.3389/fmicb.2013.00194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Moon K. H., Weber B. S., and Feldman M. F. (2017) Subinhibitory concentrations of trimethoprim and sulfamethoxazole prevent biofilm formation by Acinetobacter baumannii through inhibition of Csu pilus expression. Antimicrob. Agents Chemother. 61, e00778–17 10.1128/AAC.00778-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Patel T. S., Vanparia S. F., Patel U. H., Dixit R. B., Chudasama C. J., Patel B. D., and Dixit B. C. (2017) Novel 2,3-disubstituted quinazoline-4(3H)-one molecules derived from amino acid linked sulphonamide as a potent malarial antifolates for DHFR inhibition. Eur. J. Med. Chem. 129, 251–265 10.1016/j.ejmech.2017.02.012 [DOI] [PubMed] [Google Scholar]
- 34. Podnecky N. L., Rhodes K. A., Mima T., Drew H. R., Chirakul S., Wuthiekanun V., Schupp J. M., Sarovich D. S., Currie B. J., Keim P., and Schweizer H. P. (2017) Mechanisms of resistance to folate pathway inhibitors in Burkholderia pseudomallei: Deviation from the norm. MBio 8, e0137–17 10.1128/mBio.01357-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Thakkar S. S., Thakor P., Ray A., Doshi H., and Thakkar V. R. (2017) Benzothiazole analogues: Synthesis, characterization, MO calculations with PM6 and DFT, in silico studies and in vitro antimalarial as DHFR inhibitors and antimicrobial activities. Bioorg. Med. Chem. 25, 5396–5406 10.1016/j.bmc.2017.07.057 [DOI] [PubMed] [Google Scholar]
- 36. Sun X., Cross J. A., Bognar A. L., Baker E. N., and Smith C. A. (2001) Folate-binding triggers the activation of folylpolyglutamate synthetase. J. Mol. Biol. 310, 1067–1078 10.1006/jmbi.2001.4815 [DOI] [PubMed] [Google Scholar]
- 37. Mathieu M., Debousker G., Vincent S., Viviani F., Bamas-Jacques N., and Mikol V. (2005) Escherichia coli FolC structure reveals an unexpected dihydrofolate binding site providing an attractive target for anti-microbial therapy. J. Biol. Chem. 280, 18916–18922 10.1074/jbc.M413799200 [DOI] [PubMed] [Google Scholar]
- 38. Zhao F., Wang X. D., Erber L. N., Luo M., Guo A. Z., Yang S. S., Gu J., Turman B. J., Gao Y. R., Li D. F., Cui Z. Q., Zhang Z. P., Bi L. J., Baughn A. D., Zhang X. E., and Deng J. Y. (2014) Binding pocket alterations in dihydrofolate synthase confer resistance to para-aminosalicylic acid in clinical isolates of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 58, 1479–1487 10.1128/AAC.01775-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sheng Y., Khanam N., Tsaksis Y., Shi X. M., Lu Q. S., and Bognar A. L. (2008) Mutagenesis of folylpolyglutamate synthetase indicates that dihydropteroate and tetrahydrofolate bind to the same site. Biochemistry 47, 2388–2396 10.1021/bi701670y [DOI] [PubMed] [Google Scholar]
- 40. Babu M., Díaz-Mejía J. J., Vlasblom J., Gagarinova A., Phanse S., Graham C., Yousif F., Ding H., Xiong X., Nazarians-Armavil A., Alamgir M., Ali M., Pogoutse O., Pe'er A., Arnold R., et al. (2011) Genetic interaction maps in Escherichia coli reveal functional crosstalk among cell envelope biogenesis pathways. PLoS Genet. 7, e1002377 10.1371/journal.pgen.1002377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Forchhammer K., and Lüddecke J. (2016) Sensory properties of the PII signalling protein family. FEBS J. 283, 425–437 10.1111/febs.13584 [DOI] [PubMed] [Google Scholar]
- 42. Francis S. H., and Engleman E. G. (1978) Cascade control of E. coli glutamine synthetase. I. Studies on the uridylyl transferase and uridylyl removing enzyme(s) from E. coli. Arch. Biochem. Biophys. 191, 590–601 10.1016/0003-9861(78)90397-1 [DOI] [PubMed] [Google Scholar]
- 43. Son H. S., and Rhee S. G. (1987) Cascade control of Escherichia coli glutamine synthetase. Purification and properties of PII protein and nucleotide sequence of its structural gene. J. Biol. Chem. 262, 8690–8695 [PubMed] [Google Scholar]
- 44. Prado S., Villarroya M., Medina M., and Armengod M. E. (2013) The tRNA-modifying function of MnmE is controlled by post-hydrolysis steps of its GTPase cycle. Nucleic Acids Res. 41, 6190–6208 10.1093/nar/gkt320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kitagawa M., Ara T., Arifuzzaman M., Ioka-Nakamichi T., Inamoto E., Toyonaga H., and Mori H. (2005) Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): Unique resources for biological research. DNA Res. 12, 291–299 10.1093/dnares/dsi012 [DOI] [PubMed] [Google Scholar]
- 46. Rodionova I. A., Yang C., Li X., Kurnasov O. V., Best A. A., Osterman A. L., and Rodionov D. A. (2012) Diversity and versatility of the Thermotoga maritima sugar kinome. J. Bacteriol. 194, 5552–5563 10.1128/JB.01136-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rodionova I. A., Li X., Plymale A. E., Motamedchaboki K., Konopka A. E., Romine M. F., Fredrickson J. K., Osterman A. L., and Rodionov D. A. (2015) Genomic distribution of B-vitamin auxotrophy and uptake transporters in environmental bacteria from the Chloroflexi phylum. Environ. Microbiol. Rep. 7, 204–210 10.1111/1758-2229.12227 [DOI] [PubMed] [Google Scholar]
- 48. Butland G., Babu M., Díaz-Mejia J. J., Bohdana F., Phanse S., Gold B., Yang W., Li J., Gagarinova A. G., Pogoutse O., Mori H., Wanner B. L., Lo H., Wasniewski J., Christopolous C., et al. (2008) eSGA: E. coli synthetic genetic array analysis. Nat. Methods 5, 789–795 10.1038/nmeth.1239 [DOI] [PubMed] [Google Scholar]
- 49. Babu M., Gagarinova A., and Emili A. (2011) Array-based synthetic genetic screens to map bacterial pathways and functional networks in Escherichia coli. Methods Mol. Biol. 781, 99–126 10.1007/978-1-61779-276-2_7 [DOI] [PubMed] [Google Scholar]
- 50. de Vries S. J., and Bonvin A. M. (2011) CPORT: A consensus interface predictor and its performance in prediction-driven docking with HADDOCK. PLoS One 6, e17695 10.1371/journal.pone.0017695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. van Zundert G. C. P, Rodrigues J. P. G. L. M., Trellet M., Schmitz C., Kastritis P. L., Karaca E., Melquiond A. S. J., van Dijk M., de Vries S. J., and Bonvin A. M. J. J. (2016). The HADDOCK2.2 webserver: User-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 428, 720–725 10.1016/j.jmb.2015.09.014 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.