Abstract
c-Myc is a proto-oncogene controlling expression of multiple genes involved in cell growth and differentiation. Although the functional role of c-Myc as a transcriptional regulator has been intensively studied, targeting this protein in cancer remains a challenge. Here, we report a trimodal regulation of c-Myc function by the Ras effector, Ras-association domain family member 7 (RASSF7), a nonenzymatic protein modulating protein–protein interactions to regulate cell proliferation. Using HEK293T and HeLa cell lines, we provide evidence that RASSF7 destabilizes the c-Myc protein by promoting Cullin4B-mediated polyubiquitination and degradation. Furthermore, RASSF7 competed with MYC-associated factor X (MAX) in the formation of a heterodimeric complex with c-Myc and attenuated its occupancy on target gene promoters to regulate transcription. Consequently, RASSF7 inhibited c-Myc–mediated oncogenic transformation, and an inverse correlation between the expression levels of the RASSF7 and c-Myc genes was evident in human cancers. Furthermore, we found that RASSF7 interacts with c-Myc via its RA and leucine zipper (LZ) domains and LZ domain peptide is sufficient to inhibit c-Myc function, suggesting that this peptide might be used to target oncogenic c-Myc. These results unveil that RASSF7 and c-Myc are functionally linked in the control of tumorigenesis and open up potential therapeutic avenues for targeting the “undruggable” c-Myc protein in a subset of human cancers.
Keywords: Ras protein, Myc (c-Myc), tumor cell biology, tumor suppressor gene, ubiquitination (ubiquitination), cell-penetrating peptide (CPP), cell cycle, cell proliferation, cancer, transcription factor, colony forming assay, MAX, Peptide inhibitor, Ras effectors, RASSF7
Introduction
The RAS oncogenes are central players in many human cancers. Ras mutants are associated with poor prognosis in many cancers. Ras performs its biological functions through downstream molecules known as Ras effectors (1–3). In the past decade, a distinct class of Ras effectors has been identified known as Ras Association Domain Family of proteins (RASSF)2 that are characterized by the presence of Ras association (RA) domain (RalGDS (Ral guanine nucleotide dissociation stimulator) and AF6 (ALL-1 fusion partner from chromosome 6) (4). The RASSF family consists of 10 members and based on the location of the RA domain, they are subdivided into two groups namely classical RASSFs, also known as C-terminal RASSFs (RASSF1–6) and N-terminal RASSFs (RASSF7–10) (5). Most of the RASSF family members are known to be down-regulated in various human cancers by epigenetic modifications (6). RASSF7 was initially identified as HRC1 (H-RAS1 cluster1) located upstream of H-RAS on chromosome 11 (6). Previous reports suggest that RASSF7 plays a critical role in regulating microtubule dynamics and inhibits the pro-apoptotic signaling by promoting phosphorylation of MKK7 and regulates cell proliferation (7–10). On the other hand, RASSF7 is reported to be down-regulated by promoter hypermethylation in neuroblastoma (11) and is a key regulator of necroptosis (12). Together, these reports suggest a delicate network of pathways that RASSF7 might be regulating to control cell growth in a context-dependent manner. However, the mechanisms remain poorly understood. We therefore attempted to explore the role of RASSF7 in cell growth regulation. In the present investigation, we unveiled a novel regulatory mechanism by which RASSF7 promotes c-Myc degradation through E3-ubiquitin ligase CUL4B and alters the expression of c-Myc target genes. In addition, RASSF7 blunts c-Myc-dependent DNA synthesis and cell proliferation. Collectively, our data provide evidence that RASSF7 may be a novel regulator of oncogenic c-Myc function in human cancers.
Results
Inverse correlation of RASSF7 and c-Myc expression in human cancers
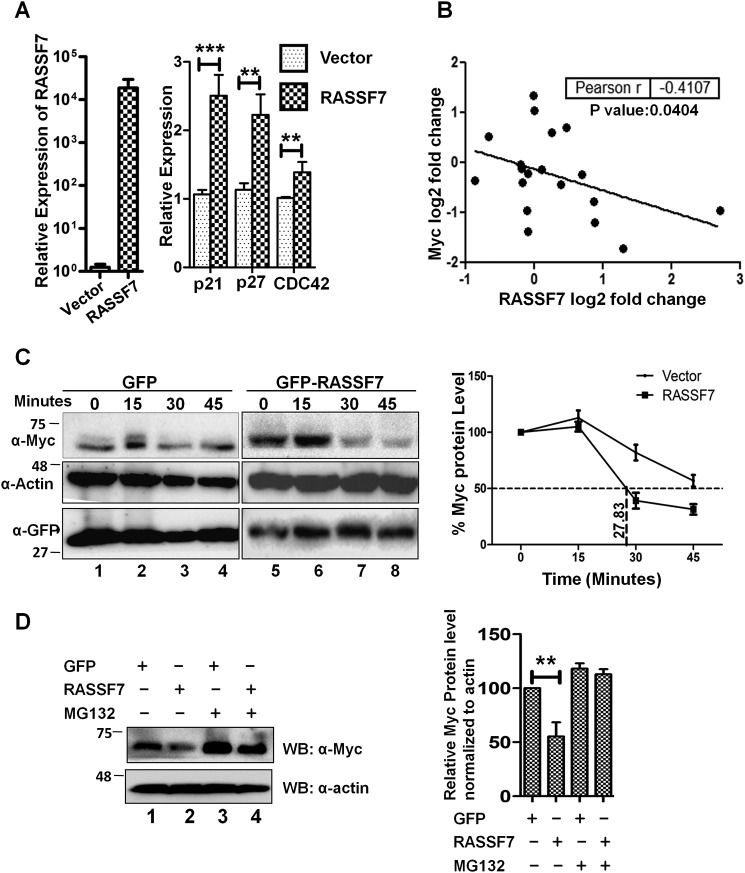
RASSF7, the first member of the N-terminal RASSF family is shown to regulate cell cycle (7–10) but the mechanisms remain unknown. Toward understanding the mechanism, we first analyzed the expression pattern of genes that are critical for cell cycle regulation such as p21 (CDKN1A), p27 (CDKN1B), and Cdc42 in the presence of RASSF7. Interestingly, results from RT-qPCR analysis indicate that ectopic expression of RASSF7 up-regulated p21, p27, and Cdc42 levels (Fig. 1A). It is worth mentioning that these genes are known to be down-regulated by c-Myc to promote cell proliferation (13, 14). This prompted us to investigate the existence of cross-talk between c-Myc and RASSF7 during tumorigenesis. Toward this, we analyzed the correlation between RASSF7 and c-Myc expression in various human cancers. Pearson correlation analysis was performed with log2 fold-change expression of RASSF7 and c-Myc transcripts across different cancer tissues retrieved from the BioXpress database (15). Analysis indicates a negative correlation (Pearson r value −0.4107) between the expression levels of RASSF7 and c-Myc in various human cancers (Fig. 1B). To further strengthen this association, expression status of RASSF7 and c-Myc were analyzed in cancer cell lines. Results from Western blot analysis suggest a negative correlation (Pearson r value −0.3147) between RASSF7 and c-Myc protein levels in tested cancer cell lines (Fig. S1). Taken together, these data suggest the existence of an inverse correlation between RASSF7 and c-Myc expression in human cancers. This leads to the hypothesis that the interplay between RASSF7 and c-Myc may be essential to regulate tumorigenesis.
Figure 1.
RASSF7 destabilizes c-Myc protein. A, RT-qPCR analysis indicates that RASSF7 up-regulates expression of p21, p27, and cdc42 in HEK293T cells (n = 3 and data are expressed as mean ± S.D.). B, correlation analysis of RASSF7 and c-Myc expression in cancer tissues available from BioXpress database. C, cycloheximide chase assay indicates that RASSF7 destabilized c-Myc protein. β-Actin served as loading control. The protein levels are normalized to percentage of protein in time zero to calculate t½ of c-Myc. D, treatment with proteosomal inhibitor rescues RASSF7 mediated degradation of c-Myc level.
RASSF7 destabilizes c-Myc through E3 ligase Cullin4B
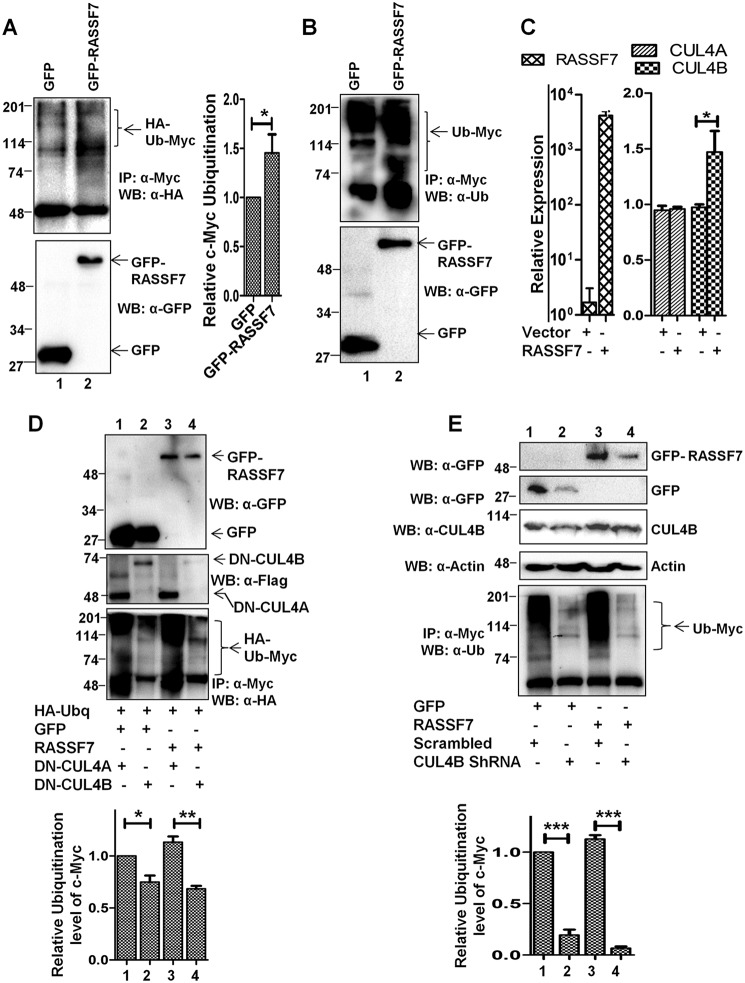
It is well established that deregulation (amplification and altered protein stability) of c-Myc oncoprotein plays a crucial role during tumorigenesis (16). To understand the mechanism and relevance of the observed negative correlation of RASSF7 and c-Myc expression in human cancers, we first investigated whether RASSF7 alters the c-Myc protein levels in cells. To prove this, HEK293T cells transfected with control or RASSF7 expression plasmids were treated with cycloheximide and the c-Myc protein levels were determined by Western blotting at different time intervals. Results from a cycloheximide chase assay suggest that the half-life of the c-Myc protein is ∼45 min (Fig. 1C, lanes 1-4). With RASSF7 expression, c-Myc protein half-life was reduced to 28 min (Fig. 1C, lanes 5-8) suggesting that RASSF7 promotes c-Myc protein destabilization. Toward understanding the mechanism, we first determined the c-Myc protein levels upon RASSF7 expression with a proteasomal inhibitor (MG132). Interestingly, MG132 is able to rescue RASSF7-mediated destabilization of c-Myc protein levels (Fig. 1D, lanes 3 and 4). These results suggest that RASSF7 destabilizes c-Myc protein levels. It is well known that the deregulated ubiquitin-proteasome pathway is critical to modulate c-Myc protein levels in cancers where amplification was not observed (17, 18). Together, these data lead to the hypothesis that RASSF7 may alter c-Myc protein levels through ubiquitination. Toward this, the status of c-Myc ubiquitination was analyzed in the presence of RASSF7 in HEK293T cells with endogenous ubiquitin as well as with HA-ubiquitin. Results from a ubiquitination assay indicate that RASSF7 promotes the polyubiquitination of c-Myc (Fig. 2, A and B, lane 2). These data suggest the possibility that RASSF7 may alter c-Myc stability. To further understand the mechanism of RASSF7-mediated degradation of c-Myc, RASSF7 was ectopically expressed in HEK293T cells and the expression levels of different E3 ligases known to be involved in c-Myc ubiquitination were analyzed by RT-qPCR. We observed a significant increase of E3 ligase Cullin4B (CUL4B) levels in the presence of RASSF7 expression (Fig. S2A). To confirm this, the expression status of CUL4 family members (CUL4A and CUL4B) was checked under RASSF7 expression and knockdown conditions. Interestingly, CUL4B not CUL4A transcript levels were altered by RASSF7 (Fig. 2C and Fig. S2B). Furthermore, CUL4B protein levels were reduced upon depletion of RASSF7 by specific shRNA (Fig. S2B). To define the role of CUL4B on c-Myc degradation, the status of c-Myc ubiquitination was checked upon co-expression of dominant-negative CUL4B (DN-CUL4B) and CUL4A (DN-CUL4A) in the presence of RASSF7. Results indicate that DN-CUL4B not DN-CUL4A was able to specifically alter RASSF7-induced ubiquitination of c-Myc (Fig. 2D, lanes 3 and 4). In addition, depletion of CUL4B by specific shRNA rescued RASSF7-induced c-Myc ubiquitination (Fig. 2E, lane 4). Together, these data provided evidence that RASSF7 up-regulates E3 ligase CUL4B expression to promote c-Myc degradation.
Figure 2.
RASSF7 induces c-Myc ubiquitination through Cullin4B. A and B, HEK293T cells expressing GFP or GFP-RASSF7 were treated with MG132 and performed ubiquitination assay. Immunoblots with HA (for HA-ubiquitin) (A) or endogenous ubiquitin (B) were performed to analyze the c-Myc ubiquitin pattern. C, RT-qPCR analysis indicates that RASSF7 induces CUL4B expression. β-Actin served as internal control (n = 3 and data are expressed as mean ± S.D.). D, ubiquitination assay performed in HEK293T cells expressing dominant-negative Cullins (DN-CUL4A or DN-CUL4B) indicate the requirement of functional CUL4B for RASSF7-induced degradation of c-Myc. E, depletion of CUL4B rescues RASSF7-induced ubiquitination of c-Myc. WB, Western blot; IP, immunoprecipitation.
RASSF7 down-regulates transcriptional activity of c-Myc
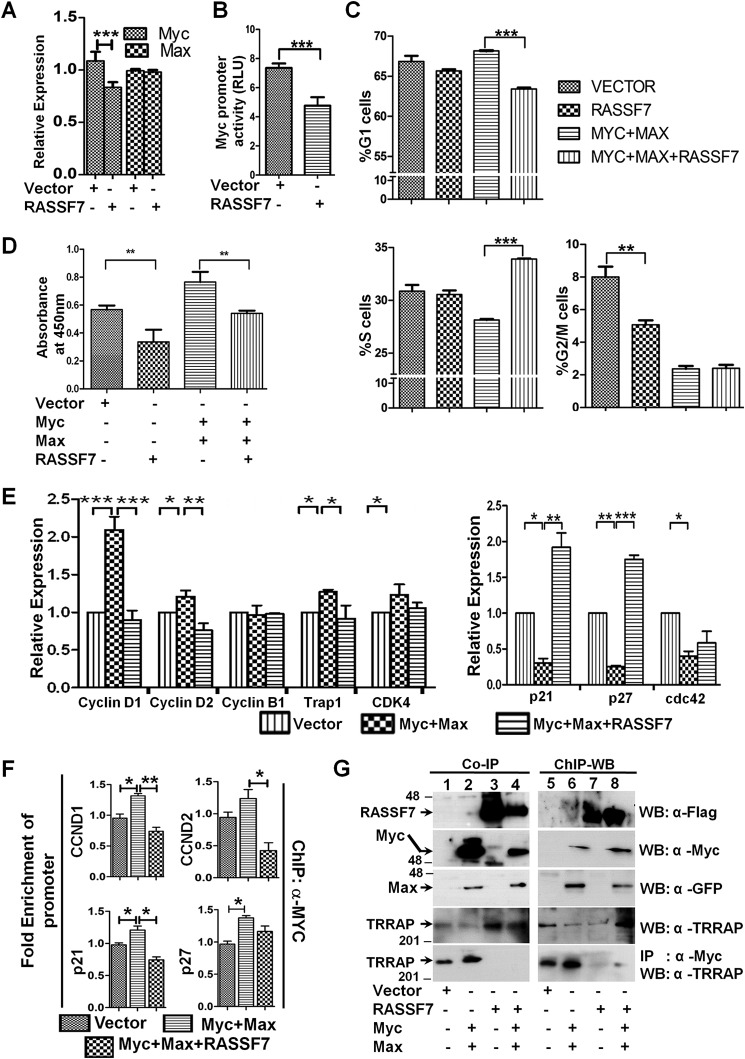
c-Myc is a well-known transcription factor controlling expression of genes that are involved in various cellular functions, including its own promoter. We next tested whether RASSF7 modulates c-Myc–mediated gene transcription. Toward this, RASSF7 was expressed in HEK293T cells and c-Myc mRNA levels were measured by RT-qPCR. Interestingly, RASSF7 reduced endogenous c-Myc mRNA levels (Fig. 3A). To understand the mechanism, c-Myc promoter activity was measured in the presence of RASSF7 expression. To our surprise, RASSF7 reduced the c-Myc promoter activity (Fig. 3B). Together, these data provide evidence that RASSF7 modulates c-Myc activity both at transcriptional and post-translational levels. To further define the effect of RASSF7 on c-Myc function, RASSF7 was co-expressed with c-Myc in HEK293T cells and the cell cycle profiles were analyzed by flow cytometry. Results in Fig. 3C indicate that RASSF7 significantly altered the status of c-Myc–mediated G1 and S phases of cell cycle. These data suggest the possibility that RASSF7 may block c-Myc–induced G1/S phase progression. To confirm this, the rate of DNA synthesis was measured in the presence of RASSF7 and c-Myc expression. RASSF7 decreased c-Myc–induced DNA synthesis, as indicated by the reduced BrdU incorporation upon co-expression of RASSF7 with c-Myc (Fig. 3D). Collectively, these data reveal that RASSF7 regulates S phase progression by blocking c-Myc–induced DNA synthesis. We next analyzed whether RASSF7 alters the expression of c-Myc target genes, which are involved in cell cycle regulation by RT-qPCR. Interestingly, RASSF7 repressed the expression of Cyclin D1, Cyclin D2, Trap1, and CDK4 (Fig. 3E) that are usually up-regulated by c-Myc. In contrast, RASSF7 rescued the c-Myc–induced repression of p21, p27, and Cdc42 expression (Fig. 3E). These data suggest that RASSF7 alters cell division by modulating the expression of c-Myc target genes that are known to play a critical role in cell proliferation. To explore the mechanism of RASSF7-mediated alteration of c-Myc transcriptional activity, we quantified the occupancy of c-Myc on the target promoters in the presence of RASSF7 by ChIP-qPCR. Interestingly, results in Fig. 3F and Fig. S2C indicate that RASSF7 attenuates the occupancy of c-Myc on target gene promoters. The RASSF8 promoter, which lacks E-box (c-Myc binding motif), served as negative control. Although, we observed a significant change in the transcriptional activity of c-Myc, a marginal decrease in the occupancy of c-Myc on the target promoters was observed. We therefore, next determined whether RASSF7 alters the c-Myc–mediated recruitment of co-factors to the target gene promoters necessary for transactivation. Earlier reports have suggested that the recruitment of co-activator, TRRAP by c-Myc is essential to activate the transcription of target genes (19). ChIP-Western blotting and co-immunoprecipitation assays confirmed that RASSF7 abrogated the c-Myc–dependent recruitment of the co-activator TRRAP to the target gene promoters (Fig. 3G, bottom panel, and lanes 3, 4, 7, and 8). Collectively, these data provide evidence that RASSF7 alters c-Myc–mediated recruitment of transcriptional co-activator to the target gene promoters and control c-Myc transcriptional activity.
Figure 3.
RASSF7 modulates c-Myc–mediated transcription. A, RT-qPCR analysis suggests that RASSF7 reduces endogenous c-Myc expression levels but not Max levels. β-Actin served as internal control (n = 3 and data are expressed as mean ± S.D.). B, promoter luciferase assay indicates RASSF7 inhibits c-Myc promoter activity. Renilla luciferase activity served as control (n = 3 and data are expressed as mean ± S.D.). C, cell cycle analysis in HEK293T cells indicates a significant change in the G1/S phase of the cell cycle when RASSF7 is co-expressed with c-Myc and Max (n = 3 and data are expressed as mean ± S.D.). D, BrdU incorporation assay reveals reduction of DNA synthesis upon co-expression of RASSF7 along with c-Myc and Max in HEK293T cells (n = 3 and data are expressed as mean ± S.D.). E, RT-qPCR analysis indicates that RASSF7 reverses expression levels of c-Myc target genes in HEK293T cells. β-Actin served as internal control (n = 3 and data are expressed as mean ± S.D.). F, ChIP-qPCR assay indicates that RASSF7 alters c-Myc occupancy on target gene promoters (n = 3 and data are expressed as mean ± S.D.). G, ChIP-Western blot (WB) and co-immunoprecipitation (IP) in HEK293T cell lysates co-expressing the indicated constructs confirms that RASSF7 inhibits the c-Myc–dependent recruitment of TRRAP to initiate the target gene transcription.
RASSF7 interacts with c-Myc and modulates its transcriptional activity
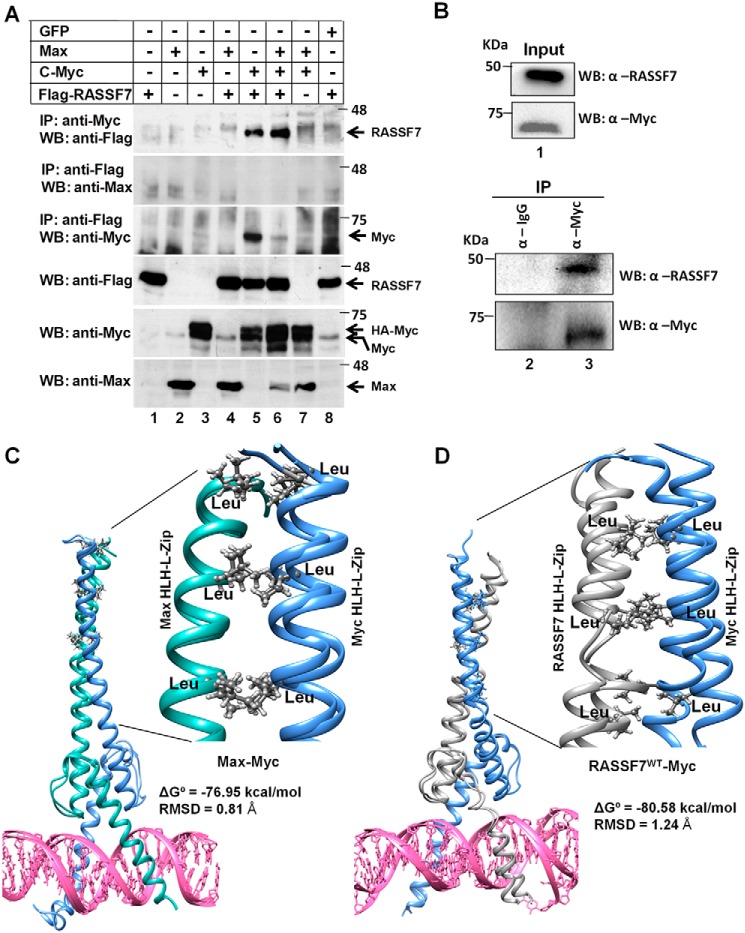
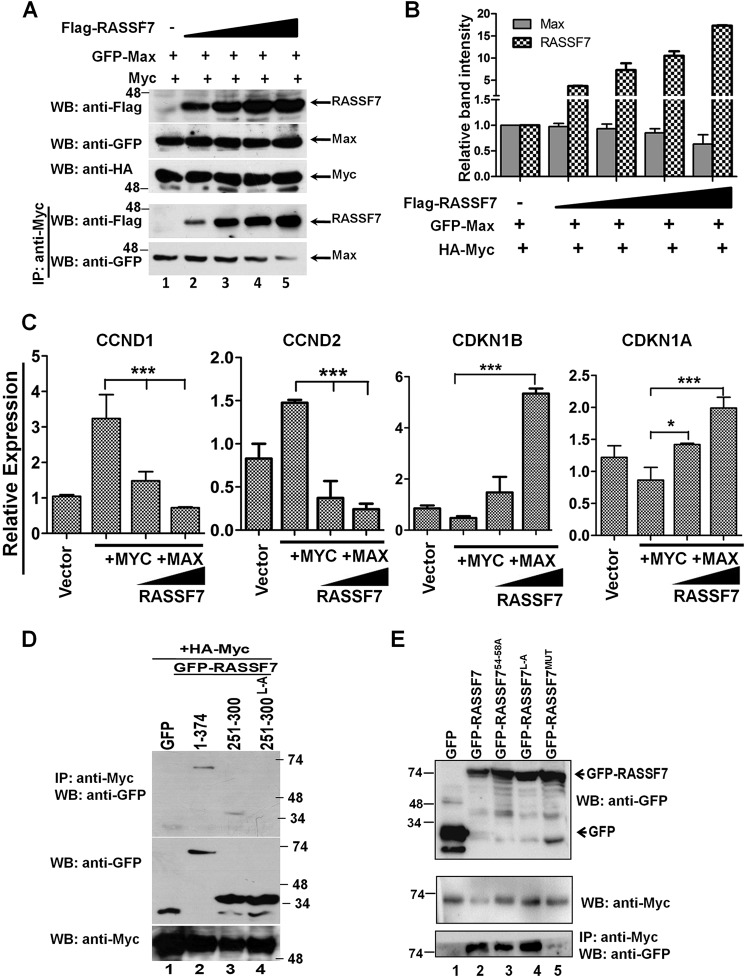
It is well documented that c-Myc forms heterodimer with Max through leucine zipper domains and occupies the E-box (c-Myc–binding motif) on the target gene promoter to regulate transcription. Based on the results from the current investigation, we hypothesized that RASSF7 blocks the recruitment of transcriptional co-activator by disrupting the formation of the heterodimeric complex between c-Myc and Max on target gene promoters. To confirm this possibility, we first tested whether RASSF7 interacts with c-Myc or Max. Immunoprecipitation experiments using HEK293T cell lysates indicate that RASSF7 specifically interacts with c-Myc (Fig. 4A, lanes 5 and 6) but not with Max (Fig. 4A, lane 4). Furthermore, co-immunoprecipitation assay confirms the existence of c-Myc and RASSF7 interaction in cells (Fig. 4B). In addition, molecular dynamics simulations revealed that the binding free energy (ΔG°) for the c-Myc/Max heterodimeric complex is −76.95 kcal/mol (Fig. 4C, Video S1), whereas it is −80.58 kcal/mol for the c-Myc/RASSF7 heterodimeric complex (Fig. 4D, Video S2). These results suggest that RASSF7 might compete with Max to form a stable heterodimeric complex with c-Myc. To further define this possibility, a competitive binding assay was performed with increasing concentrations of RASSF7 keeping the concentration of Max constant. Results indicate that a dose-dependent increase of RASSF7 levels resulted in increased formation of the c-Myc/RASSF7 complex with a gradual decrease in the c-Myc/Max complex suggest that RASSF7 competes with Max to form a heterodimeric complex with c-Myc (Fig. 5, A and B). We next checked whether the formation of a heterodimeric complex between c-Myc and RASSF7 alters c-Myc–dependent transcription. To address this, RASSF7 was co-expressed with c-Myc and Max and the mRNA levels of c-Myc–induced genes (CyclinD1 and CyclinD2) as well as repressed genes (p21 and p27) were analyzed by qPCR. Interestingly, RASSF7 inhibits the c-Myc/Max–induced expression of CyclinD1 and CyclinD2 and rescued the c-Myc/Max-repressed p21 and p27 expression in a dose-dependent manner (Fig. 5C). Taken together, these data provide evidence that RASSF7 specifically interacts with c-Myc and consequently alters c-Myc–dependent transcription.
Figure 4.
RASSF7 forms a heterodimeric complex with c-Myc. A, immunoprecipitation (IP) assay using HEK293T cell lysates reveals that RASSF7 specifically interacts with c-Myc but not Max. B, co-immunoprecipitation assay indicates interaction between endogenous RASSF7 and c-Myc. C, snapshots of heteromeric complexes obtained from molecular dynamics simulation. c-Myc/Max and (D) c-Myc/RASSF7 complexes show stable structures. The interaction of leucine pairs are showed as magnified image. WB, Western blot.
Figure 5.
RASSF7 competes with Max to form a complex with c-Myc. A, competitive binding assay suggest that RASSF7 competes with Max to interact with c-Myc. B, densitometry analysis of immunoblots from competitive binding assay. Relative band intensities of RASSF7 and Max in complex with c-Myc are represented. C, RT-qPCR analysis indicates that RASSF7 modulates the c-Myc–dependent transcription of target genes in a dose-dependent manner. β-Actin served as internal control (n = 3 and data are expressed as mean ± S.D.). D, co-immunoprecipitation (IP) assay using mutants of RASSF7 in HEK293T cells indicates that leucine residues within HLH–L-zipper domain are critical for RASSF7 interaction with c-Myc. E, both RA and HLH–L-zipper domains of RASSF7 interact with c-Myc independently. WB, Western blot.
RASSF7 interacts with c-Myc through RA and LZ domains
We next mapped the domains in RASSF7 critical for its interaction with c-Myc. Various N-terminal deletion constructs of RASSF7 were generated (Fig. S3A) and transfected in HEK293T cells. Significant reduction of RASSF7301–374 interaction with c-Myc (Fig. S3B, lane 8) indicated that the interacting domain might be located between amino acids 250 and 300 of RASSF7. Detailed analysis of amino acids within 250–300 of RASSF7 indicated the presence of a helix-loop-helix–leucine zipper domain (HLH–L-Zip), which is known to be involved in protein–protein and protein–nucleic acid interactions (20, 21). In addition, there is one more leucine-rich domain present within the N terminus of RASSF7 (Fig. S3C). To find out the involvement of RASSF7 leucine zipper domains for its interaction with c-Myc, various RASSF7 mutants were generated (Fig. S3C) and the co-immunoprecipitation assay was performed. Surprisingly, all leucine variants of RASSF7 interacted with c-Myc similar to WT RASSF7 (Fig. S3D) indicating a possibility for the involvement of an additional domain in RASSF7 on c-Myc interaction. We next generated various C-terminal deletion variants of RASSF7 (Fig. S3E) and analyzed their interaction with c-Myc. Interestingly, RASSF7 mutants without L-Zip domains retain interaction with c-Myc as like full-length RASSF7 (Fig. S3F, lanes 3–6) confirms that the presence of an additional domain within the N terminus of RASSF7 might be responsible for the observed RASSF7 interaction with c-Myc. To confirm the participation of RASSF7 HLH–L-Zip domain in the c-Myc interaction, we generated expression vector containing a RASSF7 HLH–L-Zip domain alone as fusion with GFP and exchanged all leucine residues (Fig. S4A). Interestingly, the HLH–L-Zip domain is able to interact with c-Myc, such as full-length RASSF7 and replacement of leucine residues abrogated its interaction with c-Myc (Fig. 5D, lane 4). This was further substantiated by molecular dynamics simulations indicating a weak ΔG° (ΔΔG° = +6.39 kcal/mol) for the c-Myc/RASSF7 HLH–L-ZipL-A complex (Fig. S4B and Video S3). These results suggest that leucine residues might be critical for the formation of a stable heterodimeric complex between the HLH–L-Zip domains of c-Myc and RASSF7. To identify the additional c-Myc interacting domain within the RASSF7 N terminus, we first compared the RASSF7 N-terminal sequence with known a c-Myc interaction domain of p21 (CDKN1A). The analysis indicated a conservation of amino acid motifs within the RA domain of RASSF7 with p21 (Fig. S4C). We next replaced the amino acids (indicated in box) that are conserved between p21 and the RA domain as well as the leucine residues within HLH–L-Zip domain of RASSF7 (Fig. S4D). Results from the co-immunoprecipitation experiments indicate that RASSF7 variants with mutations in either RA or the HLH–L-Zip domain retain WT interaction with c-Myc (Fig. 5E, bottom panel, lane 3 and 4). Interestingly, mutations in both RA and HLH–L-Zip domain (RASSF7MUT) completely abrogated RASSF7 interaction with c-Myc (Fig. 5E, bottom panel, lane 5). Taken together, these data suggest that RASSF7 interacts with c-Myc via both RA and leucine zipper domains independently.
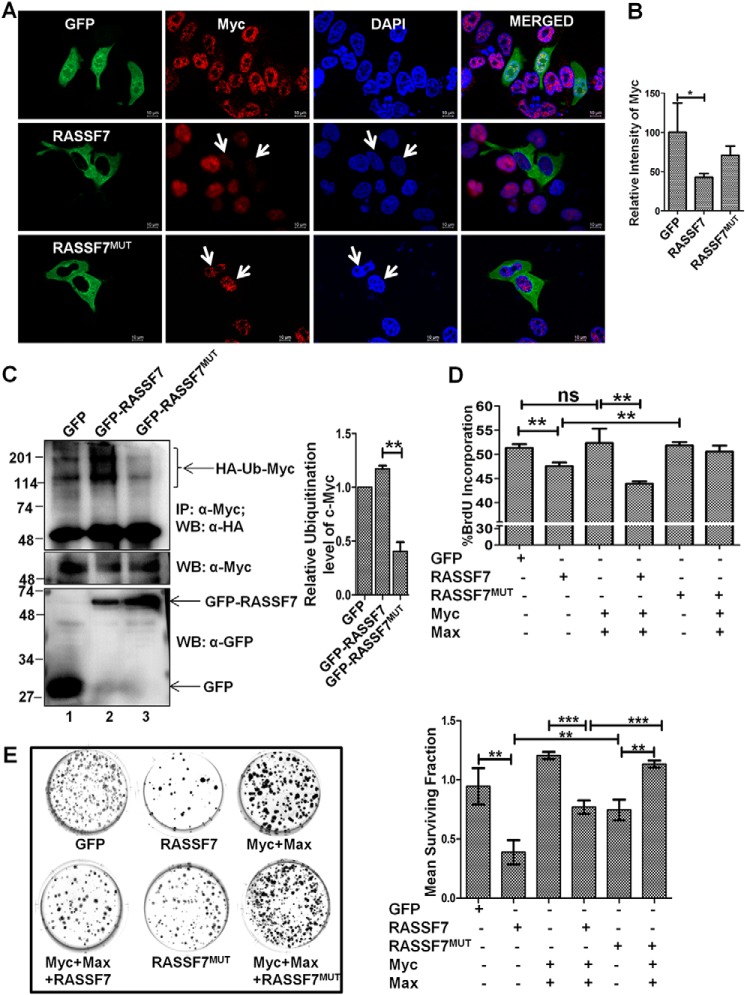
RASSF7 interaction with c-Myc is critical to inhibit cell proliferation
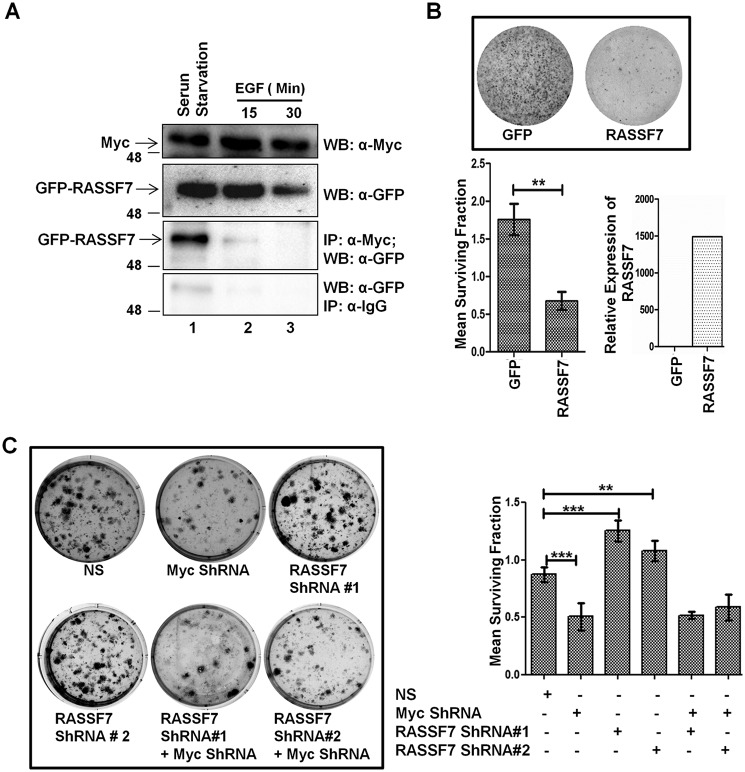
To understand the functional consequences of the RASSF7 interaction with c-Myc, we determined the cellular localization of c-Myc in the presence of RASSF7 by immunofluorescence assay. Interestingly, a c-Myc–specific signal was significantly reduced in cells expressing WT RASSF7. In contrast, c-Myc signals were not altered in cells expressing a c-Myc interaction-deficient mutant of RASSF7 (RASSF7MUT) (Fig. 6, A and B). These data lead to the hypothesis that the RASSF7 interaction with c-Myc may be critical to deplete the cellular c-Myc protein level. To confirm the specificity of RASSF7-mediated c-Myc destabilization, the status of c-Myc ubiquitination was measured in HEK293T cells ectopically expressed with RASSF7WT or RASSF7MUT. Interestingly, RASSF7MUT failed to induce polyubiquitination of c-Myc (Fig. 6C, lane 3). These data support our hypothesis that the RASSF7 interaction is critical for c-Myc polyubiquitination. We next probed the relevance of the RASSF7 interaction on c-Myc–induced cell proliferation. HEK293T cells were transiently expressed with RASSF7WT or RASSF7MUT in combination with c-Myc and Max and performed a BrdU incorporation assay. Expression of RASSF7WT not RASSF7MUT inhibited c-Myc–induced DNA synthesis (Fig. 6D, Fig. S5) indicating that the RASSF7 interaction is required for modulating the c-Myc function during the cell cycle. In addition, RASSF7 blocked c-Myc–induced cell proliferation as indicated by a lesser number of colonies in WT but not in the c-Myc interaction-deficient mutant (RASSF7MUT) expressing HEK293T cells (Fig. 6E). A similar pattern was observed in HeLa cells as well (Fig. S6A). Furthermore, RASSF7WT is able to significantly inhibit the cell migration even in the presence of c-Myc but RASSF7MUT failed to alter c-Myc–dependent cell migration (Fig. S6, B and C). Collectively, results from this investigation provide evidence that the RASSF7 interaction with c-Myc is critical to modulate c-Myc–induced cell proliferation and migration. To define the physiological relevance of the RASSF7/c-Myc heterodimeric complex formation with cell growth status, HeLa cells were subjected to serum starvation followed by treatment with epidermal growth factor (EGF) for different time points and checked their interaction. Interestingly, RASSF7 and c-Myc interaction was observed in serum-starved cells but complex formation was disrupted when cells were treated with EGF (Fig. 7A). These results suggest that the dissociation of RASSF7 from the RASSF7/c-Myc complex may be critical for c-Myc to induce cell proliferation under favorable growth conditions. Taken together, these data implicate that the cross-talk between RASSF7 and c-Myc might play a critical role in cell growth regulation.
Figure 6.
RASSF7 interaction with c-Myc is required for negative regulation of c-Myc function. A, immunofluorescence analysis indicate that WT not the c-Myc interaction-deficient mutant (RASSF7MUT) of RASSF7 deplete the total c-Myc protein in HeLa cells. RASSF7: GFP signal, green; c-Myc, Red; nuclei, blue. B, intensity of c-Myc in HeLa cells were calculated using ImageJ and plotted as a bar diagram (n = 50 cells each). C, ubiquitination assay suggests that WT not the c-Myc interaction-deficient mutant (RASSF7MUT) of RASSF7 induced ubiquitination of c-Myc in HEK293T cells. D, BrdU incorporation assay indicates that RASSF7 alters c-Myc–induced DNA synthesis (n = 3 and data are expressed as mean ± S.D.). E, RASSF7 expression blocks the c-Myc–induced colony formation ability of HEK293T cells (n = 3 and data are expressed as mean ± S.D.). WB, Western blot; IP, immunoprecipitation.
Figure 7.
RASSF7 alters c-Myc oncogenic functions in a context dependent manner. A, HeLa cells were subjected to serum starvation for 24 h followed by induction with EGF and tested the status of RASSF7 and c-Myc interaction by immunoprecipitation (IP) assay. Results suggest that interaction between RASSF7 and c-Myc is abrogated upon stimulation of cells with EGF. B, RASSF7 reduced the colony-forming ability of NIH3T3 cells. Colonies were counted using ImageJ. The surviving fraction was calculated and plotted as a bar diagram (n = 3 and data are expressed as mean ± S.D.). Expression of RASSF7 in NIH3T3 cells was quantified by RT-qPCR. β-Actin was used as internal control. C, colony-forming assay in NIH3T3 cells indicate that RASSF7 inhibits c-Myc-mediated cell transformation (n = 3 and data are expressed as mean ± S.D.). WB, Western blot.
RASSF7 inhibits c-Myc–induced cell transformation
To understand the importance of RASSF7 function during tumorigenesis, the transformation assay was performed with the immortalized mouse embryonic fibroblast cell line NIH3T3 (22) to check whether RASSF7 expression alters the c-Myc–dependent cell transformation. Results suggest that ectopic expression of RASSF7 significantly reduced the colony-forming ability of NIH3T3 (Fig. 7B). In contrast, knockdown of RASSF7 significantly increased the colony-forming ability of NIH3T3 cells (Fig. S7A). The efficiency of RASSF7 depletion was confirmed by RT-qPCR (Fig. S7B). We subsequently depleted RASSF7 and c-Myc expressions individually and in combination by specific shRNAs and tested the effect of RASSF7 on Myc-mediated transformation of immortalized NIH3T3 cells. Interestingly, an increased number of colonies were observed in the RASSF7 knockdown condition and this effect was reversed when both c-Myc and RASSF7 were depleted together (Fig. 7C). Collectively, these results suggest that RASSF7 regulates c-Myc–induced cell transformation, thereby controlling cell proliferation during tumorigenesis.
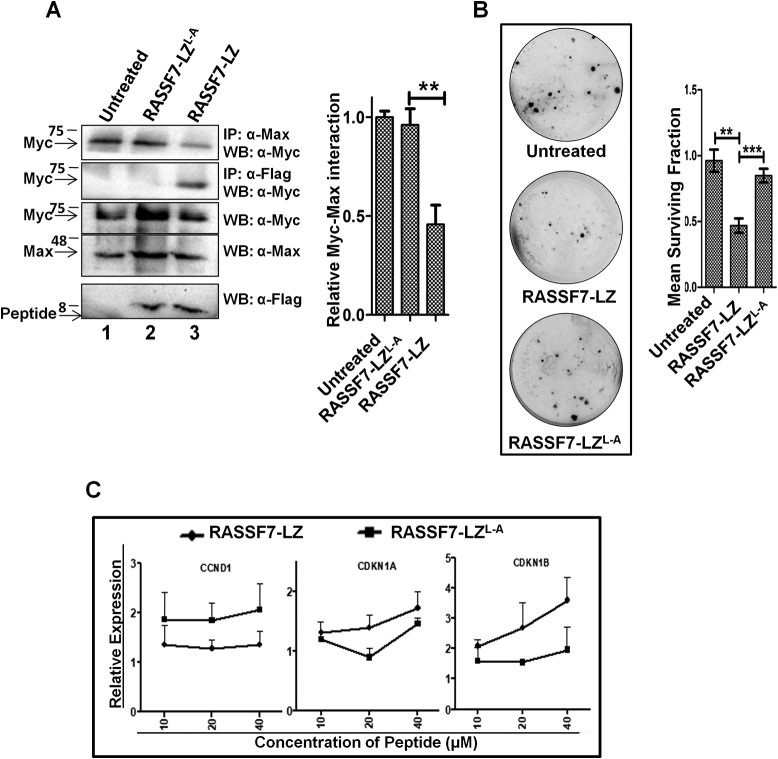
RASSF7-LZ peptide is sufficient to modulate c-Myc function
To further test the hypothesis that RASSF7 may be used to target c-Myc function, the RASSF7 leucine zipper domain (RASSF7251–300) WT (RASSF7-LZ) and mutant peptide (RASSF7-LZL-A) were designed as described under “Materials and Methods” and chemically synthesized. HEK293T cells were treated with WT and mutant RASSF7-LZ peptides and checked their interaction with Myc by co-immunoprecipitation followed by Western blot analysis with the indicated antibodies. Results indicate that WT not mutant peptide was able to interact with c-Myc (Fig. 8A, lane 3), which supports that the leucine zipper domain of RASSF7 independently interacts with c-Myc. Interestingly, the complex formation between c-Myc and Max was reduced upon treatment with WT peptide (Fig. 8A, upper panel, lane 3) compared with mutant peptide or untreated cells suggest that the RASSF7-LZ peptide might interfere with the c-Myc/Max complex formation by interacting with c-Myc. Furthermore, our results suggest that the RASSF7-LZ peptide inhibits the cell proliferation (Fig. S7C) as well as colony-forming ability of cells (Fig. 8B). To extend the ability of the RASSF7-LZ peptide to impinge on c-Myc function, transcript levels of c-Myc transcriptional targets were analyzed upon treatment of RASSF7-LZ or RASSF7-LZL-A peptide. Results indicate that WT not mutant RASSF7-LZ peptide was able to inhibit c-Myc–dependent transcription of target genes in a dose-dependent manner (Fig. 8C). In addition, RASSF7-LZ peptide treatment reduced the endogenous c-Myc level when compared with RASSF7-LZL-A peptide-treated cells (Fig. S7D). For the first time, the data from these experiments provide crucial evidence that RASSF7-LZ peptide alone is sufficient to modulate c-Myc function and may have potential therapeutic application to control c-Myc activity in a subset of human cancers.
Figure 8.
RASSF7-leucine zipper (LZ) peptide is sufficient to modulate Myc activity. A, co-immunoprecipitation (IP) suggests that RASSF7-LZ peptide interacts with c-Myc thereby reducing the c-Myc/Max complex formation, whereas mutant peptide, which lack interaction with c-Myc, failed to interfere with the c-Myc/Max complex formation. B, colony-forming assay in HEK293T cells treated with peptides reveal that the RASSF7-LZ peptide inhibits the colony-forming ability (n = 3 and data are expressed as mean ± S.D.). C, RT-qPCR analysis indicates that treatment with RASSF7-LZ peptide reverses expression levels of c-Myc target genes in a dose-dependent manner. β-Actin served as internal control (n = 3 and data are expressed as mean ± S.D.). WB, Western blot.
Discussion
With the aim to understand the mechanism of the nonenzymatic Ras effector, RASSF7 function in human cancers, the current study demonstrates that RASSF7 is a novel interactor of c-Myc and inhibits c-Myc oncogenic functions at multiple levels (Fig. 9). The steady-state level of c-Myc was altered by many E3 ligases in a context-dependent manner (23–26). Myc oncogene family members are known to be amplified in multiple human cancers and are key driver molecules regulating oncogenesis (27–30). Many approaches have been strategized to target c-Myc in human cancer (31–33). Attempts were made to engineer E3 ligases to target c-Myc for efficient degradation in cancers (34, 35). In addition, several small molecule inhibitors have been developed to inhibit c-Myc function for better management of c-Myc driven cancers (23, 36). However, c-Myc family members encode transcription factors to carry out essential functions in normal cells, rendering it a major challenge to target c-Myc function making c-Myc a prototypical example of a “undruggable target” (31). Our data provide strong evidence that RASSF7 destabilizes oncogenic c-Myc protein. Available literatures (16, 17) clearly indicate that accumulation of more oncogenic c-Myc protein in the lymphoblastic leukemia without gene amplification suggests that the stability of c-Myc protein was altered. On the other hand, RASSF7 expression was repressed in leukemia due to promoter hypermethylation (37). This interplay between RASSF7 and c-Myc may open up new avenues to exploit RASSF7 as a potential molecule for therapeutic intervention of c-Myc–driven cancers like lymphoblastic leukemia.
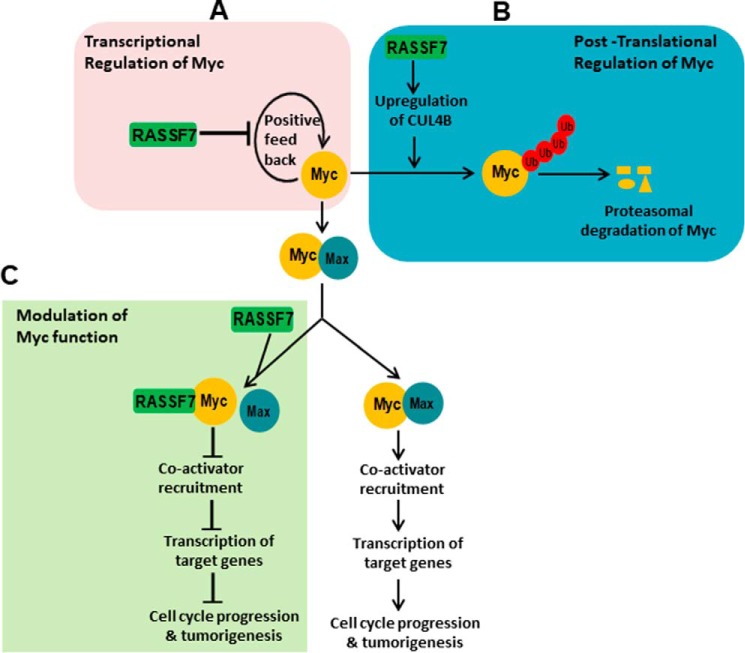
Figure 9.
Proposed model of RASSF7-mediated regulation of oncogenic c-Myc function. Results from the present investigation clearly provided evidence that RASSF7 regulates c-Myc functions at three levels. A, RASSF7 regulates c-Myc at the transcriptional level by inhibiting c-Myc promoter activity thereby reducing the mRNA levels of c-Myc. B, RASSF7 promotes polyubiquitination of c-Myc by up-regulating the expression of ligase, Cullin4B. C, RASSF7 modulates oncogenic c-Myc function by forming a heterodimeric complex with c-Myc and blocks c-Myc–mediated recruitment co-activator to the target gene promoters to induce cell cycle arrest. Taken together, our data indicate that RASSF7 impinges on expression and function of c-Myc in multiple levels to modulate oncogenic c-Myc function during tumorigenesis.
Formation of heterodimeric complex between c-Myc and Max is a prerequisite to activate a canonical c-Myc–mediated transcriptional regulatory pathway (38). c-Myc/Max heterodimeric complex occupies the E-box motif in the target gene promoters and recruits the transcriptional co-activators to trigger the transcription of target genes (21, 38). Several small molecule inhibitors have been designed to disrupt c-Myc/Max dimers and to inhibit c-Myc–induced transformation (39). Biochemical assays and molecular dynamics simulations revealed that RASSF7 forms a stable complex with c-Myc thereby inhibiting the assembly of the active c-Myc/Max heterodimer. In addition, RASSF7 inhibits the expression of c-Myc–activated target genes that are required for cell proliferation as well as the colony-forming ability of immortalized cells by altering c-Myc activity. Furthermore, under favorable growth conditions (in the presence of epidermal growth factor) we found that RASSF7 did not form a complex with c-Myc suggesting the existence of tight regulation of RASSF7 and c-Myc cross-talk during cell proliferation.
Therapeutic peptides targeting the interaction between oncogenes have seen wider applications in cancer treatment regimens. Novel peptides that have been designed to inhibit signaling cascades are proving to be a viable alternative to chemical small molecule inhibitors due to low off-target effects and more specificity (40). There have been attempts to develop a synthetic inhibitory peptide to inhibit c-Myc/Max interaction (39, 41). Applications of synthetic peptides, although superior to small molecules inhibitors, are hindered by limited knowledge about the precise molecular functions of the developed novel peptides. To circumvent such drawbacks, development of peptides from endogenous proteins could be a more practical approach. Interestingly, data from the current investigation suggest that the RASSF7-LZ peptide interacts with c-Myc and is sufficient to impinge on c-Myc activity. Although further analysis on precise molecular function of the RASSF7-LZ peptide is warranted, the data from this study indicate that RASSF7-LZ may have potential therapeutic applications to control c-Myc–dependent cancers and may be a potential molecule targeting the otherwise “undruggable” c-Myc.
A recent report suggests that RASSF7 might be a candidate target in c-Myc–driven cancers (42). Here, we report for the first time that RASSF7 inhibits c-Myc–mediated DNA synthesis and arrest the “S” phase progression of the cell cycle. In addition, RASSF7 destabilizes the c-Myc protein and modulates its oncogenic function. These results suggest that RASSF7 may be a potential candidate regulator of the cell division cycle and impinges on c-Myc–induced cell proliferation. This study is a unique demonstration of a functional link between the nonenzymatic RAS effector, RASSF7, and c-Myc to control cancer cell growth and potential application of LZ peptide as a possible drug molecule, which enables the undruggable c-Myc as a potential therapeutic target in c-Myc–driven human cancers. Conceivably, the expression status of RASSF7 and c-Myc may be used as biomarkers for designing better strategies for the management of cancers.
Materials and methods
Cell culture and transfection
HEK293T, HeLa, SCC131, AGS, IMR32, SKBR3, COLO 829, HCT 116, and NIH3T3 cell lines were maintained Dulbecco's modified Eagle's medium (Life Technologies) with 10% fetal bovine serum (Gibco) and 1% antibiotic-antimycotic (Life Technologies). Polyethylene imine (PAL Lifesciences) was used for transfection.
Plasmid construction and mutagenesis
RASSF7 and its deletion expression constructs were constructed by amplifying ORF from peripheral blood mononuclear cell cDNA library using appropriate primers and cloned between KpnI and EcoRV sites of pCDNA3-GFP plasmid. Point mutations were performed using appropriate primers with QuikChange II Site-directed Mutagenesis Kit (Agilent) as per the manufacturer's protocol. Primers used for cloning are indicated under Table S1. pcDNA3-DN-hCUL4A-FLAG (Addgene plasmid number 15821) and pcDNA3-DN-hCUL4B-FLAG (Addgene plasmid number 15822) were a gift from Wade Harper (43).
Antibodies and chemicals
Anti-GFP, anti-TRRAP, anti-RASSF7 (Santa Cruz), anti-FLAG, anti-HA, and anti-β-actin (Sigma), anti-c-Myc, anti-Max, anti-CUL4B, anti-glyceraldehyde-3-phosphate dehydrogenase, and anti-laminin (Cell Signaling Technology Inc.) antibodies were used for immunoblot analysis. Chemicals used were: MG-132 and cycloheximide (Abcam, UK).
Real Time-quantitative PCR (RT-qPCR), immunoprecipitation, and Western blotting
RT-qPCR, immunoprecipitation, and Western blotting were performed as described previously (44). Primers used for RT-qPCR are described under Table S2.
BrdU incorporation assay
For BrdU assay, cells transfected with the indicated plasmids were pulsed for 4 h with BrdU after 48 h of transfection. Cells were fixed, permeabilized, and stained with anti-BrdU antibodies conjugated with allophycocyanin), followed by total DNA staining with 7-aminoactinomycin as per the manufacturer's protocol (BD Biosciences). The amount of BrdU incorporation was analyzed in BD FACSCanto II (BD Biosciences) using FACSDIVA software (BD Biosciences).
Luciferase assay
Luciferase assay was performed as described previously (45).
Cell cycle analysis and ubiquitination assay
Cell cycle and ubiquitination assays were performed as described elsewhere (46).
ChIP
ChIP analysis was performed as described previously (47) using the indicated antibodies. For ChIP-Western blot analysis, the eluted proteins from the immune complex were resolved in SDS-PAGE (5–15% gradient) followed by Western blot analysis with indicated antibodies.
Fluorescent microscopy
Cells were transfected with the indicated plasmids. After 48 h of transfection, cells were fixed with 4% paraformaldehyde and permeabilized with 0.1% Triton X-100. Cells were then immunostained with anti-c-Myc antibodies overnight at 4 °C followed by secondary antibodies conjugated with Alexa Fluor 594 (Molecular Probes). 4′,6-Diamidino-2-phenylindole was used to stain nucleus. Images were acquired in AxioObserver.Z1 inverted microscope with Apotome (Carl Zeiss, Germany) using ZEN software (Carl Zeiss, Germany). At least 50 cells in each condition were imaged to calculate the fluorescence intensity. Myc intensity was normalized by subtracting the background intensity from the Myc-stained cells and was plotted as graph.
Database analysis
Pan cancer gene expression data from BioXpress database (https://hive.biochemistry.gwu.edu/cgi-bin/prd/bioxpress/servlet.cgi)3 was downloaded for RASSF7 and c-Myc as described previously (45) and correlation analysis was performed.
Lentivirus generation and infection
Lentiviruses expressing shRNA specific to CUL4B (TRCN 0000006532) (Sigma) and RASSF7 (clone ID: V2LHS_172434 and V3LHS_411167) (Dharmacon) were generated in HEK293T cells as per the manufacturer's protocol. Supernatant containing viral particles were collected and used to infect the indicated target cells in the presence of hexadimethrine bromide (8 mg/ml).
Colony formation assay
Colony formation assay was performed as described previously (48). Percentage survival was calculated using the following formula: plating efficiency = number of colonies counted/number of cells plated; surviving fraction = (number of colonies counted/number of cells plated)/plating efficiency of control. Each experiment was repeated three times with biological triplicates. Mean ± S.D. were plotted in the respective graphs.
In vitro cell wound-healing assay
For wound-healing assay, cells were transfected with the indicated plasmids and were subjected to serum starvation after 36 h of transfection. After 48 h of transfection, the wound was generated and wound closure was imaged at different time intervals using Cytell (GE Healthcare). The total wound area was calculated and the percentage of wound closure was plotted in a graph.
Molecular dynamics simulations
The structure of Myc/Max complex (PDB code 1NKP) was used as a template to build Myc/RASSF7HLH-L-Zip complex (49). Unrestrained simulations were performed for 40 ns in TIP3P solvent using AMBER12 (50) and ff12SB force field (51, 52). Energy minimization and equilibrations were performed as described elsewhere (53, 54). The binding free energies (ΔG°) were calculated using MM-PB(GB)SA methods (55–58).
RASSF7-LZ peptide development
RASSF7-leucine zipper domain peptide (RASSF7-LZ) comprising amino acids 251–300 was designed as a fusion protein with the TAT cell penetrating sequence and FLAG tag as mentioned below. Amino acids indicated as bold underlined letters were mutated to alanine in mutant peptide (RASSF7-LZl-A): GYGRKKRRQRRRGDYKDDDDKGQSAEVQGSLALVSRALEAAERALQAQAQELEELNRELRQCNLQQFIQQTG.
Peptides were synthesized by Synpeptide (Shanghai, China) at >95% purity and stored at −20 °C as lyophilized powder aliquots to avoid freeze-thaw cycles. Stock solution of 1 mm was prepared by dissolving in PBS and stored as aliquots in −20 °C. For cell treatment, a working concentration was achieved by diluting in Opti-MEM.
Peptide treatment of cells
In peptide-treatment assays, cells treated with peptide (at a concentration of 20 μm wherever not indicated) for 8 h were harvested, lysed in Nonidet P-40 lysis buffer and subjected to immunoprecipitation as described above under “Immunoprecipitation and Western blotting.” For RT-qPCR assays, cells were treated with peptides for 18 h with Opti-MEM containing the indicated concentration of peptide and levels of transcripts were analyzed by RT-qPCR as described above.
Statistical analysis
Statistical analyses were carried out using GraphPad Prism 5 (GraphPad Software, San Diego, CA). Data are represented as mean ± S.D. from triplicates. Statistical significance indicated by p value was calculated using Student's unpaired t test (*, p < 0.01; **, p < 0.01; ***, p < 0.001).
Author contributions
A. K., A. M., P. D., C. R., and S. M. formal analysis; A. K., A. M., P. D., A. S., L. R. P., C. R., and M. G. validation; A. K., A. M., P. D., A. S., L. R. P., and C. R. investigation; A. K., A. M., P. D., A. S., L. R. P., and C. R. methodology; A. K. and S. M. writing-original draft; M. G., K. R., and S. M. resources; S. M. conceptualization; S. M. supervision; S. M. funding acquisition; S. M. project administration; S. M. writing-review and editing.
Supplementary Material
This work was supported by Department of Science and Technology Grant VI-D&P/411/2012–2013/TDT (G) from the Government of India (to S. M.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S7, Tables S1 and S2, and Videos S1–S3.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- RASSF
- RAS association domain family of protein
- HLH–L-Zip
- helix-loop-helix–leucine zipper domain
- RA
- Ras association
- qPCR
- quantitative PCR
- HA
- hemagglutinin
- shRNA
- short hairpin RNA
- DN
- dominant-negative
- GFP
- green fluorescent protein
- EGF
- epidermal growth factor.
References
- 1. Ehrkamp A., Herrmann C., Stoll R., and Heumann R. (2013) Ras and rheb signaling in survival and cell death. Cancers (Basel) 5, 639–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Patra S. K. (2008) Ras regulation of DNA-methylation and cancer. Exp. Cell Res. 314, 1193–1201 10.1016/j.yexcr.2008.01.012 [DOI] [PubMed] [Google Scholar]
- 3. Zenonos K., and Kyprianou K. (2013) RAS signaling pathways, mutations and their role in colorectal cancer. World J. Gastrointest. Oncol. 5, 97–101 10.4251/wjgo.v5.i5.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vos M. D., and Clark G. J. (2006) RASSF family proteins and Ras transformation. Methods Enzymol. 407, 311–322 [DOI] [PubMed] [Google Scholar]
- 5. Underhill-Day N., Hill V., and Latif F. (2011) N-terminal RASSF family (RASSF7-RASSF10): a mini review. Epigenetics 6, 284–292 10.4161/epi.6.3.14108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Volodko N., Gordon M., Salla M., Ghazaleh H. A., and Baksh S. (2014) RASSF tumor suppressor gene family: biological functions and regulation. FEBS Lett. 588, 2671–2684 10.1016/j.febslet.2014.02.041 [DOI] [PubMed] [Google Scholar]
- 7. Sherwood V., Manbodh R., Sheppard C., and Chalmers A. D. (2008) RASSF7 is a member of a new family of RAS association domain-containing proteins and is required for completing mitosis. Mol. Biol. Cell 19, 1772–1782 10.1091/mbc.e07-07-0652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gulsen T., Hadjicosti I., Li Y., Zhang X., Whitley P. R., and Chalmers A. D. (2016) Truncated RASSF7 promotes centrosomal defects and cell death. Dev. Biol. 409, 502–517 10.1016/j.ydbio.2015.11.001 [DOI] [PubMed] [Google Scholar]
- 9. Recino A., Sherwood V., Flaxman A., Cooper W. N., Latif F., Ward A., and Chalmers A. D. (2010) Human RASSF7 regulates the microtubule cytoskeleton and is required for spindle formation, Aurora B activation and chromosomal congression during mitosis. Biochem. J. 430, 207–213 10.1042/BJ20100883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Takahashi S., Ebihara A., Kajiho H., Kontani K., Nishina H., and Katada T. (2011) RASSF7 negatively regulates pro-apoptotic JNK signaling by inhibiting the activity of phosphorylated-MKK7. Cell Death Differ. 18, 645–655 10.1038/cdd.2010.137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Djos A., Martinsson T., Kogner P., and Carén H. (2012) The RASSF gene family members RASSF5, RASSF6 and RASSF7 show frequent DNA methylation in neuroblastoma. Mol. Cancer 11, 40 10.1186/1476-4598-11-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hitomi J., Christofferson D. E., Ng A., Yao J., Degterev A., Xavier R. J., and Yuan J. (2008). Identification of a molecular signaling network that regulates a cellular necrotic cell death pathway by a genome wide siRNA screen. Cell 135, 1311–1323 10.1016/j.cell.2008.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kim J., Lee J., and Iyer V. R. (2008) Global identification of c-Myc target genes reveals its direct role in mitochondrial biogenesis and its E-box usage in vivo. PLoS ONE 3, e1798 10.1371/journal.pone.0001798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dang C. V. (1999) c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol. Cell Biol. 19, 1–11 10.1128/MCB.19.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wan Q., Dingerdissen H., Fan Y., Gulzar N., Pan Y., Wu T., Jung Y., Cheng Z., Zhang H., and Mazumder R. (2015) BioXpress: an integrated RNA-seq-derived gene expression database for pan-cancer analysis. Database (Oxford) 2015, bav019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Malempati S., Tibbitts D., Cunningham M., Akkari Y., Olson S., Fan G., and Sears R. C. (2006) Aberrant stabilization of c-Myc protein in some lymphoblastic leukemias. Leukemia 20, 1572–1581 10.1038/sj.leu.2404317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bahram F., von der Lehr N., Cetinkaya C., and Larsson L. G. (2000) c-Myc hot spot mutations in lymphomas result in inefficient ubiquitination and decreased proteasome-mediated turnover. Blood 95, 2104–2110 [PubMed] [Google Scholar]
- 18. Poole C. J., and van Riggelen J. (2017) MYC: master regulator of the cancer epigenome and transcriptome. Genes (Basel) 8, 142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McMahon S. B., Van Buskirk H. A., Dugan K. A., Copeland T. D., and Cole M. D. (1998) The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell 94, 363–374 10.1016/S0092-8674(00)81479-8 [DOI] [PubMed] [Google Scholar]
- 20. Landschulz W. H., Johnson P. F., and McKnight S. L. (1988) The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science 240, 1759–1764 10.1126/science.3289117 [DOI] [PubMed] [Google Scholar]
- 21. Leung I. W., and Lassam N. (1998) Dimerization via tandem leucine zippers is essential for the activation of the mitogen-activated protein kinase kinase kinase, MLK-3. J. Biol. Chem. 273, 32408–32415 10.1074/jbc.273.49.32408 [DOI] [PubMed] [Google Scholar]
- 22. Moshier J. A., Dosescu J., Skunca M., and Luk G. D. (1993) Transformation of NIH/3T3 cells by ornithine decarboxylase overexpression. Cancer Res. 53, 2618–2622 [PubMed] [Google Scholar]
- 23. Peter S., Bultinck J., Myant K., Jaenicke L. A., Walz S., Muller J., Gmachl M., Treu M., Boehmelt G., Ade C. P., Schmitz W., Wiegering A., Otto C., Popov N., Sansom O., Kraut N., and Eilers M. (2014) Tumor cell-specific inhibition of MYC function using small molecule inhibitors of the HUWE1 ubiquitin ligase. EMBO Mol. Med. 6, 1525–1541 10.1002/emmm.201303745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Amati B. (2004) c-Myc degradation: Dancing with ubiquitin ligases. Proc. Natl. Acad. Sci. U.S.A. 101, 8843–8844 10.1073/pnas.0403046101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Choi S. H., Wright J. B., Gerber S. A., and Cole M. D. (2010) c-Myc protein is stabilized by suppression of a novel E3 ligase complex in cancer cells. Genes Dev. 24, 1236–1241 10.1101/gad.1920310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Young S. Y., Herbst A., Tworkowski K. A., Salghetti S. E., and Tansey W. P. (2003) Skp2 regulates c-Myc protein stability and activity. Mol. Cell. 11, 1177–1188 10.1016/S1097-2765(03)00173-4 [DOI] [PubMed] [Google Scholar]
- 27. Dang C. V. (2012) MYC on the Path to Cancer. Cell 149, 22–35 10.1016/j.cell.2012.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gabay M., Li Y., and Felsher D. W. (2014) MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harbor Persp. Med. 4, a014241 10.1101/cshperspect.a014241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Camarda R., Williams J., and Goga A. (2017) In vivo reprogramming of cancer metabolism by MYC. Front. Cell Dev. Biol. 5, 35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nero T. L., Morton C. J., Holien J. K., Wielens J., and Parker M. W. (2014) Oncogenic protein interfaces: small molecules, big challenges. Nat. Rev. Cancer 14, 248–262 10.1038/nrc3690 [DOI] [PubMed] [Google Scholar]
- 31. Whitfield J. R., Beaulieu M.-E., and Soucek L. (2017) Strategies to inhibit c-Myc and their clinical applicability. Front. Cell Dev. Biol. 5, 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sabnis H. S., Somasagara R. R., and Bunting K. D. (2017) Targeting MYC dependence by metabolic inhibitors in cancer. Genes 8, 114 10.3390/genes8040114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hutter S., Bolin S., Weishaupt H., and Swartling F. J. (2017) Modeling and targeting MYC genes in childhood brain tumors. Genes 8, 107 10.3390/genes8040107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shin Y. J., Park S. K., Jung Y. J., Kim Y. N., Kim K. S., Park O. K., Kwon S. H., Jeon S. H., Trinh le E., Fraser A. S., Kee Y., and Hwang B. J. (2015) Nanobody-targeted E3-ubiquitin ligase complex degrades nuclear proteins. Sci. Rep. 5, 14269 10.1038/srep14269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lu J., Qia Y., Altieri M., Dong H., Wang J., Raina K., Hines J., Winkler J. D., Crew A. P., Coleman K., and Crews C. M. (2015) Hijacking the E3 ubiquitin ligase cereblon to efficiently target BRD4. Chem. Biol. 22, 755–763 10.1016/j.chembiol.2015.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jiang H., Bower K., E., Beuscher A. E., Zhou B., Bobkov A. A., Olson A. J., and Vogt P., K. (2009) Stabilizers of the Max homodimer identified in virtual ligand screening inhibit c-Myc function. Mol. Pharmacol. 76, 491–502 10.1124/mol.109.054858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shinawi T., Hill V., Dagklis A., Baliakas P., Stamatopoulos K., Agathanggelou A., Stankovic T., Maher E. R., Ghia P., and Latif F. (2012) KIBRA gene methylation is associated with unfavorable biological prognostic parameters in chronic lymphocytic leukemia. Epigenetics 7, 211–215 10.4161/epi.7.3.19222 [DOI] [PubMed] [Google Scholar]
- 38. Blackwood E. M., and Eisenman R. N. (1991) Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with c-Myc. Science 251, 1211–1217 10.1126/science.2006410 [DOI] [PubMed] [Google Scholar]
- 39. Berg T., Cohen S. B., Desharnais J., Sonderegger C., Maslyar D. J., Goldberg J., Boger D. L., and Vogt P. K. (2002) Small-molecule antagonists of c-Myc/Max dimerization inhibit c-Myc-induced transformation of chicken embryo fibroblasts. Proc. Natl. Acad. Sci. U.S.A. 99, 3830–3835 10.1073/pnas.062036999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bidwell G. L. 3rd, and Raucher D. (2009) Therapeutic peptides for cancer therapy: part I, peptide inhibitors of signal transduction cascades. Expert Opin. Drug Deliv. 6, 1033–1047 10.1517/17425240903143745 [DOI] [PubMed] [Google Scholar]
- 41. Bidwell G. L. 3rd, and Raucher D. (2005) Application of thermally responsive polypeptides directed against c-Myc transcriptional function for cancer therapy. Mol. Cancer Ther. 4, 1076–1085 10.1158/1535-7163.MCT-04-0253 [DOI] [PubMed] [Google Scholar]
- 42. Toyoshima M., Howie H. L., Imakura M., Walsh R. M., Annis J. E., Chang A. N., Frazier J., Chau B. N., Loboda A., Linsley P. S., Cleary M. A., Park J. R., and Grandori C. (2012) Functional genomics identifies therapeutic targets for MYC-driven cancer. Proc. Natl. Acad. Sci. U.S.A. 109, 9545–9550 10.1073/pnas.1121119109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jin J., Ang X. L., Shirogane T., and Wade Harper J. (2005) Identification of substrates for F-box proteins. Methods Enzymol. 399, 287–309 10.1016/S0076-6879(05)99020-4 [DOI] [PubMed] [Google Scholar]
- 44. Datta D., Anbarasu K., Rajabather S., Priya R. S., Desai P., and Mahalingam S. (2015) Nucleolar GTP-binding protein-1 (NGP-1) promotes G1 to S phase transition by activating cyclin-dependent kinase inhibitor p21 Cip1/Waf1. J. Biol. Chem. 290, 21536–21552 10.1074/jbc.M115.637280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Thoompumkal I. J., Rehna K., Anbarasu K., and Mahalingam S. (2016) Leucine zipper down-regulated in cancer-1 (LDOC1) interacts with guanine nucleotide binding protein-like 3-like (GNL3L) to modulate nuclear factor-κB (NF-κB) signaling during cell proliferation. Cell Cycle 15, 3251–3267 10.1080/15384101.2016.1242534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Suryaraja R., Anitha M., Anbarasu K., Kumari G., and Mahalingam S. (2013) The E3 ubiquitin ligase Itch regulates tumor suppressor protein RASSF5/NORE1 stability in an acetylation-dependent manner. Cell Death Dis. 4, e565 10.1038/cddis.2013.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gade P., and Kalvakolanu D. V. (2012) Chromatin immunoprecipitation assay as a tool for analyzing transcription factor activity. Methods Mol. Biol. 809, 85–104 10.1007/978-1-61779-376-9_6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Karthik I. P, Desai P., Sukumar S., Dimitrijevic A., and Rajalingam K., and Mahalingam S. (2018) E4BP4/NFIL3 modulates the epigenetically repressed Ras effector RASSF8 function through histone methyl transferases. J. Biol. Chem. 293, 5624–5635 10.1074/jbc.RA117.000623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sali A., and Blundell T. L. (1993) Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 10.1006/jmbi.1993.1626 [DOI] [PubMed] [Google Scholar]
- 50. Pearlman D. A., Case D. A., Caldwell J. W., Ross W. S., Cheatham T. E., DeBolt S., Ferguson D., Seibel G., and Kollman P. (1995) AMBER, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules. Comp. Phys. Commun. 91, 1–41 10.1016/0010-4655(95)00041-D [DOI] [Google Scholar]
- 51. Ponder J. W., and Case D. A. (2003) Force fields for protein simulations. Adv. Protein Chem. 66, 27–85 10.1016/S0065-3233(03)66002-X [DOI] [PubMed] [Google Scholar]
- 52. Duan Y., Wu C., Chowdhury S., Lee M. C., Xiong G., Zhang W., Yang R., Cieplak P., Luo R., Lee T., Caldwell J., Wang J., and Kollman P. (2003) A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem. 24,1999–2012 10.1002/jcc.10349 [DOI] [PubMed] [Google Scholar]
- 53. Ramakrishnan C., Joshi V., Joseph J. M., Vishwanath B. S., and Velmurugan D. (2014) Identification of novel inhibitors of Daboia russelli phospholipase A2 using the combined pharmacophore modeling approach. Chem. Biol. Drug Des. 84, 379–392 10.1111/cbdd.12332 [DOI] [PubMed] [Google Scholar]
- 54. Ramakrishnan C., Subramanian V., and Velmurugan D. (2010) Molecular dynamics study of secretory phospholipase A2 of Russell's viper and bovine pancreatic sources. J. Phys. Chem. B 114, 13463–13472 10.1021/jp102073f [DOI] [PubMed] [Google Scholar]
- 55. Wang J., Tingjun H., and Xiaojie X. (2006) Recent advances in free energy calculations with a combination of molecular mechanics and continuum models. Curr. Comp. Aided Drug Des. 2, 287–306 10.2174/157340906778226454 [DOI] [Google Scholar]
- 56. Wang W.,Oreola D., Reyes C. M., and Kollman P. A. (2001) Biomolecular simulations: recent developments in force fields, simulations of enzyme catalysis, protein-ligand, protein-protein, and protein-nucleic acid noncovalent interactions. Annu. Rev. Biophys. Biomol. Struct. 30, 211–243 10.1146/annurev.biophys.30.1.211 [DOI] [PubMed] [Google Scholar]
- 57. Kollman P. A., Massova I., Reyes C., Kuhn B., Huo S., Chong L., Lee M., Lee T., Duan Y., Wang W., Donini O., Cieplak P., Srinivasan J., Case D. A., and Cheatham T. E. 3rd (2000) Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc. Chem. Res. 33, 889–897 10.1021/ar000033j [DOI] [PubMed] [Google Scholar]
- 58. Miller B. R. 3rd, McGee D. T., Swails J. M., Homeyer N., Gohlke H., and Roitberg A. E. (2012) MMPBSA.py: an efficient program for end-state free energy calculations. J. Chem. Theory Comput. 8, 3314–3321 10.1021/ct300418h [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.