Abstract
PolyQ-expanded huntingtin (mHtt) variants form aggregates, termed inclusion bodies (IBs), in individuals with and models of Huntington's disease (HD). The role of IB versus diffusible mHtt in neurotoxicity remains unclear. Using a ponasterone (PA)-inducible cell model of HD, here we evaluated the effects of heat shock on the appearance and functional outcome of Htt103QExon1–EGFP expression. Quantitative image analysis indicated that 80–90% of this mHtt protein initially appears as “diffuse” signals in the cytosol, with IBs forming at high mHtt expression. A 2-h heat shock during the PA induction reduced the diffuse signal, but greatly increased mHtt IB formation in both cytosol and nucleus. Dose- and time-dependent mHtt expression suggested that nucleated polymerization drives IB formation. RNA-mediated knockdown of heat shock protein 70 (HSP70) and heat shock cognate 70 protein (HSC70) provided evidence for their involvement in promoting diffuse mHtt to form IBs. Reporter gene assays assessing the impacts of diffuse versus IB mHtt showed concordance of diffuse mHtt expression with the repression of heat shock factor 1, cAMP-responsive element-binding protein (CREB), and NF-κB activity. CREB repression was reversed by heat shock coinciding with mHtt IB formation. In an embryonic striatal neuron–derived HD model, the chemical chaperone sorbitol similarly promoted the structuring of diffuse mHtt into IBs and supported cell survival under stress. Our results provide evidence that mHtt IB formation is a chaperone-supported cellular coping mechanism that depletes diffusible mHtt conformers, alleviates transcription factor dysfunction, and promotes neuron survival.
Keywords: Huntington's disease, aggregation, heat shock protein (HSP), chaperone, heat shock factor protein 1 (HSF1), neurodegeneration, HSP chaperones, heat shock, inclusion bodies, polyQ-mHtt protein, transcription factor sequestration, CREB
Introduction
Protein misfolding and aggregation are hallmarks and contributing pathogenic mechanisms of many neurodegenerative diseases (ND)6 (1–4). Causative mutations identified in familial ND result in the aggregation of disease-related proteins: polyQ-huntingtin (mHtt) in Huntington's disease, α-synuclein in Parkinson's disease, β-amyloid in Alzheimer's disease, and prion in Creutzfeldt-Jakob disease, as examples. Although we do not know how protein aggregation may lead to neurodegeneration, the well-known association has spawned therapeutic strategies and extensive research efforts aimed at preventing and clearing such aggregated protein deposits (5).
Age is the primary risk factor in the development of ND, heritable familial cases included. That an individual harboring a mutation appears normal for decades before succumbing in mid-/later life to the pathogenic effects of the disease is confounding. In this context, our observation of an age-dependent and neuron-specific dysfunction of HSF1 for the induction of HSP chaperones is significant and relevant (6–10). HSF1 is a master transcription regulator that links stress biology to the induction of a network of HSP chaperones that collectively function as a protein quality control (PQC) mechanism to promote protein homeostasis (11, 12). The importance of PQC is underscored by the fact that mutations in HSP70 co-chaperones have been identified as causing inherited neurodegenerative and cardiac disorders, directly linking the HSP70 chaperone system to human disease (13).
Accordingly, we reason that conditions that enhance this PQC would be beneficial to counter pathologies caused by dysregulation of protein folding and aggregation in aging and ND (3, 14–19). Using an inducible cell model of HD (the 14A2.6 line of PC12 cells) (20), we evaluated the effects of heat shock and the role of HSP70 and HSC70 chaperone on the structuring and functional readout of Htt103QExon1–EGFP (mHtt) chimera protein expression. A transient and modest heat shock (HS) at 42 °C in the course of mHtt expression had a robust and reproducible effect in promoting the formation of large cytosolic inclusion bodies (IBs) and small nuclear IBs at the expense of diffusible assemblies of mHtt. Functionally, accumulation of diffuse mHtt repressed proportionately the activities of HSF1, CREB, and NF-κB as demonstrated by reporter gene analysis. The repressed CRE-reporter was reversed by HS and coincided with mHtt IB formation. Together, these results suggest that IB formation likely represents a cellular coping mechanism to rid itself of toxic mHtt conformers. This study adds to our understanding of the role of HSPs in protein homeostasis that, in addition to promoting protein folding, disaggregation, solubilization, and degradation, HSP chaperones can drive the structuring and aggregation of the intrinsically disordered mHtt protein into forming IBs for beneficial outcomes.
Results
Effect of heat shock on mHtt expression profile
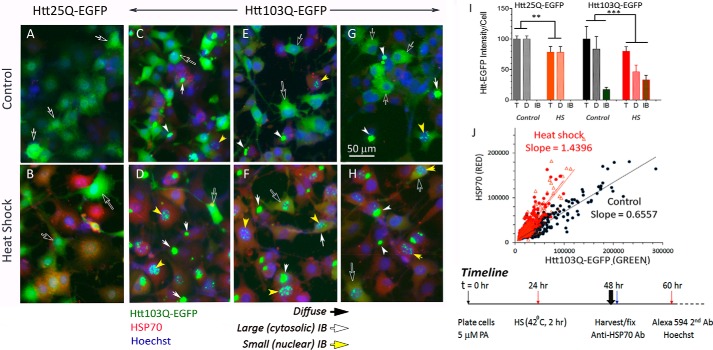
We used the 14A2.6 line of PC12 cells with stably integrated ecdysone receptor–based inducible expression of the Htt103QExon1–EGFP chimera (mHtt) as our HD cell model (20, 21). PC12 cells harboring the normal Htt construct Htt25Q–EGFP (cHtt) served as control (20). We note that transgenic mice expressing the human mHttExon1 exhibit a progressive neurological phenotype that exhibits many of the motor and nonmotor features of HD (22, 23). Fig. 1 presents images of the control and mutant Htt cells at 48 h of induction with 5 μm of ponasterone (PA), an analogue of ecdysone. To test for the effects of heat shock, cells were shifted to 42 °C for 2 h at the 24-h time point, returned to 37 °C and harvested and fixed at 48 h. Expression of HSP70 was probed with a rabbit polyclonal antibody against HSP70 followed by Alexa Fluor 594 goat anti-rabbit IgG, and nuclei were stained with Hoechst 33342. The three sets of mHtt images (Fig. 1, C and D, E and F, and G and H) were from three separate experiments and are representative of many experiments done at different times in the course of this study. Results show a robust expression of both the cHtt– and mHtt–EGFP chimera protein at the 48-h time point of PA induction, with diffusible assemblies of cytosolic Htt protein (Fig. 1, A–H, black arrows with white outline) predominant in both. In the mHtt cells, we could see occasional large cytoplasmic IBs in cells maintained at 37 °C (Fig. 1, C, E, and G; white arrows). HS decreased the diffuse cytosolic mHtt signal and conversely increased the number of IBs in both the cytosol and nucleus (Fig. 1, D, F, and H) as compared with their control counterparts (Fig. 1, C, E, and G). The cytosolic IBs were substantial in size, with some approaching the size of the 10–12 μm cell nuclei. HS also had a striking effect in increasing the number of small nuclear IBs. It should be noted that IB formation depends on polyQ expansion of the mHtt protein as this was not observed with the 25Q cHtt protein. The full-size original images presented in Fig. 1 are included as Fig. S1, A–H.
Figure 1.
Heat shock drives the remodeling of diffusible assemblies of mHtt to form IBs in a PC12-derived neuronal cell line. The 25Q cHtt and 103Q mHtt PC12 cells were plated in 96-well plate. PA was added to a final concentration of 5 μm at plating (t = 0) to induce cHtt and mHtt expression. The time line of this experiment is as indicated in the bottom right of the figure. Briefly, heat shock (HS) was at the 24-h time point by placing designated wells/strips of cells in a 42 °C incubator for 2 h and then returned to 37 °C incubator. Cells were harvested and fixed at 48 h. To track the expression of HSP70 as a response to heat shock, we used a rabbit polyclonal antibody against HSP70 followed by Alexa Fluor 594–labeled goat anti-rabbit IgG secondary antibody. Nuclei were stained with Hoechst 33342. Cells were imaged with EVOS® FL microscope with a 10× objective; all images of a given experiment were captured at the identical settings of exposure time and light intensity/contrast. A–H, cropped and contrasted areas of representative images of the 25QHtt and 103QHtt cells under control and heat shock conditions as indicated. Each of the image frames represents an area of ∼200 × 266 μm. Black arrows point to diffuse Htt–EGFP signal; white arrows point to IB; yellow arrows point to punctate nuclear IB. No IB was detected in the Htt25Q PC12 cells. I, mHtt quantitation. Averaged intensities per unit cell of total, diffuse, and IB Htt–EGFP of images from five independent experiments were done over a 2-year period with 25 independent cell samples per condition. Results shown here are relative to that of the control total of 100%. J, scatter plot of the HSP70 (red) versus Htt103Q–EGFP (green) signal of control versus HS cells. Individual cells of representative images of control (black symbol) and heat shock cells (red symbol; the two red lines represent results of two separate images) were marked as region of interest (ROI). The red (HSP70) and green (mHtt) signals of each ROI were determined by ImageJ and displayed as scatter plots. Probability of difference p >0.05 is defined as not significant, between 0.01 and 0.05 is significant (*), <0.01 is very significant (**), and <0.001 is extremely significant (***). A–H, full-frame original images are included in Fig. S1, A–H. Error bars represent S.D.
To quantify these changes in the expression and distribution of mHtt, we created “Macro” programs to measure the diffuse and IB Htt–EGFP intensity and of HSP70, and score the number and size of nuclei and IBs in ImageJ Fiji. Notably, the term diffuse is operationally defined as the microscopic diffuse appearance of the Htt protein; such diffusible assemblies can include disordered and meta-stable monomers and oligomers and low order fibrillary forms of Htt. For all IBs of varying sizes, a scatter plot of size versus green fluorescence intensity yielded a straight line with RSQ (R2) value of 0.9991 suggesting the same mHtt packing density in IBs, size notwithstanding. Diffuse and IB mHtt had significantly different mean fluorescent intensities, a result consistent with previous observations (24). This difference enabled us to analyze and score the diffuse mHtt signal separate from the IB mHtt signal. Macro programming greatly facilitated analysis of images of cells plated in 96-well plates. The Macro programs used for scoring the diffuse and IB Htt–EGFP signals and for profiling the number and size of IBs are included in Fig. S2.
The bar graph in of Fig. 1I presents our results on Htt signal quantitation. For mHtt cells at the 48-h time point of induction with 5 μm PA, the diffuse and IB forms of mHtt constituted on average 80 and 20% of the total signal intensity, respectively. HS of cells reduced the total mHtt load by about 20% and changed the relative abundance of diffuse versus IB forms of mHtt to constitute, respectively, 60 and 40% of the total. In contrast, the cHtt (25Q) protein never formed IB regardless of the level of cHtt expression, and HS reduced the total cHtt intensity by about 20%, similar to that of the mHtt cells. In sum, heat shock had two specific and notable effects on mHtt: 1) it depleted the diffuse cytosolic mHtt to form large cytosolic IBs, and 2) it greatly increased the appearance of small nuclear IBs. A scatter plot scoring individual cells for diffuse mHtt versus HSP70 (Green versus Red) in Fig. 1J shows that HS increased the slope of this scattered plot over the control. To affirm that the small, punctate-appearing IB in nuclei are indeed of nuclear localization, we imaged cells by confocal microscopy. Confocal images in Fig. S3 show that nuclear IB and the Hoechst 33342–stained nucleus are in the same focal plane.
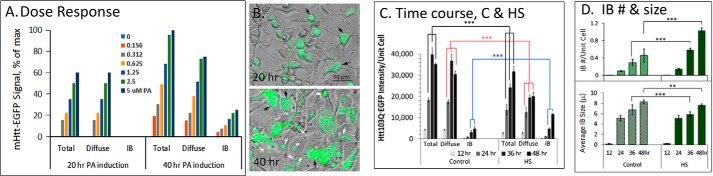
To better define conditions necessary for IB formation, we assessed their appearance vis à vis the dose of PA as well as the time to induce mHtt expression. Under control conditions and with a lower PA concentration (e.g. <1.25 μm) (Fig. 2A) or earlier time point (e.g. <24 h) (Fig. 2C) of induction, mHtt appeared exclusively or mainly as diffuse signal (Fig. 2B). Live cell images of the PA dose-dependent induction of diffuse versus IB forms of mHtt–EGFP at the 20 and 40 h time points are included as Fig. S4. HS (2 h, 42 °C) at 6 h of PA induction promoted mHtt IB formation at the expense of diffuse mHtt when assessed at subsequent time points of 12, 24, 36, and 48 h (Fig. 2, C and D). Quantitation of IB number and size of the control and HS cells showed that HS increased the number of IB per cell but reduced slightly the averaged IB size because of the increase in small nuclear IBs (Fig. 2D). These results are consistent with a nucleated polymerization process in IB formation as proposed (23).
Figure 2.
Time and ponasterone dose-dependent induction of mHtt expression and IB formation in 14A2.6 PC12 cells. A, ponasterone dose-dependent induction of mHtt expression. The 14A2.6 PC12 cells were treated with the indicated concentrations of PA to induce mHtt expression. Live cell images were captured at the 20- and 40-h time point of induction with PA. mHtt intensity was analyzed by ImageJ Fiji using the batch/Macro program protocol. Results are representative of three independent experiments. mHtt intensities are presented as % of maximum (i.e. total mHtt integrated density at 40 h with 5 μm PA = 100%). B, merged phase and EGFP images of cells. Images were captured at 20 h and 40 h of induction with 5 μm PA. The complete PA-dose dependent induction of mHtt–EGFP at the 20- and 40-h time points are included as Fig. S4. Results are representative of three separate experiments. C, time course of induction of Htt103Q–EGFP in control and HS cells. 14A2.6 cells were plated in 96-well plates and PA was added to a final concentration of 5 μm to induce the expression of the Htt103Q–EGFP protein. Cells were heat shocked at 42 °C for 2 h at the 6-h time point of induction with PA. Images of the control and HS cells at the 12-, 24-, 36-, and 48-h time points of PA induction were captured and analyzed. The bar graph represents quantitation of total, diffuse, and IB forms of mHtt intensity of control and pre-HS cells over the time course. Results indicate the average of six independent determinations under each condition. D, IB quantitation. IB count/cell (top) and average IB size (in microns; bottom) for the control and HS cells over the time course. Results are the average of six independent determinations under each condition. Probability of difference p >0.05 is defined as not significant, between 0.01 and 0.05 is significant (*), <0.01 is very significant (**), and <0.001 is extremely significant (***). Error bars represent S.D.
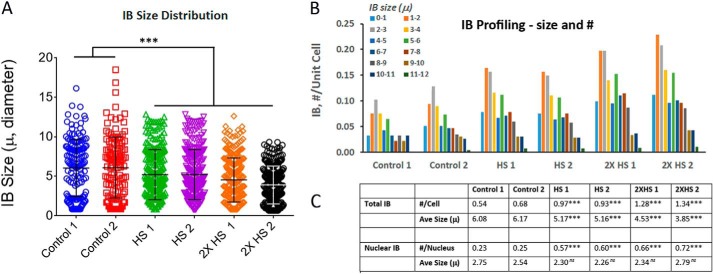
To quantitate the effects of HS on IB formation, we scored the numbers as well as size distribution of IBs of control and HS cells. For this, we selected six representative images of control and HS cells (42 °C for 2 h; 1× at 24 h; 2× at 24 and 30 h) at the 48-h time point of induction with PA. The results in Fig. 3A of IB size distribution showed that IBs ranged in size from <1 μm to >10 μm. Heat shock of cells resulted in a significant increase in the number and integrated intensity of IBs. Profiling of IB in Fig. 3B showed that there were two populations of IBs with averaged size of ∼2 and ∼6 μm in diameter, principally representing nuclear and cytosolic IBs, respectively. In the visual images, the punctate-appearing nuclear IB was a notable feature of the HS cells. Fig. 3C summarizes the quantitative results of IB in number per cell and averaged size. HS of cells resulted in statistically significant ∼2 to 3× increases in the total as well as nuclear IB number per unit cell. The averaged IB size of the HS cells was statistically smaller than the controls in large part because of the marked increase in small nuclear IB population. Because of their substantially smaller size, the nuclear IB constituted a small percentage of the total IB integrated intensity.
Figure 3.
Profiling of mHtt IB in control and heat shock 14A2.6 cells. Cells were plated and mHtt expression induced by the addition of 5 μm PA (t = 0). For heat shock (42 °C, 2 h), 1× was done at the 24-h time point and 2× was done as 24- and 30-h time points followed by recovery at 37 °C. Cells were fixed and processed for imaging at the 48-h time point of PA induction. Two images from each of the three conditions were used for this analysis. Result is representative of many images (>100 images) of experiments done over the course of 2 years. A, IB size distribution of control and HS cells. B, IB profiling. Number and size distribution of IBs of the six representative images as sorted in pivot chart. C, summary of the total and nuclear IB number/cell and average size. GraphPad InStat was used to analyze the probability of difference between the HS samples and controls with p >0.05 defined as not significant (ns), between 0.01 and 0.05 as significant (*), <0.01 as very significant (**), and <0.001 as extremely significant (***). The six original full-size images used for this analysis are shown in Fig. S5, A–F.
Effects of HSP70 and HSC70 knockdown on the restructuring of mHtt
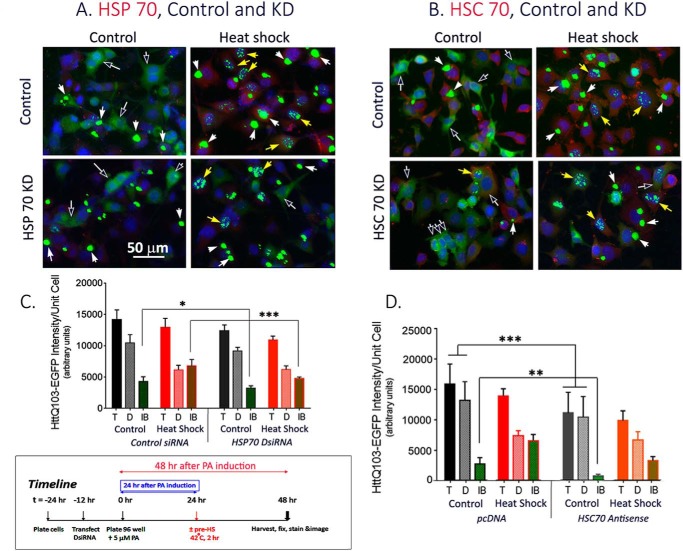
Heat shock elicits the activation of HSF1 for the induction of HSP chaperones such as HSP90, 70, 40, and others in various subcellular compartments. Collectively, the chaperones promote the folding/refolding, disaggregation, and disposition of cellular proteins for recovery after stress (19). HSP90 binds many meta-stable client proteins including the normal and the polyQ-expanded Htt (25, 26), whereas HSP70 primarily functions to chaperone exposed hydrophobic patches of nascent and nonnative proteins (27). Several of the HSP chaperones work in tandem and in cooperation for the folding versus degradation of substrate proteins (28). To probe the contribution of individual HSPs in the HS-induced dynamic change of mHtt, we asked if knocking down of HSP70 or HSC70 affected the restructuring of mHtt under either control or heat shock condition. For HSP70 KD, cells were first transfected with control and HSP70 DsiRNA (Dicer-substrate siRNA, a mixture of three Hspa1a siRNAs, Integrated DNA Technologies) and then processed for mHtt induction by PA, heat shock, and image analysis. Representative images of control and HSP70 DsiRNA-transfected cells kept under a basal versus HS condition in Fig. 4A clearly showed that HSP70 DsiRNA blunted the heat shock induction of HSP70 (Fig. 4A, red). Quantitation of the total, diffuse, and IB mHtt intensity in the control versus HSP70 DsiRNA–transfected cells showed that knocking down HSP70 reduced mHtt IB formation under both control and HS conditions, with the reduction being of greater magnitude and statistical significance in the HS cells over the controls (Fig. 4C).
Figure 4.
Effects of knocking down of HSP70 and of HSC70 expression on the restructuring of diffuse mHtt into forming IB under 37 °C control and 42 °C heat shock condition. A, HSP70 KD: Cells were transfected either with control or HSP70 DsiRNA according to methods described in the text. B, HSC70 KD: Cells were transfected with either pcDNAZeo(−) vector DNA or the hsc70 antisense vector DNA as indicated. 12 h after transfection, cells were plated in a 96-well plate in medium containing 5 μm PA to induce the expression of mHtt. HS (2 h, 42 °C) was at 24 h after plating and cells were harvested and fixed at 48 h. The time line of the experiment is illustrated in the framed box at the bottom left of the figure. Each of the image frames represents an area of ∼200 × 266 μm. Black arrows point to diffuse Htt–EGFP signal; white arrows point to IB; yellow arrows point to punctate nuclear IB. C, quantitation of the total, diffuse, and IB mHtt intensity of control and HSP70 DsiRNA–transfected cells under control and heat shock condition. Total, diffuse, and IB mHtt intensity per unit cell of three separate experiments each with four replicates per condition were determined. Result shown is the average ± S.D. of 12 representative images from each of the treatment condition (± HSP70 siRNA; ± pre-HS). D, quantitation of the total, diffuse, and IB mHtt intensity of control and HSC70 KD cells. Result shown is the average of results from three separate experiments each with 8–16 independent replicated per condition and 3–4 captured images per sample. Statistical analysis was done using the GraphPad InStat program. Probability of difference p >0.05 is defined as not significant, between 0.01 and 0.05 is significant (*), <0.01 is very significant (**), and <0.001 is extremely significant (***). The eight full frame original images shown in A and B are included in Fig. S6, A–H. Error bars represent S.D.
The heat-inducible HSP70 (HSPA1A) is a member of the HSP70 multigene family that includes the constitutively expressed HSC70 protein (HSPA8) involved in processes such as folding and trafficking of nascent polypeptide under normal conditions (29–31). To evaluate specificity of effects among members of the HSP70 protein family in the remodeling of mHtt, we determined the effects of knocking down HSC70. Representative images of control versus HSC70 KD cells under control and heat shock conditions in Fig. 4B show that HSC70 is constitutively expressed at 37 °C, and heat shock at 42 °C for 2 h gave a modest increase (<2 times) in this expression, a result consistent with previous observations (30, 31). Quantitation across multiple images of the total, diffuse, and IB mHtt intensity per unit cell in Fig. 4D shows that HSC70 KD selectively affected the restructuring of diffuse mHtt into IB under the basal 37 °C condition such that the IB signal decreased from ∼20% of the total in the control to ∼7% in the HSC70 KD cells. In HS cells, IB represented ∼46 and ∼32% of the total in control versus HSC70 KD cells, respectively. Together, the results in Fig. 4 show that KD of HSP70 selectively blunted the HS-induced restructuring of diffuse mHtt into forming IBs, whereas KD of HSC70 selectively suppressed IB formation at 37 °C.
mHtt expression and transcription factor dysregulation
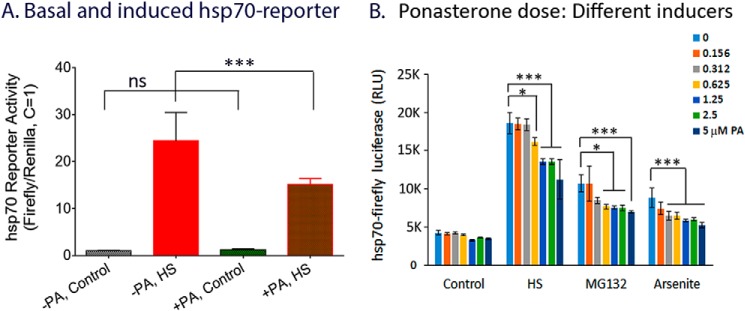
Transcription dysregulation is an important pathogenic mechanism in HD, and major changes in gene expression have been detected in postmortem HD brains and experimental model systems (32–34). Given the well-documented role of HSF1 as a driver of the HSP-dependent PQC system and its dysfunction in many model systems of aging (6–8, 11, 14, 17, 35), we asked if expression of mHtt may in turn affect the regulation and function of HSF1. For this, cells were first transfected with HSE–firefly luciferase DNA along with the internal control humanized Renilla luciferase DNA. Transfected cells were split into two halves, with and without 5 μm PA to induce mHtt expression. HSE–reporter gene activity was tested 24 h after PA induction, a time when >80% of the mHtt appeared as diffusible assemblies, by heat shock of cells at 42 °C for 2 h followed by recovery at 37 °C for 4 h. Results in Fig. 5A show that expression of mHtt significantly repressed the heat-induced HSE–reporter gene activity. Fig. 5B shows the dose-response effect of PA on HSE–firefly luciferase reporter gene activity under control and heat shock-, MG132-, and arsenite-induced conditions. HS, MG132, and arsenite were all effective in inducing the HSE–firefly luciferase reporter, with HS being the most efficacious. For each of the inducers, there was a PA dose-dependent reduction in the HSE–reporter gene activity in proportion with the increased expression of diffuse mHtt. This observation is consistent with previous demonstrations of progressive disruption of the PQC machinery in models of polyglutamine diseases (36).
Figure 5.
Expression of mHtt represses HSF1-mediated induction of hsp70 reporter gene activity. A, control and heat shock hsp70 reporter gene activity with and without mHtt induction. 14A2.6 cells were transfected with hsp70–firefly luciferase DNA and the internal control humanized Renilla luciferase DNA. Following transfection, cells were plated in 96-well plate without and with 5 μm PA as indicated. 24 h after PA induction, cells were heat shocked at 42 °C for 2 h followed by recovery incubation at 37 °C for 4 h. (Note: This HS condition of 2 h at 42 °C followed by recovery at 37 °C for 4 h is too short a time to have much of an effect on the mHtt profile.) Result of normalized hsp70 reporter gene activity, with the control firefly/Renilla luciferase ratio set at 1, is presented in bar graph format. Statistical analysis was done using GraphPad InStat. B, effect of PA dose-dependent induction of mHtt on HSE–reporter gene activity. 14A2.6 cells were transfected with the hsp70–firefly luciferase DNA and then plated into 96-well plates. Indicated concentrations of PA was added to induce the expression of mHtt protein for 24 h. hsp70–firefly luciferase activity was induced at 24 h after PA addition by HS, 2 h at 42 °C followed by recovery incubation at 37 °C for 4 h; MG132 (final concentration 10 μg/ml, 6 h); and sodium arsenite (30 μm, 6 h). The result of the firefly luciferase activity in RLU is the average of four independent determinations. Probability of difference p > 0.05 is defined as not significant, between 0.01 and 0.05 is significant (*), <0.01 is very significant (**), and <0.001 is extremely significant (***). Error bars represent S.D.
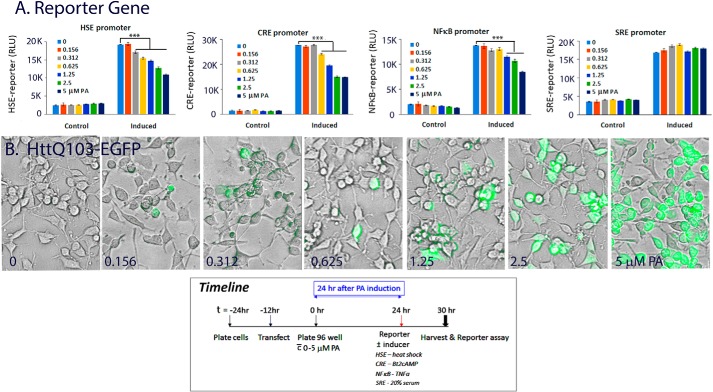
We then asked if other TF driven reporter genes may be similarly affected. In particular, we were interested in TFs implicated in memory formation and in stress response (37–41). Fig. 6A presents the result of PA dose-dependent effect on the basal and induced HSE–, CRE–, NF-κB–, and SRE–firefly luciferase reporter gene activity. Results showed that in each case, PA gave a dose-dependent repression of the induced HSE–, CRE–, and NF-κB–firefly luciferase reporter activity, whereas the serum-induced SRE–reporter gene activity was unaffected. This suppression of reporter gene activities correlates with expression of the diffuse mHtt protein as shown in fluorescent/phase images of cells from the same experiment in Fig. 6B. The basal reporter gene activity was low and it was difficult to gain statistically meaningful results of the effects of mHtt expression on the basal (uninduced) reporter gene activity. Parallel studies done with the 25Q PC12 cells did not show a PA dose-dependent effect on reporter gene activity, indicating a polyQ dependence and specificity in reporter gene repression. Expression of mHtt had little or no effect on the SRE–reporter gene under the same experimental condition. Our results are consistent with previous observation of the efficacy of polyQ and amyloid-like aggregates to sequester many meta-stable regulatory proteins, CREB, HSF1, and NF-κB included (33, 42). Together, our results in Figs. 5 and 6 show that increased expression of the diffusible assemblies of mHtt suppressed the transcriptional activity of HSF1, CREB, and NF-κB, whereas SRE-promoter was unaffected.
Figure 6.
Diffuse mHtt gives dose-dependent repression of HSE–, CRE– and NF-κB–reporter gene activity while having little or no effect on SRE-reporter. 14A2.6 PC12 cells were transfected with the HSE–, CRE–, NF-κB–, and SRE–firefly luciferase reporter DNA. After transfection, cells were plated in 96-well plated and ponasterone was added to designated wells to final concentrations as indicated. The time line of the experiment is shown at the bottom of the figure. A, reporter gene activity. 24 h after ponasterone induction, cells were tested for basal and induced reporter gene expression. HSE-reporter was induced by HS of cells for 2 h at 42 °C followed by recovery at 37 °C for 4 h, CRE-reporter induced by 1 mm dibutyryl cAMP for 6 h, NF-κB by 20 ng/ml of TNFα for 6 h, and SRE-reporter by 20% fetal bovine serum for 6 h. Reporter gene activity was assayed using the Dual-Glo Reporter gene assay kit from Promega. Results on the firefly–luciferase reporter gene activity are presented as RLU. B, merged phase and EGFP images of live cells induced with the indicated concentration of ponasterone at 24 h. Images are representative of eight independent wells of cell samples. Probability of difference p >0.05 is defined as not significant, between 0.01 and 0.05 is significant (*), <0.01 is very significant (**), and <0.001 is extremely significant (***). Error bars represent S.D.
To further define the functional readout of diffuse versus IB forms of mHtt in this paradigm of TF repression, we used a 2-h HS protocol to drive diffuse mHtt into forming IBs and tested the consequence of such manipulation on the CRE-promoter activity. The time line of the experiment is as shown in Fig. 7E. Representative images of cells at the 24-h and at the 48-h time point of PA induction, without and with pre-HS, are in Fig. 7, B–D, respectively. Results from the CRE–reporter gene assay (Fig. 7A) showed that PA-induced expression of mHtt repressed CRE–reporter gene activity at both the 24- and 48-h time points as compared with controls. Most importantly we showed that a 2-h HS of the PA-induced cells, but not of the control cells (without PA), at 24 h rescued the repressed CRE–reporter gene activity when measured at the 48-h time point (Fig. 7A). Collectively, these results demonstrate that heat shock, a condition that induces HSP chaperones to drive the structuring of diffuse mHtt into forming IBs, has beneficial consequences in the regulation and function of CREB. This also supports the notion that IB formation is an HSP-supported coping mechanism in the management and disposition of pathogenic proteins to alleviate TF dysfunction. Qualitatively similar result was observed with the NF-κB–reporter gene. Because HS affects both the remodeling of mHtt and the HSE–reporter gene, it was difficult to tease out the cause and effect relationship, and the HSE–reporter gene was not included in this analysis.
Figure 7.
Heat shock alleviates the repressed CRE-reporter gene activity concomitant with the restructuring of diffuse mHtt to form IB. Cells were transfected with the CRE–firefly luciferase DNA. Cells were then plated in 96-well plate without and with 5 μm PA to induce mHtt–EGFP expression. A, basal and dibutyryl-cAMP–induced CRE–reporter gene activity was tested at the 24- and 48-h time point after plating. Heat shock (42 °C for 2 h) was at 24 h of PA induction, and cells were returned to a 37° incubator for the remainder of the experiment. The result is the average of 24 independent determinations (eight independent samples, 3× reporter gene reading of each). B–D, representative images of cells at the 24-h time point of PA induction (B) and 48 h of induction without (C) and with a 2-h HS at the 24-h time point (D). Induction of HSP70 was tracked with a polyclonal antibody against HSP70 followed by Alexa Fluor 594 conjugated goat anti-rabbit IgG. Nuclei were stained using Hoechst 33342. E, the time line of the experiment is shown. Probability of difference p >0.05 is defined as not significant, between 0.01 and 0.05 is significant (*), <0.01 is very significant (**), and <0.001 is extremely significant (***). Error bars represent S.D.
Chemical chaperone promoted compaction of mHtt and survival of striatal neuron–derived cells
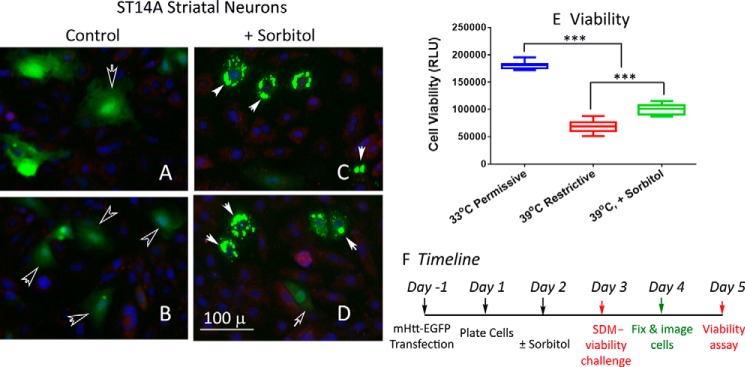
Striatal neurons are noted for their vulnerability in HD (43). To ascertain disease relevance of chaperone-dependent IB formation and the concomitant restoration of cell function, we used the ST14A line of cells derived from E14 rat striatum primordia by retroviral transduction of a temperature-sensitive SV40 large T antigen to analyze the processing and functional readout of mHtt expression (44, 45). The temperature-sensitive growth phenotype of these cells (33 °C permissive, 39 °C restrictive) negated the use of HS to induce HSP chaperones for the structuring and compaction of mHtt. Instead, we used sorbitol as a chemical chaperone to promote protein structuring (46–48). Cells were transfected with plasmid expression vectors of HttQ74–EGFP and the effects of sorbitol on the structuring and compaction of mHtt and cell viability under stress were determined. Representative images of control (Fig. 8, A and B) and sorbitol-treated (150 mm) (Fig. 8, C and D) ST14A cells show that whereas HttQ74–EGFP mostly had a diffuse appearance in control cells maintained at 33 °C, in the sorbitol-treated cells HttQ74–EGFP appeared primarily in the form of IBs. Shifting the ST14A cells from 33 °C to 39 °C under a serum-deprived condition resulted in a ∼60% decrease in cell viability at 48 h after the shift (44), whereas for the sorbitol-treated cells the decrease was ∼45%. Sorbitol had little effect on survival of the untransfected ST14A cells or cells transfected with the HttQ23–EGFP DNA. Together, the results in Fig. 8 show that sorbitol promotes the structuring and compaction of mHtt into forming IB and confers a statistically significant survival advantage to the ST14A striatal neurons under stress.
Figure 8.
Sorbitol, a chemical chaperone, promoted the structuring of diffusible assemblies of mHtt to form IBs and supported survival of the E14 rat striatal neuron–derived ST14A cell line. ST14A cells were transfected with the HttQ74–EGFP plasmid DNA and then plated in 96-well plate (day 0) according to methods described in the text. Sorbitol was added to designated wells of cells to a final concentration of 150 mm at 24 h after plating (day 1). A–D, for cell imaging, cells were fixed 72 h after plating (day 3) and processed to stain for HSP70 and nuclei according to methods described in the text. A and B are images of the control and C and D are of sorbitol-treated (150 mm, 48 h) cells. These images are representative of results from two separate experiments each with eight independent samples. E, to test for survival of the control and sorbitol-treated cells under stress, the culture medium was changed to DMEM without serum and cells were moved from a 33 °C to a 39 °C incubator on day 2 after plating. Cell viability was determined 2 days after the shift to this restrictive condition using the CellTiter-Glo reagent from Promega (G7572). Result represents the average of viability readouts (3X each sample) of 16 independent samples per condition from two separate experiments. F, the time line of the experiment is as shown. Probability of difference <0.001 is extremely significant (***). Error bars represent S.D.
Discussion
The protein deposits found in ND, including Huntington's, Alzheimer's, Parkinson's, and prion diseases, are formed of completely unrelated proteins. The presence of such protein aggregates is often the most distinct postmortem histological signs of ND, and it was hypothesized that protein aggregation is toxic to neurons and that therapeutic strategies aimed at clearing such aggregated protein deposits would be beneficial. Over time, it has become clear that the correlation of the location of protein deposits and brain regions of neuron death is weak at best (49). This and other considerations raise important questions regarding the identity of pathogenic protein conformer(s) and the cause of neuron death in ND (49–52).
Our observation that HS has a timely and reproducible effect in promoting mHtt IB formation is in general agreement with that of previous studies (53, 54), although the tenor, emphasis, and conclusion of the studies are different. In this research, we looked at changes of both diffuse and IB forms of mHtt and arrived at the conclusion that HS through induction of HSP chaperones promotes the structuring and compaction of diffuse mHtt into forming IBs with beneficial functional outcomes. Although our standard experimental protocol was to HS cells at the 24-h time point in the course of PA induction, the effective window for HS to drive the depletion of diffusible assemblies of mHtt to form IBs extended from 6 to 30 h of PA induction. Earlier than 6 h and later than 30 h, the results became less clear-cut. The significance of the size difference between cytoplasmic and nuclear IBs is not entirely clear. One possibility is a difference in mHtt concentration in the subcellular compartments. Alternatively, the presence of different HSPs and their stoichiometry in the nuclear versus cytoplasmic compartment may impart a different remodeling outcome.
mHtt and other ND-associated proteins belongs to a family of intrinsically disordered proteins (IDPs), proteins without a defined stably folded structure and with abundant interacting surfaces in their native states (55–57). Their aggregation begins with the meta-stable, intrinsically disordered conformers and ends up as IBs in Huntington's disease, β-amyloid deposits in Alzheimer's disease, and Lewy bodies in Parkinson's disease, as examples. There is a growing body of evidence that conditions that promote protein structuring guide the assembly and aggregation of IDPs, including α-synuclein, prion, and tau (48, 58, 59). Consistent with this concept, we hereby show that induction of HSP chaperones by HS greatly enhances the assembly of diffuse mHtt into forming IBs and KD of HSP70 expression blunted this process. Our observation that KD of HSC70 selectively reduced the basal rate of IB formation is consistent with the acknowledged role of HSC70 in protein folding and trafficking under normal growth conditions (29) and lends additional support to the role of protein structuring in mHtt IB formation.
The assembly of the disease protein monomer and oligomers into visually distinctive fibers and IBs is a multistep process with many ill-defined intermediates including unstructured and quasi-structured monomer and oligomers, higher order oligomers, protofibrils, and fibril bundles that may dynamically interconvert (60, 61). It is unclear which one or combinations of these may be the toxic entity that predisposes neurons to their eventual demise. An emerging concept, as exemplified by the β-amyloid protein of Alzheimer's disease, is that small intermediates including soluble oligomers adversely affect synaptic structure and plasticity (62). Studies in other ND suggest that a broadly similar process of neuronal dysfunction is caused by diffusible monomers and oligomers of disease-causing proteins (54, 62–66). In the case of HD, longitudinal tracking of individual neuron fate showed that the presence of diffuse mHtt is a prognostic indicator of neuronal death whereas IB formation is correlated with neuron survival (24, 67, 69, 70). Further, studies using an epitope-specific mAb 3B5H10 suggest that monomer or small oligomer of mHtt as opposed to higher order aggregate can best predict neuron death (71). Consistent with these observations, we showed using the ST14A embryonic striatal neuron–derived cells that an osmolyte sorbitol depleted diffuse mHtt and promoted their structuring and compaction into forming IB to support neuron survival under stress.
Identification of mHtt conformers that are directly or indirectly neurotoxic is a challenging proteomics problem with significant implications in framing therapeutics development for ND. To relate protein aggregation to disease pathogenesis, it is necessary to take into consideration the cellular components affecting protein aggregation, such as chaperones, proteases, as well as molecular crowding (72). In vitro analysis may not mimic scenarios in cells; reasons include the following: 1) intermediate conformers are likely to be meta-stable, 2) cell lysis and fractionation affect protein concentration which is a primary determinant in aggregation (73), and 3) salts and/or detergents used in the in vitro analysis would change the distribution and solubility of protein conformers (74). In this research, we relied principally on image analysis of live as well as fixed cells to track the expression and progression of diffuse versus IB forms of mHtt.
An important cause of the neurotoxicity of disease proteins implicated in ND is the anomalous interactions with many metastable regulatory proteins causing multifactorial toxicity and collapse of essential cellular functions (33, 75). In HD, there is widespread transcription dysfunction (32–34). Herein, we show that although HSF1-mediated induction of HSPs guides diffuse mHtt to form IBs and improves cell fitness, there is a negative feedback loop, namely, the expression of diffuse mHtt limits and constrains the activity of HSF1. Our observation is in agreement with previous observations which show that the expression of model β-sheet proteins leads to deficiency of the normal cytosolic stress response mediated by HSF1 and NF-κB (33), and of an impaired heat shock response in mHtt expressing cells (38, 40, 76). Likewise, a deficiency in striatal neuron proteostasis machinery likely contributes to the select demise of these neurons in HD (77).
The repression of TFs by diffusible assemblies of mHtt as a pathogenic mechanism is not limited to HSF1. We showed that the CREB- and NF-κB–driven reporter genes are similarly repressed. CREB is a TF involved in a number of processes required for memory formation (38, 40). Given that ND often involves memory compromises, it is perhaps not surprising that disruption of CREB function leads to neurodegeneration (39), that inhibition of CREB reduces neuronal excitability and plasticity associated with ND (37), and that CREB is down-regulated in Alzheimer's disease brains (79). Likewise, NF-κB functions as a crucial TF for glia and neurons, and expression of mHtt down-regulates NF-κB signaling (41, 80).
Our observation that a transient heat shock can reverse the effect of mHtt in secluding TFs has three implications: 1) IB formation is protective and represents a chaperone-supported coping mechanism; 2) part of the cyto-protective activity of heat shock is because of the reversal of TF seclusion such as HSF1, CREB, and NF-κB; and 3) for PQC purposes, the HSP chaperone network can help either by promoting the proper folding/refolding and degradation of nonnative proteins or by structuring and aggregating toxic protein conformers into IBs. Our observation is consistent with previous result on the yeast prion protein [RNQ+] in which the HSP40 chaperone protein Sis1 stimulated amyloid assembly of the otherwise toxic protein in yeast (63). These and other considerations (81, 82) suggest that chaperone-supported structuring and compaction of meta-stable IDPs can enhance cellular fitness. We do not know whether it is the remodeling of mHtt in the cytosol or the nucleus to form IB that account for the improved TF readout. HSF1 and NF-κB are located in the cytosol under normal and unstressed condition and become activated and shuttled into the nucleus upon stress (83). Teleologically, an increase in the presence of diffusible assemblies of mHtt in the cytosol can entrap HSF1 and NF-κB to constrain their translocation into the cell nucleus. For CREB, a constitutive nuclear TF, it was shown that β-amyloid blocked the nuclear translocation of activated CREB (84). Indeed, aggregation-prone artificial β-sheet proteins and authentic disease proteins have been shown to cause a pronounced impairment of nucleocytoplasmic transport and a redistribution of nuclear shuttle factors to the cytosol (85, 86).
We further note that independent of the mHtt-dependent seclusion of key TFs involved in cell fitness and survival, there is also age-dependent attenuation of various prosurvival and adaptive responses, a decreased activation of HSF1 in various aging models being a prime example (8, 14). Together, these two changes likely would have a snowballing effect to progressively limit and constrain the HSF1-dependent PQC machinery leading eventually to PQC collapse. In the context of a therapeutics framework for HD, this would mean that an earlier intervention to enhance the chaperone network in the course of disease progression would have a significantly greater efficacy, whereas treatment effectiveness would greatly diminish as the disease progresses.
Experimental procedures
Materials
siRNAs of rat HSP70 (rn.Ri.Hspa1a13.1, 13.2, and 13.3) were from Integrated DNA Technologies. Hsc70 antisense vector was from Dr. Janice S. Blum's laboratory of Indiana University School of Medicine (78). The hsp70–firefly luciferase reporter gene construct was described previously (13). The other three reporter gene constructs were cloned in the pGL4 vector (Promega): pCRE–firefly luciferase (E8471), pNF-κB–firefly luciferase (E8491), and pSRE–firefly luciferase (E1340), humanized Renilla luciferase DNA (phRLSV40), the Dual-Glo Luciferase Assay Reagent (E2920), and the CellTiter-Glo reagent (G7572) were all obtained from Promega. Plasmid expression vectors for the Q23 (plasmid no. 40261) and Q74mHtt–EGFP (plasmid no.40262) were from Addgene. The ST14A rat E14 striatal neuron–derived cell line was from Coriell Institute (CH00066; CHDI-90000066). Lipofectamine 2000 reagent used for DNA transfection was from Invitrogen. Rabbit polyclonal antibody against HSP70–RTG76 antibody that we generated against a recombinant histidine-tagged human HSP70 protein (9) or from Enzo Life Sciences, ADI-SPA-812, was used at 1:200 dilution for probing of the heat-inducible HSP70. The SPA-816 rabbit polyclonal antibody (Enzo Life Sciences, ADI-SPA-816) was used at 1:40 dilution for probing of HSC70. All other biochemical and chemical reagents were of molecular biology or reagent grade.
Cell culture
This study uses PC12-derived cell lines with stably integrated DNA of the normal (25Q) and polyQ-expanded (103Q; 14A2.6 line) HttExon1 sequences under the control of the inducible VgRxR promoter. Cells were cultured and maintained in DMEM supplemented with 5% fetal bovine serum, 5% horse serum, 50 μg/ml streptomycin, 50 units/ml of penicillin, 100 μg/ml G418, and 200 μg/ml zeocin essentially as described previously (20). For imaging experiments, cells were plated in 96 Stripwell plate at a density of ∼1–2 × 104 cells/well. For reporter gene assay, cells were plated at ∼5–6 × 104cells/well. Ponasterone (Invitrogen) was added to induce the expression of Htt–EGFP and the induction was monitored by live-cell imaging using an EVOS® FL microscope. Unless indicated otherwise, heat shock was for 2 h at 42° at 24 h after PA addition followed by recovery at 37 °C for the remainder of the experiment. Culturing of the ST14A rat E14 striatal neuron–derived cell line was done according to instructions provided by the HD Community Biorepository via Coriell Institute for Medical Research in DMEM with 10% fetal bovine serum.
Image acquisition and quantification
Cells were fixed with 4% paraformaldehyde in PBS for 15 min followed by blocking with 10% fetal bovine serum in PBS and 0.1% Triton X-100. All subsequent washes and incubation were in PBS with 0.1% Triton X-100. Fixed cells were incubated overnight at 4 °C with α-HSP70 antibody, followed by washing three times for10 min each. Cells were then incubated for 2 h at room temperature with Alexa Fluor 594 goat anti-rabbit IgG (Invitrogen, A11037) and Hoechst 33342 for nuclear stain (300–500 ng/ml). Cells were imaged with an EVOS® fluorescent microscope at an exposure setting to optimize the difference between the diffuse and IB mHtt signals. In a given experiment, all images were captured under identical conditions.
For quantification of Htt intensities in the diffuse versus IB format, we wrote Macro programs to quantitate Htt intensity and scoring of individual cells for diffuse mHtt signal versus HSP70 intensity. These Macro programs are included in Fig. S2.
DsiRNA knockdown of HSP70 expression and antisense knockdown of HSC70 expression
A rat HSP70 RNAi kit, TriFECTaRNAi, was obtained from Integrated DNA Technologies. For our experiment, we used the three Dicer-substrate siRNAs of rat HSP70 (rn.Ri.Hspa1a13.1, 13.2, and 13.3), a negative control DsiRNA, and a Tye563 transfection control DsiRNA, all at 20 pmol siRNA/transfection of a 35-mm plate of cells. All siRNA was prepared in Opti-MEM and mixed with Lipofectamine 2000 according to protocols provided by Invitrogen. The hsc70 antisense vector was generated by inserting hsc70 cDNA (1042–2187) in an inverted orientation into pcDNAZeo(−) (78). Freshly plated 14A2.6 cells were transfected with the designated DNA– or oligoRNA–lipid mixture. After transfection, cells were plated in 96-well plate and mHtt was induced by the addition of 5 μm PA. HS was done at 24 h after plating at 42 °C for 2 h followed by recovery at 37 °C. The time line of the experiment is as indicated in Fig. 4.
Reporter gene and cell viability assays
The four reporter genes used were HSE–, CRE–, NF-κB–, and SRE–firefly luciferase. Reporter gene activity was assayed using standard transient transfection protocol as reported previously (68). Briefly, freshly plated 80–90% confluent cells were used, the amount of each DNA was 0.5 μg/35 mm plate or 1.5 μg/60 mm plate. The amount of Lipofectamine 2000 (in microliters) was three times that of the total amount of DNA (in micrograms). 6 h after DNA transfection, cells were trypsinized and plated into individual wells of a 96 StripwellTM plate at a density of ∼5–6 × 104 cells/well (Corning/Costar 9102). The identically transfected cells provided a very stable baseline to assess the effects of mHtt expression and of treatment conditions (e.g. heat shock or dibutyryl cAMP) on reporter gene activity. When specified, the reporter DNA was transfected along with the internal control of phRLSV40 (synthetic humanized Renilla luciferase DNA) to normalize for possible nonspecific effects of treatment conditions on transcription and translation processes. Conditions to induce the promoters of reporter genes were heat shock (2 h at 42 °C plus 4 h of recovery at 37 °C) for HSE, dibutyryl cAMP (1 mm, 6 h) for CRE, TNFα (20 ng/ml, 6 h) for NF-κB, and fetal bovine serum for SRE (20%, 6 h). Results are the average ± S.D. of four separate determinations of each sample/condition. Within each experiment, the reporter gene signal was tight, with a <10% sample to sample variation.
The Dual-Glo Luciferase Assay Reagent from Promega (E2920) was used to assay for first the firefly then the Renilla luciferase activity according to manufacturer's instructions. Luciferase activity was measured using the PerkinElmer VICTOR 2 Multiplate Reader equipped with dual injectors. Where indicated, result of the Hsp70–firefly luciferase activity (in relative luminescence units, RLU) was normalized against that of the Renilla luciferase (RLU), and to facilitate comparison across experiments for statistical analysis across experiments this ratio was set at 1 for the control (without heat shock) (see Fig. 5A). By normalizing the Hsp70–firefly luciferase activity against that of the Renilla luciferase internal control, we effectively negated experimental variables such as differences in cell number, as well as nonselective effects of the treatment conditions/reagents on gene expression. Cell viability assay was done using the CellTiter-Glo regent from Promega (G7572) as described previously (9, 10).
Data and statistical analysis
ImageJ Fiji and Macro programs (Fig. S2) were used for image analysis and quantitation. Statistical analyses were done using GraphPad InStat or GraphPad Prism 6. Data shown are the mean ± S.D. The significance of difference between groups of data were determined using ANOVA followed by post hoc Tukey-Kramer multiple comparisons test. Probability of difference p >0.05 is defined as not significant, between 0.01 and 0.05 is significant (*), <0.01 is very significant (**), and <0.001 is extremely significant (***).
Author contributions
J. Y. C., M. P., H. S., D. B., K. K., and A. Y. C. L. data curation; J. Y. C., M. P., H. S., and A. Y. C. L. software; J. Y. C., M. P., H. S., and A. Y. C. L. validation; J. Y. C., M. P., H. S., and A. Y. C. L. visualization; J. Y. C., M. P., H. S., D. B., K. K., and A. Y. C. L. methodology; M. P., H. S., D. B., and A. Y. C. L. formal analysis; M. P., H. S., K. K., and A. Y. C. L. investigation; W. D., K. Y. C., and A. Y. C. L. resources; K. K., K. Y. C., and A. Y. C. L. funding acquisition; K. K., K. Y. C., and A. Y. C. L. writing-review and editing; K. Y. C. and A. Y. C. L. conceptualization; K. Y. C. and A. Y. C. L. supervision; A. Y. C. L. writing-original draft; A. Y. C. L. project administration.
Supplementary Material
Acknowledgments
We thank Dr. Long Jun Wu of the Department of Cell Biology and Neuroscience of Rutgers University and now of the Mayo Clinic (Minnesota) for the generous support in making the EVOS® fluorescence microscope available for this work. We thank Dr. Lori Covey of the Department of Cell Biology and Neuroscience of Rutgers University for providing us with TNFα to test for the induction of NF-κB–firefly luciferase. We thank Dr. Janice Blum and the then postdoctoral research Dr. Victoria L. Crotzer for the HSC70 antisense vector. We are grateful for the help of current students Samantha Chen, Hannaan Choudhry, Irene Nicholas, and Shreyaas Aravindun in image analysis and reporter gene assays. We thank Dr. Samuel Gunderson of Rutgers University for help in the planning of the HSP70 siRNA experiment.
This work was supported by internal University funding (to A. Y. C. L.), the Chemistry and Chemical Biology Fund 800177 of Rutgers (to K. Y. C.), and National Institutes of Health Grant R01 5DC15000 (to K. K.). M. P., H. S., and D. B. were supported by Aresty Research Fellowships from Rutgers University while working on this research project. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
We dedicate this work to the memory of Dr. J Fred “Paulo” Dice. It was through his generosity and introduction that we secured the HSC70 antisense construct from Dr. Janice Blum's laboratory of Indiana University School of Medicine in 2006.
This article contains supporting Figs. S1–S6.
- ND
- neurodegenerative disease
- ANOVA
- analysis of variance
- CRE
- cAMP-responsive element
- CREB
- CRE-binding protein
- HD
- Huntington's disease
- HS
- heat shock
- HSE
- heat shock element
- IB
- inclusion body
- IDP
- intrinsically disordered protein
- KD
- knockdown (of expression)
- PA
- ponasterone
- polyQ
- poly-glutamine
- PQC
- protein quality control
- RLU
- relative luminescence units
- SRE
- serum-response element
- TF
- transcription factor.
References
- 1. Forman M. S., Trojanowski J. Q., and Lee V. M. (2004) Neurodegenerative diseases: A decade of discoveries paves the way for therapeutic breakthroughs. Nat. Med. 10, 1055–1063 10.1038/nm1113 [DOI] [PubMed] [Google Scholar]
- 2. Knowles T. P., Vendruscolo M., and Dobson C. M. (2014) The amyloid state and its association with protein misfolding diseases. Nat. Rev. Mol. Cell Biol. 15, 384–396 10.1038/nrm3810 [DOI] [PubMed] [Google Scholar]
- 3. Hipp M. S., Park S.-H., and Hartl F. U. (2014) Proteostasis impairment in protein-misfolding and -aggregation diseases. Trends Cell Biol. 24, 506–514 10.1016/j.tcb.2014.05.003 [DOI] [PubMed] [Google Scholar]
- 4. Ross C. A., and Poirier M. A., (2004) Protein aggregation and neurodegenerative disease. Nat. Med. 10 (suppl.) S10–S17 10.1038/nm1066 [DOI] [PubMed] [Google Scholar]
- 5. Wolozin B., and Behl C. (2000) Mechanisms of neurodegenerative disorders: Part 1: Protein aggregates. Arch. Neurol. 57, 793–796 10.1001/archneur.57.6.793 [DOI] [PubMed] [Google Scholar]
- 6. Liu A. Y., Choi H.-S., Lee Y.-K., and Chen K. Y. (1991) Molecular events involved in transcriptional activation of heat shock genes become progressively refractory to heat stimulation during aging of human diploid fibroblasts. J. Cell Physiol. 149, 560–566 10.1002/jcp.1041490327 [DOI] [PubMed] [Google Scholar]
- 7. Liu A. Y., Lee Y. K., Manalo D., and Huang L. E. (1996) Attenuated heat shock transcriptional response in aging: molecular mechanism and implication in the biology of aging. EXS 77, 393–408 [DOI] [PubMed] [Google Scholar]
- 8. Liu A. Y., Lin Z., Choi H. S., Sorhage F., and Li B. (1989) Attenuated induction of heat shock gene expression in aging diploid fibroblasts. J. Biol. Chem. 264, 12037–12045 [PubMed] [Google Scholar]
- 9. Oza J., Yang J., Chen K. Y., and Liu A. Y. (2008) Changes in the regulation of heat shock gene expression in neuronal cell differentiation. Cell Stress Chaperones 13, 73–84 10.1007/s12192-008-0013-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yang J., Oza J., Bridges K., Chen K. Y., and Liu A. Y. (2008) Neural differentiation and the attenuated heat shock response. Brain Res. 1203, 39–50 10.1016/j.brainres.2008.01.082 [DOI] [PubMed] [Google Scholar]
- 11. Akerfelt M., Morimoto R. I., and Sistonen L. (2010) Heat shock factors: Integrators of cell stress, development and lifespan. Nat. Rev. Mol. Cell Biol. 11, 545–555 10.1038/nrm2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Labbadia J., and Morimoto R. I. (2015) The biology of proteostasis in aging and disease. Annu. Rev. Biochem. 84, 435–464 10.1146/annurev-biochem-060614-033955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Duncan E. J., Cheetham M. E., Chapple J. P., and van der Spuy J. (2015) The role of HSP70 and its co-chaperones in protein misfolding, aggregation and disease. Subcell. Biochem. 78, 243–273 10.1007/978-3-319-11731-7_12 [DOI] [PubMed] [Google Scholar]
- 14. Gomez-Pastor R., Burchfiel E. T., and Thiele D. J. (2018) Regulation of heat shock transcription factors and their roles in physiology and disease. Nat. Rev. Mol. Cell Biol. 19, 4–19 10.1038/nrm.2017.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Carman A., Kishinevsky S., Koren J. 3rd, Lou W., and Chiosis G. (2013) Chaperone-dependent neurodegeneration: A molecular perspective on therapeutic intervention. J. Alzheimers Dis. Parkinsonism 2013, Suppl. 10, 007 10.4172/2161-0460.S10-007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lindberg I., Shorter J., Wiseman R. L., Chiti F., Dickey C.A., and McLean P. J. (2015) Chaperones in neurodegeneration. J. Neurosci. 35, 13853–13859 10.1523/JNEUROSCI.2600-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Morimoto R. I. (2011) The heat shock response: Systems biology of proteotoxic stress in aging and disease. Cold Spring Harb. Symp. Quant. Biol. 76, 91–99 10.1101/sqb.2012.76.010637 [DOI] [PubMed] [Google Scholar]
- 18. Sontag E. M., Vonk W. I. M., and Frydman J. (2014) Sorting out the trash: The spatial nature of eukaryotic protein quality control. Curr. Opin. Cell Biol. 26, 139–146 10.1016/j.ceb.2013.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Saibil H. (2013) Chaperone machines for protein folding, unfolding and disaggregation. Nat. Rev. Mol. Cell Biol. 14, 630–642 10.1038/nrm3658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Apostol B. L., Kazantsev A., Raffioni S., Illes K., Pallos J., Bodai L., Slepko N., Bear J. E., Gertler F. B., Hersch S., Housman D. E., Marsh J. L., and Thompson L. M. (2003) A cell-based assay for aggregation inhibitors as therapeutics of polyglutamine-repeat disease and validation in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 100, 5950–5955 10.1073/pnas.2628045100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tan Z., Dai W., van Erp T. G., Overman J., Demuro A., Digman M. A, Hatami A., Albay R., Sontag E. M., Potkin K. T., Ling S., Macciardi F., Bunney W. E., Long J. D., Paulsen J. S., et al. (2015) Huntington's disease cerebrospinal fluid seeds aggregation of mutant huntingtin. Mol. Psychiatry 20, 1286–1293 10.1038/mp.2015.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sathasivam K., Neueder A., Gipson T. A., Landles C., Benjamin A. C., Bondulich M. K., Smith D. L., Faull R. L., Roos R. A., Howland D., Detloff P.J., Housman D. E., and Bates G. P. (2013) Aberrant splicing of HTT generates the pathogenic exon 1 protein in Huntington disease. Proc. Natl. Acad. Sci. U.S.A. 110, 2366–2370 10.1073/pnas.1221891110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Scherzinger E., Sittler A., Schweiger K., Heiser V., Lurz R., Hasenbank R., Bates G. P., Lehrach H., and Wanker E. E. (1999) Self-assembly of polyglutamine-containing huntingtin fragments into amyloid-like fibrils: Implications for Huntington's disease pathology. Proc. Natl. Acad. Sci. U.S.A. 96, 4604–4609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Arrasate M., Mitra S., Schweitzer E. S., Segal M. R., and Finkbeiner S. (2004) Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature 431, 805–810 10.1038/nature02998 [DOI] [PubMed] [Google Scholar]
- 25. Karagöz G. E., and Rüdiger S. G. (2015) Hsp90 interaction with clients. Trends Biochem. Sci. 40, 117–125 10.1016/j.tibs.2014.12.002 [DOI] [PubMed] [Google Scholar]
- 26. Baldo B., Weiss A., Parker C. N., Bibel M., Paganetti P., and Kaupmann K. (2012) A screen for enhancers of clearance identifies huntingtin as a heat shock protein 90 (Hsp90) client protein. J. Biol. Chem. 287, 1406–1414 10.1074/jbc.M111.294801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Clerico E. M., Tilitsky J. M., Meng W., and Gierasch L. M. (2015) How hsp70 molecular machines interact with their substrates to mediate diverse physiological functions. J. Mol. Biol. 427, 1575–1588 10.1016/j.jmb.2015.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kundrat L., and Regan L. (2010) Balance between folding and degradation for Hsp90-dependent client proteins: A key role for CHIP. Biochemistry 49, 7428–7438 10.1021/bi100386w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Daugaard M., Rohde M., and Jäättelä M. (2007) The heat shock protein 70 family: Highly homologous proteins with overlapping and distinct functions. FEBS Lett. 581, 3702–3710 10.1016/j.febslet.2007.05.039 [DOI] [PubMed] [Google Scholar]
- 30. O'Malley K., Mauron A., Barchas J. D., and Kedes L. (1985) Constitutively expressed rat mRNA encoding a 70-kilodalton heat-shock-like protein. Mol. Cell Biol. 5, 3476–3483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sorger P. K., and Pelham H. R. (1987) Cloning and expression of a gene encoding hsc73, the major hsp70-like protein in unstressed rat cells. EMBO J. 6, 993–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hodges A., Strand A. D., Aragaki A. K., Kuhn A., Sengstag T., Hughes G., Elliston L. A., Hartog C., Goldstein D.R., Thu D., Hollingsworth Z. R., Collin F., Synek B., Holmans P. A., Young A. B., et al. (2006) Regional and cellular gene expression changes in human Huntington's disease brain. Hum. Mol. Genet. 15, 965–977 10.1093/hmg/ddl013 [DOI] [PubMed] [Google Scholar]
- 33. Olzscha H., Schermann S. M., Woerner A. C., Pinkert S., Hecht M. H., Tartaglia G. G., Vendruscolo M., Hayer-Hartl M., Hartl F. U., and Vabulas R. M. (2011) Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell 144, 67–78 10.1016/j.cell.2010.11.050 [DOI] [PubMed] [Google Scholar]
- 34. van Hagen M., Piebes D. G. E., de Leeuw W. C., Vuist I. M., van Roon-Mom W. M. C., Moerland P. D., and Verschure PJ (2017) The dynamics of early-state transcriptional changes and aggregate formation in a Huntington's disease cell model. BMC Genomics 18, 373 10.1186/s12864-017-3745-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Morimoto R. I.,. (2006) Stress, aging, and neurodegenerative disease. N. Engl. J. Med. 355, 2254–2255 10.1056/NEJMcibr065573 [DOI] [PubMed] [Google Scholar]
- 36. Gidalevitz T., Ben-Zvi A., Ho K. H., Brignull H. R., and Morimoto R. I. (2006) Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science 311, 1471–1474 10.1126/science.1124514 [DOI] [PubMed] [Google Scholar]
- 37. Jancic D., Lopez de Armentia M., Valor L. M., Olivares R., and Barco A. (2009) Inhibition of cAMP response element-binding protein reduces neuronal excitability and plasticity, and triggers neurodegeneration. Cereb. Cortex 19, 2535–2547 10.1093/cercor/bhp004 [DOI] [PubMed] [Google Scholar]
- 38. Kandel E. R. (2012) The molecular biology of memory: cAMP, PKA, CRE, CREB-1, CREB-2, and CPEB. Mol. Brain 5, 14 10.1186/1756-6606-5-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mantamadiotis T., Lemberger T., Bleckmann S. C., Kern H., Kretz O., Martin Villalba A., Tronche F., Kellendonk C., Gau D., Kapfhammer J., Otto C., Schmid W., Schütz G. (2002) Disruption of CREB function in brain leads to neurodegeneration. Nat. Genet. 31, 47–54 10.1038/ng882 [DOI] [PubMed] [Google Scholar]
- 40. Silva A. J., Kogan J. H., Frankland P. W., and Kida S. (1998) CREB and memory. Annu. Rev. Neurosci. 21, 127–148 10.1146/annurev.neuro.21.1.127 [DOI] [PubMed] [Google Scholar]
- 41. O'Neill L. A., and Kaltschmidt C. (1997) NF-κB: A crucial transcription factor for glial and neuronal cell function. Trends Neurosci. 20, 252–258 10.1016/S0166-2236(96)01035-1 [DOI] [PubMed] [Google Scholar]
- 42. Shimohata T., Nakajima T., Yamada M., Uchida C., Onodera O., Naruse S., Kimura T., Koide R., Nozaki K., Sano Y., Ishiguro H., Sakoe K., Ooshima T., Sato A., Ikeuchi T., et al. (2000) Expanded polyglutamine stretches interact with TAFII130, interfering with CREB-dependent transcription. Nat. Genet. 26, 29–36 10.1038/79139 [DOI] [PubMed] [Google Scholar]
- 43. Ehrlich M. E. (2012) Huntington's disease and the striatal medium spiny neuron: Cell-autonomous and non-cell-autonomous mechanisms of disease. Neurotherapeutics 9, 270–284 10.1007/s13311-012-0112-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rigamonti D., Bauer J. H., De-Fraja C., Conti L., Sipione S., Sciorati C., Clementi E., Hackam A., Hayden M. R., Li Y., Cooper J. K., Ross C. A., Govoni S., Vincenz C., and Cattaneo E. (2000) Wild-type huntingtin protects from apoptosis upstream of caspase-3. J. Neurosci. 20, 3705–3713 10.1523/JNEUROSCI.20-10-03705.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cattaneo E., and Conti L. (1998) Generation and characterization of embryonic striatal conditionally immortalized ST14A cells. J. Neurosci. Res. 53, 223–234 10.1002/(SICI)1097-4547(19980715)53:2%3C223::AID-JNR11%3E3.0.CO%3B2-7 [DOI] [PubMed] [Google Scholar]
- 46. Howard M., Fischer H., Roux J., Santos B. C., Gullans S. R., Yancey P. H., and Welch W. J. (2003) Mammalian osmolytes and S-nitrosoglutathione promote ΔF508 cystic fibrosis transmembrane conductance regulator (CFTR) protein maturation and function. J. Biol. Chem. 278, 35159–35167 10.1074/jbc.M301924200 [DOI] [PubMed] [Google Scholar]
- 47. Street T. O., Bolen D. W., and Rose G. D. (2006) A molecular mechanism for osmolyte-induced protein stability. Proc. Natl. Acad. Sci. U.S.A. 103, 13997–14002 10.1073/pnas.0606236103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Naik V., Kardani J., and Roy I. (2016) Trehalose-induced structural transition accelerates aggregation of α-synuclein. Mol. Biotechnol. 58, 251–255 10.1007/s12033-016-9923-4 [DOI] [PubMed] [Google Scholar]
- 49. Treusch S., Cyr D. M., and Lindquist S. (2009) Amyloid deposits: Protection against toxic protein species? Cell Cycle 8, 1668–1674 10.4161/cc.8.11.8503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. DiFiglia M., Sapp E., Chase K. O., Davies S. W., Bates G. P., Vonsattel J. P., and Aronin N. (1997) Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science 277, 1990–1993 [DOI] [PubMed] [Google Scholar]
- 51. Todd T. W., and Lim J. (2013) Aggregation formation in the polyglutamine diseases: Protection at a cost? Mol. Cells 36, 185–194 10.1007/s10059-013-0167-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wolfe K. J., and Cyr D. M. (2011) Amyloid in neurodegenerative diseases: Friend or foe? Semin. Cell Dev. Biol. 22, 476–481 10.1016/j.semcdb.2011.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bersuker K., Hipp M. S., Calamini B., Morimoto R. I., and Kopito R. R. (2013) Heat shock response activation exacerbates inclusion body formation in a cellular model of Huntington disease. J. Biol. Chem. 288, 23633–23638 10.1074/jbc.C113.481945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wyttenbach A., Carmichael J., Swartz J., Furlong R. A., Narain Y., Rankin J., and Rubinsztein D. C. (2000) Effects of heat shock, heat shock protein 40 (HDJ-2), and proteasome inhibition on protein aggregation in cellular models of Huntington's disease. Proc. Natl. Acad. Sci. U.S.A. 97, 2898–2903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dyson H. J. (2016) Making sense of intrinsically disordered proteins. Biophys. J. 110, 1013–1016 10.1016/j.bpj.2016.01.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Fink A. L. (2005) Natively unfolded proteins. Curr. Opin. Struct. Biol. 15, 35–41 10.1016/j.sbi.2005.01.002 [DOI] [PubMed] [Google Scholar]
- 57. Uversky V. N. (2015) Intrinsically disordered proteins and their (disordered) proteomes in neurodegenerative disorders. Front. Aging Neurosci. 7, 18 10.3389/fnagi.2015.00018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Levine Z. A., Larini L., LaPointe N. E., Feinstein S. C., and Shea J. E. (2015) Regulation and aggregation of intrinsically disordered peptides. Proc. Natl. Acad. Sci. U.S.A. 112, 2758–2763 10.1073/pnas.1418155112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sabareesan A. T., and Udgaonkar J. B. (2016) Pathogenic mutations within the disordered palindromic region of the prion protein induce structure therein and accelerate the formation of misfolded oligomers. J. Mol. Biol. 428, 3935–3947 10.1016/j.jmb.2016.08.015 [DOI] [PubMed] [Google Scholar]
- 60. Poirier M. A., Li H., Macosko J., Cai S., Amzel M., and Ross C. A. (2002) Huntingtin spheroids and protofibrils as precursors in polyglutamine fibrilization. J. Biol. Chem. 277, 41032–41037 10.1074/jbc.M205809200 [DOI] [PubMed] [Google Scholar]
- 61. Laganowsky A., Liu C., Sawaya M. R., Whitelegge J. P., Park J., Zhao M., Pensalfini A., Soriaga A. B., Landau M., Teng P. K., Cascio D., Glabe C., and Eisenberg D. (2012) Atomic view of a toxic amyloid small oligomer. Science 335, 1228–1231 10.1126/science.1213151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Haass C., and Selkoe D. J. (2007) Soluble protein oligomers in neurodegeneration: Lessons from the Alzheimer's amyloid β-peptide. Nat. Rev. Mol. Cell Biol. 8, 101–112 10.1038/nrm2101 [DOI] [PubMed] [Google Scholar]
- 63. Douglas P. M., Treusch S., Ren H. Y., Halfmann R., Duennwald M. L., Lindquist S., and Cyr D. M. (2008) Chaperone-dependent amyloid assembly protects cells from prion toxicity. Proc. Natl. Acad. Sci. U.S.A. 105, 7206–7211 10.1073/pnas.0802593105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Duennwald M. L. (2011) Polyglutamine misfolding in yeast: Toxic and protective aggregation. Prion 5, 285–290 10.4161/pri.18071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Saudou F., Finkbeiner S., Devys D., and Greenberg M. E. (1998) Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell 95, 55–66 [DOI] [PubMed] [Google Scholar]
- 66. Bodner R. A., Outeiro T. F., Altmann S., Maxwell M. M., Cho S. H., Hyman B. T., McLean P. J., Young A. B., Housman D. E., and Kazantsev A. G. (2006) Pharmacological promotion of inclusion formation: A therapeutic approach for Huntington's and Parkinson's diseases. Proc. Natl. Acad. Sci. U.S.A. 103, 4246–4251 10.1073/pnas.0511256103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Arrasate M., and Finkbeiner S. (2012) Protein aggregates in Huntington's disease. Exp. Neurol. 238, 1–11 10.1016/j.expneurol.2011.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Liu D. J., Hammer D., Komlos D., Chen K. Y., Firestein B. L., and Liu A. Y. (2014) SIRT1 knockdown promotes neural differentiation and attenuates the heat shock response. J. Cell Physiol. 229, 1224–1235 10.1002/jcp.24556 [DOI] [PubMed] [Google Scholar]
- 69. Finkbeiner S. (2011) Huntington's disease. Cold Spring Harb. Perspect. Biol. 3, a007476 10.1101/cshperspect.a007476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Miller J., et al. (2010) Quantitative relationships between huntingtin levels, polyglutamine length, inclusion body formation, and neuronal death provide novel insight into Huntington's disease molecular pathogenesis. J. Neurosci. 30, 10541–10550 10.1523/JNEUROSCI.0146-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Miller J., Arrasate M., Brooks E., Libeu C. P., Legleiter J., Hatters D., Curtis J., Cheung K., Krishnan P., Mitra S., Widjaja K., Shaby B. A., Lotz G. P., Newhouse Y., Mitchell E. J., et al. (2011) Identifying polyglutamine protein species in situ that best predict neurodegeneration. Nat. Chem. Biol. 7, 925–934 10.1038/nchembio.694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ami D., Natalello A., Lotti M., and Doglia S. M. (2013) Why and how protein aggregation has to be studied in vivo. Microb. Cell Fact. 12, 17 10.1186/1475-2859-12-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Scherzinger E., Lurz R., Turmaine M., Mangiarini L., Hollenbach B., Hasenbank R., Bates G. P., Davies S. W., Lehrach H., and Wanker E. E. (1997) Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell 90, 549–558 10.1016/S0092-8674(00)80514-0 [DOI] [PubMed] [Google Scholar]
- 74. Schmit J. D., Ghosh K., and Dill K. (2011) What drives amyloid molecules to assemble into oligomers and fibrils? Biophys. J. 100, 450–458 10.1016/j.bpj.2010.11.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Halliday M., Radford H., and Mallucci G. R. (2014) Prions: Generation and spread versus neurotoxicity. J. Biol. Chem. 289, 19862–19868 10.1074/jbc.R114.568477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Chafekar S. M., and Duennwald M. L. (2012) Impaired heat shock response in cells expressing full-length polyglutamine-expanded huntingtin. PLoS One 7, e37929 10.1371/journal.pone.0037929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Margulis J., and Finkbeiner S. (2014) Proteostasis in striatal cells and selective neurodegeneration in Huntington's disease. Front. Cell Neurosci. 8, 218 10.3389/fncel.2014.00218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zhou D., Li P., Lin Y., Lott J. M., Hislop A. D., Canaday D. H., Brutkiewicz R. R., and Blum J. S. (2005) Lamp-2a facilitates MHC class II presentation of cytoplasmic antigens. Immunity 22, 571–581 10.1016/j.immuni.2005.03.009 [DOI] [PubMed] [Google Scholar]
- 79. Pugazhenthi S., Wang M., Pham S., Sze C. I., and Eckman C. B. (2011) Downregulation of CREB expression in Alzheimer's brain and in Aβ-treated rat hippocampal neurons. Mol. Neurodegener. 6, 60 10.1186/1750-1326-6-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Reijonen S., Kukkonen J. P., Hyrskyluoto A., Kivinen J., Kairisalo M., Takei N., Lindholm D., and Korhonen L. (2010) Downregulation of NF-κB signaling by mutant huntingtin proteins induces oxidative stress and cell death. Cell Mol. Life Sci. 67, 1929–1941 10.1007/s00018-010-0305-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Escusa-Toret S., Vonk W. I. M., and Frydman J. (2013) Spatial sequestration of misfolded proteins by a dynamic chaperone pathway enhances cellular fitness during stress. Nat. Cell Biol. 15, 1231–1243 10.1038/ncb2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Wolfe K. J., Ren H. Y., Trepte P., and Cyr D. M. (2013) The Hsp70/90 cochaperone, Sti1, suppresses proteotoxicity by regulating spatial quality control of amyloid-like proteins. Mol. Biol. Cell 24, 3588–3602 10.1091/mbc.E13-06-0315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Vujanac M., Fenaroli A., and Zimarino V. (2005) Constitutive nuclear import and stress-regulated nucleocytoplasmic shuttling of mammalian heat-shock factor 1. Traffic 6, 214–229 10.1111/j.1600-0854.2005.00266.x [DOI] [PubMed] [Google Scholar]
- 84. Arvanitis D. N., Ducatenzeiler A., Ou J. N., Grodstein E., Andrews S. D., Tendulkar S. R., Ribeiro-da-Silva A., Szyf M., and Cuello A. C. (2007) High intracellular concentrations of amyloid-β block nuclear translocation of phosphorylated CREB. J. Neurochem. 103, 216–228 10.1111/j.1471-4159.2007.04704.x [DOI] [PubMed] [Google Scholar]
- 85. Woerner A. C., Frottin F., Hornburg D., Feng L. R., Meissner F., Patra M., Tatzelt J., Mann M., Winklhofer K. F., Hartl F. U., and Hipp M. S. (2016) Cytoplasmic protein aggregates interfere with nucleocytoplasmic transport of protein and RNA. Science 351, 173–176 10.1126/science.aad2033 [DOI] [PubMed] [Google Scholar]
- 86. Zhang Y. J., Gendron T. F., Grima J. C., Sasaguri H., Jansen-West K., Xu Y. F., Katzman R. B., Gass J., Murray M. E., Shinohara M., Lin W. L., Garrett A., Stankowski J. N., Daughrity L., Tong J., et al. (2016) C9ORF72 poly(GA) aggregates sequester and impair HR23 and nucleocytoplasmic transport proteins. Nat. Neurosci. 19, 668–677 10.1038/nn.4272 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.