Abstract
Pseudomonas aeruginosa is a Gram-negative bacterium responsible for a large number of nosocomial infections. The P. aeruginosa respiratory chain contains the ion-pumping NADH:ubiquinone oxidoreductase (NQR). This enzyme couples the transfer of electrons from NADH to ubiquinone to the pumping of sodium ions across the cell membrane, generating a gradient that drives essential cellular processes in many bacteria. In this study, we characterized P. aeruginosa NQR (Pa-NQR) to elucidate its physiologic function. Our analyses reveal that Pa-NQR, in contrast with NQR homologues from other bacterial species, is not a sodium pump, but rather a completely new form of proton pump. Homology modeling and molecular dynamics simulations suggest that cation selectivity could be determined by the exit ion channels. We also show that Pa-NQR is resistant to the inhibitor 2-n-heptyl-4-hydroxyquinoline N-oxide (HQNO). HQNO is a quinolone secreted by P. aeruginosa during infection that acts as a quorum sensing agent and also has bactericidal properties against other bacteria. Using comparative analysis and computational modeling of the ubiquinone-binding site, we identified the specific residues that confer resistance toward this inhibitor. In summary, our findings indicate that Pa-NQR is a proton pump rather than a sodium pump and is highly resistant against the P. aeruginosa–produced compound HQNO, suggesting an important role in the adaptation against autotoxicity. These results provide a deep understanding of the metabolic role of NQR in P. aeruginosa and provide insight into the structural factors that determine the functional specialization in this family of respiratory complexes.
Keywords: Pseudomonas aeruginosa (P. aeruginosa), ubiquinone, proton pump, respiratory chain, bacterial metabolism, inhibition mechanism, quorum sensing, antibacterial compound, autotoxicity, HQNO, Na+-NQR, quinolone, respiratory enzyme
Introduction
Pseudomonas aeruginosa is a facultative anaerobic Gram-negative γ-proteobacterium that colonizes a diverse range of habitats, including surgical equipment and catheters (1–3). Once established, this pathogen can rapidly manipulate its environment, both physically and chemically, through biofilm and bactericidal agent production (4–7). P. aeruginosa is the primary cause of infection in cystic fibrosis patients (8–11) and one of the main causes, along with Escherichia coli and Enterococci, of nosocomial catheter-associated urinary tract infections (UTIs)3 (2, 4, 12, 13). Due to high infection recurrence, which primarily results from bacterial resistance against multiple antibiotics (5, 14–16), the treatment of UTIs in the United States costs more than $450 million per year (http://www.cdc.gov/hai/pdfs/hai/scott_costpaper.pdf).
P. aeruginosa contains a highly branched respiratory chain that is regulated by the availability of substrates and oxygen, which supports energy production and contributes to pathogenesis (9, 18). The respiratory chain of P. aeruginosa is composed of a number of ubiquinol oxidases, such as the bc1, bo3, and bd complexes (8, 19, 20). Cytochromes bo3 and bd transfer electrons directly to oxygen, whereas the bc1 complex transfers electrons to cytochrome c (9). From cytochrome c, electrons can travel to any of the three different cytochrome c oxidases, which play major roles in lung and micro-aerobic environment colonization (18). Moreover, this microorganism has 17 respiratory dehydrogenases, including three different types of NADH dehydrogenases: complex I, NDH-2, and NQR (8, 9). Although these three enzymes transfer electrons from NADH to ubiquinone, their composition and mechanisms are completely different (21–23). Complex I is a proton-pumping, multisubunit enzyme complex, found in the respiratory chains of both mitochondria and bacteria (22, 24). NDH-2 is a single-subunit flavoenzyme that does not participate in ion pumping (25). In all previously reported cases, NQR is a sodium-pumping complex found only in prokaryotes, which plays a major role in the metabolism and pathogenicity of numerous bacterial species (26–29). The sodium gradient generated by NQR supplies energy for essential processes, such as ATP synthesis, flagellar rotation, drug extrusion, and nutrient transport (29–33). Indeed, for the obligate intracellular parasite Chlamydia trachomatis, NQR plays a vital role during infection and intracellular multiplication (34). A recent report by our group shows that the energetic metabolism of C. trachomatis is sustained by the sodium gradient generated by NQR (34). Because of its fundamental importance to pathogenic bacteria and its absence in mammalian cells, this complex is a promising target for drug design (34, 35). However, the specific role of NQR in the physiology of P. aeruginosa, its catalytic properties, and the biochemical adaptations to the diverse microorganism's habitats are unknown.
The well-characterized V. cholerae NQR (Vc-NQR) complex is composed of six subunits (subunits A–F) and couples the transfer of electrons from NADH to ubiquinone to the pumping of sodium across the cell membrane (27, 28, 36). Five confirmed redox cofactors are involved in electron transfer through the enzyme: FAD, a 2Fe-2S center, two covalently bound FMN molecules, and riboflavin (27, 28, 36). NQR is the only known enzyme to utilize riboflavin as a redox cofactor (37–40). Crystallographic data suggest the participation of an additional iron cofactor, located in subunits D and E (41). However, no direct evidence is currently available to support the role of this cofactor.
To understand the role of NQR in P. aeruginosa physiology, structural and kinetic analyses were conducted. In this work, we found that the Pa-NQR complex is monomeric and is composed of six subunits, similar to previously studied homologues. Additionally, our data show the expected cofactor composition, with around four flavin cofactors, and comparable functional properties. However, significant differences were found, such as the absence of the neutral radical in riboflavin, which is a hallmark of the family (37–40). Our results also show that the cation specificity of Pa-NQR differs significantly compared with other homologues. Whereas the characterized NQR complexes (mostly from Vibrio species) are sodium-specific transporters that are regulated by potassium (42–44), Pa-NQR is a proton-specific pump that is regulated by several monovalent cations. Structural analysis indicates that the ion channels in subunit B of Pa-NQR show significant differences in their location, size, and depth compared with those of the sodium-pumping complexes, providing information to understand the structural basis that determines cation selectivity in this family.
Moreover, experiments were performed to characterize the effects of HQNO on Pa-NQR activity. HQNO is a quinolone compound actively produced by P. aeruginosa that forms part of quorum sensing pathways (7, 14, 45), promotes biofilm formation (1, 7), and exhibits strong antibiotic effects by inhibiting the respiratory chain of competing bacteria (7, 10, 46, 47). Our results show that Pa-NQR is 5–10 times more resistant to HQNO compared with other NQR homologues and that the HQNO inhibition is partial, allowing P. aeruginosa to survive in the presence of this toxin. Molecular modeling was carried out to understand the structural basis for the differences in the behavior of this enzyme. Our models show that the ubiquinone-binding site has specific sequence differences that confer resistance toward HQNO, while maintaining ubiquinone binding activity. These predictions were corroborated by mutating residues 151 and 155 of Vc-NQR subunit B, which turn it into an HQNO-resistant enzyme. The data presented in this paper provide a better understanding of the NQR family, its role in P. aeruginosa, and the biochemical adaptations that allow the colonization and survival of this microorganism.
Results
Protein complex characterization
To understand the kinetic and structural properties of Pa-NQR, the recombinant protein complex was purified and compared with Vc-NQR, which is the best-characterized member of the family (29–33, 48, 49). The Pa-NQR operon was cloned into the pBAD HisB plasmid, and the protein complex was expressed (using arabinose as a gene expression inducer) in Δnqr attenuated Vibrio cholerae O395N1 cells, previously obtained by our group (26). Induced cells were harvested at early stationary phase and disrupted by sonication. Cytoplasmic membranes were obtained by ultracentrifugation, and the Pa-NQR complex was purified using two chromatographic steps: Ni-NTA and DEAE FPLC chromatography, as reported before (26, 38). Following chromatographic purification, a purity of ∼80% was obtained for Pa-NQR, comparable with 95% for Vc-NQR (26, 42). Purity was assessed densitometrically using ImageJ (50), following urea SDS-PAGE (Fig. 1A). The six subunits of Pa-NQR were identified based on estimated mass derived from their corresponding amino acid sequences (subunit A, 48 kDa; B, 44 kDa; C, 28 kDa; D, 24 kDa; E, 28 kDa; and F, 46 kDa). Additionally, the covalently attached FMN cofactors of subunits B and C were visualized by exposing the SDS-polyacrylamide gel to UV light (Fig. 1A), which makes these bands glow (43, 51).
Figure 1.
Urea SDS-acrylamide gel of Vc-NQR and Pa-NQR and blue native/second-dimension urea SDS-PAGE of Pa-NQR. A, subunit and flavin cofactor identification following Ni-NTA and DEAE FPLC chromatography purification. Left, Coomassie Blue–stained gel lanes of Vc-NQR and Pa-NQR. Right, unstained gel lanes carrying Vc-NQR (Vc) and Pa-NQR (Pa) exposed to UV light, using a transilluminator. B, Coomassie Blue–stained blue native gel of Pa-NQR (i); in-gel NADH dehydrogenase activity assay (ii); Coomassie Blue–stained second-dimension SDS-PAGE (iii); second-dimension SDS-PAGE exposed to UV light (iv).
Blue native gel electrophoresis (BN-PAGE) (52, 53) was conducted on the purified Pa-NQR sample, followed by an in-gel NADH dehydrogenase activity assay (53), which allowed us to characterize the molecular weight of the complex. Fig. 1B (i) shows the blue native electrophoresis of Pa-NQR with a main band of ∼220 kDa, corresponding to the expected molecular mass of the monomeric complex. This band exhibits NADH dehydrogenase activity, as shown by in-gel activity assays (Fig. 1B, ii). The BN-PAGE was incubated with NADH and nitro blue tetrazolium (NBT), an electrophilic dicationic compound, which readily accepts electrons and forms diformazan precipitates (52). NBT reduction solely occurred at the band corresponding to Pa-NQR. To confirm the presence of all six subunits in the functional Pa-NQR complex, a second-dimension 30% urea, 15% acrylamide SDS-PAGE electrophoresis was run using a BN-PAGE lane, and the six bands with the corresponding molecular weights of subunits A–F were observed (Fig. 1B, iii). Additionally, the fluorescent flavin molecules of subunits B and C were visualized under UV light, as mentioned previously (Fig. 1B, iv).
Spectra analysis and cofactor composition
To determine the cofactor composition of Pa-NQR, spectrophotometric analyses were conducted under denaturing conditions and with the native complex under reducing and oxidizing conditions (Fig. 2). Concentrated Pa-NQR (>100 μm) samples were dissolved (1:20 dilution) in 7 m guanidine chloride to completely denature the sample and oxidize the riboflavin cofactor, which in Vc-NQR is found as a neutral radical (37–39). This method allows the quantification of the total flavin content in the sample. According to our data, the flavin/protein ratio of Pa-NQR (∼3.3) is close to the expected value of 4 (i.e. FAD, riboflavin, and two covalently bound FMNs) (27, 28, 36, 39), especially considering that our sample has 80% purity.
Figure 2.
Reduced-minus-oxidized spectra of Vc-NQR and Pa-NQR. A, Vc-NQR (continuous) and Pa-NQR (dashed) reduced-minus-oxidized spectra. B, double difference spectra of Vc-NQR minus Pa-NQR. C, spectral component corresponding to the one-electron reduction of the riboflavin neutral radical (RibH• → RibH2). The spectrum was obtained from previously published data (37, 54).
Spectrophotometric analyses were also carried on the native sample under reducing and oxidizing conditions. Full reduction of the sample was achieved by adding a few grains of dithionite to the solution and was compared with the air-oxidized sample. The reduced-minus-oxidized spectrum of the sample would reveal the presence and stoichiometry of the different cofactors, because they possess characteristic absorbance spectra, which can be used to quantify their concentrations (54–57). The reduced-minus-oxidized spectra of Pa-NQR and Vc-NQR (Fig. 2A) were similar, with about the same minima and maxima. However, some differences were evident, especially in the region of 400–430 and 525–700 nm. The double difference spectrum (Vc-NQR minus Pa-NQR) shows a missing spectral component in Pa-NQR that is very similar to the one-electron reduction of the riboflavin flavin radical (RibH• → RibH2) (Fig. 2, B and C) (27, 28, 36, 37, 39). This result indicates that whereas the cofactor composition and redox transitions of both complexes are similar, the riboflavin cofactor of Pa-NQR seems to be mostly found in the fully oxidized state, which is very different compared with Vc-NQR, in which it is found as a neutral radical (37, 54).
Pa-NQR cation specificity
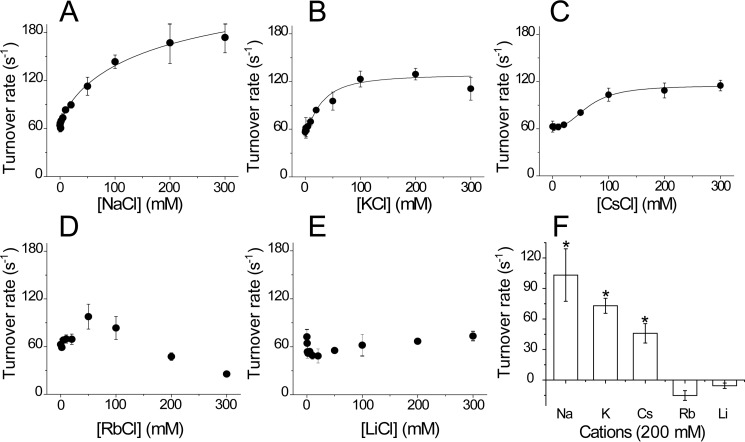
Previous reports have shown that the activity of Vc-NQR and other homologues depends on the presence of sodium and other monovalent cations (42). In Vc-NQR, it has been shown that the rate-limiting step of the reaction, the one-electron reduction of FMN in subunit C, is specifically stimulated by sodium (54, 58). To investigate cation specificity for Pa-NQR, the NADH-dependent ubiquinone reductase activity was measured in the presence of different concentrations of monovalent cations, such as sodium, lithium, potassium, rubidium, and cesium. Sodium increased the activity by 3 times (compared with when no cation is present) with an activation constant (Ka) of 90 mm (Fig. 3A and Table 1). Potassium and cesium produced a similar stimulation, doubling the activity, with Ka values of 30 and 65 mm, respectively (Fig. 3 (B and C) and Table 1). The enzyme was also slightly stimulated by rubidium (60%), but this cation also showed inhibitory effects at concentrations above 50 mm (Fig. 2D). Lithium had an inhibitory effect at low concentrations and produced minimal effects at higher concentrations. These effects differ drastically from the results reported previously with Vc-NQR, which shows 8- and 3-fold stimulation of the activity with sodium and lithium, respectively, activation with potassium, and inhibition by rubidium (42). The results indicate that Pa-NQR has a very different cation selectivity compared with other studied homologues. It should be pointed out that the Km values for NADH of Pa-NQR (KmNADH =9 ± 1 μm) and Vc-NQR (KmNADH = 25 μm) were similar (59). The Km values for ubiquinone-1 were also similar between the two complexes (KmUQ = 5 ± 1.3 μm for Pa-NQR and 3 μm for Vc-NQR) (59). Thus, the changes that we are reporting are not due to general enzyme misfolding or lack of functionality of the complex.
Figure 3.
Pa-NQR ubiquinone reductase activity in the presence of different monovalent cations. Activity was measured in the presence of 250 μm NADH and 50 μm ubiquinone-1, at different concentrations of NaCl (A), KCl (B), CsCl (C), RbCl (D), and LiCl (E). F, comparison of the activity obtained at a 200 mm concentration of each cation. *, p < 0.05. Error bars, S.D.
Table 1.
Effect of monovalent cations on Pa-NQR activity
Activity measurements were conducted in the presence of 250 μm NADH and 50 μm ubiquinone-1, with different concentrations of monovalent cations (NaCl, KCl, and CsCl). The saturation kinetics were fitted to the Michaelis–Menten equation to calculate the kcat and Ka values.
| Cation | kcat | Ka |
|---|---|---|
| s−1 | mm | |
| NaCl | 210 ± 21 | 92 ± 25 |
| KCl | 129 ± 7 | 30 ± 6 |
| CsCl | 115 ± 2 | 65 ± 3 |
Pa-NQR ion pumping
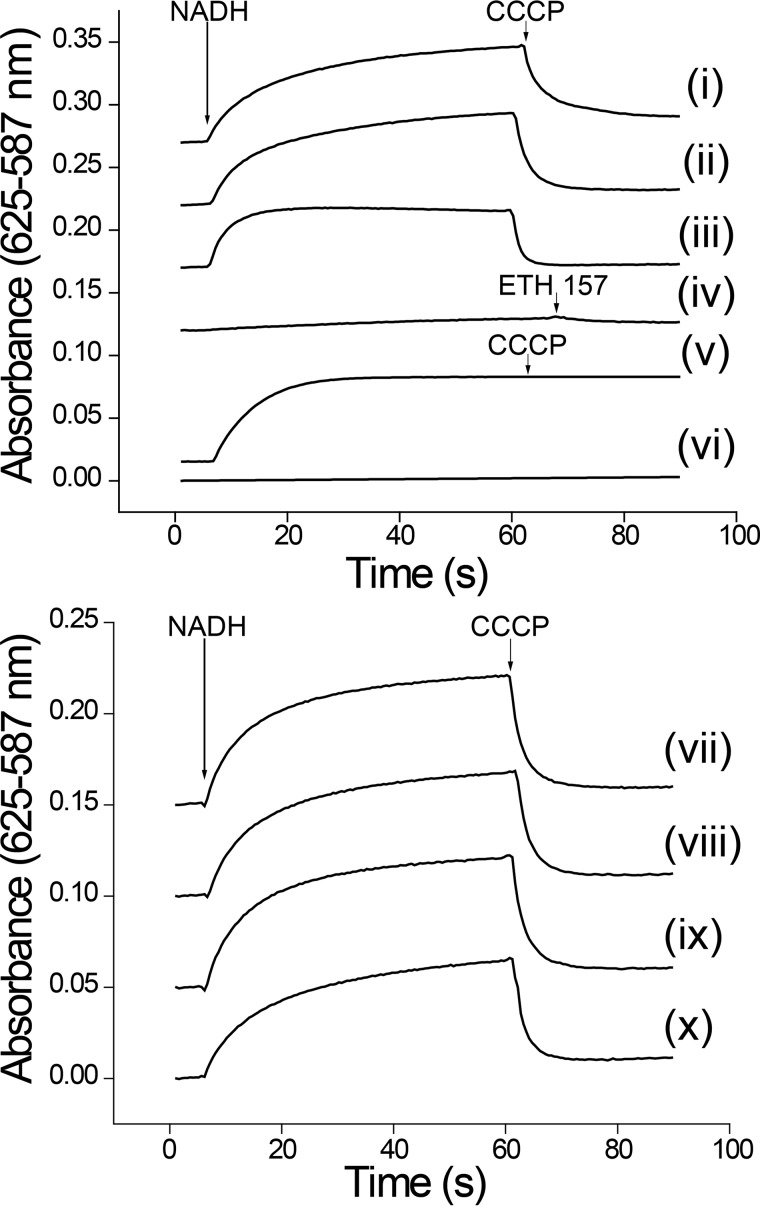
Due to the significant differences in the cation specificity of Pa-NQR, we decided to study the ion-pumping activity of the reconstituted enzyme into proteoliposomes. The purified Pa-NQR complex was incorporated into E. coli phospholipid proteoliposomes following a method reported by Juarez et al. (38, 42), based on the protocol from Rigaud et al. (60). Cation transport was measured indirectly, through the generation of membrane potential in reconstituted proteoliposomes, using the anionic dye Oxonol VI (61). The results show that membrane potential is formed in the presence of all of the tested cations: sodium, potassium, rubidium, cesium, and lithium (Fig. 4, traces ii, vii, viii, ix, and x), which could indicate that Pa-NQR is a nonspecific pump that is able to transport any monovalent cation. However, the data also indicate that the membrane potential formed in all of these cases is almost completely abolished by the proton ionophore CCCP (62) and that it is also formed in the absence of any added cation (Fig. 4, trace i) (the pH of the reconstitution and assay buffers were adjusted with Tris base; see “Experimental Procedures”). The data demonstrate that the membrane potential is formed through proton pumping. Indeed, the use of CCCP from the start of the reaction eliminated any significant generation of membrane potential (Fig. 4, trace iv). To further corroborate that sodium is not transported by the reconstituted Pa-NQR complex, experiments were performed in the presence of the sodium ionophore ETH 157 (38, 63). Fig. 4 (trace iii) shows that the sodium ionophore has no effect on membrane potential formation. To corroborate that the proton pumping activity of Pa-NQR is not due to any factors that could affect the reconstitution process, the experiments were also carried out in Vc-NQR proteoliposomes. The results obtained corroborate that Vc-NQR is a sodium-specific ion pump and that the gradient produced with this ion is not dissipated by CCCP (trace v) and cannot be established in the absence of sodium (trace vi). Thus, unlike all other studied NQR homologues, Pa-NQR does not function as a sodium transporter, but rather as a proton-specific pump. The ability of P. aeruginosa NQR to transport protons is a novel characteristic previously unknown in the NQR family.
Figure 4.
Cation transport in reconstituted artificial proteoliposome membranes containing Pa-NQR. Ion transport was measured indirectly using Oxonol VI (5 μm). The buffer contained 50 μm ubiquinone-1, and the reaction was started with the addition of NADH (250 μm). The proton ionophore CCCP (2 μm) or the sodium ionophore ETH 157 (5 μm) was added after 60 s. Top, Pa-NQR membrane potential generation in the absence of cations (i), in the presence of sodium (ii), sodium and ETH 157 (iii), and sodium with CCCP (iv). Shown is Vc-NQR membrane potential generation in the presence (v) or absence (vi) of sodium. Bottom, membrane potential generation by Pa-NQR in the presence of KCl (vii), CsCl (viii), RbCl (ix), and LiCl (x).
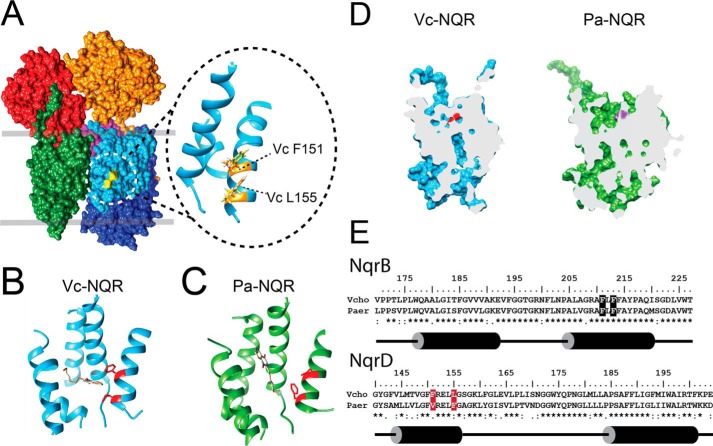
Sequence comparison between Vc-NQR and Pa-NQR shows that the negatively charged residues associated with sodium uptake and transport (i.e. B-Asp-397, B-Asp-346, and E-Glu-95) (38, 58, 64, 65) are conserved in Pa-NQR (Fig. S1). To understand the structural factors that determine the cation selectivity of the family, we performed homology modeling of subunit B of Pa-NQR, using the crystallographic data previously published for Vc-NQR (41). Moreover, molecular dynamics were performed for 50 ns to relax the model. Analysis of the van der Waals surface unveils significant differences in the structure of subunit B between Vc-NQR and Pa-NQR (Fig. 6D). Subunit B plays a critical role in electron transfer and in sodium transport. In particular, it contains one of the two sodium-binding sites (66) and carries the binding site for one FMN cofactor (51, 67), riboflavin (37, 39, 54, 68), and ubiquinone (59). In both cases, an ion entry channel is formed that would deliver the ion to Asp-346 (V. cholerae numbering). This residue had been identified as a key element that controls the release of sodium to the external side of the membrane (38). Whereas some differences are apparent regarding the size and depth of the entry channel, the exit channel of Vc-NQR and Pa-NQR are completely different. Vc-NQR has a channel running almost perfectly vertically, with Asp-346 being at the very top. On the other hand, Pa-NQR carries an L-shaped channel that is completely different compared with the one found in Vc-NQR. We are currently investigating the role of these two channels in the ion specificity of the NQR family.
Figure 6.
Structural analysis of the ubiquinone-binding site and ion channels of Vc-NQR crystal and Pa-NQR homology model. A, location of the ubiquinone-binding site in Vc-NQR. Inset, enlarged view of the residues (Phe-151 and Leu-155) in subunit D of Vc-NQR that contribute to HQNO resistance in Pa-NQR. Docking analysis of HQNO to Vc-NQR (B) and Pa-NQR (C) is shown. The figure shows the HQNO mode with the lowest energy. Residues involved in HQNO resistance are highlighted in red. D, cross-section of Vc-NQR and Pa-NQR subunit B showing the locations of the entry and exit channels for the ion. Aspartate 346 (V. cholerae numbering) is shown for Vc-NQR (red) and Pa-NQR (purple). E, sequence alignment of NqrB and NqrD of V. cholerae and P. aeruginosa. Residues involved in ubiquinone binding are highlighted in black. Residues involved in HQNO resistance are highlighted in red.
Effect of HQNO on Pa-NQR activity
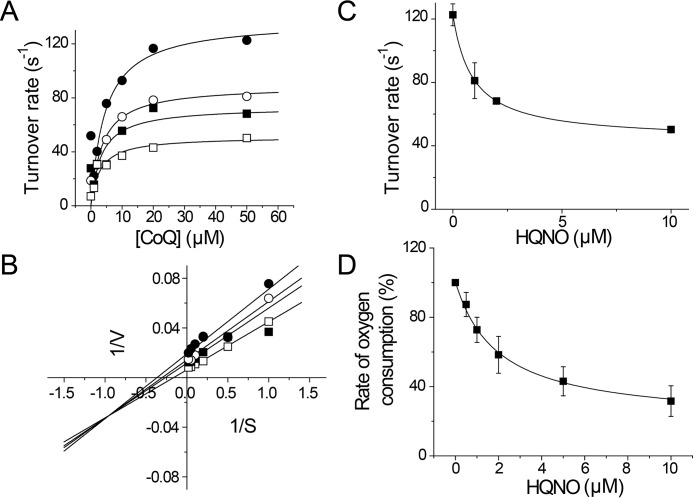
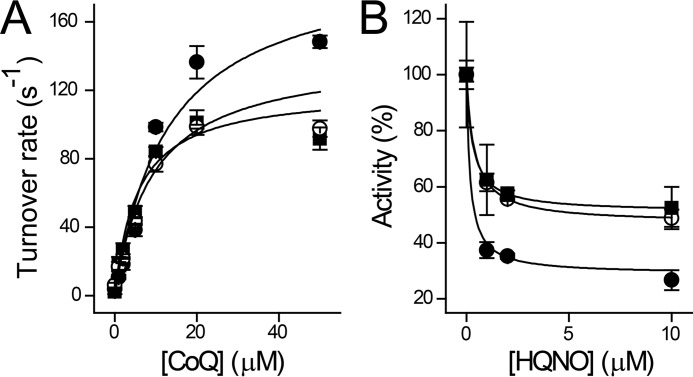
P. aeruginosa actively produces several types of quinolone compounds, including 3,4-dihydroxy-2-heptylquinoline and HQNO (7, 46, 69, 70). These quinolones fulfill different roles, such as quorum sensing, promoting biofilm formation, and acting as bactericidal agents (7, 11, 70, 71). Previous reports have shown that HQNO inhibits the activity of NQR, acting as a mixed type inhibitor versus ubiquinone (26, 72, 73). The production of HQNO by P. aeruginosa seems paradoxical, because it would trigger the autoinhibition of the respiratory chain, which would be deleterious for this microorganism, compromising its survival. To understand the role of NQR and HQNO in the physiology of P. aeruginosa, the effect of this inhibitor was tested on both purified Pa-NQR and in P. aeruginosa membranes (Fig. 5). The mechanism of inhibition was studied by measuring the effects of HQNO on the saturation kinetics of ubiquinone-1 (Fig. 5A). The data were best fitted to Equation 1, which corresponds to the behavior of a partial mixed inhibitor. Indeed, the double reciprocal plot shows lines intersecting in the third quadrant, characteristic of this type of inhibition (Fig. 5B). From the data fitting, we obtained two inhibition constants, for the competitive and uncompetitive components of the inhibition (Kic and Kiu), 1.6 and 1.3 μm, respectively. These Ki values are significantly higher (5–16 times) compared with the Ki values for HQNO of other members of the family, 0.1–0.3 μm (26, 73, 74). Moreover, the HQNO-resistant activity (kcatR), corresponds to around 40% of the activity. These two properties are key factors that would allow P. aeruginosa to survive its own production of HQNO. This analysis was also conducted over the NADH-dependent respiratory activity of P. aeruginosa membranes. Fig. 5D shows that the respiratory activity is inhibited by relatively high concentrations of HQNO, with a Ki of 2.0 μm, which roughly corresponds to the Ki obtained in the isolated enzyme (Fig. 5C). Because the Ki values of HQNO for other respiratory enzymes, such as succinate dehydrogenase, complex I, NDH-2, and quinol oxidases, are higher (47), our data suggest that most of the NADH-dependent respiratory activity could be due to NQR activity.
Figure 5.
HQNO inhibition mechanism of Pa-NQR and P. aeruginosa membranes. A, Pa-NQR activity was tested at different concentrations of ubiquinone-1 (CoQ) in the presence of increasing concentrations of HQNO (0 μm (●), 1 μm (○), 2 μm (■), and 10 μm (□)). B, Lineweaver–Burk plot of the ubiquinone versus HQNO titration data. The data shown in A and B were fitted to the equation corresponding to a partial mixed inhibitor (Equation 1). Shown is HQNO titration of Pa-NQR activity obtained at a fixed ubiquinone concentration (50 μm) (C) and of NADH (200 μm)-dependent oxygen consumption of P. aeruginosa membranes (D). Error bars, S.D.
Modeling of Pa-NQR ubiquinone-binding site
To understand the structural factors involved in HQNO resistance, we performed an analysis of the sequence and the structure of the proposed ubiquinone-binding sites. Fig. 6E shows sequence alignment of the helices of Vc-NQR and Pa-NQR NqrB (residues 170–225) and NqrD (residues 140–200) that form the ubiquinone-binding pocket (Fig. 6A). The critical residues in subunit B for ubiquinone binding are conserved in both complexes, which is consistent with the ability of Pa-NQR to bind and reduce ubiquinone, especially considering that the Km values are similar. However, the analysis shows that Phe-151 and Leu-155 in subunit D of Vc-NQR are switched to leucine and phenylalanine, respectively, in Pa-NQR (Fig. 6E). The two amino acid sites are located in the same α helix of subunit D, on the adjacent side that faces the binding site for ubiquinone (Fig. 6A), which may modify the affinity or stability of HQNO in the binding site of Pa-NQR. Molecular dynamics simulations showed a loop located at the bottom of the associated helix in Pa-NQR that may favor rotation of the helix carrying these residues. These residue and chain flexibility differences between Vc-NQR and Pa-NQR potentially account for decreased binding of HQNO. Docking analysis was performed to understand the role of these changes in the resistance toward HQNO. Fig. 6 (B and C) shows the docking of the inhibitor to the crystal structure of Vc-NQR and the model of Pa-NQR. The calculations predict that in Vc-NQR, HQNO interacts with the ubiquinone-binding site proposed by our group and establishes contacts with Phe-151 (with a docking score of −35.7 kcal/mol). In the case of Pa-NQR, the HQNO's aromatic rings are oriented in a completely different way (with a comparable docking score of −37.5 kcal/mol). The HQNO pose predicted for the Vc-NQR structure interacts relatively weakly with the Pa-NQR model (docking score of −23.2 kcal/mol). On the other hand, the HQNO pose predicted for the Pa-NQR model is unlikely to bind to the Vc-NQR structure (docking score of −2.9 kcal/mol). The ability of HQNO to be bound by Pa-NQR in a crevice near the ubiquinone site would explain the partial-type inhibition, in which both HQNO and ubiquinone would be able to be bound by the enzyme, without producing complete inhibition. Fig. 6 (B and C) shows the modes with the lowest energies, and other modes (Fig. S2) showed the same behavior. The docking calculations suggest that relative to Vc-NQR, Pa-NQR has an alternative binding site for HQNO.
Designing of HQNO-resistant mutants
To test whether the amino acid residues identified through molecular docking participate in HQNO resistance, mutagenesis analysis was carried out in Vc-NQR. Three mutations were performed making the substitutions found in NqrD of Pa-NQR: F151I, L155F, and the double mutant F151I/L155F. These proteins were characterized kinetically to understand the effect of the mutation. Activities were measured at different concentrations of ubiquinone-1 to calculate the Km for this substrate (KmUQ). Fig. 7A shows that the mutant F151I is the most active, but this mutant shows a significantly higher KmUQ than WT Vc-NQR (3.5 μm (59)) (Table 2). The mutant L155F shows small decreases in the kcat and KmUQ. Remarkably, the double mutant has a behavior that is almost identical to Pa-NQR (Table 2). A characterization of the inhibitor sensitivity of the mutants was carried out, performing a titration with HQNO at a fixed concentration of ubiquinone (50 μm) (Fig. 7B). The mutant F151I has the same Kiapp as WT Vc-NQR. The mutation in residue 155 increased Kiapp by a factor of 2 in both the single and double mutant, but did not reach the values found in Pa-NQR. Interestingly, the HQNO-resistant component (kcatR) is increased by the mutation in residue 155, and in the double mutant, it resembled the behavior of Pa-NQR. Taken together, the data indicate that residue 155 confers the resistance to HQNO. Our results show the structural basis that allows Pa-NQR to resist HQNO inhibition and further confirm our previous data regarding the location of the ubiquinone-binding site in subunit B.
Figure 7.
Kinetic characterization of Vc-NQR mutants F151I (●), L155F (○), and F151I/L155F double mutant (■). A and B, activity titrations varying the concentration of ubiquinone-1 (CoQ) (A) or HQNO (B) at a fixed concentration of ubiquinone (50 μm). Error bars, S.D.
Table 2.
Kinetic properties of Pa-NQR and Vc-NQR mutants
Activity measurements were conducted in the presence of 250 μm NADH, with different concentrations of ubiquinone-1 or HQNO. The saturation kinetic curves were fitted to Equation 1 to calculate kcat, kcatR, KmUQ, or Ki.
| Mutant | kcat | KmUQ | Kiapp | kcatR |
|---|---|---|---|---|
| s−1 | mm | mm | s−1 | |
| F151I | 196 ± 40 | 13 ± 6 | 0.2 ± 0.15 | 29 ± 5 |
| L155F | 141 ± 16 | 9 ± 5 | 0.4 ± 0.02 | 47 ± 5 |
| 151/155 | 120 ± 15 | 5 ± 2 | 0.5 ± 0.03 | 51 ± 3 |
| Pa-NQR | 143 ± 11 | 5 ± 1.3 | 0.9 ± 0.04 | 43.6 ± 3 |
Discussion
P. aeruginosa is a Gram-negative multidrug-resistant bacterium with a highly branched metabolism that can survive in almost any environment (9, 14, 18). Previous studies have shown that this bacterium uses oxidative phosphorylation, rather than fermentation, to support ATP production and maintain the redox balance within the cell (8, 9, 19). The respiratory chain of this pathogen is complex and contains three different NADH dehydrogenases (8, 21): complex I (proton pumping) (22, 24), NQR (sodium pumping, in other bacteria) (26–29), and NDH-2 (not linked to energy production) (25). Although the physiologic function of the three different NADH dehydrogenases is unknown, it has been reported that complex I is essential for anaerobic growth in presence of nitrate (75). On the other hand, NQR could be involved in ion homeostasis and in the development of the pathogenic phenotype (as previously reported for V. cholerae) (30–32, 76, 77). To understand the physiologic role of this complex, we performed a structural and enzymatic characterization of Pa-NQR.
Subunit and cofactor composition of Pa-NQR
According to our data, the subunit and cofactor composition of Pa-NQR is similar to other studied homologues: a monomeric complex of six subunits and four flavin cofactors, with two covalently bound flavins (presumably FMN) in subunits B and C. However, Pa-NQR shows significant differences. In Vc-NQR, riboflavin has been identified as the final electron acceptor of the protein, delivering electrons to ubiquinone (37, 54). Interestingly, riboflavin is found as a neutral semiquinone radical that is stable even in the presence of strong oxidants (37–39). In Pa-NQR, however, the neutral radical of riboflavin is absent or greatly diminished, and riboflavin seems to be found in the fully oxidized state. A more detailed investigation of the cofactor composition of Pa-NQR is needed to verify the redox state of this cofactor and its role in the significant functional differences of Pa-NQR.
Cation specificity
Pa-NQR as a new proton pump
Our work demonstrates that the membrane potential generation by Pa-NQR reconstituted into proteoliposomes is independent of monovalent cations. Whereas the rate of membrane potential formation in the presence of sodium is significantly higher compared with when it is absent (Fig. 4), the ion-pumping activity can be sustained in the absence of any added cations. Moreover, the membrane potential is completely abolished by the protonophore CCCP, demonstrating that Pa-NQR is a proton pump. To corroborate that Pa-NQR does not pump the traces of sodium found in solution (a few μm is expected, even when the buffers are prepared in double-distilled water and the pH is adjusted with Tris base), the sodium ionophore ETH 157 was added to the assays. The addition of ETH 157 had no effect on the formation of membrane potential, which together with the effect of CCCP demonstrates that Pa-NQR is a proton pump, in contrast with all other NQR homologues that are sodium-specific transporters (30, 48, 49). This finding indicates that the NQR family (formerly Na+-NQR) is more diverse than previously anticipated. Most of the previous studies have been carried out with Vibrio species, and our understanding of other bacteria is limited.
Regulatory sites
Previous studies involving Vc-NQR have shown that the ubiquinone reductase activity is regulated by different monovalent cations (38, 42). In these studies, sodium and lithium were able to stimulate the enzyme's activity, whereas potassium and rubidium act on a separate regulatory site. Ubiquinone reductase activity was stimulated 8–9 times in the presence of sodium (with a Km of 2.5 mm) and 3 times with lithium (Km = 3.4 mm) (42). Interestingly, these two cations are transported by Vc-NQR and in this case can be considered as co-substrates. Moreover, the enzyme also carries a regulatory site that is stimulated by potassium and inhibited by rubidium (42). This behavior differs significantly from what we obtained for Pa-NQR. The steady-state ubiquinone reductase activity of Pa-NQR is stimulated by the monovalent cations sodium, potassium, and cesium, with the highest activity observed with sodium. Even though Pa-NQR enzyme activity was stimulated in the presence of sodium, its effect was only a third of that of Vc-NQR. Additionally, rubidium showed a biphasic behavior and at 50 mm showed the same activatory effect as potassium, followed by an inhibitory effect at higher concentrations. On the other hand, negligible inhibitory activity was observed for lithium. Because Pa-NQR is a proton-specific pump, our results indicate that the stimulation of the activity is due to the interactions of the cations with the regulatory site. This site seems to be less specific in Pa-NQR compared with Vc-NQR, because the latter is specific for potassium (42). This site has not been located in the structure of the complex, but we can speculate that it might contain 6–8 ligands, which would allow the binding of potassium and cesium (79–81). It is possible that the geometry of the site would also allow the binding of sodium, through six ligands in an octahedral array (79–81), which is consistent with the relatively high Ka for sodium compared with larger cations (Table 1). Of all the tested cations, lithium has the smallest ionic radius (0.76 Å) and requires five ligands for binding (79, 81). These two factors might explain the lack of stimulation by the smaller cation. Rubidium has an ionic radius between that of potassium and cesium (1.64 versus 1.46 and 1.73 Å, respectively) and is bound by eight ligands (79). Whereas the activation phase can be explained as the interaction of rubidium with the regulatory site, having an effect similar to potassium, the inhibitory phase could be mediated by nonspecific interactions of the cation with other parts of the protein.
In a recent report, we showed that V. cholerae ApbE, a flavin transferase involved in NQR assembly, is positively regulated by potassium (82). We proposed that V. cholerae production of the cholera toxin and hemolysin would lead to the release of potassium from the epithelial cells, increasing NQR assembly and enhancing V. cholerae pathogenesis. The regulation of Pa-NQR by cations could follow a similar mechanism. Whereas sodium produces the maximum stimulation of the activity (3 times), the Ka is very high, which makes this ion an unlikely regulator. The regulation by potassium seems to be the physiologic function of this regulatory site, especially due to the Ka of 30 mm. Indeed, it has been reported that P. aeruginosa induces the release of potassium from epithelial cells (83), which would have an activatory effect on NQR, enhancing the infection, analogous to the regulation of ApbE.
Mechanisms of resistance against HQNO autopoisoning
P. aeruginosa produces HQNO, a component of its quinolone signal system, for quorum sensing (7, 14, 45). HQNO is a bactericidal agent that acts on competing bacteria and inhibits different complexes of the respiratory chain (7, 10, 46, 47). This compound has a strong inhibitory effect on Vc-NQR, with submicromolar inhibition constants (0.1–0.3 μm). Our group recently reported the inhibition mechanism of HQNO (26). HQNO is a ubiquinone analog that acts as a mixed-type inhibitor, instead of the expected competitive behavior. Our studies show that HQNO is bound to the ubiquinone-binding site in two different redox states of the enzyme (26). Although HQNO acts on a single site, it exhibits mixed-type inhibition because it is an analog of both ubiquinone and ubiquinol.
The production of HQNO by P. aeruginosa appears paradoxical, because the inhibitor would have autoinhibitory properties that might compromise the survival of this microorganism, especially considering that >70% of the rate of oxygen consumption linked to the NADH dehydrogenase activity is sensitive to this inhibitor, indicating an important role of NQR in the aerobic respiratory chain of this bacteria. Moreover, the concentration of HQNO produced by P. aeruginosa is very high and could shut down the respiratory activity. HQNO concentration can reach 4 μm in cystic fibrosis sputum (45), but it can be as high as 40 μm in cell culture (7, 69). To understand P. aeruginosa physiology and the metabolic adaptations that allow the bacteria to avoid or survive autopoisoning, we performed a characterization of the mechanism of inhibition by HQNO. In the case of Pa-NQR, HQNO exhibits a partial mixed inhibitory effect versus ubiquinone, which is different compared with the simple mixed inhibition reported in Vc-NQR (26). The partial component (40%) indicates that even at saturating concentrations of HQNO (tens of micromolar), Pa-NQR would be able to carry out its function. Moreover, the inhibition constants (Kic and Kiu, 1.6 and 1.3 μm) are 5–16 times higher compared with the Ki values of other members of the family (26). These differences would allow P. aeruginosa not only to outcompete other bacteria in its environment, but to survive its own production of HQNO and establish the infection. This resistance mechanism can work together with other mechanisms to protect the bacteria against HQNO autopoisoning, such as the use of efflux pumps, the use of HQNO as precursor to produce other quorum sensing molecules, and biofilm formation (69, 70). Although no studies have been conducted to elucidate the role of NQR in P. aeruginosa physiology, the data indicate that it has important functions in disease and in biofilm formation. Indeed, P. aeruginosa strains isolated from cystic fibrosis patients show an increase in NQR expression (84, 85). Moreover, Pa-NQR subunits have been shown to be expressed by P. aeruginosa during biofilm formation (86, 87), especially in the oxygen-rich, metabolically active regions of the biofilm, where bacteria are exposed to competing bacteria, antibacterial metabolites and antibiotics, and other varying environmental factors.
Ito et al. (88) proposed that a binding site for ubiquinone is located in subunit A of Vc-NQR. Using photoaffinity labeling, the group identified ubiquinone-binding regions in an aqueous cavity of this subunit. Given the importance of ubiquinone for NQR function, the binding site must be conserved across species. However, the regions identified by the authors are highly variable. One of the peculiar characteristics of these regions is that they are rich in positively charged residues, such as arginine and lysine, which are uncommon in ubiquinone-binding motifs (59), and could react easily with the reactive ubiquinone analogs. Another factor that indicates that this putative site does not participate in the catalytic cycle is its location. This site is completely cytosolic, and the ubiquinone molecule would need to be pulled out 25–30 Å from the membrane environment, exposing a large portion of ubiquinone's hydrophobic isoprene chain. Our group has recently located the catalytic ubiquinone-binding site of Vc-NQR, which is found in the interface of subunits B and D, in the core of the phospholipid bilayer (59), and is composed of absolutely conserved residues. Mutations of these residues not only inactivate the enzyme but specifically knock out ubiquinone and HQNO binding. Moreover, this site is located within 20 Å of the FMN molecule in subunit B, which according to functional data must be in close proximity to riboflavin, the electron donor of ubiquinone (37, 39, 54, 68). Thus, the site proposed by our group is a more likely candidate to carry out this important function. Indeed, the mutations that we performed in subunit D, which is adjacent to the site that we proposed, turned Vc-NQR into an HQNO-resistant enzyme.
Conclusion
Taken together, the data demonstrate that Pa-NQR functions unlike any of the other previously characterized NQR homologues: not as a sodium pump but rather as a proton pump with a high resistance to HQNO. These findings raise important questions about the physiologic role of NQR and the redundancy of the three different NADH dehydrogenases that P. aeruginosa carries. It is possible that the expression of these enzymes would vary, depending on the environment of the niches that this bacterium colonizes, including the human body. The differential expression of these complexes may allow the survival of the bacterium by reducing the sensitivity to allelopathic and antibiotic molecules produced by plants, fungi, and other bacteria. For instance, NQR could be the main NADH dehydrogenase during the colonization of plants, because they produce a high amount of rotenone and other flavones, which are inhibitors of complex I and NDH-2 (89). It has been recently proposed that HQNO autopoisoning induces the secretion of diverse factors that favors biofilm formation, while inhibiting the growth of other bacteria (7). However, the bacteria must be able to survive its own production of toxins and virulence factors. Further studies are necessary to characterize the role of Pa-NQR in the physiology of P. aeruginosa.
Experimental procedures
Cloning
The operon encoding P. aeruginosa (strain PAO1) NQR was amplified by PCR, with the forward primer 5′-ATGATCAAGATAAAACGTGGCCTG-3′ and reverse primer 5′-TCAATGATGATGATGATGATGTGCTCCTGCTCCGCCACCGAAATCGTCCAGCAG-3′. On the reverse primer, a triplicate glycine-alanine repeat spacer and a six-histidine coding sequence were included. The operon was inserted in-frame into the 5′-EcoRI and 3′-BglII restriction site of pBAD/His B plasmid. The Pa-NQR construct was verified by sequencing (Operon MWG) and subsequently used to transform V. cholerae O395N1 Δnqr for protein expression.
Site-directed mutagenesis
F151I and L155F mutations in Vc-NQR were obtained with a site-directed mutagenesis kit (Agilent Technologies), using sense primer 5′-TGATGACGGTTGGTTTCATCCGTGAGCTTTTAGGC-3′ and antisense primer 5′-GCCTAAAAGCTCACGGATGAAACCAACCGTCATCA-3′ for F151I and sense primer 5′-GGTTTCTTCCGTGAGCTTTTTGGCTCAGGTAAGCTATTTGG-3′ and antisense primer 5′-CCAAATAGCTTACCTGAGCCAAAAAGCTCACGGAAGAAACC-3′ for L155F. The mutations were subcloned in-frame into the 5′-KpnI and 3′-EcoRI restriction enzyme sites in the Vc-NQR pBAD/HisB construct. A construct carrying both mutations, F151I and L155F, was produced by using the Vc-NQR-F151I construct as a template, inducing the L155F mutation using primers 5′-GGTTTCATCCGTGAGCTTTTTGGCTCAGGTAAGCTATTTGG-3′ and 5′-CCAAATAGCTTACCTGAGCCAAAAAGCTCACGGATGAAACC-3′ (sense and antisense, respectively).
Protein expression and purification
V. cholerae cells carrying the WT and mutant Vc-NQR-pBAD/HisB construct, as described previously by Tuz et al. (26), or the Pa-NQR-pBAD/HisB construct were grown in Luria broth medium (59). NQR genetic expression was induced with arabinose. The induced cells were harvested, washed, and disrupted via sonication (60-s pulsed sonication, 50% duty cycle), and then the cell membranes were obtained by differential centrifugation. The membranes were solubilized in buffer containing 0.05% n-dodecyl-β-d-maltoside (DDM), 5 mm imidazole, 50 mm Na2HPO4, 300 mm NaCl, 5% glycerol, pH 8.0. The NQR complex was purified by Ni-NTA affinity chromatography, followed by cation-exchange chromatography using DEAE-Sepharose.
Urea SDS-polyacrylamide gel analysis
Purified Vc-NQR and Pa-NQR complexes were run in urea (30%) SDS-PAGE 15% acrylamide gels to determine enzyme subunit composition and protein purity. After electrophoresis, the gel was exposed to UV light for the detection of the fluorescent FMN cofactors previously identified in subunits NqrB and NqrC (43, 44, 67). The gel was stained with Coomassie Blue to identify the six different subunits of NQR. The intensity of the bands was analyzed densitometrically in a gel image, using ImageJ (50), and was compared with the total pixel intensity in the lane to calculate purity.
Blue native gel electrophoresis
BN-PAGE of Pa-NQR was performed as described by Wittig et al. (53). Briefly, 10 μg of protein per lane were separated in a 5–16% gradient gel acrylamide/bisacrylamide (38.5%:1.5%), using cathode buffer I (10 mm Tricine, 3 mm Bistris, pH 7, and 0.02% Coomassie Blue G-250), cathode buffer II (10 mm Tricine, 3 mm Bistris, and 0.002% Coomassie Blue G-250), and anode buffer (10 mm Bistris, pH 7). Lanes were cut to perform in-gel NADH dehydrogenase activity, Coomassie Blue staining, and second-dimension SDS-PAGE.
NADH dehydrogenase activity in-gel
In-gel activity of Pa-NQR was performed as described previously (90). After BN-PAGE, gel lanes were incubated at room temperature for 30 min in 10 mm Tris, 0.2 mm NADH, 0.5 mg/ml NBT, pH 7.0 (91). The excess of Coomassie Blue was removed by incubating the gel in 0.1% SDS.
Second-dimension gel electrophoresis
The second-dimension gel electrophoresis was performed using a lane obtained from the first-dimension BN-PAGE, placed on top of an SDS-polyacrylamide gel (30% urea, 15% acrylamide) (52, 53). Following the second-dimension SDS electrophoresis, the gel was exposed to UV light to identify the two covalently bound FMN molecules and stained with Coomassie Blue, as described previously (43, 67).
UV-visible spectra analysis
Cofactor composition of Pa-NQR was assessed spectrophotometrically (300–700 nm) using the denatured complex or the native complex under reducing and oxidizing conditions. Protein samples were denatured using 7 m guanidine chloride. Spectrophotometric analyses with the native complex were carried out in buffer containing 20 mm Tris, 1 mm EDTA, 5% glycerol, 0.05% DDM, pH 8.0. Protein samples were fully reduced by the addition of sodium dithionite.
Activity measurements
Enzymatic activity was measured spectrophometrically, following ubiquinone reductase activity of NQR. Ubiquinone reductase was measured at 282 nm, using a molar extinction coefficient of 10.2 mm−1 cm−1 (42). Enzymatic assays were carried out in buffer containing 250 μm K2-NADH, 50 mm Tris-HCl, 1 mm EDTA, 5% glycerol, 0.05% DDM, pH 8.0. Activity measurements were conducted in the presence of varying concentrations of ubiquinone, monovalent cations (NaCl, LiCl, KCl, RbCl, and CsCl), or HQNO.
Experiments to characterize the inhibition mechanism of HQNO were carried out, performing a ubiquinone-1 titration at several fixed concentrations of HQNO. The resulting curves were fitted to Equation 1, which describes mixed partial inhibition, where v is the turnover rate, kcat is the maximum turnover rate in the absence of the inhibitor, kcatR is the turnover rate obtained at saturating concentrations of HQNO, [S] is the concentration of ubiquinone-1, [I] is the concentration of HQNO, Km is the Km, and Kic and Kiu are the inhibition constants of the competitive and uncompetitive components. This equation was obtained from the model shown in Scheme 1 (92).
| (Eq. 1) |
Scheme 1.

NQR reconstitution in proteoliposomes and membrane potential measurement
Pa-NQR was reconstituted into proteoliposomes following the protocol previously reported by Juárez et al. (38, 60). Briefly, purified Pa-NQR was added to a solution containing E. coli phospholipids (20 mm, lipid/protein ratio of 10) and 0.5 m n-octyl glucoside in reconstitution buffer (250 mm sucrose, 25 mm HEPES, 1 mm EDTA, pH 7.5-adjusted with Tris base). The detergent was gradually removed from the sample by the addition of SM Bio-Beads (60). Ion transport was assayed in reconstituted proteoliposome membranes containing Pa-NQR or Vc-NQR, following membrane potential generation in the presence of different monovalent cations or ionophores. The transmembrane potential was measured spectrophotometrically using Oxonol VI, at 625 minus 587 nm (61). The assays were carried out in reconstitution buffer containing 100 μm ubiquinone-1, 5 μm oxonol VI, 250 μm NADH, and a 50 mm concentration of the monovalent cation (NaCl, KCl, CsCl, RbCl, or LiCl). The protonophore CCCP (2 μm) and the sodium ionophore ETH 157 (5 μm) were added at different time points to determine proton or sodium pumping.
P. aeruginosa membrane preparation
P. aeruginosa (PAO1 strain) was cultured in Luria broth medium at 37 °C under agitation (250 rpm). The cells were harvested by centrifugation at early stationary phase of growth. Cells were washed twice with KHE buffer (50 mm HEPES, 1 mm EDTA, 100 mm KCl, 0.05% DDM, pH 7.5) and stored at −80 °C. The cells were thawed, and 1 mm phenylmethylsulfonyl fluoride was added before cell disruption. The cell suspension was sonicated three times using a Sonifier® Cell Disruptor 350 (Branson) at 50% duty cycle, output control 6, for 1 min in ice. Cell debris was removed by centrifugation at 7,000 rpm for 30 min in a Beckman centrifuge using a JA-20 rotor at 4 °C. The supernatant was collected and centrifuged at 20,000 rpm in a Beckman JA-20 rotor at 4 °C. The resulting supernatant was ultracentrifuged at 30,000 rpm in a Beckman ultracentrifuge at 4 °C for 5 h. The pellet containing the membranes was resuspended in KHE buffer and stored at −80 °C.
Oximetry
Respiratory activity of P. aeruginosa membranes (200 μg/ml) was measured using a Clark type electrode (YSI 5300), in a 2-ml custom-made chamber at 36.5 °C (34). Oxygen consumption was evaluated in buffer containing 150 mm KCl, 20 mm HEPES, 1 mm EDTA, 5 mm MgCl2, 10 mm K2HPO4, pH 7.5, in the presence of 200 μm NADH at different concentrations of HQNO (0, 0.5, 1, 2, 5, and 10 μm).
Molecular modeling
The sequence and crystallographic structure of Na+-NQR from V. cholerae were obtained from the RCSB Protein Data bank (entry 4P6V) Because several loops were missing from the crystallographic structure, MODELLER version 9.14 was used to construct complete models of subunits B and D (93). A template search with BLAST and PSI-BLAST did not find three-dimensional structures homologous to the missing loops (94). Thus, the loops were modeled based on a template from the pGenTHREADER server, which contains a method for fold recognition and identification of distant homologues (95). Ubiquinone-1 was downloaded from the ZINC database (accession number 1559692) (96). The atomic charges of all molecules in ZINC database were calculated by the semiempirical quantum mechanical program AMSOL (97). The P. aeruginosa homology model was built by first using protein BLAST to align the sequences to the structure of V. cholerae NQR (94). MODELLER 9.14 was again used to create homologous secondary and tertiary structure for each protein subunit. The best alignment was chosen based on MODELLER DOPE scoring function. The subunits were assembled identically to the conformation of Vc-NQR (93). A model of P. aeruginosa NQR subunits B, D, and E in a membrane, based on the CHARMM36 force field (98), was built using the CHARMM-GUI membrane builder with the following settings. Terminal group patching was activated for all subunits, and the membrane selection was homologous DOPC (99–101). The system was built using the replacement method, and 0.15 m NaCl ions were added to the explicit solvent. Molecular dynamics simulations were performed at 303.15 K in the NPT ensemble with a Langevin dynamics integrator with a 2-fs time step using OpenMM version 7.0.1 (17). The simulation ran for a total of 50 ns.
To prepare the model for docking with UCSF DOCK version 6.6 (78), hydrogen atoms were stripped using UCSF Chimera version 1.9. A molecular surface was prepared using DMS, a tool within DOCK 6.6. Another DOCK 6.6 tool, Sphgen, was used to generate vacancy spheres surrounding the entire protein. Spheres had a minimum and maximum radius of 1.0 and 5.0 Å, respectively. HQNO was docked into all spheres using flexible docking. Docking orientations were ranked based on grid score, a molecular mechanics-like scoring function.
Author contributions
D. A. R., M. R.-L., W. M. M., C. L., X. F., P. L., and K. T. conducted the experiments. D. A. R., M. R.-L., D. D. L. M., and O. J. designed the experiments. D. A. R., M. R.-L., W. M. M., K. T., D. D. L. M., and O. J. analyzed the data and wrote the manuscript. All authors participated in the final review of the manuscript.
Supplementary Material
This work was supported by National Institutes of Health Grant R15GM114781 (to D. D. L. M.), startup funds from the Illinois Institute of Technology (to O. J.), and CONACYT (to M. R.-L.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1 and S2.
- UTI
- urinary tract infection
- NQR
- ion-pumping NADH:ubiquinone oxidoreductase
- Vc-NQR
- V. cholerae NQR
- Pa-NQR
- P. aeruginosa NQR
- CCCP
- carbonyl cyanide 3-chlorophenylhydrazone
- DDM
- n-dodecyl β-d-maltoside
- HQNO
- 2-n-heptyl-4-hydroxyquinoline N-oxide
- ETH 157
- N,N′-dibenzyl-N,N′-diphenyl-1,2-phenylenedioxydiacetamide
- Ni-NTA
- nickel-nitrilotriacetic acid
- BN-PAGE
- blue native PAGE
- NBT
- nitro blue tetrazolium
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- Bistris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
References
- 1. Rasamiravaka T., Labtani Q., Duez P., and El Jaziri M. (2015) The formation of biofilms by Pseudomonas aeruginosa: a review of the natural and synthetic compounds interfering with control mechanisms. Biomed Res. Int. 2015, 759348 10.1155/2015/759348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cole S. J., Records A. R., Orr M. W., Linden S. B., and Lee V. T. (2014) Catheter-associated urinary tract infection by Pseudomonas aeruginosa is mediated by exopolysaccharide-independent biofilms. Infect. Immun. 82, 2048–2058 10.1128/IAI.01652-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Warren J. W., Steinberg L., Hebel J. R., and Tenney J. H. (1989) The prevalence of urethral catheterization in Maryland nursing homes. Arch. Intern. Med. 149, 1535–1537 10.1001/archinte.1989.00390070073009 [DOI] [PubMed] [Google Scholar]
- 4. Jarvis W. R., and Martone W. J. (1992) Predominant pathogens in hospital infections. J. Antimicrob. Chemother. 29, 19–24 10.1093/jac/29.suppl_A.19 [DOI] [PubMed] [Google Scholar]
- 5. Høiby N., Bjarnsholt T., Givskov M., Molin S., and Ciofu O. (2010) Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 35, 322–332 10.1016/j.ijantimicag.2009.12.011 [DOI] [PubMed] [Google Scholar]
- 6. Parsek M. R., and Singh P. K. (2003) Bacterial biofilms: an emerging link to disease pathogenesis. Annu. Rev. Microbiol. 57, 677–701 10.1146/annurev.micro.57.030502.090720 [DOI] [PubMed] [Google Scholar]
- 7. Hazan R., Que Y. A., Maura D., Strobel B., Majcherczyk P. A., Hopper L. R., Wilbur D. J., Hreha T. N., Barquera B., and Rahme L. G. (2016) Auto poisoning of the respiratory chain by a quorum-sensing-regulated molecule favors biofilm formation and antibiotic tolerance. Curr. Biol. 26, 195–206 10.1016/j.cub.2015.11.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Williams H. D., Zlosnik J. E. A., and Ryall B. (2007) Oxygen, cyanide and energy generation in the cystic fibrosis pathogen Pseudomonas aeruginosa. Adv. Microb. Physiol. 52, 1–71 10.1016/S0065-2911(06)52001-6 [DOI] [PubMed] [Google Scholar]
- 9. Arai H. (2011) Regulation and function of versatile aerobic and anaerobic respiratory metabolism in Pseudomonas aeruginosa. Front. Microbiol. 2, 103 10.3389/fmicb.2011.00103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Orazi G., and O'Toole G. A. (2017) Pseudomonas aeruginosa alters Staphylococcus aureus sensitivity to vancomycin in a biofilm model of cystic fibrosis infection. MBio 8, e00873–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mitchell G., Séguin D. L., Asselin A.-E., Déziel E., Cantin A. M., Frost E. H., Michaud S., and Malouin F. (2010) Staphylococcus aureus σ B-dependent emergence of small-colony variants and biofilm production following exposure to Pseudomonas aeruginosa 4-hydroxy-2-heptylquinoline-N-oxide. BMC Microbiol. 10, 33 10.1186/1471-2180-10-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mittal R., Aggarwal S., Sharma S., Chhibber S., and Harjai K. (2009) Urinary tract infections caused by Pseudomonas aeruginosa: a minireview. J. Infect. Public Health 2, 101–111 10.1016/j.jiph.2009.08.003 [DOI] [PubMed] [Google Scholar]
- 13. Gaynes R., Edwards J. R., and National Nosocomial Infections Surveillance System (2005) Overview of nosocomial infections caused by Gram-negative bacilli. Clin. Infect. Dis. 41, 848–854 10.1086/432803 [DOI] [PubMed] [Google Scholar]
- 14. Moradali M. F., Ghods S., and Rehm B. H. A. (2017) Pseudomonas aeruginosa lifestyle: a paradigm for adaptation, survival, and persistence. Front. Cell Infect. Microbiol. 7, 39 10.3389/fcimb.2017.00039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lister P. D., Wolter D. J., and Hanson N. D. (2009) Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin. Microbiol. Rev. 22, 582–610 10.1128/CMR.00040-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Drenkard E. (2003) Antimicrobial resistance of Pseudomonas aeruginosa biofilms. Microbes Infect. 5, 1213–1219 10.1016/j.micinf.2003.08.009 [DOI] [PubMed] [Google Scholar]
- 17. Eastman P., Swails J., Chodera J. D., McGibbon R. T., Zhao Y., Beauchamp K. A., Wang L. P., Simmonett A. C., Harrigan M. P., Stern C. D., Wiewiora R. P., Brooks B. R., and Pande V. S. (2017) OpenMM 7: rapid development of high performance algorithms for molecular dynamics. PLoS Comput. Biol. 13, e1005659 10.1371/journal.pcbi.1005659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Alvarez-Ortega C., and Harwood C. S. (2007) Responses of Pseudomonas aeruginosa to low oxygen indicate that growth in the cystic fibrosis lung is by aerobic respiration. Mol. Microbiol. 65, 153–165 10.1111/j.1365-2958.2007.05772.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Matsushita K., Yamada M., Shinagawa E., Adachi O., and Ameyama M. (1983) Membrane-bound respiratory chain of Pseudomonas aeruginosa grown aerobically: a KCN-insensitive alternate oxidase chain and its energetics. J. Biochem. 93, 1137–1144 10.1093/oxfordjournals.jbchem.a134239 [DOI] [PubMed] [Google Scholar]
- 20. Matsushita K., Patel L., and Kaback H. R. (1984) Cytochrome o type oxidase from Escherichia coli: characterization of the enzyme and mechanism of electrochemical proton gradient generation. Biochemistry 23, 4703–4714 10.1021/bi00315a028 [DOI] [PubMed] [Google Scholar]
- 21. Kerscher S., Dröse S., Zickermann V., and Brandt U. (2008) The three families of respiratory NADH dehydrogenases. Results Probl. Cell Differ. 45, 185–222 10.1007/400_2007_028 [DOI] [PubMed] [Google Scholar]
- 22. Brandt U. (2011) A two-state stabilization-change mechanism for proton-pumping complex i. Biochim. Biophys. Acta 1807, 1364–1369 10.1016/j.bbabio.2011.04.006 [DOI] [PubMed] [Google Scholar]
- 23. Yagi T. (1991) Bacterial NADH-quinone oxidoreductases. J. Bioenerg. Biomembr. 23, 211–225 10.1007/BF00762218 [DOI] [PubMed] [Google Scholar]
- 24. Friedrich T., Steinmüller K., and Weiss H. (1995) The proton-pumping respiratory complex-I of bacteria and mitochondria and its homolog in chloroplasts. FEBS Lett. 367, 107–111 10.1016/0014-5793(95)00548-N [DOI] [PubMed] [Google Scholar]
- 25. Heikal A., Nakatani Y., Dunn E., Weimar M. R., Day C. L., Baker E. N., Lott J. S., Sazanov L. A., and Cook G. M. (2014) Structure of the bacterial type II NADH dehydrogenase: a monotopic membrane protein with an essential role in energy generation. Mol. Microbiol. 91, 950–964 10.1111/mmi.12507 [DOI] [PubMed] [Google Scholar]
- 26. Tuz K., Mezic K. G., Xu T., Barquera B., and Juárez O. (2015) The kinetic reaction mechanism of the Vibrio cholerae sodium-dependent NADH dehydrogenase. J. Biol. Chem. 290, 20009–20021 10.1074/jbc.M115.658773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Juárez O., and Barquera B. (2012) Insights into the mechanism of electron transfer and sodium translocation of the Na+-pumping NADH:quinone oxidoreductase. Biochim. Biophys. Acta 1817, 1823–1832 10.1016/j.bbabio.2012.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Verkhovsky M. I., and Bogachev A. V. (2010) Sodium-translocating NADH:quinone oxidoreductase as a redox-driven ion pump. Biochim. Biophys. Acta 1797, 738–746 10.1016/j.bbabio.2009.12.020 [DOI] [PubMed] [Google Scholar]
- 29. Reyes-Prieto A., Barquera B., and Juárez O. (2014) Origin and evolution of the sodium-pumping NADH:ubiquinone oxidoreductase. PLoS One 9, e96696 10.1371/journal.pone.0096696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Häse C. C., and Barquera B. (2001) Role of sodium bioenergetics in Vibrio cholerae. Biochim. Biophys. Acta 1505, 169–178 10.1016/S0005-2728(00)00286-3 [DOI] [PubMed] [Google Scholar]
- 31. Kojima S., Yamamoto K., Kawagishi I., and Homma M. (1999) The polar flagellar motor of Vibrio cholerae is driven by an Na+ motive force. J. Bacteriol. 181, 1927–1930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Häse C. C., Fedorova N. D., Galperin M. Y., and Dibrov P. A. (2001) Sodium ion cycle in bacterial pathogens: evidence from cross-genome comparisons. Microbiol. Mol. Biol. Rev. 65, 353–370, table of contents 10.1128/MMBR.65.3.353-370.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Skulachev V. P. (1984) Sodium bioenergetics. Trends Biochem. Sci. 9, 483–485 10.1016/0968-0004(84)90317-7 [DOI] [Google Scholar]
- 34. Liang P., Rosas-Lemus M., Patel D., Fang X., Tuz K., and Juárez O. (2018) Dynamic energy dependency of Chlamydia trachomatis on host cell metabolism during different stages of intracellular growth: possible role of sodium-based energetics in chlamydial ATP generation. J. Biol. Chem. 293, 510–522 10.1074/jbc.M117.797209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dibrov P., Dibrov E., Maddaford T. G., Kenneth M., Nelson J., Resch C., and Pierce G. N. (2017) Development of a novel rationally designed antibiotic to inhibit a nontraditional bacterial target. Can. J. Physiol. Pharmacol. 95, 595–603 10.1139/cjpp-2016-0505 [DOI] [PubMed] [Google Scholar]
- 36. Steuber J., Vohl G., Muras V., Toulouse C., Claussen B., Vorburger T., and Fritz G. (2015) The structure of Na+-translocating of NADH:ubiquinone oxidoreductase of Vibrio cholerae: implications on coupling between electron transfer and Na+ transport. Biol. Chem. 396, 1015–1030 [DOI] [PubMed] [Google Scholar]
- 37. Juárez O., Nilges M. J., Gillespie P., Cotton J., and Barquera B. (2008) Riboflavin is an active redox cofactor in the Na+-pumping NADH:quinone oxidoreductase (Na+-NQR) from Vibrio cholerae. J. Biol. Chem. 283, 33162–33167 10.1074/jbc.M806913200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Juárez O., Athearn K., Gillespie P., and Barquera B. (2009) Acid residues in the transmembrane helices of the Na+-pumping NADH:quinone oxidoreductase from Vibrio cholerae involved in sodium translocation. Biochemistry 48, 9516–9524 10.1021/bi900845y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Barquera B., Zhou W., Morgan J. E., and Gennis R. B. (2002) Riboflavin is a component of the Na+-pumping NADH-quinone oxidoreductase from Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 99, 10322–10324 10.1073/pnas.162361299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Barquera B., Morgan J. E., Lukoyanov D., Scholes C. P., Gennis R. B., and Nilges M. J. (2003) X- and W-band EPR and Q-band ENDOR studies of the flavin radical in the Na+-translocating NADH:quinone oxidoreductase from Vibrio cholerae. J. Am. Chem. Soc. 125, 265–275 10.1021/ja0207201 [DOI] [PubMed] [Google Scholar]
- 41. Steuber J., Vohl G., Casutt M. S., Vorburger T., Diederichs K., and Fritz G. (2014) Structure of the V. cholerae Na+-pumping NADH:quinone oxidoreductase. Nature 516, 62–67 10.1038/nature14003 [DOI] [PubMed] [Google Scholar]
- 42. Juárez O., Shea M. E., Makhatadze G. I., and Barquera B. (2011) The role and specificity of the catalytic and regulatory cation-binding sites of the Na+-pumping NADH:quinone oxidoreductase from Vibrio cholerae. J. Biol. Chem. 286, 26383–26390 10.1074/jbc.M111.257873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Barquera B., Hellwig P., Zhou W., Morgan J. E., Häse C. C., Gosink K. K., Nilges M., Bruesehoff P. J., Roth A., Lancaster C. R. D., and Gennis R. B. (2002) Purification and characterization of the recombinant Na+-translocating NADH:quinone oxidoreductase from Vibrio cholerae. Biochemistry 41, 3781–3789 10.1021/bi011873o [DOI] [PubMed] [Google Scholar]
- 44. Zhou W., Bertsova Y. V., Feng B., Tsatsos P., Verkhovskaya M. L., Gennis R. B., Bogachev A. V., and Barquera B. (1999) Sequencing and preliminary characterization of the Na+-translocating NADH:ubiquinone oxidoreductase from Vibrio harveyi. Biochemistry 38, 16246–16252 10.1021/bi991664s [DOI] [PubMed] [Google Scholar]
- 45. Barr H. L., Halliday N., Cámara M., Barrett D. A., Williams P., Forrester D. L., Simms R., Smyth A. R., Honeybourne D., Whitehouse J. L., Nash E. F., Dewar J., Clayton A., Knox A. J., and Fogarty A. W. (2015) Pseudomonas aeruginosa quorum sensing molecules correlate with clinical status in cystic fibrosis. Eur. Respir. J. 46, 1046–1054 10.1183/09031936.00225214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kang J., and Kim Y. J. (2007) HQNO-sensitive NADH:quinone oxidoreductase of Bacillus cereus KCTC 3674. J. Biochem. Mol. Biol. 40, 53–57 [DOI] [PubMed] [Google Scholar]
- 47. Meunier B., Madgwick S. A., Reil E., Oettmeier W., and Rich P. R. (1995) New inhibitors of the quinol oxidation sites of bacterial cytochromes bo and bd. Biochemistry 34, 1076–1083 10.1021/bi00003a044 [DOI] [PubMed] [Google Scholar]
- 48. Barquera B. (2014) The sodium pumping NADH:quinone oxidoreductase (Na+-NQR), a unique redox-driven ion pump. J. Bioenerg. Biomembr. 46, 289–298 10.1007/s10863-014-9565-9 [DOI] [PubMed] [Google Scholar]
- 49. Minato Y., Fassio S. R., Kirkwood J. S., Halang P., Quinn M. J., Faulkner W. J., Aagesen A. M., Steuber J., Stevens J. F., and Häse C. C. (2014) Roles of the sodium-translocating NADH:quinone oxidoreductase (Na+-NQR) on Vibrio cholerae metabolism, motility and osmotic stress resistance. PLoS One 9, e97083 10.1371/journal.pone.0097083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Abràmofff M. D., and Magalhães P. J., and Ram S. J. (2005) Image processing with ImageJ Part II. Biophotonics Int. 11, 36–43 [Google Scholar]
- 51. Barquera B., Häse C. C., and Gennis R. B. (2001) Expression and mutagenesis of the NqrC subunit of the NQR respiratory Na+ pump from Vibrio cholerae with covalently attached FMN. FEBS Lett. 492, 45–49 10.1016/S0014-5793(01)02224-4 [DOI] [PubMed] [Google Scholar]
- 52. Schägger H., and von Jagow G. (1991) Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199, 223–231 10.1016/0003-2697(91)90094-A [DOI] [PubMed] [Google Scholar]
- 53. Wittig I., Braun H. P., and Schägger H. (2006) Blue native PAGE. Nat. Protoc. 1, 418–428 10.1038/nprot.2006.62 [DOI] [PubMed] [Google Scholar]
- 54. Juárez O., Morgan J. E., and Barquera B. (2009) The electron transfer pathway of the Na+-pumping NADH:quinone oxidoreductase from Vibrio cholerae. J. Biol. Chem. 284, 8963–8972 10.1074/jbc.M809395200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bogachev A. V., Bertsova Y. V., Ruuge E. K., Wikström M., and Verkhovsky M. I. (2002) Kinetics of the spectral changes during reduction of the Na+-motive NADH:quinone oxidoreductase from Vibrio harveyi. Biochim. Biophys. Acta 1556, 113–120 10.1016/S0005-2728(02)00342-0 [DOI] [PubMed] [Google Scholar]
- 56. Neehaul Y., Juárez O., Barquera B., and Hellwig P. (2012) Thermodynamic contribution to the regulation of electron transfer in the Na+-pumping NADH:quinone oxidoreductase from Vibrio cholerae. Biochemistry 51, 4072–4077 10.1021/bi300343u [DOI] [PubMed] [Google Scholar]
- 57. Bogachev A. V., Kulik L. V., Bloch D. A., Bertsova Y. V., Fadeeva M. S., and Verkhovsky M. I. (2009) Redox properties of the prosthetic groups of Na+-translocating NADH:quinone oxidoreductase. 1. Electron paramagnetic resonance study of the enzyme. Biochemistry 48, 6291–6298 10.1021/bi900524m [DOI] [PubMed] [Google Scholar]
- 58. Juárez O., Morgan J. E., Nilges M. J., and Barquera B. (2010) Energy-transducing redox steps of the Na+-pumping NADH:quinone oxidoreductase from Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 107, 12505–12510 10.1073/pnas.1002866107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tuz K., Li C., Fang X., Raba D. A., Liang P., Minh D. D. L., and Juárez O. (2017) Identification of the catalytic ubiquinone-binding site of Vibrio cholerae sodium-dependent NADH dehydrogenase: a novel ubiquinone-binding motif. J. Biol. Chem. 292, 3039–3048 10.1074/jbc.M116.770982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rigaud J. L., Pitard B., and Levy D. (1995) Reconstitution of membrane proteins into liposomes: application to energy-transducing membrane proteins. Biochim. Biophys. Acta 1231, 223–246 10.1016/0005-2728(95)00091-V [DOI] [PubMed] [Google Scholar]
- 61. Apell H. J., and Bersch B. (1987) Oxonol-Vi as an optical indicator for membrane-potentials in lipid vesicles. Biochim. Biophys. Acta 903, 480–494 10.1016/0005-2736(87)90055-1 [DOI] [PubMed] [Google Scholar]
- 62. Parker V. H. (1965) Uncouplers of rat-liver mitochondrial oxidative phosphorylation. Biochem. J. 97, 658–662 10.1042/bj0970658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Schaffar B. P. H., and Wolfbeis O. S. (1989) A sodium-selective optrode. Mikrochim. Acta 99, 109–116 10.1007/BF01242796 [DOI] [Google Scholar]
- 64. Neehaul Y., Juárez O., Barquera B., and Hellwig P. (2013) Infrared spectroscopic evidence of a redox-dependent conformational change involving ion binding residue NqrB-D397 in the Na+-pumping NADH:quinone oxidoreductase from Vibrio cholerae. Biochemistry 52, 3085–3093 10.1021/bi4000386 [DOI] [PubMed] [Google Scholar]
- 65. Shea M. E. M. E., Juárez O., Cho J., and Barquera B. (2013) Aspartic acid 397 in subunit B of the Na+-pumping NADH:quinone oxidoreductase from Vibrio cholerae forms part of a sodium-binding site, is involved in cation selectivity, and affects cation-binding site cooperativity. J. Biol. Chem. 288, 31241–31249 10.1074/jbc.M113.510776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Shea M. E., Mezic K. G., Juárez O., and Barquera B. (2015) A mutation in Na+-NQR uncouples electron flow from Na+ translocation in the presence of K+. Biochemistry 54, 490–496 10.1021/bi501266e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Nakayama Y., Yasui M., Sugahara K., Hayashi M., and Unemoto T. (2000) Covalently bound flavin in the NqrB and NqrC subunits of Na-translocating NADH-quinone reductase from Vibrio alginolyticus. FEBS Lett. 474, 165–168 10.1016/S0014-5793(00)01595-7 [DOI] [PubMed] [Google Scholar]
- 68. Casutt M. S., Huber T., Brunisholz R., Tao M., Fritz G., and Steuber J. (2010) Localization and function of the membrane-bound riboflavin in the Na+-translocating NADH:quinone oxidoreductase (Na+-NQR) from Vibrio cholerae. J. Biol. Chem. 285, 27088–27099 10.1074/jbc.M109.071126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Déziel E., Lépine F., Milot S., He J., Mindrinos M. N., Tompkins R. G., and Rahme L. G. (2004) Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc. Natl. Acad. Sci. U.S.A. 101, 1339–1344 10.1073/pnas.0307694100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zemke A. C., and Bomberger J. M. (2016) Microbiology: social suicide for a good cause. Curr. Biol. 26, R80–R82 10.1016/j.cub.2015.12.018 [DOI] [PubMed] [Google Scholar]
- 71. Häussler S., and Becker T. (2008) The Pseudomonas quinolone signal (PQS) balances life and death in Pseudomonas aeruginosa populations. PLoS Pathog. 4, e1000166 10.1371/journal.ppat.1000166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Nakayama Y., Hayashi M., Yoshikawa K., Mochida K., and Unemoto T. (1999) Inhibitor studies of a new antibiotic, korormicin, 2-n-heptyl-4-hydroxyquinoline N-oxide and Ag+ toward the Na+-translocating NADH-quinone reductase from the marine Vibrio alginolyticus. Biol. Pharm. Bull. 22, 1064–1067 10.1248/bpb.22.1064 [DOI] [PubMed] [Google Scholar]
- 73. Juárez O., Neehaul Y., Turk E., Chahboun N., DeMicco J. M., Hellwig P., and Barquera B. (2012) The role of glycine residues 140 and 141 of subunit B in the functional ubiquinone binding site of the Na+-pumping NADH:quinone oxidoreductase from Vibrio cholerae. J. Biol. Chem. 287, 25678–25685 10.1074/jbc.M112.366088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yoshikawa K., Nakayama Y., Hayashi M., Unemoto T., and Mochida K. (1999) Korormicin, an antibiotic specific for Gram-negative marine bacteria, strongly inhibits the respiratory chain-linked Na+-translocating NADH:quinone reductase from the marine Vibrio alginolyticus. J. Antibiot. 52, 182–185 10.7164/antibiotics.52.182 [DOI] [PubMed] [Google Scholar]
- 75. Filiatrault M. J., Picardo K. F., Ngai H., Passador L., and Iglewski B. H. (2006) Identification of Pseudomonas aeruginosa genes involved in virulence and anaerobic growth. Infect. Immun. 74, 4237–4245 10.1128/IAI.02014-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Muras V., Dogaru-Kinn P., Minato Y., Häse C. C., and Steuber J. (2016) The Na+-translocating NADH:quinone oxidoreductase (Na+-NQR) enhances oxidative stress in the cytoplasm of Vibrio cholerae. J. Bacteriol. 198, 2307–2317 10.1128/JB.00342-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Merrell D. S., Hava D. L., and Camilli A. (2002) Identification of novel factors involved in colonization and acid tolerance of Vibrio cholerae. Mol. Microbiol. 43, 1471–1491 10.1046/j.1365-2958.2002.02857.x [DOI] [PubMed] [Google Scholar]
- 78. Lang P. T., Brozell S. R., Mukherjee S., Pettersen E. F., Meng E. C., Thomas V., Rizzo R. C., Case D. A., James T. L., and Kuntz I. D. (2009) DOCK 6: combining techniques to model RNA-small molecule complexes. RNA 15, 1219–1230 10.1261/rna.1563609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Mähler J., and Persson I. (2012) A study of the hydration of the alkali metal ions in aqueous solution. Inorg. Chem. 51, 425–438 10.1021/ic2018693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Bucher D., Guidoni L., Carloni P., and Rothlisberger U. (2010) Coordination numbers of K+ and Na+ ions inside the selectivity filter of the KcsA potassium channel: insights from first principles molecular dynamics. Biophys. J. 98, L47–L49 10.1016/j.bpj.2010.01.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Nayal M., and Di Cera E. (1996) Valence screening of water in protein crystals reveals potential Na+ binding sites. J. Mol. Biol. 256, 228–234 10.1006/jmbi.1996.0081 [DOI] [PubMed] [Google Scholar]
- 82. Fang X., Liang P., Raba D. A., Rosas-Lemus M., Chakravarthy S., Tuz K., and Juárez O. (2017) Kinetic characterization of Vibrio cholerae ApbE: substrate specificity and regulatory mechanisms. PLoS One 12, e0186805 10.1371/journal.pone.0186805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Arlehamn C. S. L., Pétrilli V., Gross O., Tschopp J., and Evans T. J. (2010) The role of potassium in inflammasome activation by bacteria. J. Biol. Chem. 285, 10508–10518 10.1074/jbc.M109.067298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kamath K. S., Pascovici D., Penesyan A., Goel A., Venkatakrishnan V., Paulsen I. T., Packer N. H., and Molloy M. P. (2016) Pseudomonas aeruginosa cell membrane protein expression from phenotypically diverse cystic fibrosis isolates demonstrates host-specific adaptations. J. Proteome Res. 15, 2152–2163 10.1021/acs.jproteome.6b00058 [DOI] [PubMed] [Google Scholar]
- 85. Guina T., Purvine S. O., Yi E. C., Eng J., Goodlett D. R., Aebersold R., and Miller S. I. (2003) Quantitative proteomic analysis indicates increased synthesis of a quinolone by Pseudomonas aeruginosa isolates from cystic fibrosis airways. Proc. Natl. Acad. Sci. U.S.A. 100, 2771–2776 10.1073/pnas.0435846100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Babin B. M., Atangcho L., van Eldijk M. B., Sweredoski M. J., Moradian A., Hess S., Tolker-Nielsen T., Newman D. K., and Tirrell D. A. (2017) Selective proteomic analysis of antibiotic-tolerant cellular subpopulations in pseudomonas aeruginosa biofilms. MBio 8, e01593–17 10.1128/mBio.01593-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Crouzet M., Claverol S., Lomenech A. M., Le Sénéchal C., Costaglioli P., Barthe C., Garbay B., Bonneu M., and Vilain S. (2017) Pseudomonas aeruginosa cells attached to a surface display a typical proteome early as 20 minutes of incubation. PLoS One 12, e0180341 10.1371/journal.pone.0180341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Ito T., Murai M., Ninokura S., Kitazumi Y., Mezic K. G. G., Cress B. F. F., Koffas M. A. G. G. A. G., Morgan J. E. E., Barquera B., and Miyoshi H. (2017) Identification of the binding sites for ubiquinone and inhibitors in the Na+-pumping NADH-ubiquinone oxidoreductase from Vibrio cholerae by photoaffinity labeling. J. Biol. Chem. 292, 7727–7742 10.1074/jbc.M117.781393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Lümmen P. (1998) Complex I inhibitors as insecticides and acaricides. Biochim. Biophys. Acta 1364, 287–296 10.1016/S0005-2728(98)00034-6 [DOI] [PubMed] [Google Scholar]
- 90. Wittig I., and Schägger H. (2005) Advantages and limitations of clear-native PAGE. Proteomics 5, 4338–4346 10.1002/pmic.200500081 [DOI] [PubMed] [Google Scholar]
- 91. Schägger H. (2002) Respiratory chain supercomplexes of mitochondria and bacteria. Biochim. Biophys. Acta 1555, 154–159 10.1016/S0005-2728(02)00271-2 [DOI] [PubMed] [Google Scholar]
- 92. Segel I. H. (1993) Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady-state Enzyme Systems, 2nd Ed., p. 957, John Wiley & Sons, Inc., New York [Google Scholar]
- 93. Sali A., and Blundell T. L. (1993) Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 10.1006/jmbi.1993.1626 [DOI] [PubMed] [Google Scholar]
- 94. Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., and Lipman D. J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Lobley A., Sadowski M. I., and Jones D. T. (2009) pGenTHREADER and pDomTHREADER: new methods for improved protein fold recognition and superfamily discrimination. Bioinformatics 25, 1761–1767 10.1093/bioinformatics/btp302 [DOI] [PubMed] [Google Scholar]
- 96. Irwin J. J., Sterling T., Mysinger M. M., Bolstad E. S., and Coleman R. G. (2012) ZINC: a free tool to discover chemistry for biology. J. Chem. Inf. Model. 52, 1757–1768 10.1021/ci3001277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Cramer C. J., and Truhlar D. G. (1992) AM1-SM2 and PM3-SM3 parameterized SCF solvation models for free energies in aqueous solution. J. Comput. Aided Mol. Des. 6, 629–666 10.1007/BF00126219 [DOI] [PubMed] [Google Scholar]
- 98. Lee J., Cheng X., Swails J. M., Yeom M. S., Eastman P. K., Lemkul J. A., Wei S., Buckner J., Jeong J. C., Qi Y., Jo S., Pande V. S., Case D. A., Brooks C. L., MacKerell A. D., et al. (2016) CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput. 12, 405–413 10.1021/acs.jctc.5b00935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Jo S., Lim J. B., Klauda J. B., and Im W. (2009) CHARMM-GUI membrane builder for mixed bilayers and its application to yeast membranes. Biophys. J. 97, 50–58 10.1016/j.bpj.2009.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Jo S., Kim T., and Im W. (2007) Automated builder and database of protein/membrane complexes for molecular dynamics simulations. PLoS One 2, e880 10.1371/journal.pone.0000880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Jo S., Kim T., Iyer V. G., and Im W. (2008) CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 10.1002/jcc.20945 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.