Abstract
The small ubiquitin-like modifier (SUMO) post-translationally modifies lysine residues of transcription factors and co-regulators and thereby contributes to an important layer of control of the activities of these transcriptional regulators. Likewise, deSUMOylation of these factors by the sentrin-specific proteases (SENPs) also plays a role in gene regulation, but whether SENPs functionally interact with other regulatory factors that control gene expression is unclear. In the present work, we focused on SENP1, specifically, on its role in activation of gene expression investigated through analysis of the SENP1 interactome, which revealed that SENP1 physically interacts with the chromatin remodeler chromodomain helicase DNA-binding protein 3 (CHD3). Using several additional methods, including GST pulldown and co-immunoprecipitation assays, we validated and mapped this interaction, and using CRISPR-Cas9–generated CHD3- and SENP1-KO cells (in the haploid HAP1 cell line), we investigated whether these two proteins are functionally linked in regulating chromatin remodeling and gene expression. Genome-wide ATAC-Seq analysis of the CHD3- and SENP1-KO cells revealed a large degree of overlap in differential chromatin openness between these two mutant cell lines. Moreover, motif analysis and comparison with ChIP-Seq profiles in K562 cells pointed to an association of CHD3 and SENP1 with CCCTC-binding factor (CTCF) and SUMOylated chromatin–associated factors. Lastly, genome-wide RNA-Seq also indicated that these two proteins co-regulate the expression of several genes. We propose that the functional link between chromatin remodeling by CHD3 and deSUMOylation by SENP1 uncovered here provides another level of control of gene expression.
Keywords: small ubiquitin-like modifier (SUMO), sumoylation, chromatin remodeling, chromatin regulation, transcription, transcription coregulator, ATAC-seq, CHD3, HAP1, SENP1, SUMO protease
Introduction
SUMOylation is a fundamental post-translational modification (PTM)3 involved in a broad range of cellular processes such as transcription, nuclear transport, signal transduction, and maintenance of genome integrity (1–4). A large fraction of nuclear proteins has been found to be SUMO-conjugated, in particular transcription factors and chromatin modifiers. In fact, 60–80% of all proteins involved in key nuclear functions (transcription factors, chromatin regulators, DNA damage-response factors, the spliceosome, or cell cycle regulators) were recently identified as SUMO target proteins (5). In the majority of studies of transcription, SUMOylation has been linked to repression (1, 6–8), although exceptions exist (9). Therefore, removal of SUMO conjugation is expected largely to be associated with transcriptional activation, implicating SUMO-proteases as putative players in gene activation. Our knowledge is limited of how they exert this role and within which interaction networks they operate.
The SUMO family (SUMO1–4) consists of small 10-kDa proteins that are conjugated to lysine residues in their target proteins using an enzymatic pathway consisting of E1, E2, and E3 enzymes, which resembles that of ubiquitinylation. SUMO1 is only 45% identical to SUMO2 and SUMO3, whereas the latter two are almost identical and are functionally undistinguishable. The vast majority of SUMO1 exists in conjugated species, whereas a free pool of SUMO2/3 species is usually found in cells. SUMO2/3 conjugation is strongly induced in response to in vivo heat shock and oxidative stress (1–4).
SUMOylated proteins may be part of dynamic and complex interaction networks. SUMO modifications are recognized, or “read,” by proteins harboring one or several SUMO interaction motifs (SIM). This noncovalent interaction depends on a short hydrophobic core flanked by acidic amino acids ((V/I)X(V/I)(V/I)aaa) (10).
In the SUMO system, SUMOylation is finely balanced by deSUMOylation (11–13). Proteolytic enzymes are responsible for the maturation of the SUMO precursor, for substrate deconjugation, and for depolymerizing SUMO2/3 chains (12). Six mammalian SUMO proteases have been described, SENP1–3 and SENP5–7 (12, 13). The C-terminal regions of SENP proteins encode a highly conserved catalytic domain, whereas their N-terminal domains are variable and appear to direct subcellular localization and substrate specificity (11–13).
The SUMO system is essential. Deletion of either SUMO-conjugating or -deconjugating genes resulted in embryonal lethality (14–16). Dysregulation of the SUMO system has been increasingly implicated in cancer and other diseases (17–19).
SENP1 is an essential gene; its knockout in mice is embryonic lethal, causing anemia between embryonic day 13.5 and postnatal day 1 (20). This fetal anemia probably stems from deficient erythropoietin production appearing because SENP1-mediated GATA1 deSUMOylation is critical for definitive erythropoiesis (15). SENP1 was also found to be essential for the development of early T and B cells (21). However, the full function of the SENP/ULP family within the cell has yet to be determined (22–24).
Transcription depends on a dynamic chromatin structure that can respond to changes in the regulatory inputs. This dynamic is controlled through PTMs of histones and other chromatin-associated factors. Among these, SUMOylation has gained increasing importance, playing a key role in the regulation of transcription and chromatin dynamics through a variety of mechanisms (6). However, despite extensive studies, our understanding of the role of SUMOylation in chromatin dynamics remains incomplete. Several chromatin-related mechanisms have been proposed to explain the repressive effect observed for a large number of factors as a consequence of their SUMOylation. SUMO appears to recruit a diversity of chromatin-modifying enzymes and chromatin-associated proteins, including the histone deacetylase HDAC2, the histone demethylase LSD1, the histone methyltransferase SETDB1, the nucleosome remodeling ATPase Mi-2, and chromatin-associated proteins HP1, L3MBTL1, and L3MBTL2 (25–27). It has been proposed that SUMO sets up a local repressive heterochromatin (26). Even if this process is only partially understood, we know even less when it comes to how this repression is abolished or relieved. The dampening effect of SUMOylation implies that in many cases relief from SUMO repression may represent an important contribution to the transcriptional activation caused by a transcription factor.
In this work, we focus on SUMO-proteases as putative players in gene activation. We reasoned that their role in gene activation might be clarified through their interactome. By screening for interaction partners of SENP1, we identified the chromodomain helicase DNA–binding protein 3 (CHD3), implicating a novel link between deSUMOylation and chromatin remodeling. We used CRISPR-Cas9–generated KO cells in combination with ATAC-Seq and RNA-Seq to uncover a functional link between the two proteins.
Results
SENP1 interacts with CHD3
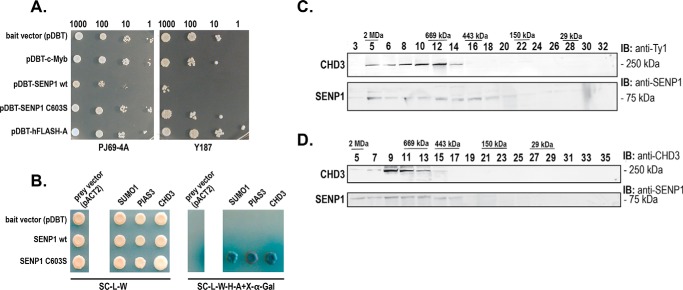
As a first step toward finding novel gene activation mechanisms dependent on SENP1, we performed a yeast two-hybrid screening of a human thymus cDNA library using a centromeric bait-plasmid to identify partners of SENP1. Even at the low expression level achieved with our single copy CEN-plasmid approach, SENP1 turned out to be toxic to yeast cells precluding ordinary screening (Fig. 1A). However, by using a protease-dead mutant of SENP1 (C603S), the toxic effects were minor, and we were able to perform a screening of 6 × 107 clones, of which 92 positive grew on various selective media. We sequenced the cDNA of 40 of the clones with the strongest blue color on 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal) plates. The interaction between SENP1 and selected prey was verified in yeast by retransformation and growth on reporter-selective media (Fig. 1B). Among the cDNAs with highest frequency in this screening, we found the chromatin remodeler CHD3 (Figs. 1B and 2A), which we selected for further studies because we thought CHD3 may represent an interesting novel link between the SUMO system, chromatin remodeling, and transcription.
Figure 1.
CHD3 is a candidate interaction partner of SENP1. A, toxicity test of the bait plasmid pDBT–hSENP1-WT and its C603S mutant derivative in the yeast strains Y187 and PJ69-4A on SC/−trp medium. Controls used were pDBT (empty vector), pDBT-hcM (encoding c-Myb (65)), and pDBT-hFlashA (66). The cells were incubated for 48 h at 30 °C. Number of cells/μl are indicated. Each spot represents 5 μl plated. B, validation by remating of selected positive cDNAs in the pACT2 vector (SUMO1, PIAS3, and CHD3), crossed with the indicated bait plasmids (in the pDBT vector). The left panel shows growth on the control plate (SC/−trp/−leu medium) selecting only for diploid a/α-cells containing both pDBT and pACT2 plasmids. The right panel shows growth on SC/−trp/−leu/−his/−ade/+X-α-Gal medium where growth and color depend on interaction. C, Superose-6 fractions of 3×Ty1–CHD3 K562 nuclear extract. The fractions were revealed with a rabbit polyclonal anti-SENP1 antibody and with a mouse anti-Ty1 mAb. D, Superose-6 fractions of 3×Ty1-Empty nuclear extract. The fractions were revealed with a rabbit polyclonal anti-CHD3 antibody and with a rabbit polyclonal anti-SENP1 antibody.
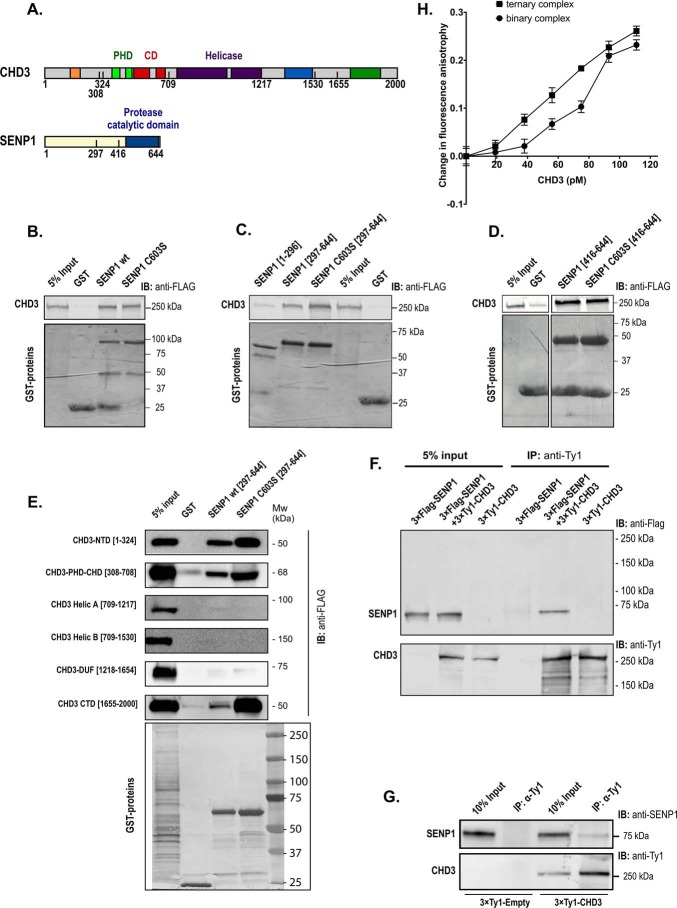
Figure 2.
CHD3 interacts with SENP1. A, human CHD3 and SENP1 are depicted with their domain structures. B–E, GST pulldown binding assays were performed with different GST protein domains and 3×FLAG–CHD3 from transfected COS-1 cells. The GST fusion proteins used were full length versions of SENP1 (panel B), and the indicated deletions of SENP1 (panels C, D, and E). In panels B, C, and D, binding of the GST proteins to full-length CHD3 was monitored, while binding to deletions of 3×FLAG-CHD3 was monitored in panel E. 24 h after transfection, the COS-1 cells were lysed in KAc-interaction buffer, and the lysates were incubated with comparable amounts of the different GST fusion proteins bound to GSH beads. The bound proteins were separated by SDS-PAGE, and the immunoblot was analyzed using anti-FLAG antibody (1:10,000) and LI-COR IRDye 680RD anti-mouse secondary antibody (1:10,000). 5% of total cell extract used for each pulldown was loaded as reference. The upper panels show the Western blots (anti-FLAG) for CHD3, and the lower panel shows the Coomassie-stained gel of the indicated GST fusion proteins. B and C are derived from the same experiment and the same gel with the common input and GST controls placed in the middle. Therefore, the controls of B are re-used in C. F, co-immunoprecipitation of SENP1 with CHD3. COS-1 cells were transfected with the indicated combinations of pCIneo-3×FLAG-SENP1 and pEF1–3×Ty1–CHD3. Whole-cell lysates were immunoprecipitated (IP) with anti-Ty1 antibody and separated by SDS-PAGE, and SENP1 was revealed by immunoblotting (IB) using anti-FLAG antibody. 5% of total transfected cell lysate was loaded as input reference. G, co-immunoprecipitation at endogenous levels of SENP1 with 3×Ty1–CHD3. The K562 nuclear extract from 3×Ty1–CHD3 (clone H6) was incubated with protein A magnetic beads coupled to an anti-Ty1 mAb (right). A nuclear extract from a 3×Ty1–Empty stable K562 cell line served as control (left). From a measure of the pixels in the bands in the upper panel, we estimated that in the right part from 3×Ty1–CHD3-expressing cells, 1.7% of the total input with endogenous SENP1 was found in the anti-Ty1 precipitate (pixels in 4th lane = 17% in 3rd lane). This is in contrast to the left control pair from 3×Ty1 empty cells where the co-IP band was only 0.01% of the total input. H, change in fluorescence anisotropy of SUMO1–AMC in complex with increasing amounts of recombinant full-length CHD3–(1994-SIMmutant) in the absence (binary complex) or presence (ternary complex) of recombinant SENP1-(C603S)(297–644). The anisotropy values were measured in the presence and absence of added protein, and the difference was plotted as indicated. In the ternary complex binding curve, the fixed concentration of SENP1-(C603S)(297–644) was 580 pm. This fixed concentration of SENP1 used for the ternary complex curve was based on a separate titration of SENP1-(C603S)(297–644) to SUMO1–AMC, where a concentration of SENP1 well below saturation was selected.
But before we embarked on a further analysis, given that the interaction was observed in yeast cells, we performed size fractionation of nuclear extracts from K562 cells. Using Superose-6 fractionation of nuclear extracts from a derived K562 cell line expressing 3×Ty1-tagged CHD3 and a control cell line, we observed in both cases that endogenous SENP1 migrated in high-molecular weight fractions supporting its association with larger proteins or complexes (Fig. 1, C and D). CHD3 eluted as a high-molecular weight complex as expected from its association with NuRD-type complexes (28).
To confirm the interaction between SENP1 and CHD3 by independent methods, we first performed GST-pulldown assays using full-length GST–SENP1 WT and C603S mutant. The purified fusion proteins were incubated with lysate from COS-1 cells transfected with an expression plasmid for full-length CHD3. As shown in Fig. 2B, CHD3 was retained on GST–SENP1 (WT and mutant) but not on the GST control. To map in more detail the interacting regions, we performed GST pulldown assays with deletion constructs of both SENP1 and CHD3. The catalytic domain of SENP1(297–644) bound more strongly to CHD3 than its N-terminal regions (amino acids 1–296) (Fig. 2C). It appears that the minimal catalytic domain of SENP1(416–644) is sufficient for strong interaction of SENP1 with CHD3 (Fig. 2D). When GST fusions of SENP1(297–644) were used to retain subdomains of CHD3 expressed in COS-1 cells, we observed more than one region of CHD3 binding to SENP1. Both the N-terminal domain (CHD3(1–324)), the PHD-chromodomain region (CHD3(308–708)) and the C-terminal CTD-region (CHD3(1655–2000)) were retained by GST–SENP1(297–644) (Fig. 2E). In contrast, the central ATPase domain of CHD3(709–1217) and the DUF region of CHD3(1218–1654) showed no affinity for SENP1.
An additional line of evidence for the CHD3-SENP1 interaction was provided by co-immunoprecipitation assays, first using lysates from COS-1 cells transfected with 3×FLAG-SENP1 and 3×Ty1–CHD3. As shown in Fig. 2F, SENP1 was co-immunoprecipitated with CHD3 (anti-Ty1 antibody). Control transfections indicated that this interaction is specific. We then used a K562 cell line stably expressing 3×Ty1–CHD3 to perform co-immunoprecipitation assays under more stringent, semi-endogenous conditions. In K562 nuclear extracts from this stable cell line, endogenous SENP1 was co-immunoprecipitated with CHD3 as revealed by a polyclonal rabbit anti-SENP1 antibody (Fig. 2G). The level of expression of 3×Ty1–CHD3 in this stable cell line was close to the endogenous level of CHD3 in the parental K562 cells, supporting an interaction at physiological levels of both SENP1 and CHD3 (Fig. S1).
Fluorescence anisotropy experiments with a fluorescently labeled SUMO1 (SUMO1–AMC) gave evidence for the formation of a CHD3–SENP1–SUMO1 ternary complex in solution (Fig. 2H). CHD3 has been reported to bind SUMO1 (29). Hence, we monitored the binding of SUMO1–AMC to CHD3 as fluorescence anisotropy. Upon increasing concentrations of CHD3, we observed the expected change in anisotropy (Fig. 2H). If CHD3 in addition were binding to SENP1, we expected an increase in anisotropy because of the increased size of the ternary complex. To avoid too strong a binding of SUMO1 to CHD3, we made use of a CHD3 mutant where its SUMO interaction motif close to its C terminus was mutated (amino acid positions 1994–1997 changed from VICI to AACA). We also used a catalytic dead mutant of SENP1 (C603S) to avoid cleavage of the AMC fluorophore under these conditions, and we used only the catalytic domain of SENP1(297–644), binding CHD3 strongly (Fig. 2C). The shift in the CHD3-titration curve upon addition of a fixed amount of SENP1 suggests that SUMO1–AMC was binding to a larger complex when CHD3 and SENP1 were combined compared to when CHD3 was titrated alone, supporting the interaction of the two proteins in solution.
Because CHD3 operates on chromatin, we tested whether the remodeler was able to recruit SENP1 to chromatin. For this purpose, we performed ChIP experiments in the HEK293-c1 cell line (26) transfected with a Gal–CHD3 fusion and monitored the occupancy of SENP1 both at the Gal–CHD3-bound site (5×Gal4 response-element promoter) and at a more distant locus (NCOA5 intron) serving as background reference. SENP1 was not recruited to any of these sites in the presence of the control Gal4 DNA-binding domain (GBD) only. However, in presence of the Gal–CHD3 fusion protein, which becomes directed to the Gal4-responsive promoter, the recruitment of SENP1 was evident (Fig. 3). These observations are in line with the interaction experiments and consistent with the hypothesis that SENP1 and CHD3 interact in a chromatin context where SENP1 becomes recruited to chromatin-associated CHD3. Taken together, these independent interaction assays all supported the validity of an interaction between SENP1 and CHD3 both in vitro and in vivo involving the catalytic domain of SENP1 and regions of CHD3.
Figure 3.
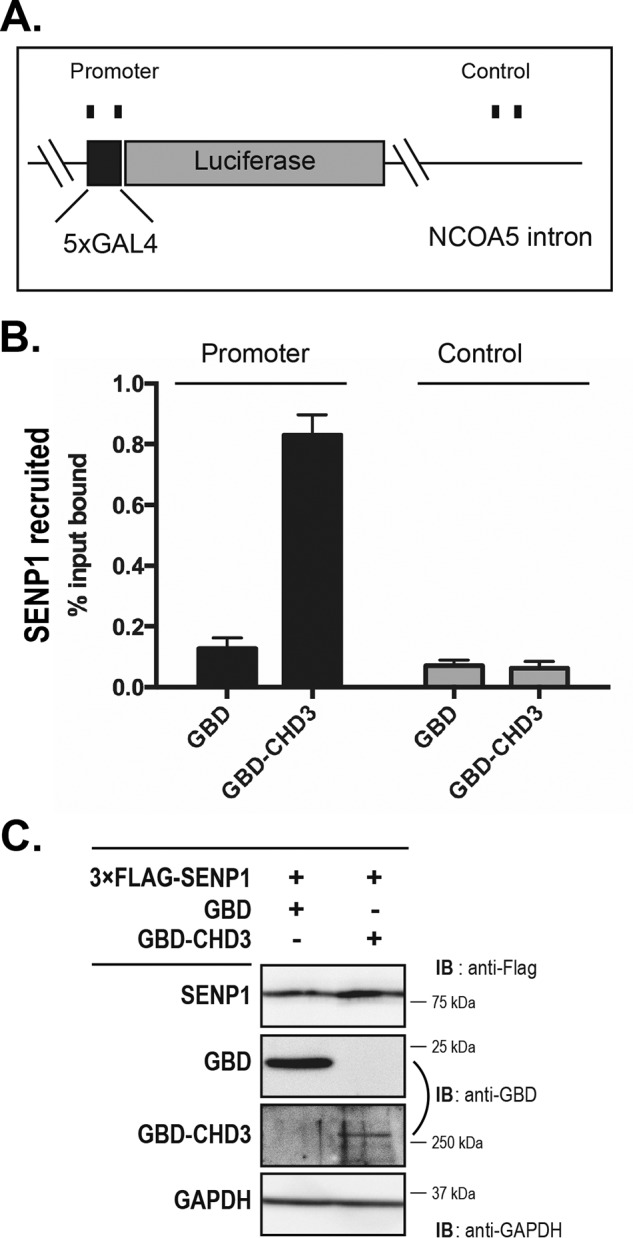
CHD3 recruits SENP1 to the chromatin. ChIP assays were performed in the HEK293-c1 cell line containing an integrated 5×GAL4-Luciferase gene (26). Cells were transfected with plasmids encoding the Gal4 DNA-binding domain (GBD), GBD–CHD3, and 3×FLAG–SENP1 in the combinations indicated. A, illustration showing the genomic loci in the NCOA5 gene where the 5×GAL4-Luciferase gene is integrated. B, ChIP assay against 3×FLAG-SENP1. The mean of three biological replicates is shown, with error bars showing standard error of the mean. C, representative immunoblot of the transfections.
SENP1 and CHD3 affect shared loci in the genome
To address the functional implications of the interaction between SENP1 and CHD3, we focused first on the effects on chromatin features. We reasoned that because CHD3 is a chromatin remodeler, the accessibility of some loci should be affected by loss of CHD3. Furthermore, if CHD3 and SENP1 interact in a way that affects the function of CHD3, there should also be some shared loci where accessibility is affected by loss of each of the two proteins. For this purpose, we made use of HAP1 cells in which the genes for either SENP1 or CHD3 were inactivated by CRISPR-Cas9–generated frameshift mutations into the coding sequence of the respective genes. HAP1 is a near-haploid human cell line that was derived from the male chronic myelogenous leukemia (CML) cell line KBM-7 (30). The characterization of the KO cell lines showed that they both lacked the full-length proteins (Fig. S2). However, they appear to express a reduced amount of a truncated protein (20% for the truncated CHD3, less for the truncated SENP1), suggesting that there is an alternative start codon downstream of the primary ATG or that an alternatively spliced transcript is formed that does not include the edited exon. Hence, the KO cell lines used here may be regarded as a KO for the full-length protein but a knockdown for the total pool of proteins caused by expressed isoforms.
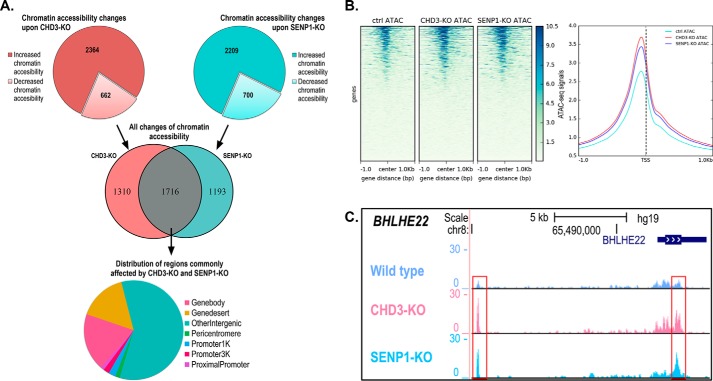
The WT reference cell line and the two KO HAP1 cell lines (CHD3-KO and SENP1-KO) were analyzed genome-wide for chromatin accessibility using the ATAC-Seq method. A total of about 60,000 ATAC signals were found in each of the three cell lines. (For details on statistical correlation of ATAC-Seq replicates, see Fig. S3.) Fig. 4A shows an overview of the differential chromatin accessibility based on the number of sites that exhibit a ≥50% fold change in ATAC signals in the KO cell lines relative to the control WT cell line. Using this threshold, the CHD3-KO cells showed differential peaks at 3026 sites, a majority of which showed increased chromatin accessibility after knockdown (2364), in line with a main role of CHD3 as a repressor of transcription. However, a significant fraction showed decreased chromatin accessibility (662), suggesting that CHD3 may also contribute to open chromatin at specific sites. Because the role of SENP1 on chromatin is less well characterized, the expected profile for the SENP1-KO was not obvious. However, it turned out to be very similar to that of CHD3-KO. The SENP1-KO cells showed differential peaks at 2909 sites, a majority of which showed increased chromatin accessibility after knockdown (2209), suggesting a main role of SENP1 at chromatin as restricting openness. However, also in this case, a significant fraction showed decreased chromatin accessibility (700). Finally, and most important, was the observation of a significant overlap between the differentially accessible regions (1716 sites) between SENP1-KO (both increased and decreased) and CHD3-KO (both increased and decreased). The most frequent type of genomic region affected by both deletions was “other intergenic” in which most enhancers are found, but also regions in the “gene body” and the different “promoter” categories were significant, representing together about 24% of the sites and being associated with specific genes. When we centered all the differential ATAC peaks according to the transcription start sites (TSS) of the closest gene, the resulting heatmap of the average ATAC-Seq signals showed an increased openness in the two KO strains around the TSS with a peak just upstream of the TSS (Fig. 4B). This suggests that for the subfraction of ATAC-Seq peaks that are close to a TSS, both CHD3 and SENP1 appear to affect the nucleosome-free regions of promoters. We show examples of this effect, where in the CHD3-KO and SENP1-KO cell lines chromatin is more open next to a TSS (BHLHE22, SERTAD4, CTGF, and TFPI2), and one case represents the less frequent opposite effect (RHOB) (Fig. 4C and Fig. S4). This suggests that CHD3 is involved in keeping a significant fraction of loci closed in a way that also depends on associated SENP1.
Figure 4.
Chromatin accessibility changes assayed by ATAC-Seq. A, diagrams show the regions that exhibit ≥50% fold change upon CHD3-KO and SENP1-KO with increased and decreased chromatin accessibility indicated. The Venn diagram shows the overlapping differentially accessible regions (n = 1716) between SENP1-KO (both increased and decreased) and CHD3-KO (both increased and decreased). The pie chart shows the distribution of genomic regions for the shared changes in chromatin accessibility (n = 1716). Differential chromatin accessibility was obtained by comparing with the HAP1 control cell lines. B, heatmap showing the average ATAC-Seq signal centered on the TSS of the nearest genes. Ensembl human reference genome annotation (GRCh37 release 87) was used as regions for calculating enrichment of the ATAC signal at and around the TSS. The line plot shows the intensity of the centered average ATAC signal at and ±1 kb around the TSS. The heatmap and line plot were made using deepTools2 (67). C, example ATAC track for the BHLHE22 locus showing ATAC-Seq signals for CHD3-KO and SENP1-KO as well as control cell lines. Visualization of the tracks were made using the UCSC genome browser (68).
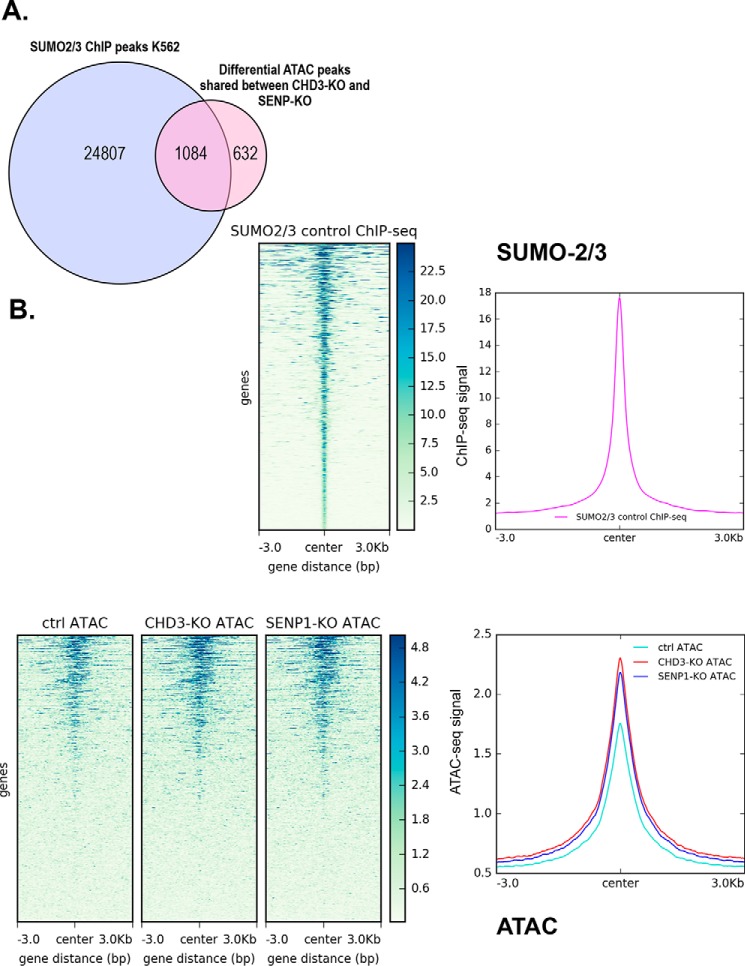
Unfortunately, there are no public genome-wide ChIP-Seq data on HAP1 cells. To evaluate the relevance of our ATAC profiling, we took advantage of the fact that both HAP1 cells and K562 cells are CML-derived cell lines, so we reasoned that ChIP-Seq data from K562 might be used as a proxy to evaluate whether the ATAC sites found in HAP1 cells correlate with relevant ChIP-Seq profiles. Unfortunately, genome-wide ChIP-Seq data for CHD3 and SENP1 are lacking. However, we reasoned that an enzyme needs a substrate, so the location of SENP1 along chromatin might show a correlation with the location of SUMO associated with chromatin. Niskanen et al. (31) analyzed chromatin occupancy of SUMO-2/3–modified proteins in K562 cells using ChIP-Seq and concluded that heat-shock SUMOylation targets promoters and enhancers of actively transcribed genes, thereby restricting the transcriptional activity of the induced genes. When we compared the 1716 shared ATAC peaks with the SUMO-2/3 peaks from K562 (control, not heat shock), a majority (1084 peaks, i.e. 63%) of the regions overlap with SUMO-2/3 peaks in K562 cells (Fig. 5). The overlap shown in Fig. 5A is based on shared peak annotations localizing to specific genes. A comparable result was obtained with another calculation based on physical overlaps of peaks (Fig. S5). When we used the SUMO data and centered all the ATAC peaks according to the SUMO2/3 peaks found in K562 cells, the resulting heatmap of the average ATAC-Seq signals showed an increase in the ATAC profile centered over the SUMO peaks (Fig. 5). Given that HAP1 cells and K562 cells are not identical, we find this overlap indicative of a link between the fraction of SENP1 and of SUMO associated with chromatin.
Figure 5.
Correlation between open regions affected by CHD3 and SENP1 and chromatin-associated SUMO. A, Venn diagram showing number of overlapping regions (n = 1084) between the ATAC-Seq subgroup with shared differentially accessible regions (n = 1716) and SUMO2/3 ChIP-Seq peaks from K562 (GSE66448). The overlapping regions were obtained by annotating SUMO2/3 peaks and the jointly affected differential ATAC regions using the HOMER program annotatePeaks.pl (32). The annotated files were then used to obtain intersections of peak regions that are annotated to promoter ID of the nearest gene. B, heatmap showing the average ATAC-Seq signal centered on SUMO2/3 binding sites along with line plot, showing the intensity of the centered average ATAC signal in HAP1 cells. Similarly, a heatmap showing average SUMO2/3 ChIP-Seq signal from K562 centered on SUMO2/3-binding sites along with a line plot showing the intensity of the centered average ChIP-Seq signal is displayed. Public ChIP-Seq data for SUMO2/3 in K562 (GSE66448) (31) was used as regions for calculating enrichment of the ATAC signal at and ±3 kb around SUMO2/3-binding sites. Heatmaps and line plots were made using deepTools2 (67).
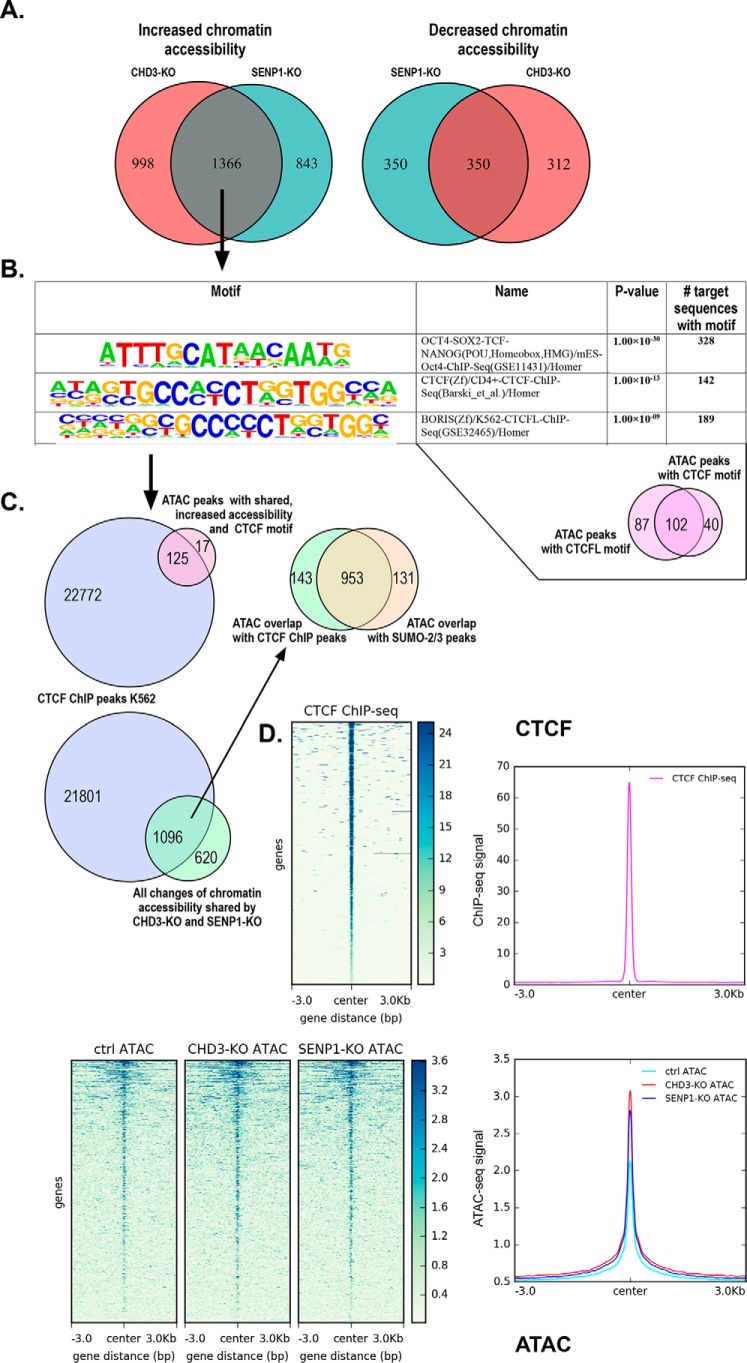
As expected from the differential effect of each KO (Fig. 4A), also the shared 1716 peaks were dominated by regions showing increased chromatin accessibility after knockdown (1366 sites) compared with decreased chromatin accessibility (350 peaks) (Fig. 6A). To better characterize the regions dependent on both CHD3 and SENP1 for being closed, we performed a motif analysis of the 1366 sites with increased chromatin accessibility using the HOMER motif discovery algorithm, searching for enrichment of known motifs (32). The argument is that because neither CHD3 nor SENP1 are sequence-specific DNA-binding proteins; they probably are recruited to specific sites through association with transcription factors. Among the list of enriched motifs (Fig. 6B), the two with the highest p values (estimated from the frequency of the found known motif in the target set relative to the background frequency of the same motif in the background set) were the following: 1) motifs for the pluripotency factors OCT4–SOX2–TCF–NANOG (328 sites, p = 1 × 10−30), and 2) motifs for CTCF (142 sites, p = 2 × 10−13) and for CTCFL/BORIS (189 sites, p = 1 × 10−9), a paralog of CTCF having a DNA-binding specificity identical to that of CTCF (33). The latter is reflected in 102 sites being shared between CTCF and CTCFL in our dataset. An extended list is shown in Fig. S6.
Figure 6.
Correlation between open regions affected by CHD3 and SENP1 and chromatin-bound CTCF. A, Venn diagram showing the number of overlapping peaks (n = 1366) between differential ATAC peak regions in SENP1-KO and CHD3-KO having increased chromatin accessibility (left). The right Venn diagram shows the same overlap for differential ATAC peak regions with decreased chromatin accessibility (n = 350). B, motif enrichment of the subgroup with shared increased chromatin-accessible regions (n = 1366). The specific binding motifs are arranged in descending order based on their p value. Motif analysis around peak regions of the shared differential ATAC subgroup with increased chromatin accessibility (n = 1366) was done using the HOMER program (32). The small Venn diagram below shows the overlap of motif instances (n = 102) between two of the most significantly known motifs enriched among the shared differential ATAC subgroup (n = 1366). Motif regions with CTCF binding (n = 142) and with CTCFL binding (n = 189) was used to calculate the overlapping regions. C, Venn diagram showing the number of overlapping regions (n = 125) between motif regions with CTCF binding (n = 142), extracted from enriched motifs above, and CTCF ChIP-Seq peaks from K562 (ENCSR000EGM). The lower Venn diagram shows overlapping regions (n = 1096) between the same CTCF ChIP-Seq peaks from K562 and the ATAC-Seq subgroup with shared differentially accessible regions (n = 1716). The upper right Venn diagram shows the overlapping regions (n = 953) between the subset that is shared between ATAC differential accessible regions and CTCF peaks above (n = 1096) and that of SUMO2/3 peaks (GSE66448) (n = 1084) (from Fig. 5A). The overlapping regions were obtained by annotating the relevant files (i.e. extracted motif instances from ATAC peaks, public CTCF peaks, and the jointly affected differential ATAC regions) using the HOMER program annotatePeaks.pl (32). The annotated files were then used to obtain the corresponding pairwise intersections of peak regions that are annotated to promoter ID of the nearest gene. A separate analysis of the pairwise intersection using a different tool representing physical overlaps is shown in Fig. S5. D, heatmap showing the average ATAC-Seq signal centered on CTCF-binding sites along with a line plot showing the intensity of the centered average ATAC signal. Similarly, a heatmap showing average CTCF ChIP-Seq signal centered on CTCF-binding sites along with a line plot showing the intensity of the centered average ChIP-Seq signal. Public ChIP-Seq data for CTCF in K562 (ENCSR000EGM) from ENCODE was used as regions for calculating enrichment of the ATAC signal at and ±3 kb around CTCF-binding sites. Heatmaps and line plots were made using deepTools2 (67).
To evaluate the relevance of these motifs, we again took advantage of the fact that both HAP1 cells and K562 cells are CML-derived cell lines and that CTCF occupancy often is conserved between different cell types (34–36). Hence, we compared the 142 sites affected by both SENP1 and CHD3 KO in HAP1 cells (and harboring a CTCF motif) with available ChIP-Seq profiles for CTCF from K562 cells. As shown in the Venn diagram in Fig. 6C (upper left) and illustrated by example profiles in Fig. S7, the vast majority (i.e. 88%) of the 142 regions are in fact occupied by CTCF in K562 cells. When all the 1716 shared ATAC peaks were compared with CTCF ChIP peaks in K562 (Fig. 6C, lower diagram), again a high percentage (1096 peaks, 64% of the total) overlapped, suggesting a significant association between CTCF and the sites where CHD3 and SENP1 cooperate. The overlap shown in Fig. 6C is based on shared peak annotations localizing to specific genes. A comparable result was obtained with another calculation based on physical overlaps of peaks (Fig. S5). Of note, 87% of the 1096 overlapping peaks also shared a SUMO-2/3 ChIP peak in K562 cells (Fig. 6C, upper right), in line with an association between SENP1 and SUMO at chromatin. When all the ATAC peaks were centered according to the CTCF peaks found in K562 cells, the resulting heatmap of the average ATAC-Seq signals showed an increase in the ATAC profile centered over the CTCF peaks (Fig. 6D). We also performed a “functional term analysis” of this subgroup (i.e. genes closest to the ATAC peak regions of the 142 sites) and saw an enrichment of various processes, in particular linked to immature B cell differentiation and regulation of mRNA splicing (Fig. S8). Taken together, this suggests that one important group of regions where CHD3 and SENP1 operate together is CTCF-occupied sites.
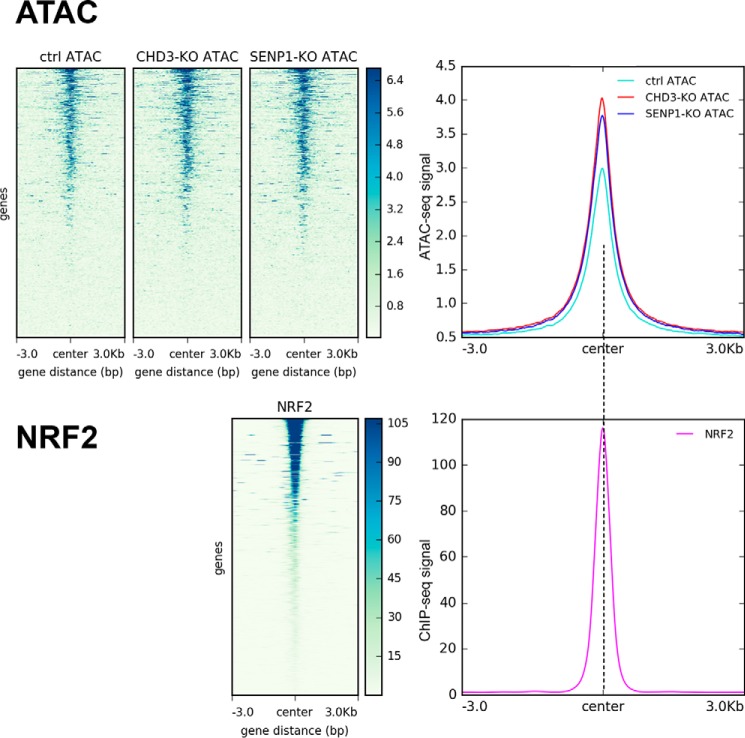
A similar analysis of the link to the motifs with the highest p value was not possible because the associated factors are not expressed in K562 cells, although at least SOX2 is expressed in HAP1. OCT4, SOX2, and NANOG are pluripotency factors expressed in embryonic stem cells. However, we compared some factors on the list that are well expressed in K562, NRF2, NF-E2, and Bach1. The HOMER analysis revealed 58 peaks (p = 0.007) with motifs for NRF2/GABPA (EntrezGene ID:2551) (70). The ATAC peaks centered clearly around regions enriched for binding of this factor (Fig. 7). A similar comparison with NF-E2 (59 sites, p = 0.02), also highly expressed in K562 (EntrezGene ID:4778) (70), however, showed only a weak, less convincing correlation (Fig. S9). The same poor correlation was true for Bach1 (63 peaks, p = 0.002) (Fig. S10). The latter illustrates that the ATAC peak regions are found enriched only at a selection of open sites.
Figure 7.
Correlation between open regions affected by CHD3 and SENP1 and chromatin-bound NRF2. Upper panel, heatmap showing average ATAC-Seq signals centered on NRF2-binding sites along with a line plot showing the intensity of the centered average ATAC signal. Lower panel, similar heatmap showing average NRF2 ChIP-Seq signals centered on NRF2-binding sites along with a line plot showing the intensity of the centered average ChIP-Seq signal. Public ChIP-Seq data for NRF2 in K562 (ENCSR290MUH) from ENCODE was used as regions for calculating enrichment of the ATAC signal at and ±3 kb around NRF2-binding sites. Heatmaps and line plots were made using deepTools2 (67).
In addition to genome-wide comparison of our ATAC-Seq data to publicly available ChIP-Seq data for different factors, we also took advantage of DNase-Seq data in HAP1 cells (ENCSR620QNS), which is the only public data set to date that is generated using HAP1 cells. Similar comparisons were done, where we centered ATAC-Seq signals on DNase-Seq peaks from HAP1 cells. The resulting comparison revealed as expected enrichment of ATAC-Seq signals on DNase-Seq peaks (Fig. S11).
SENP1 and CHD3 affect the expression of shared genes in the genome
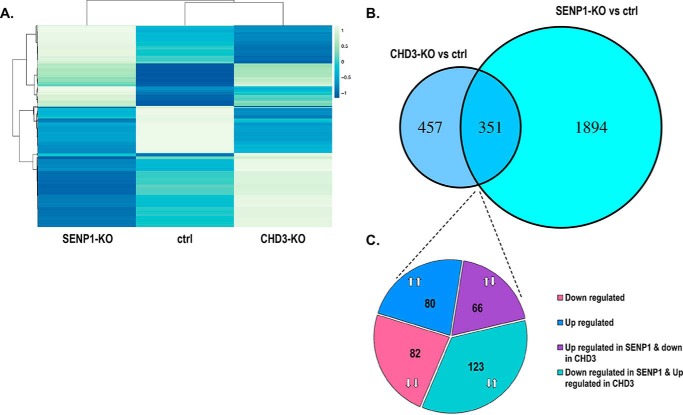
To further analyze the functional implications of the interaction between SENP1 and CHD3, we reasoned that their shared effect on accessibility of many regions would imply a shared effect on the expression of some genes. We therefore performed RNA-Seq analysis of the same variants of the HAP1 cells as used for the ATAC-Seq analysis. Hence, RNA was isolated from three independent biological replicates of SENP1-KO, CHD3-KO, and WT reference HAP1 cells, delivered to high-throughput sequencing and subjected to downstream statistical processing. Transcriptome-wide fold change of differentially expressed genes for the knockout cell lines relative to the control is presented in volcano plots (Fig. S12). Analysis of the sequencing data revealed that SENP1-KO had a significant change in the expression of 2245 genes, whereas the CHD3-KO had a significant change in the expression of 808 genes (Fig. 8). 43% of the latter (351 genes) were also among those affected in SENP1-KO cells, supporting the hypothesis that SENP1 affects the function of CHD3 at a significant fraction of genes. SENP1 clearly also affects a large number of genes through other mechanisms than through CHD3. We noticed that all combinations of up- and down-regulation were found in comparable numbers among the shared genes, probably because both direct and indirect effects are to be expected in this type of analysis. We assume that this is the reason why the RNA-Seq analysis gave a more diverse pattern than the ATAC-seq. One may argue that opposite expression patterns are more likely to be caused by indirect effects. In support of this, we observed a higher correlation between genes associated with ATAC-Seq peaks and genes with altered expression when we compared genes that were similarly regulated (both up- or both down-regulated) than for oppositely regulated genes. In the first case, visual inspection of the ATAC tracks close to each gene with altered expression identified that 40% of the latter had a significantly altered ATAC profile. The same analysis in the oppositely regulated group of genes showed only 13% correlation. However, because the majority of the differential ATAC peaks were found in locations (“other intergenic”) at larger distances from genes, it is difficult to assess precisely which genes are affected by the changes in chromatin openness.
Figure 8.
RNA-Seq changes in CHD3-KO and SENP1-KO HAP1 cells. A, heatmap showing hierarchical clustering of significant differentially regulated genes shared between SENP1-KO and CHD3-KO (n = 351), with each row representing a gene. The cluster heatmap was generated using ClustVis webtool (41). B, significant differentially regulated genes upon CHD3-KO and SENP1-KO and the intersection between them (n = 351). C, proportion of different subgroups among the shared affected genes by CHD3-KO and SENP1-KO; the number of shared genes that were both down-regulated (n = 82), both up-regulated (n = 80), up-regulated in SENP1-KO, and down-regulated in CHD3-KO (n = 66), and vice versa (n = 123). Differential gene expression was calculated using cuffdiff (69).
Finally, to get an idea of which functions were involved, we performed a gene ontology (GO) analysis of the shared genes. A quite diverse pattern of functions was found in the groups with opposite effects of the two knockouts. In the group of genes where SENP1-KO and CHD3-KO both were associated with down-regulation, we observed an enrichment for functions linked to growth and signaling. In the group of genes where SENP1-KO and CHD3-KO both were associated with up-regulation, we observed an enrichment for a signaling pathway, but also to histone modifications (Fig. S13).
Discussion
SUMO proteome analysis has revealed the involvement of SUMO in a wide range of nuclear functions, including transcription and chromatin remodeling (5, 37). As is well known from extensive studies of phosphorylation, acetylation, and methylation of nuclear proteins, the processes of de-modifications are equally important as the modification processes. Our objective in this study was to focus on the deSUMOylation process and seek a better understanding through the identification of novel interaction partners of the SENP1 enzyme. We observed that SENP1 from a nuclear extract of K562 cells migrates as a rather high-molecular weight entity (Fig. 1, C and D). Using a stringent version of the yeast two-hybrid method and a nontoxic mutant of SENP1, we identified the chromatin remodeler CHD3 as a novel interaction partner of SENP1 (Fig. 1B). The interaction was validated in vitro by GST-pulldown assays, co-immunoprecipitations, and fluorescence anisotropy titrations (Fig. 2). The interaction was also validated in vivo by a ChIP assay where we directed CHD3 to a specific locus through fusion to a Gal4–DNA-binding domain and observed that SENP1 became enriched at the same locus (Fig. 3). We also mapped the domains involved as the catalytic domain of SENP1 and at least two regions of CHD3, its C-terminal region and the N-terminal region, including its PHD and CHD domains (Fig. 2).
To test the functional relevance of this novel interaction, we took a global approach and used ATAC-Seq profiling of HAP1 cells in which either CHD3 or SENP1 were knocked out. The sites where the accessibility of chromatin depended on CHD3 largely overlapped with those where the accessibility of chromatin depended on SENP1, strongly supporting the functional relevance of their interaction (Fig. 4). A more indirect analysis of the transcriptomes of the KO cells still showed that a significant fraction of the genes affected by loss of CHD3 were also affected by loss of SENP1 (Fig. 8). We conclude that the SUMO system and the chromatin-remodeling system involving CHD3 are linked and operate together at specific chromatin loci.
Several links between the SUMO system and chromatin remodeling have already been reported for other remodelers, such as the yeast chromatin–remodeling Isw1a complex (38) and the INO80 chromatin–remodeling complex (39), both of which appear to be regulated by SUMOylation.
Our findings raise several questions on how SUMOylation and deSUMOylation processes cooperate with chromatin-remodeling processes. The simplest model for explaining our observations would be that CHD3 was SUMOylated and associated with SENP1 because of being a substrate. CHD3 is well known to be involved in SUMO binding, acting as a SUMO-dependent corepressor and contributing to the repressive effect often observed after SUMOylation of transcription factors (26, 27, 41). A well-studied example is the repressor protein KAP1, the SUMOylation of which is required for KAP1-mediated gene silencing. KAP1 seems to function by directly recruiting the SETDB1 histone methyltransferase and CHD3 via SUMO-interacting motifs (40). Less is known about SUMO modification of CHD3 itself. However, according to the proteome studies of Hendricks et al. (41), three SUMOylation sites were identified in CHD3 (Lys-627, Lys-1238, and Lys-1308), whereas eight sites were found in the paralog CHD4. If and how the activity of CHD3 is modulated through these modifications is not known. Recently, another SUMO conjugation site in CHD3 (Mi2α) was identified in Lys-1971 (42). Based on various functional studies, the authors concluded that the transcription factor hDREF might incite transcriptional activation by SUMOylating CHD3, resulting in its dissociation from chromatin. Combined with our observations, an attractive hypothesis would be that SENP1 through deSUMOylation of CHD3 would enhance its chromatin association. This hypothesis also implies that CHD3 would be less chromatin-associated in the absence of SENP1.
However, several of our interaction assays were performed with recombinant proteins that are unmodified, so the SENP1–CHD3 interaction is clearly not dependent on SUMO modification. It appears that CHD3 and SENP1 interact more strongly and specifically than a transient enzyme–substrate interaction. Another attractive hypothesis is based on the concept of “group SUMOylation” (43), assuming that SUMOylation controls the activity not only of individual proteins but also of entire complexes. Here, the scenario would be that when a locus needs to be remodeled, as it occurs during changes in gene programs, one may imagine a coordinated action where CHD3 is recruited to SUMOylated clusters in chromatin (possibly enhanced by SENP1), followed by remodeling the local chromatin. At the same time, SENP1 recruited by CHD3 will remove SUMO modifications of several proteins or complexes in the same region, leading to coordinated reprogramming.
The latter scenario is also interesting in light of our finding of a link to CTCF, where a significant subfraction of the chromatin sites that were affected by loss of both SENP1 and CHD3 were also enriched in CTCF-binding motifs. Combined with a correlation with K562 CTCF ChIP-Seq data, supporting actual binding of CTCF to these sites, this suggests that CHD3 and SENP1 operate together at some CTCF-occupied sites. Because CTCF is a key component of the structures that determine chromatin organization, one may even imagine a cooperation between CHD3 and SENP1 contributing to the regulation of chromatin architecture during processes where gene programs need to be modulated. Whether CHD3 or SENP1 is directly recruited to chromatin by CTCF is not known, but it has been reported that CHD7 and CHD8 in fact associate with CTCF (44, 45). Other chromatin remodelers have also been observed to operate and organize arrays of nucleosomes adjacent to CTCF-bound sites, promoting CTCF binding (46). Moreover, CTCF is itself SUMOylated at two major sites (47), affecting its transcriptional and chromatin opening activity (49, 50). This makes CTCF a putative substrate for SENP1. The functional implications of these interactions and cooperation require further studies.
The link between SUMO, CTCF, and chromatin architecture, including enhancer–promoter looping, will be a highly interesting focus for future research. A recent study by Whalen et al. (48), where they developed a computational method for reconstructing regulatory landscapes from diverse features along the genome, concluded that most of this signature is not proximal to the enhancers and promoters but instead decorates the looping DNA. In particular, they found that SUMOylation in the window between an enhancer and a promoter was a top predictor of interactions, nearly as important as CTCF (48).
We also found it intriguing that the most significant motif enrichment indicated a link to pluripotency factors or other transcription factors playing a role in development. The model here would be that a specific transcription factor recruits CHD3 to assist in remodeling the local chromatin, whereas SENP1 performs its group deSUMOylation leading to a coordinated action. Interestingly, the pluripotency factors are also pioneer factors with a key role in making a region accessible to other transcription factors (49). It is still a debate whether they perform this only by themselves or by recruiting chromatin remodelers (50). In the latter case, recruiting CHD3 would, according to our findings, also imply recruitment of SENP1 leading to deSUMOylation. An example of a transcription factor operating both as a pioneer factor and associated with CHD3 is c-Myb (51, 52).
In summary, the interaction between SENP1 and CHD3 studied in this work suggests a novel level of coordinated control of gene expression where chromatin remodeling and deSUMOylation cooperate at specific genomic loci. Our studies have pointed to unexplored links with deSUMOylation, chromatin remodeling, CTCF, SUMO, and pioneer factors. Future studies will be necessary to understand the molecular details of the cooperation between these nuclear systems.
Experimental procedures
Cell cultures
The following four cell lines were used: COS-1 (ATCC® CRL-1650TM Cercopithecus aethiops kidney); HEK293 (ATCC® CRL-1573TM Homo sapiens embryonic kidney); K562 (ATCC® CCL-243TM H. sapiens bone marrow, chronic myelogenous leukemia); and HAP1 cells (near-haploid cell line derived from the male CML cell line KBM-7, Horizon). HAP1 knockout (KO) cell lines for CHD3 and SENP1, and a parental control cell line were obtained from Horizon Discovery. The cells were grown at 37 °C and 5% CO2. The COS-1 and K562 cells were grown and transfected with the indicated plasmids as described previously (53, 54). The HAP1 cell cultures were maintained in Iscove's modified Dulbecco's medium containing 10% fetal calf serum and 1% penicillin/streptomycin. K562 cells stably expressing a tagged CHD3 variant (3×Ty1–CHD3) were selected by G418 for 2 weeks before selecting single clones for 1 week. Stable expression was confirmed by Western blotting.
Plasmid construction
The pDBT–hSENP1 WT and C603S derivative were constructed by subcloning a human SENP1 cDNA (53) into the pDBT vector. We discovered four point mutations in our original SENP1 cDNA that were repaired prior to designing the various SENP1-containing plasmids. The C603S mutant derivative of SENP1 was generated by QuikChange site-directed mutagenesis. The various GST–SENP1 fusion proteins were PCR-cloned into pGEX-6P2 vector using BamHI and NotI. Human CHD3 cDNA from the construct pCIneoB-3×FLAG–CHD3 (52) was transferred using SacII and NotI to the pEF1neo-3×Ty1 vector to express a 3×Ty1-tagged version of CHD3. The plasmid expressing a Gal4 fusion to CHD3 has been described (52). All constructs were verified by DNA sequencing.
Antibodies
For immunoblot detection, the following antibodies were used: mouse anti-FLAG M2 mAb (F3165, Sigma); mouse anti-proliferating cell nuclear antigen (PC-10, sc-56 Santa Cruz Biotechnology, Inc.); mouse anti-GAPDH (AM4300, Invitrogen); rabbit anti-GBD (06-62, Upstate Biotechnology); rabbit anti-CHD3 (ab114100 and ab109195, Abcam); rabbit anti-SENP1 (ab108981, Abcam); and from Li-Cor Biosciences, IRDye 800CW anti-mouse (926-32212); IRDye 680RD anti-mouse (926-68072); IRDye 680RD anti-rabbit (92668073); and IRDye 800CW anti-rabbit (926-32213). The anti-Ty1 monoclonal mouse antibody was produced in our laboratory from a hybridoma cell line (55).
Yeast two-hybrid screening
The two-hybrid screening was performed by mating as described previously (52, 56–58), using as bait the CEN plasmid pDBT–hSENP1–C603S pretransformed in the Y187 yeast strain and mated with the PJ69-4A strain pretransformed with a human thymus cDNA library. The interactions were confirmed by transformation of pDBT, pDBT–hSENP1, and pDBT–hSENP1–C603S and the rescued interaction candidates in pACT2 into yeast strains of opposite mating type followed by mating. Diploids were grown on yeast minimal medium lacking the selection amino acids or adenine.
GST-binding assay
GST and GST fusion proteins were expressed in Escherichia coli, as described previously (59). GST pulldown assays were performed essentially as described (52) but with the F-buffer replaced with a KAc-interaction buffer (150 mm KAc, 20 mm HEPES, pH 7.4, 10% glycerol, 0.2% Triton X-100, 1 mm DTT, 1× Complete protease inhibitor, Roche Applied Science) or with a NaCl-interaction buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 5 mm EDTA, 1% (v/v) Triton X-100, 1 mm DTT, 1 mm PMSF, and 1×Complete protease inhibitor), using total cell extracts from COS-1 cells transfected with the indicated expression constructs. The proteins retained on the GST-Sepharose beads were analyzed on a 4–15% Criterion SDS-polyacrylamide gradient gel. The membrane was blocked with LI-COR blocking solution (product no. 927-50010) for 1 h, probed with anti-FLAG antibody in LI-COR blocking solution overnight at 4 °C, and washed before being probed with LI-COR IRDye 680RD anti-mouse secondary antibody in LI-COR blocking solution for 1 h. After washing with TBS-T, the PVDF membrane was scanned with the LI-COR Odyssey® CLx Imaging System.
Co-immunoprecipitation
Co-immunoprecipitations were performed essentially as described (58) using the KAc-interaction buffer (150 mm KAc, 20 mm HEPES, pH 7.4, 10% glycerol, 0.2% Triton X-100, 1 mm DTT, 1× Complete protease inhibitor) and lysates from transfected COS-1 cells for immunoprecipitation. Proteins were separated by SDS-PAGE and detected with immunoblotting. SENP1 and CHD3 were detected with anti-FLAG and anti-Ty1 antibodies, respectively.
The endogenous co-immunoprecipitation of SENP1 was done with a nuclear extract of the K562 3×Ty1–CHD3 cell line. 500 μg of nuclear extract was incubated with 5 μg of anti-Ty1 antibody (mouse monoclonal), 10 μl of 10 mg/ml BSA, and 20 μl of protein A Dynabeads for 2 h at 4 °C with rotation. The beads were washed three times with 1 ml of BC300 + 0.05% Nonidet P-40 and once with 1 ml of BC100 + 0.05% Nonidet P-40. Proteins were separated by SDS-PAGE and detected with immunoblotting. SENP1 was detected with an anti-SENP1 rabbit polyclonal antibody and CHD3 with an anti-Ty1 mouse mAb.
Preparation of K562 nuclear extract
The methodology described in Wright et al. (60) was followed. K562 cells were grown to confluency and processed essentially as described. The clear supernatant after the ammonium sulfate precipitation was subjected to dialysis (rather than column desalting), against Buffer C (25 mm HEPES, pH 7.6, 150 mm NaCl, 12.5 mm MgCl2, 0.1 mm EDTA, 10% (v/v) glycerol, 1 mm DTT, 0.2 mm PMSF, and Complete protease inhibitors). The extract was finally centrifuged at 16,000 rpm for 5 min at 4 °C and flash-frozen in liquid nitrogen.
Superose-6 fractionation of K562 nuclear extracts
The Superose-6 column was equilibrated in BC200 buffer (25 mm HEPES, pH 7.6, 200 mm NaCl, 1 mm MgCl2, 0.5 mm EGTA, 0.1 mm EDTA, 10% (v/v) glycerol, 1 mm DTT, and 2 mm PMSF) filtered through a 0.2-μm membrane. 500 μl of the K562 nuclear extract were injected (flow rate 0.3 ml/min). The 0.5-ml fractions were frozen in liquid nitrogen and stored at −80 °C. Before analysis of the fractions by SDS-PAGE, they were precipitated with TCA to a final concentration of 20%, incubated for 10 min on ice, and centrifuged at 13,000 rpm at 4 °C for 10 min. The pellets were washed once with 1500 μl of ice-cold acetone. The protein pellets were dried at room temperature, resuspended in 1× SDS-PAGE loading buffer before separation on a 4–15% Criterion Gel, transferred to PVDF, and immunoblotted with the desired primary antibodies.
Expression and purification of 3×FLAG–CHD3 protein from COS-1 cells
Two 15-cm plates with COS-1 cells were transfected with 132 μg of pCIneoB–3×FLAG–CHD3 or pCIneoB–3×FLAG–CHD3–SIM1994–AACA constructs using polyethyleneimine (23966, Polysciences, Inc.). 24 h after transfection, the cells were washed and resuspended in 1.5 ml of BC100 + 0.05% Nonidet P-40. The cell suspension was sonicated (Up400s, Hielscher Ultrasonics GmbH) four times at 50% intensity for 15 s. The cell debris was removed (13,400 rpm for 20 min), and the supernatant was flash-frozen in liquid nitrogen and stored at −80 °C until purification.
The FLAG–CHD3 proteins were purified by affinity chromatography. First, 125 μl of FLAG M2-magnetic beads (M8823, Sigma) were added and incubated at 4 °C with rotation. The beads were washed once with 2 ml of BC1000 + 0.05% Nonidet P-40 and once with 2 ml of BC100 + 0.05% Nonidet P-40. The FLAG–CHD3 proteins were eluted with 100 μl of BC100 + 0.05% Nonidet P-40 + 0.5 mg/ml FLAG peptide (GenScript HK Ltd.). The eluted proteins were separated on a 4–15% Criterion Gel, transferred to a PVDF membrane, and detected with anti-FLAG antibody (mouse monoclonal).
Expression and purification of SENP1
Recombinant SENP1 was expressed as a GST fusion protein in E. coli BL21 cells using standard methods. The GST fusion tag was cleaved on column with 40 units of PreScission protease (27-0843-01, GE Healthcare) in the following buffer: 50 mm Tris-HCl, pH 7.0, 150 mm NaCl, 1 mm EDTA, and 1 mm DTT. The cleavage was done overnight at 4 °C. The cleaved protein was further purified by gel-filtration chromatography on a Superdex 75 column.
Protein–protein interaction studies using fluorescence anisotropy
The binding of SENP1 and CHD3 to SUMO1–AMC (UL-551, Boston Biochem) was followed with fluorescence anisotropy in the following buffer: 25 mm HEPES, 200 mm NaCl, 1 mm DTT, pH 7.6, using a PerkinElmer Life Sciences LS-50 fluorescence spectrometer with excitation at 380 nm and emission recorded at 460 nm. For binding of SENP1-(297–640) to SUMO1–AMC, the catalytic dead C603S mutant was used. For CHD3, we used the full-length FLAG-tagged protein (affinity-purified) but harboring a mutant in the SUMO1-interaction motif (SIM, at amino acids 1994–1997), with weaker SUMO binding that facilitated the titration. The binding of FLAG–CHD3 1994 SIM mutant proteins to SUMO1–AMC was measured at a SUMO1–AMC concentration of 200 pm.
ChIP
A HEK293 reporter cell line harboring a 5×GRE Gal4-luciferase reporter integrated transgene (26) was co-transfected with an HA-tagged Gal-fusion derivative of CHD3 and a 3×FLAG-tagged SENP1. The ChIP analysis was performed as described previously (61).
RNA-Seq and analysis
Total RNA was isolated from HAP1 cell lines using an RNA isolation kit (Qiagen-RNeasy mini kit). The quality and quantity of total RNA were determined using NanoDrop (ThermoFisher Scientific) and agarose gel electrophoresis, respectively. RNA samples were delivered for sequencing to the Norwegian sequencing center, Oslo, Norway, where libraries were prepared using strand-specific TruSeqTM library preparation kit. Transcriptomic data for the three cell lines with three biological replicates were generated using Illumina HiSeq 4000 sequencer, where 150-bp paired-end reads were obtained. The subsequent bioinformatics analysis is detailed in the supporting Materials and methods. Data have been made publicly available under GEO (accession number GSE111272).
ATAC sequencing and analysis
ATAC libraries for the HAP1 cell lines were made as described previously (62). Ad1 noMX and Ad2.1–2.12 bar-coded primers were used from Ref. 63. Crude nuclei were prepared from 150,000 cells per cell line, and the amount of all reagents used in the transposition assay were doubled relative to Ref. 62.
Background regions that are normally Tn5 transposase-accessible were obtained by isolating genomic DNA from HAP1 control cell lines using GenEluteTM mammalian genomic DNA miniprep kit (Sigma). This was followed by the transposition reaction and ATAC library preparation as described above. ATAC libraries were delivered to the Norwegian sequencing center, Oslo, Norway, where library quality was determined and sequenced. Three biological replicates were used to generate 40-bp paired-end reads using the Illumina NextSeq 500 sequencer. The bioinformatics analysis is detailed in the supporting Materials and methods. Data have been publicly available through GEO (accession number GSE111047).
Public datasets
ChIP-Seq datasets from K562 cells that were mapped to hg19 for CTCF (ENCSR000EGM), NRF2 (ENCSR290MUH), NFE2 (ENCSR552YGL), and BACH1 (ENCSR740NPG) were obtained from the ENCODE project (64). HAP1-derived DNase-Seq data (ENCSR620QNS) that are mapped to hg19 were obtained from the ENCODE project (64). K562-derived raw SUMO2/3 ChIP-Seq data (GSE66448) (31) were obtained from GEO. The SUMO2/3 ChIP-Seq sequencing files were processed the same way as the ATAC-Seq files, where peak calling was made using MACS2 with the parameters “-m 5 50 -bw 500 -fix-bimodal -extsize 200 call summits -B -q 0.01”. Peak refinement was performed, and the resulting peak calls were used to generate heatmaps and profile (line) plots compared with the ATAC-Seq data.
Author contributions
F. R.-C., R. B. L., I. C., M. B., L. M. M., M. L., and O. S. G. investigation; F. R.-C., R. B. L., and R. E. methodology; F. R.-C., R. B. L., and R. E. writing-review and editing; R. B. L. data curation; R. B. L. formal analysis; R. E. resources; R. E. and O. S. G. supervision; R. E. and O. S. G. funding acquisition; O. S. G. conceptualization; O. S. G. writing-original draft; O. S. G. project administration.
Supplementary Material
Acknowledgments
We thank E. T. Yeh for the SENP1 cDNA-expressing plasmid and Keith Gull for the TY1 hybridoma cell line (BB2). We thank Bettina Maria Fuglerud for helpful discussions and guidance on the ATAC experiments. We also thank Vilborg Matre for co-supervising L. M. M. during screening and data analysis. The sequencing service was provided by the Norwegian Sequencing Centre, a national technology platform hosted by the University of Oslo and supported by the “Functional Genomics” and “Infrastructure” programs of the Research Council of Norway and the South-Eastern Regional Health Authorities. All genome-wide analyses were made on the Abel computer cluster (Project nn9374k), which is owned by the University of Oslo and the Norwegian metacenter for high-performance computing (NOTUR). The Abel Cluster is operated by the Department for Research Computing at USIT and the University of Oslo IT-department. We thank Arvind Sundaram for guidance on RNA-Seq analysis.
This work was supported by Norwegian Cancer Society Grant 3485238-2013 (to R. E.) and Research Council of Norway Grants 240768 (to O. S. G.) and 231217 (to R. E.). The authors declare that they have no conflicts of interest with the contents of this article.
This article was selected as one of our Editors' Picks.
This article contains Figs. S1–S13, supporting Materials and Methods, and supporting Refs. 1–19.
- PTM
- post-translational modification
- SUMO
- small ubiquitin-like modifier
- SENP
- sentrin-specific protease
- Seq
- sequencing
- CTCF
- CCCTC-binding factor
- PVDF
- polyvinylidene difluoride
- GBD
- Gal4 DNA-binding domain
- CML
- chronic myelogenous leukemia
- TSS
- transcription start site
- SIM
- SUMO interaction motif
- AMC
- amido-4-methylcoumarin
- mAb
- monoclonal antibody
- GST
- glutathione S-transferase
- PMSF
- phenylmethylsulfonyl fluoride.
References
- 1. Gareau J. R., and Lima C. D. (2010) The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 11, 861–871 10.1038/nrm3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Flotho A., and Melchior F. (2013) Sumoylation: a regulatory protein modification in health and disease. Annu. Rev. Biochem. 82, 357–385 10.1146/annurev-biochem-061909-093311 [DOI] [PubMed] [Google Scholar]
- 3. Geiss-Friedlander R., and Melchior F. (2007) Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 8, 947–956 10.1038/nrm2293 [DOI] [PubMed] [Google Scholar]
- 4. Seeler J.-S., and Dejean A. (2003) Nuclear and unclear functions of SUMO. Nat. Rev. Mol. Cell Biol. 4, 690–699 10.1038/nrm1200 [DOI] [PubMed] [Google Scholar]
- 5. Hendriks I. A., Lyon D., Young C., Jensen L. J., Vertegaal A. C., and Nielsen M. L. (2017) Site-specific mapping of the human SUMO proteome reveals co-modification with phosphorylation. Nat. Struct. Mol. Biol. 10.1038/nsmb.3366 [DOI] [PubMed] [Google Scholar]
- 6. Cubeñas-Potts C., and Matunis M. J. (2013) SUMO: a multifaceted modifier of chromatin structure and function. Dev. Cell 24, 1–12 10.1016/j.devcel.2012.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gill G. (2003) Post-translational modification by the small ubiquitin-related modifier SUMO has big effects on transcription factor activity. Curr. Opin. Genet. Dev. 13, 108–113 10.1016/S0959-437X(03)00021-2 [DOI] [PubMed] [Google Scholar]
- 8. Núñez-O'Mara A., and Berra E. (2013) Deciphering the emerging role of SUMO conjugation in the hypoxia-signaling cascade. Biol. Chem. 394, 459–469 10.1515/hsz-2012-0319 [DOI] [PubMed] [Google Scholar]
- 9. Chymkowitch P., Nguéa P. A., Enserink J. M. (2015) SUMO-regulated transcription: challenging the dogma. Bioessays 37, 1095–1105 10.1002/bies.201500065 [DOI] [PubMed] [Google Scholar]
- 10. Kerscher O. (2007) SUMO junction-what's your function? New insights through SUMO-interacting motifs. EMBO Rep. 8, 550–555 10.1038/sj.embor.7400980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yeh E. T. (2009) SUMOylation and De-SUMOylation: wrestling with life's processes. J. Biol. Chem. 284, 8223–8227 10.1074/jbc.R800050200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hickey C. M., Wilson N. R., and Hochstrasser M. (2012) Function and regulation of SUMO proteases. Nat. Rev. Mol. Cell Biol. 13, 755–766 10.1038/nrm3478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huang C.-J., Wu D., Khan F. A., and Huo L.-J. (2015) DeSUMOylation: an important therapeutic target and protein regulatory event. DNA Cell Biol. 34, 652–660 10.1089/dna.2015.2933 [DOI] [PubMed] [Google Scholar]
- 14. Nacerddine K., Lehembre F., Bhaumik M., Artus J., Cohen-Tannoudji M., Babinet C., Pandolfi P. P., and Dejean A. (2005) The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev. Cell 9, 769–779 10.1016/j.devcel.2005.10.007 [DOI] [PubMed] [Google Scholar]
- 15. Cheng J., Kang X., Zhang S., and Yeh E. T. (2007) SUMO-specific protease 1 is essential for stabilization of HIF1α during hypoxia. Cell 131, 584–595 10.1016/j.cell.2007.08.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kang X., Qi Y., Zuo Y., Wang Q., Zou Y., Schwartz R. J., Cheng J., and Yeh E. T. (2010) SUMO-specific protease 2 is essential for suppression of polycomb group protein-mediated gene silencing during embryonic development. Mol. Cell 38, 191–201 10.1016/j.molcel.2010.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bartek J., and Hodny Z. (2010) SUMO boosts the DNA damage response barrier against cancer. Cancer Cell 17, 9–11 10.1016/j.ccr.2009.12.030 [DOI] [PubMed] [Google Scholar]
- 18. Bettermann K., Benesch M., Weis S., and Haybaeck J. (2012) SUMOylation in carcinogenesis. Cancer Lett. 316, 113–125 10.1016/j.canlet.2011.10.036 [DOI] [PubMed] [Google Scholar]
- 19. Sarge K. D., and Park-Sarge O.-K. (2011) SUMO and its role in human diseases. Int. Rev. Cell Mol. Biol. 288, 167–183 10.1016/B978-0-12-386041-5.00004-2 [DOI] [PubMed] [Google Scholar]
- 20. Yu L., Ji W., Zhang H., Renda M. J., He Y., Lin S., Cheng E.-C., Chen H., Krause D. S., and Min W. (2010) SENP1-mediated GATA1 deSUMOylation is critical for definitive erythropoiesis. J. Exp. Med. 207, 1183–1195 10.1084/jem.20092215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Van Nguyen T., Angkasekwinai P., Dou H., Lin F.-M., Lu L.-S., Cheng J., Chin Y. E., Dong C., and Yeh E. T. (2012) SUMO-specific protease 1 is critical for early lymphoid development through regulation of STAT5 activation. Mol. Cell 45, 210–221 10.1016/j.molcel.2011.12.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hay R. T. (2007) SUMO-specific proteases: a twist in the tail. Trends Cell Biol. 17, 370–376 10.1016/j.tcb.2007.08.002 [DOI] [PubMed] [Google Scholar]
- 23. Mukhopadhyay D., and Dasso M. (2007) Modification in reverse: the SUMO proteases. Trends Biochem. Sci. 32, 286–295 10.1016/j.tibs.2007.05.002 [DOI] [PubMed] [Google Scholar]
- 24. Wang Y., and Dasso M. (2009) SUMOylation and deSUMOylation at a glance. J. Cell Sci. 122, 4249–4252 10.1242/jcs.050542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ouyang J., and Gill G. (2009) SUMO engages multiple corepressors to regulate chromatin structure and transcription. Epigenetics 4, 440–444 10.4161/epi.4.7.9807 [DOI] [PubMed] [Google Scholar]
- 26. Stielow B., Sapetschnig A., Wink C., Krüger I., and Suske G. (2008) SUMO-modified Sp3 represses transcription by provoking local heterochromatic gene silencing. EMBO Rep. 9, 899–906 10.1038/embor.2008.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stielow B., Sapetschnig A., Krüger I., Kunert N., Brehm A., Boutros M., and Suske G. (2008) Identification of SUMO-dependent chromatin-associated transcriptional repression components by a genome-wide RNAi screen. Mol. Cell 29, 742–754 10.1016/j.molcel.2007.12.032 [DOI] [PubMed] [Google Scholar]
- 28. Lai A. Y., and Wade P. A. (2011) Cancer biology and NuRD: a multifaceted chromatin remodeling complex. Nat. Rev. Cancer 11, 588–596 10.1038/nrc3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Goodarzi A. A., Kurka T., and Jeggo P. A. (2011) KAP-1 phosphorylation regulates CHD3 nucleosome remodeling during the DNA double-strand break response. Nat. Struct. Mol. Biol. 18, 831–839 10.1038/nsmb.2077 [DOI] [PubMed] [Google Scholar]
- 30. Carette J. E., Raaben M., Wong A. C., Herbert A. S., Obernosterer G., Mulherkar N., Kuehne A. I., Kranzusch P. J., Griffin A. M., Ruthel G., Dal Cin P., Dye J. M., Whelan S. P., Chandran K., and Brummelkamp T. R. (2011) Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature 477, 340–343 10.1038/nature10348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Niskanen E. A., Malinen M., Sutinen P., Toropainen S., Paakinaho V., Vihervaara A., Joutsen J., Kaikkonen M. U., Sistonen L., and Palvimo J. J. (2015) Global SUMOylation on active chromatin is an acute heat stress response restricting transcription. Genome Biol. 16, 153 10.1186/s13059-015-0717-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Heinz S., Benner C., Spann N., Bertolino E., Lin Y. C., Laslo P., Cheng J. X., Murre C., Singh H., and Glass C. K. (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lobanenkov V. V., and Zentner G. E. (2018) Discovering a binary CTCF code with a little help from BORIS. Nucleus 9, 33–41 10.1080/19491034.2017.1394536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dixon J. R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J. S., and Ren B. (2012) Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 10.1038/nature11082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schmitt A. D., Hu M., Jung I., Xu Z., Qiu Y., Tan C. L., Li Y., Lin S., Lin Y., Barr C. L., and Ren B. (2016) A compendium of chromatin contact maps reveals spatially active regions in the human genome. Cell Rep. 17, 2042–2059 10.1016/j.celrep.2016.10.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ji X., Dadon D. B., Powell B. E., Fan Z. P., Borges-Rivera D., Shachar S., Weintraub A. S., Hnisz D., Pegoraro G., Lee T. I., Misteli T., Jaenisch R., and Young R. A. (2016) 3D chromosome regulatory landscape of human pluripotent cells. Cell Stem Cell 18, 262–275 10.1016/j.stem.2015.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hendriks I. A., and Vertegaal A. C. (2016) A comprehensive compilation of SUMO proteomics. Nat. Rev. Mol. Cell Biol. 17, 581–595 10.1038/nrm.2016.81 [DOI] [PubMed] [Google Scholar]
- 38. Shen Q., Beyrouthy N., Matabishi-Bibi L., and Dargemont C. (2017) The chromatin remodeling Isw1a complex is regulated by SUMOylation. Biochem. J. 474, 3455–3469 10.1042/BCJ20170172 [DOI] [PubMed] [Google Scholar]
- 39. Cox E., Hwang W., Uzoma I., Hu J., Guzzo C. M., Jeong J., Matunis M. J., Qian J., Zhu H., and Blackshaw S. (2017) Global analysis of SUMO-binding proteins identifies SUMOylation as a Key regulator of the INO80 chromatin remodeling complex. Mol. Cell. Proteomics 16, 812–823 10.1074/mcp.M116.063719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ivanov A. V., Peng H., Yurchenko V., Yap K. L., Negorev D. G., Schultz D. C., Psulkowski E., Fredericks W. J., White D. E., Maul G. G., Sadofsky M. J., Zhou M.-M., and Rauscher F. J. 3rd. (2007) PHD domain-mediated E3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Mol. Cell 28, 823–837 10.1016/j.molcel.2007.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hendriks I. A., D'Souza R. C., Yang B., Verlaan-de Vries M., Mann M., and Vertegaal A. C. (2014) Uncovering global SUMOylation signaling networks in a site-specific manner. Nat. Struct. Mol. Biol. 21, 927–936 10.1038/nsmb.2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yamashita D., Moriuchi T., Osumi T., and Hirose F. (2016) Transcription factor hDREF is a novel SUMO E3 ligase of Mi2α. J. Biol. Chem. 291, 11619–11634 10.1074/jbc.M115.713370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Psakhye I., and Jentsch S. (2012) Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell 151, 807–820 10.1016/j.cell.2012.10.021 [DOI] [PubMed] [Google Scholar]
- 44. Ishihara K., Oshimura M., and Nakao M. (2006) CTCF-dependent chromatin insulator is linked to epigenetic remodeling. Mol. Cell 23, 733–742 10.1016/j.molcel.2006.08.008 [DOI] [PubMed] [Google Scholar]
- 45. Allen M. D., Religa T. L., Freund S. M., and Bycroft M. (2007) Solution structure of the BRK domains from CHD7. J. Mol. Biol. 371, 1135–1140 10.1016/j.jmb.2007.06.007 [DOI] [PubMed] [Google Scholar]
- 46. Wiechens N., Singh V., Gkikopoulos T., Schofield P., Rocha S., and Owen-Hughes T. (2016) The chromatin remodeling enzymes SNF2H and SNF2L position nucleosomes adjacent to CTCF and other transcription factors. PLoS Genet. 12, e1005940 10.1371/journal.pgen.1005940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. MacPherson M. J., Beatty L. G., Zhou W., Du M., and Sadowski P. D. (2009) The CTCF insulator protein is post-translationally modified by SUMO. Mol. Cell. Biol. 29, 714–725 10.1128/MCB.00825-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Whalen S., Truty R. M., and Pollard K. S. (2016) Enhancer-promoter interactions are encoded by complex genomic signatures on looping chromatin. Nat. Genet. 48, 488–496 10.1038/ng.3539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Soufi A., Garcia M. F., Jaroszewicz A., Osman N., Pellegrini M., and Zaret K. S. (2015) Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell 161, 555–568 10.1016/j.cell.2015.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Swinstead E. E., Paakinaho V., Presman D. M., and Hager G. L. (2016) Pioneer factors and ATP-dependent chromatin remodeling factors interact dynamically: a new perspective: multiple transcription factors can effect chromatin pioneer functions through dynamic interactions with ATP-dependent chromatin remodeling factors. Bioessays 38, 1150–1157 10.1002/bies.201600137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fuglerud B. M., Lemma R. B., Wanichawan P., Sundaram A. Y. M., Eskeland R., and Gabrielsen O. S. (2017) A c-Myb mutant causes deregulated differentiation due to impaired histone binding and abrogated pioneer factor function. Nucleic Acids Res. 45, 7681–7696 10.1093/nar/gkx364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Saether T., Berge T., Ledsaak M., Matre V., Alm-Kristiansen A. H., Dahle O., Aubry F., and Gabrielsen O. S. (2007) The chromatin remodeling factor Mi-2α acts as a novel co-activator for human c-Myb. J. Biol. Chem. 282, 13994–14005 10.1074/jbc.M700755200 [DOI] [PubMed] [Google Scholar]
- 53. Molværsmyr A.-K., Saether T., Gilfillan S., Lorenzo P. I., Kvaløy H., Matre V., and Gabrielsen O. S. (2010) A SUMO-regulated activation function controls synergy of c-Myb through a repressor-activator switch leading to differential p300 recruitment. Nucleic Acids Res. 38, 4970–4984 10.1093/nar/gkq245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bengtsen M., Klepper K., Gundersen S., Cuervo I., Drabløs F., Hovig E., Sandve G. K., Gabrielsen O. S., and Eskeland R. (2015) c-Myb binding sites in haematopoietic chromatin landscapes. PLoS ONE 10, e0133280 10.1371/journal.pone.0133280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bastin P., Bagherzadeh Z., Matthews K. R., and Gull K. (1996) A novel epitope tag system to study protein targeting and organelle biogenesis in Trypanosoma brucei. Mol. Biochem. Parasitol. 77, 235–239 10.1016/0166-6851(96)02598-4 [DOI] [PubMed] [Google Scholar]
- 56. Alm-Kristiansen A. H., Saether T., Matre V., Gilfillan S., Dahle O., and Gabrielsen O. S. (2008) FLASH acts as a co-activator of the transcription factor c-Myb and localizes to active RNA polymerase II foci. Oncogene 27, 4644–4656 10.1038/onc.2008.105 [DOI] [PubMed] [Google Scholar]
- 57. Matre V., Nordgård O., Alm-Kristiansen A. H., Ledsaak M., and Gabrielsen O. S. (2009) HIPK1 interacts with c-Myb and modulates its activity through phosphorylation. Biochem. Biophys. Res. Commun. 388, 150–154 10.1016/j.bbrc.2009.07.139 [DOI] [PubMed] [Google Scholar]
- 58. Alm-Kristiansen A. H., Lorenzo P. I., Molværsmyr A.-K., Matre V., Ledsaak M., Sæther T., and Gabrielsen O. S. (2011) PIAS1 interacts with FLASH and enhances its co-activation of c-Myb. Mol. Cancer 10, 21 10.1186/1476-4598-10-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Dahle Ø., Bakke O., and Gabrielsen O. S. (2004) c-Myb associates with PML in nuclear bodies in hematopoietic cells. Exp. Cell Res. 297, 118–126 10.1016/j.yexcr.2004.03.014 [DOI] [PubMed] [Google Scholar]
- 60. Wright K. J., Marr M. T. 2nd., and Tjian R. (2006) TAF4 nucleates a core subcomplex of TFIID and mediates activated transcription from a TATA-less promoter. Proc. Natl. Acad. Sci. U.S.A. 103, 12347–12352 10.1073/pnas.0605499103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ledsaak M., Bengtsen M., Molværsmyr A.-K., Fuglerud B. M., Matre V., Eskeland R., and Gabrielsen O. S. (2016) PIAS1 binds p300 and behaves as a coactivator or corepressor of the transcription factor c-Myb dependent on SUMO-status. Biochim. Biophys. Acta 1859, 705–718 10.1016/j.bbagrm.2016.03.011 [DOI] [PubMed] [Google Scholar]
- 62. Buenrostro J. D., Wu B., Chang H. Y., and Greenleaf W. J. (2015) ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 109, 21.29.1–9 10.1002/0471142727.mb2129s109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Buenrostro J. D., Giresi P. G., Zaba L. C., Chang H. Y., and Greenleaf W. J. (2013) Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 10.1038/nmeth.2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. ENCODE Project Consortium. (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Dahle Ø., Andersen T. Ø., Nordgård O., Matre V., Del Sal G., and Gabrielsen O. S. (2003) Transactivation properties of c-Myb are critically dependent on two SUMO-1 acceptor sites that are conjugated in a PIASy enhanced manner. Eur. J. Biochem. 270, 1338–1348 10.1046/j.1432-1033.2003.03504.x [DOI] [PubMed] [Google Scholar]
- 66. Alm-Kristiansen A. H., Norman I. L., Matre V., and Gabrielsen O. S. (2009) SUMO modification regulates the transcriptional activity of FLASH. Biochem. Biophys. Res. Commun. 387, 494–499 10.1016/j.bbrc.2009.07.053 [DOI] [PubMed] [Google Scholar]
- 67. Ramírez F., Ryan D. P., Grüning B., Bhardwaj V., Kilpert F., Richter A. S., Heyne S., Dündar F., and Manke T. (2016) deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kent W. J., Sugnet C. W., Furey T. S., Roskin K. M., Pringle T. H., Zahler A. M., and Haussler D. (2002) The human genome browser at UCSC. Genome Res. 12, 996–1006 10.1101/gr.229102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D. R., Pimentel H., Salzberg S. L., Rinn J. L., and Pachter L. (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Abugessaisa I., Shimoji H., Sahin S., Kondo A., Harshbarger J., Lizio M., Hayashizaki Y., Carninci P., FANTOM consortium, Forrest A., Kasukawa T., and Kawaji H. (2016) FANTOM5 transcriptome catalog of cellular states based on Semantic MediaWiki. Database (Oxford) 2016, baw105 10.1093/database/baw105 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.