Abstract
The RNA-binding protein Musashi 2 (MSI2) has emerged as an important regulator in cancer initiation, progression, and drug resistance. Translocations and deregulation of the MSI2 gene are diagnostic of certain cancers, including chronic myeloid leukemia (CML) with translocation t(7;17), acute myeloid leukemia (AML) with translocation t(10;17), and some cases of B-precursor acute lymphoblastic leukemia (pB-ALL). To better understand the function of MSI2 in leukemia, the mRNA targets that are bound and regulated by MSI2 and their MSI2-binding motifs need to be identified. To this end, using photoactivatable ribonucleoside cross-linking and immunoprecipitation (PAR-CLIP) and the multiple EM for motif elicitation (MEME) analysis tool, here we identified MSI2's mRNA targets and the consensus RNA-recognition element (RRE) motif recognized by MSI2 (UUAG). Of note, MSI2 knockdown altered the expression of several genes with roles in eukaryotic initiation factor 2 (eIF2), hepatocyte growth factor (HGF), and epidermal growth factor (EGF) signaling pathways. We also show that MSI2 regulates classic interleukin-6 (IL-6) signaling by promoting the degradation of the mRNA of IL-6 signal transducer (IL6ST or GP130), which, in turn, affected the phosphorylation statuses of signal transducer and activator of transcription 3 (STAT3) and the mitogen-activated protein kinase ERK. In summary, we have identified multiple MSI2-regulated mRNAs and provided evidence that MSI2 controls IL6ST activity that control oncogenic signaling networks. Our findings may help inform strategies for unraveling the role of MSI2 in leukemia to pave the way for the development of targeted therapies.
Keywords: post-transcriptional regulation, RNA-binding protein, leukemia, interleukin 6 (IL-6), Janus kinase (JAK), Janus kinase (JAK)/STAT signaling, mitogen-activated protein kinase (MAPK), mitogen-activated protein kinase (MAPK) signaling, phosphorylation, cancer, Musashi 2, cancer and Musashi2, PAR-CLIP
Introduction
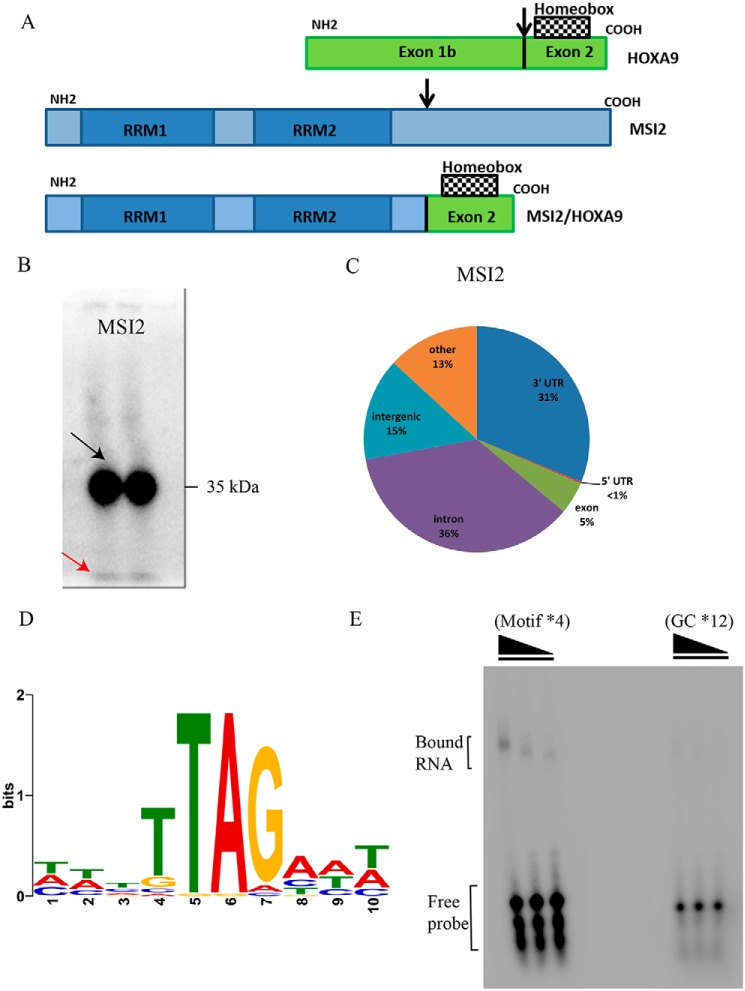
Cell division is orchestrated by signal transduction pathways composed of multiple proteins including transcription factors, receptor G proteins, and protein kinases. Genetic alterations such as chromosomal translocations in these proteins turn them into oncogenes, leading to loss of function and uncontrolled cell growth. Recurrent (balanced) chromosomal translocations in oncogenes like MLL, ETV6, BCL2, and c-Myc lead to multiple hematological malignancies (1). Recently, a rare but recurrent new translocation in gene MSI2 was identified at disease progression stage of chronic myeloid leukemia (CML)7 (2). MSI2 forms a fusion product with HOXA9, thus forming a cryptic in-frame MSI2-HOXA9 with translocation t(7;17) fusion transcript (Fig. 1A). The breakpoint on HOXA9 occurs in exon 1b and in MSI2 after exon 9, which results in a fusion transcript that retains both the RNA-recognition elements (RRE) of MSI2 and the homeobox domain of HOXA9. HOXA9 belongs to family of homeobox-containing transcription factors that have roles in morphogenesis, proliferation, and hematopoietic stem cell expansion (3). HOXA9 is commonly deregulated in acute leukemia and is a strong predictor of poor prognosis. Almost 50% of AML cases have 2- to 8-fold higher expression of HOXA9 compared with healthy individuals (4, 5). Although HOXA9 family of genes has been well-studied, less is known about the fusion partner MSI2. Musashi RNA-binding protein 2 (MSI2) is one of the two Musashi family of RNA-binding proteins (RBPs) containing two RNA recognition motifs (Fig. S1A) (6). MSI2 was found be highly expressed in multiple hematological malignancies including AML, CML, and B-ALL (2, 7, 8). MSI2 plays a major role as cell cycle regulator and maintainer of stem cell phenotype by balancing self-renewal and differentiation in HSCs, thus balancing HSC homeostasis (9–11). Despite some overall progress, the RNA-binding protein properties and the actually regulated mRNAs of MSI2 have not been studied in detail. Few publications have previously searched for the binding partners using techniques such as HITS-CLIP and RIP-CHIP, and although these techniques provide good insights into the interactome of the investigated RBP, they have several drawbacks related to the cross-linking efficiency and the use of UV-254 (12–15). Therefore, we used PAR-CLIP (16) to gain insight into the transcriptome-wide mRNA targets and the RNA-binding motif of MSI2. PAR-CLIP utilizes a photoactivatable nucleoside, 4-S-uridine, to achieve higher cross-linking efficiency and determine binding sites with higher resolution compared with other techniques.
Figure 1.
RNA–protein interactions and RNA-binding motif of the Musashi 2. A, schematic representation of HOXA9, MSI2, and the chimeric MSI2/HOXA9. The arrows represent break points of translocation. B, phosphorimage of SDS-PAGE gel that resolved 32P-labeled RNA-FLAG-HA MSI2 immunoprecipitate in two lanes. The black arrow points to the radioactive bands corresponding to the expected size of the MSI2 (running at 35 kDa), and the red arrow to the radioactive running front. C, distribution of PAR-CLIP–obtained clusters within introns, exons, and UTRs. Numbers indicate the total percentage of CCs within mRNA gene bodies. D, nucleotide sequence of RNA motif inferred from MSI2-binding sites. E, phosphorimage of native PAGE resolving complexes of MSI2 protein with RNA oligonucleotides: GC × 12 as control and motif × 4. Bound RNA and free probe are indicated.
We here report targets regulated by MSI2 post-transcriptionally, which play roles in functions including DNA replication, DNA repair, and cell death and survival. Pathways regulated by MSI2 included EGF, HGF signaling, and eIF2 signaling pathways. The EGF and HGF pathways in turn regulate PI3K/AKT, JAK/STAT, and ERK/MAPK pathways. We investigated one of the most highly regulated targets from our pathway analysis, IL6ST, a transmembrane signal transducer belonging to the IL-6 signaling cascade entity. IL-6 signaling includes multiple cytokines like IL-6, IL-11, LIF (leukemia inhibitory factor), OSM (oncostatin M) which together are called IL-6 type cytokines. These cytokines activate the receptor IL6ST which in turn phosphorylates downstream targets, thereby activating JAK/STAT, ERK, and PI3K/AKT signaling pathways (17, 18). The list of targets and the pathways they regulate increase our understanding of the role of MSI2 in leukemia, and thus may pave the way for the development of targeted therapies.
Results
Identification of transcriptome-wide direct mRNA targets of MSI2 by PAR-CLIP
We performed PAR-CLIP to identify the transcriptome-wide targets of MSI2 in human embryonic kidney cells (HEK293, Flp-In TREx) (Fig. 1B). The analysis of the MSI2 PAR-CLIP datasets revealed a total of 20,531 RBP-bound genomic regions after filtering for confident clusters (CCs). CCs contained two or more T-C conversion sites (cross-linked sites) and ≥10 overlapping reads (for cluster lengths and cluster site distances between CCs, see Fig. S1, B and C). Using gene annotation information for GRCh38 we identified 7378 distinct genes targeted by MSI2. We then examined the distribution of CCs across intronic and exonic regions of mRNA primary transcripts. MSI2 bound to intronic regions (39%) and 3′UTR regions (40%) equally, whereas only 7% bound to exonic regions (Fig. 1C).
Identification of the RNA-recognition element of the MSI2 protein
Next, we used the data from PAR-CLIP to identify the actual RNA sequence motif that was bound by the RBPs. Many RBPs bind RNA in a sequence-specific manner, with the recognized binding region usually referred to as the RNA-recognition element. This motif prediction was performed using MEME (multiple EM for motif elicitation). To elucidate the RRE of MSI2, we restricted our analysis to CCs with a T-C mutation frequency of ≥20% and two or more T-C positional changes. The genomic reference sequences of the 100 most abundantly sequenced clusters located in introns, exons, or 3′UTRs were used. MSI2 datasets revealed the motif as TTAG (E-value (MSI2) = 9.3E-44) (Fig. 1D). We performed EMSAs to test a selection of synthetic RNAs representing the MSI2-binding sites using recombinant MSI2 protein. We tested a tetranucleotide repeat of the motif (UUAG) (length of 24 nucleotides) as well as a control with GC repeated 12 times to match the length of the motif probe. We observed a shift in the protein-RNA complex containing the predicted MSI2 motif, whereas a shift is completely absent using the control probe, indicating no binding between the RBP and the control probe (Fig. 1E). The exact motif was present in 76% of the top 100 MSI2 CCs, whereas the remaining clusters contained slight variations of the given motif.
Data integration of PAR-CLIP and RNA-Seq reveals direct mRNA target spectrum of MSI2
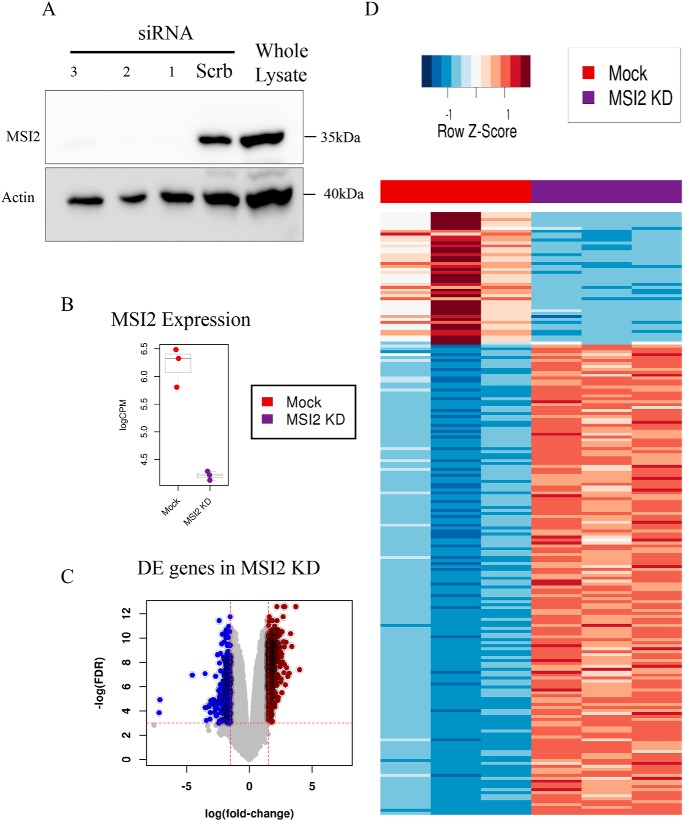
Next, we investigated which transcripts were preferentially bound and directly regulated by MSI2. Therefore, we performed siRNA-mediated knockdown (KD) of MSI2 in HEK293T cells (Fig. 2A) and investigated the gene expression profiles with high-throughput RNA-sequencing (RNA-Seq) (Fig. 2B). We identified differentially expressed genes with stringent thresholds having a logFC (logarithm of fold change) >1.5 or <−1.5 and false discovery rate (FDR) ≤0.05 (Fig. 2C). Next, we integrated the PAR-CLIP data for MSI2 to identify physically bound and differentially regulated genes, which we term as the direct targets of MSI2. In total, we found 230 genes to be directly under regulation of MSI2 (Fig. 2D; Table S1). Among those, 82% of the genes showed a down-regulation versus 18% to be up-regulated upon MSI2 knockdown. Previous RIP-CHIP analysis for MSI2 also revealed a similar pattern of more down-regulated genes versus up-regulated ones upon MSI2 knockdown (13).
Figure 2.
Targets bound and regulated by MSI2. A, Western blotting showing the down-regulation of MSI2 after siRNA knockdown. Antibodies are indicated. Beta-actin was used as loading control and molecular size markers are indicated. B, expression changes of MSI2 in MSI2 KD (logFC = −2.02; FDR = 2.59E-29) measured by RNA-Seq data. C, volcano plots showing transcriptome-wide differences in RNA-Seq expression data upon MSI2 knockdown, with a logFC threshold set to −1.5/1.5 and FDR threshold set to 0.05 (red dotted lines). D, heat map representing the relative difference in the expression of genes between mock and MSI2 knockdown samples.
Several growth signaling pathways are regulated by MSI2
Using our results from direct MSI2 targets, we evaluated the biological processes that are regulated by MSI2 by applying Ingenuity Pathway Analysis (IPA). The entire output of differentially regulated genes for MSI2 KD (FDR ≤0.05 and logFC ≥ ± 1) as well as only the genes, which were differentially expressed and showed at least one confident PAR-CLIP cluster (FDR ≤0.05, logFC ≥ ± 1.5, T-C conversion ≥0.25), were selected to be analyzed by IPA.
The entirety of differentially regulated genes revealed six canonical pathways to be significantly affected (FDR ≤0.05) by MSI2 (Table S2). These included but were not limited to growth factor pathways such as HGF and EGF signaling, which were activated upon MSI2 KD (z-scores of 2.4 and 1.7, respectively). The eIF2 signaling pathway was the most significantly affected pathway upon MSI2 KD and showed a total overlap of 43.6% genes of the pathway with the differentially expressed genes of MSI2 KD. It was predicted to be highly inhibited upon MSI2 knockdown with z-score of −5.217 for MSI2. The other pathways, including eIF4 (eukaryotic translation initiation factor 4) and p70S6 kinase pathway, actin cytoskeleton pathway and EGF, HGF growth factor pathways were all highly activated upon MSI2KD with positive z-scores. To investigate the downstream effects of MSI2 further, we searched for upstream targets in the direct targets list of MSI2 that are associated with these signaling pathways, and found IL6ST to be bound and transcriptionally regulated by MSI2 in a negative fashion as proposed by our genome-wide pathway analysis.
MSI2 regulates the expression of IL6ST
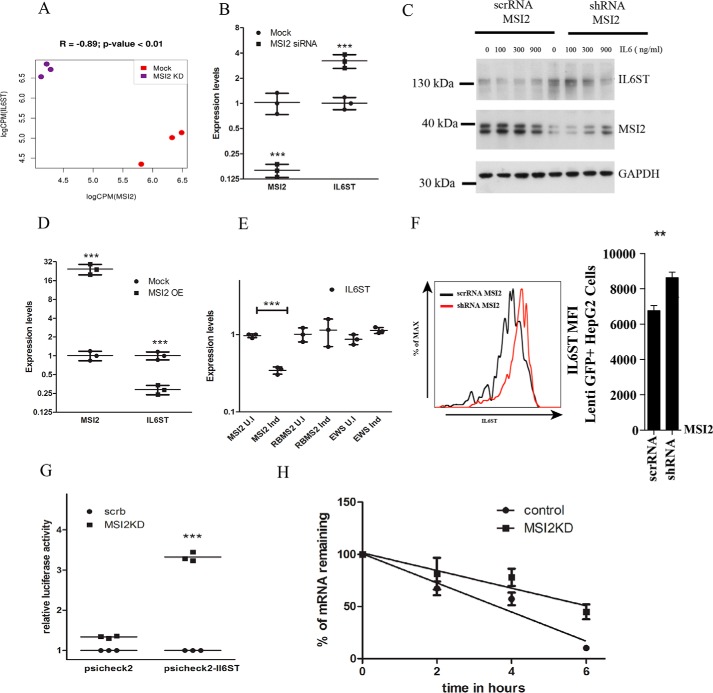
We eventually focused our attention onto interleukin 6 signal transducer, or IL6ST, which was also one of the most highly regulated targets. IL6ST expression was significantly increased (logFC = 1.98; FDR = 6.4E-17) upon MSI2 knockdown and thus negatively correlated with MSI2 expression (Fig. 3A). To confirm the result, we performed quantitative PCR (qPCR) on MSI2 knockdown cells and observed increased expression of IL6ST (Fig. 3B). In addition, IL6ST contained an MSI2-binding site in its 3′UTR, which we validated by EMSA (Fig. S1D). We also created stable knockdown of MSI2 in HEK293T cells using shRNA construct and observed an increase in the levels of IL6ST protein levels (Fig. 3C). To further confirm that MSI2 is a negative regulator of IL6ST, a control vector and a vector expressing MSI2 stably were transfected into HEK293T cells. The stable expression of MSI2 significantly decreased IL6ST mRNA levels (Fig. 3D). Similarly, we tested this regulatory link in cells with MSI2 expression under the control of an inducible tetracycline promoter. As controls, we used cells in which RBMS2 and EWSR1 expression were under the control of tetracycline promoter. Upon induction of these three cell lines with doxycycline, levels of IL6ST decreased in MSI2 cells, whereas no effect was observed in cells expressing RBMS2 and EWSR1 (Fig. 3E). To avoid cell-specific impact of MSI2 on IL6ST expression, we chose to examine Lenti GFP+ Hep G2 cells, which also have high native IL6ST surface expression. We knocked down MSI2 in these cells and checked for surface expression of IL6ST using FACS and observed an increased surface expression of IL6ST (Fig. 3F). We also analyzed RNA-Seq data of 72 AML patient datasets and found that this reciprocal relationship between IL6ST and MSI2 holds true in AML context as well (see supporting information). Together, these data suggest that overexpression of MSI2 decreases IL6ST transcript and protein levels and vice versa. These effects on mRNA abundance of IL6ST by differential expression of MSI2 suggest that MSI2 likely regulates the stability of the IL6ST transcript.
Figure 3.
MSI2 regulates IL6ST transcript levels. A, expression correlation measured by RNA-Seq between IL6ST and MSI2 in Mock and MSI2 KD samples. B, quantitative real-time PCR of differential gene regulation of IL6ST upon MSI2 siRNA–treated cells and untreated cells. qPCR data were analyzed using two-way ANOVA and presented as mean ± S.D. (***, p < 0.001; n = 3 biological replicates). Data were normalized to actin levels. C, Western blotting showing the protein levels of IL6ST, MSI2, and GAPDH in cells stably expressing MSI2 shRNA and scrambled RNA upon IL-6 treatment. Concentration of IL-6 and antibodies are indicated. Molecular size markers are indicated. D, qPCR of relative mRNA levels of IL6ST in cells expressing MSI2 stably and HEK293T cells as control. Data were analyzed using two-way ANOVA and presented as mean ± S.D. (***, p < 0.001; n = 3 biological replicates). Data were normalized to actin levels. E, qPCR of relative mRNA levels of IL6ST in induced and uninduced cells where MSI2, RBMS2, and EWSR1 are under inducible tetracycline promoter. Data were analyzed using two-way ANOVA and presented as mean ± S.D. (***, p < 0.001; n = 3 biological replicates). Data were normalized to actin levels. F, to the left is flow cytometric analysis of IL6ST surface expression on Lenti GFP+ Hep G2 cells which were stably transfected with scrambled and MSI2 shRNA. The same is represented as bar diagram on the right side. Data were analyzed with the Student's t test and presented as mean ± S.D. (**, p < 0.01; n = 3 biological replicates). G, MSI2 silencing increased the reporter activity in cells treated with psiCHECK2-IL6ST compared with those treated with vector control (psiCHECK2). Data are shown as the -fold increase in luciferase activity (RLU). Data were analyzed with the Student's t test and presented as the mean ± S.D. (***, p < 0.001; n = 3 biological replicates). H, IL6ST mRNA transcript levels in cells treated with scrambled or MSI2 siRNAs followed by actinomycin D treatment were plotted against time after actinomycin D addition and fitted to a linear regression model. Data were analyzed with the Student's t test and the data points are presented as the mean ± S.E. The solid line represents the fitted line through the data points. Experiment was carried out in biological triplicates.
MSI2 regulates IL6ST mRNA stability
To gain understanding of how MSI2 binding to the IL6ST 3′UTR affects IL6ST mRNA translation, we cloned the 3′UTR region of IL6ST that contains multiple MSI2 binding motifs into a luciferase reporter construct (psiCHECK2) bearing the Renilla luciferase coding region along with firefly luciferase, which served as an internal control for transfection (Fig. S2A). psiCHECK2-IL6ST (3′UTR) and the parent vector psiCHECK2 were transfected into HEK293Ts that expressed either normal or reduced levels of MSI2. As shown in Fig. 3G, MSI2 silencing clearly increased the luciferase expression of psiCHECK2-IL6ST, whereas parent vector levels did not change, indicating that the binding of MSI2 to IL6ST 3′UTR was responsible for promoting the degradation of the IL6ST.
To further test if the binding of MSI2 to IL6ST mRNA affects its stability, we performed mRNA stability assays by treating the cells with actinomycin D to inhibit de novo RNA synthesis. We found that the mRNA levels of IL6ST were higher in cells treated with MSI2 siRNA when compared with those in control cells (Fig. 3H). Together, these findings indicate that binding of MSI2 to the 3′UTR of IL6ST mRNA enhances the degradation of the IL6ST by destabilizing it.
MSI2 expression affects JAK/STAT and MAPK pathways
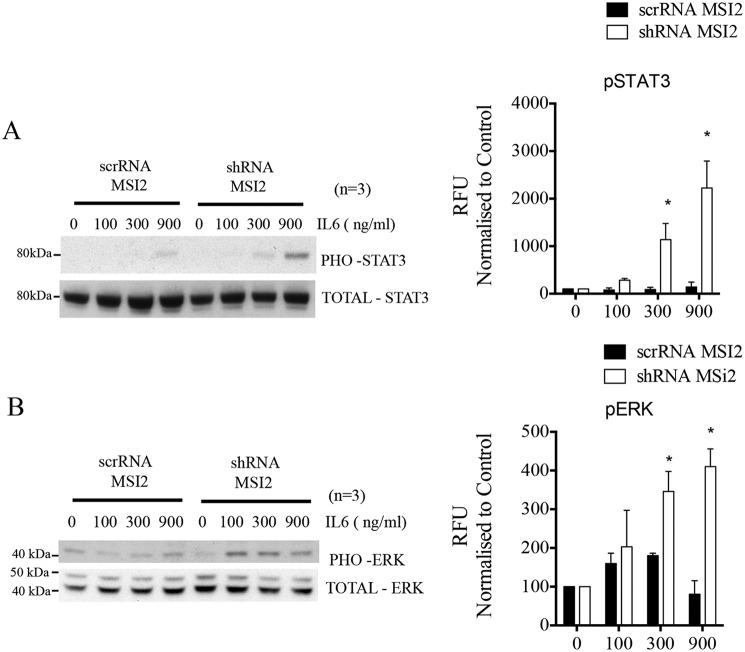
Having established the regulatory role of MSI2 on IL6ST stability and expression, we aimed to understand the molecular mechanisms modulated by this regulation. We therefore evaluated the pathways, STAT3 phosphorylation, and ERK phosphorylation, which are induced by signal transduction through IL6ST as well as affected by EGF and HGF signaling. We checked for the phosphorylation status of STAT3 and ERK upon IL6ST overexpression in the stable MSI2 knockdown cells. For this, we stimulated the stable knockdown cells with different concentrations of IL-6 cytokine (0–900 ng/μl). IL-6 activates IL6ST which in turn leads to initiation of JAK/STAT signaling. The protein levels of IL6ST, total STAT3, and phosphorylated STAT3 were examined after IL-6 stimulation. We noticed a constant hyperphosphorylation of STAT3 upon increased IL-6 concentration whereas total protein content remained unchanged (Fig. 4A). Similarly, we also checked the protein levels of IL6ST, total ERK, and phosphorylated ERK after IL-6 stimulation and noticed similar hyperphosphorylation in ERK (Fig. 4B). These results indicated that post-transcriptional gene regulation of IL6ST by MSI2 affects the phosphorylation of STAT3 and ERK proteins, which in turn affects JAK/STAT and MAPK signaling pathways.
Figure 4.
MSI2 effects phosphorylation of STAT3 and ERK. A, Western blotting showing the protein levels of phosphorylated STAT3 and total STAT3 in cells stably expressing scrambled (control) and MSI2 shRNA upon IL-6 treatment. Antibodies and concentration of IL-6 are indicated. On the right is the densitometric quantification of phosphorylated STAT3 protein band, which is normalized to total STAT3 protein from three independent blots. Data were analyzed with the Student's t test and presented as the mean with S.D. (*, p < 0.05; n = 3 independent blots). B, Western blotting showing the protein levels of phosphorylated ERK and total ERK in cells stably expressing scrambled (control) and MSI2 shRNA upon IL-6 treatment. Antibodies and concentration of IL-6 are indicated. On the right is the densitometric quantification of phosphorylated ERK protein band normalized to total ERK protein from three independent blots. Data were analyzed with the Student's t test and presented as the mean with S.D. (*, p < 0.05; n = 3 independent blots).
Discussion
In this study we investigated the RNA-binding partners of the RNA-binding protein MSI2 and confirmed the RRE by both PAR-CLIP and EMSA. Furthermore, we identified that IL6ST, a signal transducer for multiple cytokines, is bound and regulated by MSI2. We also showed that stimulating MSI2 knockdown cells with IL-6 led to hyperphosphorylation of STAT3 and ERK1/2. Thus, we identified targets bound and regulated by MSI2 and put forth a novel mechanism by which IL6ST is post-transcriptionally regulated by an RNA-binding protein. Because IL6ST is part of IL-6 signaling mechanism, we described a novel pathway, by which this signaling mechanism can be regulated.
Although MSI2 has been implicated in the development of several aggressive cancers as well as exerting important cellular functions, the RNA-binding partners of this RNA-binding protein have not been thoroughly investigated. Applying PAR-CLIP and RNA-Seq to MSI2, we found several relevant RNA-binding partners and their respective RNA-binding motif. The high degree of conservation among MSI family members across broad ranges of animal phyla indicates that their motifs might be similar. Okano and co-workers identified a similar motif in Drosophila MSI and mouse MSI1 as (G/A)U1–3AGU (19). Also, the motif in mouse MSI1 was found to be 87% identical to human MSI2 (20). Although the motifs are identical, there is a differential binding specificity among Drosophila, mouse, and human MSI RRM family domains, suggesting a functional divergence. Despite this divergence, the core element driving the binding among MSI family has been evolutionarily conserved and was established by Zearfoss et al. as UAG using quantitative fluorescence anisotropy assay (20). Consistent with this study, our data also revealed a similar motif in human MSI2 to be U followed by UAG (UUAG) which was present in 76% of the top 100 binding sites in our PAR-CLIP datasets.
We have identified several novel targets of MSI2 which were previously shown to play an important role in hematopoiesis. Among them is KMT2A (alias ALL1, MLL), which is a transcriptional co-activator that plays an important role in hematopoiesis and is also known to be frequently translocated in many leukemias (21). Another target, EGR1, was shown to be often deregulated in hematological malignancies such as AML, CML, multiple myelomas and B cell lymphomas (22). ASXL2, another target of MSI2 is a tumor suppressor which maintains normal HSC function (23, 24). The functional link between MSI2 and these targets gives us a better idea about the role of MSI2 in hematopoiesis and leukemia. Similarly, targets such as SMG1 and RUNX1, which regulate cell growth in AML (25, 26), might be affected upon overexpression of MSI2 in AML. This regulation therefore could explain the increased cell expansion seen in AML patients (27). Consistent with the previous finding that MSI2 modulates the TGF-β pathway by direct binding to genes such as TGFBR1 and SMAD3, our data add additional targets of MSI2 involved in this pathway such as RAF1, p300, CBP, SMAD5, and SMAD9 (13).
Our genome-wide pathway analysis revealed targets of MSI2 that regulate important cellular pathways such as eIF2, eIF4/p70S6K, EGF, and HGF. Both the eukaryotic translation initiation factor pathways (eIF2, eIF4 and p70S6K) attribute a translational role to MSI2 protein. The genes included in these pathways, that are bound and regulated by MSI2, are ribosomal proteins including RPL11, RPS7, RPL19, and RPS20, which suggests that MSI2 might be an essential factor in recruiting these proteins for protein translation. Additionally, the two growth factor pathways HGF and EGF in turn regulate central signaling cascades including PI3K-Akt, JAK/STAT, or MAPK (28–30), which have increased activity after MSI2 KD, thus being negatively regulated. The EGF and HGF signaling pathways activate the PI3K-Akt and MAPK/ERK pathways by phosphorylation/dephosphorylation of downstream targets, indicating that the phosphorylation in general seems to be likely affected by MSI2 (31, 32). Additionally, to support this idea, we have identified many targets in the pathway analysis, including JAK1, PIK3CA, ITPR2, and MAP3K1. Among several targets of MSI2, we here described a regulation of IL6ST, which is one of the upstream targets of these signaling pathways. We showed that regulation of IL6ST by MSI2 affected the phosphorylation of downstream targets such as ERK1/2 and STAT3 upon IL-6 stimulation. Because MSI2 was found to be up-regulated whereas IL6ST was down-regulated in AML patients, we hypothesize that STAT3 is hypophosphorylated upon IL-6 stimulation thus rendering the cells less susceptible to respond to immune modulatory attacks. We showed that ERK1/2 was hyperphosphorylated in stable MSI2 knockdown cells upon IL-6 stimulation, which is not in line with the previous findings that ERK phosphorylation decreased, and leads to inactivation of MAPK pathway (33). This ambiguity might be because of the cellular context in which the study was undertaken or that IL-6–stimulated cells might react differently from nonstimulated cells.
Taken together, our study identified multiple novel target mRNAs regulated by human Musashi protein MSI2. We describe a novel mode of regulation of IL6ST (GP130) by MSI2. Our findings show that MSI2 broadly controls the post-transcriptional regulation of multiple mRNAs that control many oncogenic signaling networks, but a precise understanding of the underlying mechanisms will be vital to be able to translate this knowledge to clinical therapy.
Experimental procedures
Plasmids
Plasmid pENTR4_MSI2 was generated by PCR amplification of the respective coding sequence (CDS) followed by restriction digestion with SalI and NotI and ligation into pENTR4 (Invitrogen). Sequences were as follows: MSI2_Not_Rev, ATAGTTTAGCGGCCGCTCAATGGTATCCATTTGTAAAGGC and MSI2_Sal_For, ACGCGTCGACATGGAGGCAAATGGGAGCCAAGGC. PENTR4_MSI2 was recombined into pFRT_DESTFLAGHA modified destination vector to allow for expression of FLAG/HA-tagged protein in stably transfected Flp-In TREx HEK293 cells (Invitrogen) (34).
PAR-CLIP
PAR-CLIP from Flp-In TREx HEK293 cells (Invitrogen) stably overexpressing FLAG/HA-tagged MSI2 protein was performed as described previously (16, 35). Briefly, cells were grown in medium supplemented with 100 μm 4SU (4-S-uridine) (Sigma-Aldrich) for 12 h prior to cross-linking. The medium was decanted and cells were washed in PBS and irradiated with 0.15 J per cm2 total energy of 365 nm UV light (Spectrolinker XL-1500). Cells were then harvested and lysed and the cleared lysate was treated with RNase T1 (Thermo Fisher). FLAG/HA-tagged MSI proteins were immunoprecipitated with an anti-FLAG antibody (Sigma-Aldrich, F3165) conjugated to Dynabeads (Thermo Fisher). After second RNase T1 digestion, beads were washed in immunoprecipitation wash buffer and resuspended in dephosphorylation buffer and incubated with calf intestinal alkaline phosphatase beads. The cross-linked RNA was then radiolabeled with T4 polynucleotide kinase (Thermo Fisher) and washed, and the protein-RNA complex was released from the beads by incubating at 90 °C and separated on SDS gels (NuPAGE Bis-Tris Midi Gels and XCell4 SureLock Midi-Cell chamber from Thermo Fisher Scientific). The protein-RNA complex was excised from the gel followed by proteinase K digestion. The leftover RNA was extracted using phenol-chloroform (Sigma-Aldrich) extraction and ethanol precipitation. The 5′-32P-phosphorylated RNA14C was then subjected to a standard cDNA library preparation protocol (36). The RNA was ligated with adaptors at 3′ and 5′ ends followed by reverse transcription. The resulting cDNA was amplified by adaptor-specific primers and sequenced with an Illumina HiSeq 2000 (50 cycles).
siRNA knockdown, library preparation, and RNA-Seq
siRNA transfections (50 nm final concentration; duration of knockdown 72 h) of HEK293 cells were performed in 24-well format using Lipofectamine RNAiMAX (Invitrogen) as described by the manufacturer. The sequences of three different MSI2 siRNAs are given in the supporting information. For other knockdown experiments (apart from RNA-Seq), we used a mixture of these three different siRNAs. Total RNA was extracted using TRIzol (Invitrogen) and further purified using the RNeasy purification kit (Qiagen). Libraries were prepared according to manufacturer's instructions using TruSeq Total RNA sample preparation kit (Illumina) and further sequenced using Illumina HiSeq 2500 platform (single read, 100 cycles).
Stable knockdown of MSI2
Subconfluent (50–60%) HEK293T cells were transfected with 10 μg of plasmid expressing MSI2 shRNA and 10 μg of scramble control (Origene, TF303128) along with 5 μg of lentiviral packaging plasmids (pMDG1.vsvg, pRSV-Rev, and pMDLg/pRRE). After 48 h, high titer lentiviral particles were collected and filtered through a 0.45 μm filter. Subsequently, HEK293T cells were transduced with lentiviral particles expressing the MSI2 shRNAs and scramble controls containing 0.8 μg/ml of Polybrene. After 48 h, cells were selected with puromycin selection (0.75 μg/ml of puromycin). Lenti GFP+ Hep G2 cells, which have high IL6ST surface expression, were used to knockdown MSI2.
qRT-PCR
Total RNA was extracted using the RNeasy Mini kit (Qiagen). cDNA was synthesized using SuperScript III Reverse Transcriptase kit (Invitrogen) according to the manufacturer's instructions (oligo dT). qRT-PCR was carried out using Power SYBR Green Kit (Applied Biosystems). All reactions were run on an ABI 7500 Real-Time PCR System (Applied Biosystems) in triplicate. Data were acquired using the ABI SDS 2.0.1 software package. RNA isolated from the samples was tested for the expression levels of the chosen targets and their ΔCt values were subtracted from the respective beta-actin expression levels.
Western blotting
Cells were harvested, resuspended in Nonidet P-40 lysis buffer and cOmplete EDTA-free Protease Inhibitor mixture (Roche) and lysed. A 10% SDS-PAGE gel was run in Tris-glycine-SDS buffer. A semi-dry transfer procedure (Bio-Rad) was carried out onto cellulose membrane. After transfer, the membrane was blocked in TBS with Tween 20 and 5% milk. The membrane was probed with monoclonal antibodies detecting beta-actin (Sigma Aldrich, A2228), MSI2 (Abcam, ab76148), IL6ST (BD Biosciences, 564151), STAT3 (Cell Signaling Technology, 9139), phosphorylated STAT3 (Cell Signaling Technology, 9131), Total ERK (Cell Signaling Technology, 4695), phosphorylated ERK (Cell Signaling Technology, 9101), GAPDH (Cell Signaling Technology, 14C10) and FLAG M2 (Sigma, F3165). The antibodies were used at a concentration of 1:1000 for MSI2, IL6ST, STAT3, pSTAT3, ERK, pERK, and GAPDH and 1:10,000 for beta-actin. HRP-conjugated anti-mouse and anti-rabbit antibodies (Santa Cruz Biotechnology) were used as secondary antibodies at 1:10,000 dilution. Super Signal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific) was used to detect the signal using Image Gauge software (Fujifilm).
Recombinant protein expression and purification
A synthetized gene encoding the Musashi 2 protein containing optimized codon usages for Escherichia coli was purchased from GeneArt. Following digestion with PmlI and AvrII restriction enzymes (New England Biolabs), the gene was ligated into pET302/NT-His vector and transformed into E. coli BL21 (DE3) cells. Protein expression was induced by addition of isopropyl β-d-1-thiogalactopyranoside (IPTG) (to a final concentration of 1 mm) to the cells grown at 37 °C, with A600 of 0.7. After 4 h, cells were harvested by centrifugation at 5000 × g. Cell pellets were resuspended in lysis buffer (500 mm NaCl, 20 sodium phosphate, 20 mm imidazol, pH 7.5, containing 5 mm DTT and EDTA-free protease inhibitor tablets (Roche Applied Sciences)) and lysed using a cell disrupter (Constant Systems). After a mild sonication step, insoluble materials were removed by centrifugation at 28,000 × g. The soluble fraction was loaded onto a HisTrap FF ml column (GE Healthcare Life Sciences). The His-tagged Musashi protein was eluted with 200 mm imidazol in 20 mm sodium phosphate, 500 mm NaCl, pH 7.5 buffer containing 5 mm DTT. Eluted fractions were pooled and dialyzed against 20 mm Tris HCl (pH 7.5) containing 5 mm DTT and 1 m NaCl (molecular weight cut-off, 3 kDa) to remove RNA impurities. After an additional dialysis step against the same buffer without NaCl, the protein sample was loaded onto a cation exchange column (HiTrap SP FF 5 ml, GE Healthcare). Next, the purified protein was further purified by size-exclusion chromatography using a HiLoad 16/60 Superdex 75 (GE Healthcare), equilibrated in 20 mm Tris HCl, 150 mm NaCl, pH 8.0, containing 5 mm DTT. All purification steps were carried out with an Äkta purifier system and monitored by measuring the absorbance at 215 nm, 254 nm, and 280 nm. Protein sample analysis by 15% PAGE (SDS-PAGE) did not detect any impurities. Protein concentration was determined using UV spectroscopy.
Electrophoretic mobility shift assays (EMSA)
1 nm radioactively labeled (γ-ATP) RNA was incubated with protein concentrations varying from 0–1 μm for 1 h in 1.5 ml passivated (50 μl 1 mg/ml acetylated BSA, 27 °C, 1 h) prelubricated reaction tubes. The reaction was separated by native PAGE for 2 h at 300 V. The radioactive signal was visualized by phosphorimaging and the signal was quantified using Image Gauge software (Fujifilm).
Luciferase assays
200 ng of control plasmid psiCHECK2 (Promega) or psiCHECK2 bearing the 3′UTR of IL6ST mRNA containing multiple binding motifs were transfected into HEK293T cells together with siRNAs for MSI2 and control using Lipofectamine 2000 (Invitrogen). 48 h later, Renilla luciferase and firefly luciferase were measured using Dual-Glo Luciferase Assay System (Promega) using Tecan 10 m Spark Luminometer.
Half-life assays
The mRNA half-life was determined by treating the cells with 3 μg/μl actinomycin D (Sigma-Aldrich). Cells were collected at different time points (0–6 h) and total RNA was isolated followed by cDNA synthesis. Real-time PCR, to measure the percentage of RNA remaining for MSI2, IL6ST along with GAPDH as an internal control, was performed.
Statistical analysis
All experiments were performed at least in triplicates (biological triplicates). Numerical data were expressed as mean ± S.D. Group comparisons were analyzed by two-way ANOVA or unpaired t test. p values were calculated and a p value of <0.05 was considered significant.
Bioinformatic analysis
Processing RNA-Seq reads
We obtained a total of 138,315, 336 sequencing reads for the regular RNA-Seq before and after MSI2 knockdown (each sequenced in biological triplicates). First, adapter sequences and low-quality ends of the raw sequencing reads were trimmed using seqtk (https://github.com/lh3/seqtk)8 and cutadapt (37). All sequencing reads shorter than 25 bases after trimming were discarded. The trimmed and filtered sequencing reads were then aligned against the human genome sequence GRCh38 (38) with TopHat2 version 2.0.13 (39). Next, read counts on a gene basis were estimated by applying HTSeq (40) using gene annotations downloaded from Ensembl Genes version 77 (41). A total of 94,422,196 sequencing reads were mapped to annotated genes. The read counts represented an estimate of the respective transcript abundance and were normalized according to the sequencing library size (resulting in the so-called counts per million or cpm) using the Bioconductor package edgeR (42). Genes with low (no discernible) expression were discarded, as these were not accessible for statistical testing. Subsequently, statistical testing was performed with edgeR to identify differentially expressed genes between the controls and the MSI2 knockdown samples. The rate of differential expression is represented by the logarithm of the -fold change (logFC). To adjust for multiple testing, we used the FDR and chose genes as significantly differentially expressed if the FDR was below 5%. All sequencing data were deposited at European Nucleotide Archive (ENA), accession number PRJEB25085.
Processing of PAR-CLIP reads
The PAR-CLIP sequencing reads were preprocessed similarly to the RNA-Seq reads. However, because of some intrinsic properties compared with RNA-Seq reads (43), we kept sequencing reads ≥18 bases after trimming with cutadapt for further analyses. Next, the sequencing reads were aligned against the reference sequence of GRCh38 using Burrows-Wheeler Aligner (BWA) allowing for up to two mismatches between a single read and the reference sequence (44). All aligned reads were piled up into clusters by the PARA-suite (45) applying a hierarchical clustering algorithm. CCs were identified by filtering the obtained clusters for containing at least 10 reads, being shorter than 75 bases, showing at least 25% T-C conversions; 100% T-C conversion sites or T-C conversions reported as SNPs in the dbSNP version 142 (46) were excluded. To identify the genomic RBP-binding sites of the respective RBP, the CCs were overlapped with gene annotations downloaded from Ensembl Genes V77 using the Homer annotation tool (47).
Motif discovery
To predict the binding motifs from the PAR-CLIP data, we considered the 100 most abundant CCs in each dataset defined by the highest number of reads. The detection of the sequence motif of the RBP-binding sites was performed using MEME (48). We assumed a single occurrence of the sequence motif per cluster sequence, so we chose the one occurrence per sequence (OOPS) motif model with a minimum motif length of four nucleotides.
Ingenuity pathway analysis (IPA)
Pathway analysis for MSI2 knockdown was performed using Qiagen's Ingenuity® Pathway Analysis. Only genes with a significant differential expression (logFC ≥ ± 1, FDR ≤0.05) were used as input.
Author contributions
S. D., A. K., and J. I. H. data curation; S. D., A. K., and P. C. M. software; S. D. formal analysis; S. D., A. K., P. C. M., and A. C. M. validation; S. D., S. K. M., H. S., and J. I. H. investigation; S. D. visualization; S. D., S. K. M., and K. H. methodology; S. D. writing-original draft; W. H., P. A. L., A. B., and J. I. H. supervision; A. C. M., A. B., and J. I. H. writing-review and editing; A. B. and J. I. H. project administration; J. I. H. conceptualization; J. I. H. funding acquisition.
Supplementary Material
Acknowledgments
We thank Michael Gombert and Katayoun Alemazkour-Fathayan for help with Illumina sequencing. We also thank Ute Fischer for helpful comments on the manuscript.
This work was supported by Heinrich-Heine University Düsseldorf Forschungskommission 17/2013, the Düsseldorf School of Oncology (funded by the Comprehensive Cancer Center Düsseldorf/Deutsche Krebshilfe and the Medical Faculty HHU Düsseldorf), and the Deutsche Forschungsgemeinschaft HO 5456/3-1. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supporting information, Tables S1 and S2, and Figs. S1 and S2.
All sequencing data were deposited at the European Nucleotide Archive (ENA) with accession number PRJEB25085.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- CML
- chronic myeloid leukemia
- RRE
- RNA recognition elements
- AML
- acute myeloid leukemia
- RBP
- RNA-binding protein
- ALL
- acute lymphoblastic leukemia
- HSC
- hematopoietic stem cell
- PAR-CLIP
- photoactivatable ribonucleoside cross-linking and immunoprecipitation
- HGF
- hepatocyte growth factor
- CC
- confident cluster
- T-C
- thymidine to cytidine
- KD
- knockdown
- FDR
- false discovery rate
- qPCR
- quantitative PCR
- ANOVA
- analysis of variance
- RLU
- relative light unit.
References
- 1. Aplan P. D. (2006) Causes of oncogenic chromosomal translocation. Trends Genet. 22, 46–55 10.1016/j.tig.2005.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barbouti A., Höglund M., Johansson B., Lassen C., Nilsson P.-G., Hagemeijer A., Mitelman F., and Fioretos T. (2003) A novel gene, MSI2, encoding a putative RNA-binding protein is recurrently rearranged at disease progression of chronic myeloid leukemia and forms a fusion gene with HOXA9 as a result of the cryptic t(7;17)(p15;q23). Cancer Res. 63, 1202–1206 [PubMed] [Google Scholar]
- 3. Collins C. T., and Hess J. L. (2016) Role of HOXA9 in leukemia: Dysregulation, cofactors and essential targets. Oncogene 35, 1090–1098 10.1038/onc.2015.174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Collins C., Wang J., Miao H., Bronstein J., Nawer H., Xu T., Figueroa M., Muntean A. G., and Hess J. L. (2014) C/EBPα is an essential collaborator in Hoxa9/Meis1-mediated leukemogenesis. Proc. Natl. Acad. Sci. U.S.A. 111, 9899–9904 10.1073/pnas.1402238111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Figueroa M. E., Lugthart S., Li Y., Erpelinck-Verschueren C., Deng X., Christos P. J., Schifano E., Booth J., van Putten W., Skrabanek L., Campagne F., Mazumdar M., Greally J. M., Valk P. J., Löwenberg B., Delwel R., and Melnick A. (2010) DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell 17, 13–27 10.1016/j.ccr.2009.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sakakibara S., Nakamura Y., Satoh H., and Okano H. (2001) RNA-binding protein Musashi2: Developmentally regulated expression in neural precursor cells and subpopulations of neurons in mammalian CNS. J. Neurosci. 21, 8091–8107 10.1523/JNEUROSCI.21-20-08091.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Saleki R., Christensen T., Liu W., Wang X., Chen Q. C., Aakre M., Gomes N. M., Alexiev B. A., Schappert J., Baer M. R., and Zou Y. S. (2015) A novel TTC40-MSI2 fusion in de novo acute myeloid leukemia with an unbalanced 10;17 translocation. Leuk. Lymphoma 56, 1137–1139 10.3109/10428194.2014.947611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Aly R. M., and Ghazy H. F. (2015) Prognostic significance of MSI2 predicts unfavorable outcome in adult B-acute lymphoblastic leukemia. Int. J. Lab. Hematol. 37, 272–278 10.1111/ijlh.12284 [DOI] [PubMed] [Google Scholar]
- 9. Sutherland J. M., McLaughlin E. A., Hime G. R., and Siddall N. A. (2013) The Musashi family of RNA binding proteins: Master regulators of multiple stem cell populations. Adv. Exp. Med. Biol. 786, 233–245 10.1007/978-94-007-6621-1_13 [DOI] [PubMed] [Google Scholar]
- 10. Kharas M. G., Lengner C. J., Al-Shahrour F., Bullinger L., Ball B., Zaidi S., Morgan K., Tam W., Paktinat M., Okabe R., Gozo M., Einhorn W., Lane S. W., Scholl C., Fröhling S., et al. (2010) Musashi-2 regulates normal hematopoiesis and promotes aggressive myeloid leukemia. Nat. Med. 16, 903–908 10.1038/nm.2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kudinov A. E., Deneka A., Nikonova A. S., Beck T. N., Ahn Y. H., Liu X., Martinez C. F., Schultz F. A., Reynolds S., Yang D. H., Cai K. Q., Yaghmour K. M., Baker K. A., Egleston B. L., Nicolas E., et al. (2016) Musashi-2 (MSI2) supports TGF-β signaling and inhibits claudins to promote non-small cell lung cancer (NSCLC) metastasis. Proc. Natl. Acad. Sci. U.S.A. 113, 6955–6960 10.1073/pnas.1513616113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bennett C. G., Riemondy K., Chapnick D. A., Bunker E., Liu X., Kuersten S., and Yi R. (2016) Genome-wide analysis of Musashi-2 targets reveals novel functions in governing epithelial cell migration. Nucleic Acids Res. 44, 3788–3800 10.1093/nar/gkw207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Park S. M., Deering R. P., Lu Y., Tivnan P., Lianoglou S., Al-Shahrour F., Ebert B. L., Hacohen N., Leslie C., Daley G. Q., Lengner C. J., and Kharas M. G. (2014) Musashi-2 controls cell fate, lineage bias, and TGF-β signaling in HSCs. J. Exp. Med. 211, 71–87 10.1084/jem.20130736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang S., Li N., Yousefi M., Nakauka-Ddamba A., Li F., Parada K., Rao S., Minuesa G., Katz Y., Gregory B. D., Kharas M. G., Yu Z., and Lengner C. J. (2015) Transformation of the intestinal epithelium by the MSI2 RNA-binding protein. Nat. Commun. 6, 6517 10.1038/ncomms7517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ascano M., Hafner M., Cekan P., Gerstberger S., and Tuschl T. (2012) Identification of RNA-protein interaction networks using PAR-CLIP. Wiley Interdiscip. Rev. RNA 3, 159–177 10.1002/wrna.1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hafner M., Landthaler M., Burger L., Khorshid M., Hausser J., Berninger P., Rothballer A., Ascano M. Jr., Jungkamp A. C., Munschauer M., Ulrich A., Wardle G. S., Dewell S., Zavolan M., and Tuschl T. (2010) Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141, 129–141 10.1016/j.cell.2010.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Heinrich P. C., Behrmann I., Haan S., Hermanns H. M., Müller-Newen G., and Schaper F. (2003) Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem. J. 374, 1–20 10.1042/bj20030407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Heinrich P. C., Behrmann I., Muller-Newen G., Schaper F., and Graeve L. (1998) Interleukin-6-type cytokine signalling through the gp130/Jak/STAT pathway. Biochem. J. 334, 297–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Imai T., Tokunaga A., Yoshida T., Hashimoto M., Mikoshiba K., Weinmaster G., Nakafuku M., and Okano H. (2001) The neural RNA-binding protein Musashi1 translationally regulates mammalian numb gene expression by interacting with its mRNA. Mol. Cell. Biol. 21, 3888–3900 10.1128/MCB.21.12.3888-3900.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zearfoss N. R., Deveau L. M., Clingman C. C., Schmidt E., Johnson E. S., Massi F., and Ryder S. P. (2014) A conserved three-nucleotide core motif defines Musashi RNA binding specificity. J. Biol. Chem. 289, 35530–35541 10.1074/jbc.M114.597112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rao R. C., and Dou Y. (2015) Hijacked in cancer: the KMT2 (MLL) family of methyltransferases. Nat. Rev. Cancer 15, 334–346 10.1038/nrc3929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tian J., Li Z., Han Y., Jiang T., Song X., and Jiang G. (2016) The progress of early growth response factor 1 and leukemia. Intractable Rare Dis. Res. 5, 76–82 10.5582/irdr.2015.01049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Micol J. B., Pastore A., Inoue D., Duployez N., Kim E., Lee S. C., Durham B. H., Chung Y. R., Cho H., Zhang X. J., Yoshimi A., Krivtsov A., Koche R., Solary E., Sinha A., Preudhomme C., and Abdel-Wahab O. (2017) ASXL2 is essential for haematopoiesis and acts as a haploinsufficient tumour suppressor in leukemia. Nat. Commun. 8, 15429 10.1038/ncomms15429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li J., He F., Zhang P., Chen S., Shi H., Sun Y., Guo Y., Yang H., Man N., Greenblatt S., Li Z., Guo Z., Zhou Y., Wang L., Morey L., et al. (2017) Loss of Asxl2 leads to myeloid malignancies in mice. Nat. Commun. 8, 15456 10.1038/ncomms15456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Du Y., Lu F., Li P., Ye J., Ji M., Ma D., and Ji C. (2014) SMG1 acts as a novel potential tumor suppressor with epigenetic inactivation in acute myeloid leukemia. Int. J. Mol. Sci. 15, 17065–17076 10.3390/ijms150917065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ichikawa M., Yoshimi A., Nakagawa M., Nishimoto N., Watanabe-Okochi N., and Kurokawa M. (2013) A role for RUNX1 in hematopoiesis and myeloid leukemia. Int. J. Hematol. 97, 726–734 10.1007/s12185-013-1347-3 [DOI] [PubMed] [Google Scholar]
- 27. Byers R. J., Currie T., Tholouli E., Rodig S. J., and Kutok J. L. (2011) MSI2 protein expression predicts unfavorable outcome in acute myeloid leukemia. Blood 118, 2857–2867 10.1182/blood-2011-04-346767 [DOI] [PubMed] [Google Scholar]
- 28. Madonna R., Bolli R., Rokosh G., and De Caterina R. (2013) Targeting phosphatidylinositol 3-kinase-Akt through hepatocyte growth factor for cardioprotection. J. Cardiovasc. Med. 14, 249–253 10.2459/JCM.0b013e3283542017 [DOI] [PubMed] [Google Scholar]
- 29. Zhang Y. W., and Vande Woude G. F. (2003) HGF/SF-met signaling in the control of branching morphogenesis and invasion. J. Cell. Biochem. 88, 408–417 10.1002/jcb.10358 [DOI] [PubMed] [Google Scholar]
- 30. Normanno N., De Luca A., Bianco C., Strizzi L., Mancino M., Maiello M. R., Carotenuto A., De Feo G., Caponigro F., and Salomon D. S. (2006) Epidermal growth factor receptor (EGFR) signaling in cancer. Gene 366, 2–16 10.1016/j.gene.2005.10.018 [DOI] [PubMed] [Google Scholar]
- 31. Yu C. F., Liu Z. X., and Cantley L. G. (2002) ERK negatively regulates the epidermal growth factor-mediated interaction of Gab1 and the phosphatidylinositol 3-kinase. J. Biol. Chem. 277, 19382–19388 10.1074/jbc.M200732200 [DOI] [PubMed] [Google Scholar]
- 32. Yu C. F., Roshan B., Liu Z. X., and Cantley L. G. (2001) ERK regulates the hepatocyte growth factor-mediated interaction of Gab1 and the phosphatidylinositol 3-kinase. J. Biol. Chem. 276, 32552–32558 10.1074/jbc.M104493200 [DOI] [PubMed] [Google Scholar]
- 33. Zhang H., Tan S., Wang J., Chen S., Quan J., Xian J., Zhang S., He J., and Zhang L. (2014) Musashi2 modulates K562 leukemic cell proliferation and apoptosis involving the MAPK pathway. Exp. Cell Res. 320, 119–127 10.1016/j.yexcr.2013.09.009 [DOI] [PubMed] [Google Scholar]
- 34. Spitzer J., Landthaler M., and Tuschl T. (2013) Rapid creation of stable mammalian cell lines for regulated expression of proteins using the Gateway® recombination cloning technology and Flp-In T-REx® lines. Methods Enzymol. 529, 99–124 10.1016/B978-0-12-418687-3.00008-2 [DOI] [PubMed] [Google Scholar]
- 35. Spitzer J., Hafner M., Landthaler M., Ascano M., Farazi T., Wardle G., Nusbaum J., Khorshid M., Burger L., Zavolan M., and Tuschl T. (2014) PAR-CLIP (photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation): A step-by-step protocol to the transcriptome-wide identification of binding sites of RNA-binding proteins. Methods Enzymol. 539, 113–161 10.1016/B978-0-12-420120-0.00008-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hafner M., Lianoglou S., Tuschl T., and Betel D. (2012) Genome-wide identification of miRNA targets by PAR-CLIP. Methods 58, 94–105 10.1016/j.ymeth.2012.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Martin M. (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- 38. Lander E. S., Linton L. M., Birren B., Nusbaum C., Zody M. C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Funke R., Gage D., Harris K., Heaford A., Howland J., et al. (2001) Initial sequencing and analysis of the human genome. Nature 409, 860–921 10.1038/35057062 [DOI] [PubMed] [Google Scholar]
- 39. Trapnell C., Pachter L., and Salzberg S. L. (2009) TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Anders S., Pyl P. T., and Huber W. (2015) HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cunningham F., Amode M. R., Barrell D., Beal K., Billis K., Brent S., Carvalho-Silva D., Clapham P., Coates G., Fitzgerald S., Gil L., Girón C. G., Gordon L., Hourlier T., Hunt S. E., et al. (2015) Ensembl 2015. Nucleic Acids Res. 43, D662–D669 10.1093/nar/gku1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Robinson M. D., McCarthy D. J., and Smyth G. K. (2010) edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kloetgen A., Münch P. C., Borkhardt A., Hoell J. I., and McHardy A. C. (2015) Biochemical and bioinformatic methods for elucidating the role of RNA-protein interactions in posttranscriptional regulation. Brief. Funct. Genomics 14, 102–114 10.1093/bfgp/elu020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li H., and Durbin R. (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kloetgen A., Borkhardt A., Hoell J. I., and McHardy A. C. (2016) The PARA-suite: PAR-CLIP specific sequence read simulation and processing. PeerJ 4, e2619 10.7717/peerj.2619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sherry S. T., Ward M. H., Kholodov M., Baker J., Phan L., Smigielski E. M., and Sirotkin K. (2001) dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 29, 308–311 10.1093/nar/29.1.308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Heinz S., Benner C., Spann N., Bertolino E., Lin Y. C., Laslo P., Cheng J. X., Murre C., Singh H., and Glass C. K. (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bailey T. L., and Elkan C. (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28–36 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.