Key Points
Genomic analysis of 1193 donor–recipient samples found no association with autosomal minor histocompatibility antigens and acute GVHD.
Y-chromosome–encoded minors that mismatch paralogous sites in female donors associate with acute GVHD.
Abstract
Allogeneic hematopoietic stem cell transplantation (allo-HCT) is a curative option for blood cancers, but the coupled effects of graft-versus-tumor and graft-versus-host disease (GVHD) limit its broader application. Outcomes improve with matching at HLAs, but other factors are required to explain residual risk of GVHD. In an effort to identify genetic associations outside the major histocompatibility complex, we conducted a genome-wide clinical outcomes study on 205 acute myeloid leukemia patients and their fully HLA-A–, HLA-B–, HLA-C–, HLA-DRB1–, and HLA-DQB1–matched (10/10) unrelated donors. HLA-DPB1 T-cell epitope permissibility mismatches were observed in less than half (45%) of acute GVHD cases, motivating a broader search for genetic factors affecting clinical outcomes. A novel bioinformatics workflow adapted from neoantigen discovery found no associations between acute GVHD and known, HLA-restricted minor histocompatibility antigens (MiHAs). These results were confirmed with microarray data from an additional 988 samples. On the other hand, Y-chromosome–encoded single-nucleotide polymorphisms in 4 genes (PCDH11Y, USP9Y, UTY, and NLGN4Y) did associate with acute GVHD in male patients with female donors. Males in this category with acute GVHD had more Y-encoded variant peptides per patient with higher predicted HLA-binding affinity than males without GVHD who matched X-paralogous alleles in their female donors. Methods and results described here have an immediate impact for allo-HCT, warranting further development and larger genomic studies where MiHAs are clinically relevant, including cancer immunotherapy, solid organ transplant, and pregnancy.
Visual Abstract
Introduction
Allogeneic hematopoietic cell transplantation (allo-HCT) can cure certain inherited diseases and acquired malignancies of the blood, yet biological mechanisms that provide beneficial effects, such as graft-versus-tumor (GVT),1-3 also contribute to life-threatening graft-versus-host disease (GVHD).4 Outcomes improve dramatically with donor–recipient matching of HLAs, but GVHD still occurs at frequencies of up to 70% in fully matched unrelated pairs, and to a lesser degree in related, HLA-identical transplant recipients,5 suggesting unaccounted-for genetic factors impact clinical responses.
Minor histocompatibility antigens (MiHAs) are germline-encoded immunogenic peptides presented by specific HLA molecules on the surface of cancer cells or normal tissues. Although donor and recipient mismatching at the major histocompatibility complex (MHC) confers the highest proportional risk of GVHD, many clinically relevant MiHAs with defined HLA restriction have been identified,6 including Y-chromosome–encoded antigens that affect outcomes in sex-mismatched HCT.7 In other nonmalignant conditions, such as solid organ transplant or pregnancy, MiHAs carry risk of rejection8-10 or miscarriage,11,12 respectively.
In leukemia, there is evidence for tumor-specific antigenicity by exogenous activation of gene expression,13-15 gene fusion,16 and alternative splicing.17 In all cancers, driver and passenger mutations mark tumor progression,18-20 which may guide biomarker discovery21,22 and individualized treatment.23-25 A subset of cancer variants give rise to immunoreactive neoantigens encoded by somatic changes in tumor DNA, and these changes are presented exclusively by tumors and targeted by patients’ normal immune systems.26 In a clinical setting, this effect may theoretically be exploited for GVT in allo-HCT27 or precision medicine approaches to cancer immunotherapy.
Despite the physiological connection between MiHAs and neoantigens, there are important differences that should guide genomic analysis. Neoantigen discovery from DNA or RNA sequences requires high sensitivity to detect rare or private variants from heterogenous tumor tissue,28 which is often chemically preserved.29 Sequencing patient and adjacent normal samples adds cost but also reduces false positives.30,31 MiHAs, on the other hand, arise from heritable germline polymorphisms that may be common in populations and accessible with less expensive microarrays or lower-coverage sequencing panels.32 In both cases, antigens are only immunoreactive if they are displayed by patient HLA in affected tissues. Therefore, it is important to annotate variants with predicted MHC restriction, binding affinity, and tissue-specific expression.
We sought a controlled, clinical-outcomes–based study in HLA-matched donor–recipient pairs to discover genetic variation outside the MHC that may contribute to the risk of acute GVHD following allo-HCT.
Methods
Study design
The study population consisted of high-resolution HLA-A–, HLA-B–, HLA-C–, HLA-DRB1–, and HLA-DQB1–matched (10/10) unrelated donor and recipient allo-HCT pairs. Patients were selected to obtain equal numbers with and without clinical evidence of grade II-IV acute GVHD, which was assessed as described based on severity or degree of organ involvement before day 100 after transplant.33 All patients received myeloablative conditioning for acute myeloid leukemia (AML) or other blood cancers in complete remission (CR1 or CR2). After quality control and other filtering (see “Whole-genome sequencing”), acute GVHD positive and negative cohorts were balanced for age, disease status, self-reported race or ethnicity, GVHD prophylaxis, and other factors (Table 1).
Table 1.
Counts of patients and donors in the GVHD and non-GVHD groups
| Variable | Acute GVHD | No acute GVHD | P |
|---|---|---|---|
| Number of recipients | 102 | 103 | |
| Number of centers | 47 | 50 | |
| Patient related | |||
| Recipient age at transplant, y | .09 | ||
| 0-9 | 3 (3) | 1 (1) | |
| 10-19 | 10 (10) | 6 (6) | |
| 20-29 | 16 (16) | 17 (17) | |
| 30-39 | 21 (21) | 15 (15) | |
| 40-49 | 23 (23) | 42 (41) | |
| 50-59 | 29 (28) | 22 (21) | |
| Median (range) | 41 (1-66) | 44 (9-66) | .58 |
| Recipient race/ethnicity | .79 | ||
| White, non-Hispanic | 87 (92) | 94 (92) | |
| African American, non-Hispanic | 1 (1) | 2 (2) | |
| Asian, non-Hispanic | 2 (2) | 3 (3) | |
| Hispanic, white | 5 (5) | 3 (3) | |
| Unknown | 7 (N/A) | 1 (N/A) | |
| Recipient sex | .009 | ||
| Male | 64 (63) | 46 (45) | |
| Female | 38 (37) | 57 (55) | |
| Karnofsky score | .47 | ||
| 10-80 | 24 (24) | 32 (31) | |
| 90-100 | 72 (71) | 66 (64) | |
| Missing | 6 (6) | 5 (5) | |
| Disease related | |||
| Disease status at transplant | .61 | ||
| Early (CR1) | 71 (70) | 75 (73) | |
| Intermediate (CR2+) | 31 (30) | 28 (27) | |
| Transplant related | |||
| Stem cell source | .24 | ||
| Marrow | 27 (26) | 35 (34) | |
| PBSCs | 75 (74) | 68 (66) | |
| TCE nonpermissiveness | .22 | ||
| Ambiguous DPB1 allele | 0 | 1 (1) | |
| Permissive DPB1 | 54 (54) | 62 (63) | |
| Nonpermissive DPB1 | 46 (46) | 35 (36) | |
| Data missing | 2 (N/A) | 5 (N/A) | |
| GVHD prophylaxis | .80 | ||
| Tacrolimus + MMF ± others | 22 (22) | 20 (19) | |
| Tacrolimus + MTX ± others (except MMF) | 56 (55) | 57 (55) | |
| Tacrolimus + others (except MTX, MMF) | 5 (5) | 2 (2) | |
| Tacrolimus alone | 3 (3) | 1 (1) | |
| CSA + MMF ± others (except tacrolimus) | 2 (2) | 3 (3) | |
| CSA + MTX ± others (except tacrolimus, MMF) | 12 (12) | 18 (17) | |
| CSA + others (except tacrolimus, MTX, MMF) | 1 (1) | 1 (1) | |
| CSA alone | 1 (1) | 1 (1) | |
| Donor–recipient sex matching | .007 | ||
| Male–male | 44 (42) | 40 (39) | |
| Male–female | 24 (25) | 38 (37) | |
| Female–male | 21 (21) | 6 (6) | |
| Female–female | 13 (13) | 19 (18) | |
| Donor–recipient CMV serostatus | .68 | ||
| Negative–negative | 33 (32) | 29 (28) | |
| Negative–positive | 37 (36) | 36 (35) | |
| Positive–negative | 10 (10) | 14 (14) | |
| Positive–positive | 19 (19) | 23 (22) | |
| Unknown | 3 (3) | 1 (1) | |
| Donor age at donation, y | .18 | ||
| 18-19 | 4 (4) | 1 (1) | |
| 20-29 | 45 (44) | 54 (52) | |
| 30-39 | 29 (28) | 31 (30) | |
| 40-49 | 16 (16) | 15 (15) | |
| ≥50 | 8 (8) | 2 (2) | |
| Median (range) | 30 (19-56) | 28 (20-52) | .11 |
| Donor–recipient ethnicity matching | |||
| African American–African American | 0 (0) | 2 (2) | |
| Asian-Pacific Islander–Asian-Pacific Islander | 2 (2) | 2 (2) | |
| Asian-Pacific Islander–white | 0 (0) | 2 (2) | |
| White–African American | 2 (2) | 1 (1) | |
| White–white | 84 (82) | 85 (83) | |
| White–deceased | 3 (3) | 2 (2) | |
| White–Native American | 0 (0) | 2 (2) | |
| White–unknown | 4 (4) | 2 (2) | |
| Hispanic–white | 1 (1) | 2 (2) | |
| Hispanic–deceased | 1 (1) | 0 (0) | |
| Hispanic–unknown | 1 (1) | 1 (1) | |
| Multiple–white | 3 (3) | 1 (1) | |
| Unknown–white | 1 (1) | 1 (1) | |
| Year of transplant | .61 | ||
| 2000 | 0 | 3 (3) | |
| 2001 | 2 (2) | 4 (4) | |
| 2002 | 1 (1) | 0 | |
| 2003 | 1 (1) | 3 (3) | |
| 2004 | 5 (5) | 8 (8) | |
| 2005 | 15 (15) | 11 (11) | |
| 2006 | 16 (16) | 15 (15) | |
| 2007 | 17 (17) | 17 (17) | |
| 2008 | 11 (11) | 8 (8) | |
| 2009 | 14 (14) | 9 (9) | |
| 2010 | 18 (18) | 22 (21) | |
| 2011 | 2 (2) | 3 (3) | |
| Follow-up among survivors, mo | |||
| Number evaluated | 44 | 50 | |
| Median (range) | 60 (33-99) | 61 (30-123) | .93 |
Data are expressed as n (%), except as noted. P values were calculated using the Pearson χ2 test to compare discrete variables or the Kruskal-Wallis test to compare continuous variables.
CMV, cytomegalovirus; CSA, cyclosporine; MMF, mycophenolate mofetil; MTX, methotrexate; PBSCs, peripheral blood stem cells.
Clinical data collection
Clinical data were collected by the Center for International Blood and Bone Marrow Transplant Research (CIBMTR), a collaboration between the National Marrow Donor Program and the Medical College of Wisconsin representing a worldwide network of transplant centers that contribute detailed data on HCT. The CIBMTR conducts research in compliance with all applicable federal regulations pertaining to the protection of human research participants. All participants provided informed consent for participation in the CIBMTR research program, including submission of biological samples to the Research Repository, and this study was approved by the National Marrow Donor Program Institutional Review Board.
HLA typing and histocompatibility matching
HLA matching was determined at high resolution for HLA-A, HLA-B, HLA-C, HLA-DRB1, and HLA-DQB1 through retrospective typing of stored pretransplant samples and/or reported by the transplant center and match assessment performed per CIBMTR criteria as previously described.34 Five-locus haplotype matching was performed with the HapLogic algorithm.35
Whole-genome sequencing
Two hundred fifty donor and 250 HCT recipient samples (500 samples total) were sequenced at Human Longevity, Inc. (San Diego, CA) to a mean coverage depth of 30× with 2 × 150 bp paired reads using Illumina HiSeq X instruments. One hundred twenty-five pairs came from transplants with clinical evidence of acute GVHD; 125 pairs came from transplants without evidence of GVHD. Ten recipient samples did not produce adequate sequencing data. A further 2 recipient samples and 1 donor sample failed the heterozygosity test that was applied to remove contaminated samples. An additional 32 samples were missing data for their paired donor or recipient and were removed from analysis. The final set included 205 pairs of donor–recipient samples (102 acute GVHD and 103 non-GVHD). Secondary analysis with Isaac alignment and variant calling pipeline36 resulted in 1 binary alignment map37 and 1 variant call format38 file per sample using the human genome reference assembly hg38. Variants with below average read depth (30×) were excluded from analysis.
Microarray data and analysis
The microarray data and primary analysis for supplemental Table 1 have been described previously.39
Bioinformatics
Genomic similarity was measured using identity-by-descent (IBD) sequencing with default parameters.40 This technique determines phase for donor and patient genotypes to form haplotype segments of varying lengths, which indicate common ancestry. Normalizing the lengths of these segments to those of specific genomic features (including the whole genome itself) gives a relative measure of genetic similarity for each feature (Figure 1). For comparison, the null distribution of normalized IBD in each region is simulated from an all-by-all pairing of donors and recipients (excluding actual HLA-matched pairs). X and Y chromosomes were excluded from analysis. Removal of low-quality variants due to read misalignment resulted in small broken intervals in the ARS and MHC, explaining lower than expected genetic similarity for HLA-matched donor–recipient pairs within these regions.
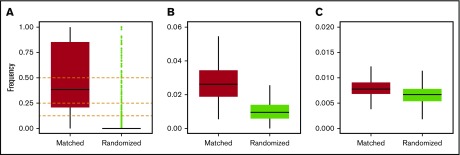
Figure 1.
Measurement of genetic similarity. Frequency distributions of IBD segments normalized by the total lengths of regions of interest for the following: (A) MHC, including HLA-A, HLA-B, HLA-C, HLA-DR, and HLA-DQ; (B) chromosome 6; (C) and the whole genome. Horizontal black bars represent median values. Outliers (green dots) are shown only for panel A (see text for details).
Comparisons of donor–recipient variant call format files (Figures 3-5; supplemental Figure 2) was performed with RTG tools41 to generate patient-specific variants, which were functionally annotated with snpEff.42 Sex-mismatched pairs were considered as special cases with Y-chromosome–specific variants in male recipients aligned to paralogous sites on the X chromosome. In all samples, missense and nonsense variants were mapped to their corresponding primary transcript and translated into amino acid sequences for proteasomal cleavage site prediction with netChop 3.1.43 MHC binding prediction was performed with netMHCpan 3.044 using patient HLA typing to determine MHC restriction. Ranked peptides were further annotated with minor allele frequencies from dbSNP45 build 147. Acute GVHD usually affects the skin, liver, and gastrointestinal tract.46 While patient-specific MiHA expression is most informative, collecting these data requires invasive tissue biopsy specimens. Therefore, we opted to corroborate our results with public data from the Genotype-Tissue Expression Project47-49 using previously described methods50 to associate MiHAs with a measure of broad tissue-specific gene expression. The entire workflow is freely available at https://github.com/wwang-nmdp/MiHAIP.
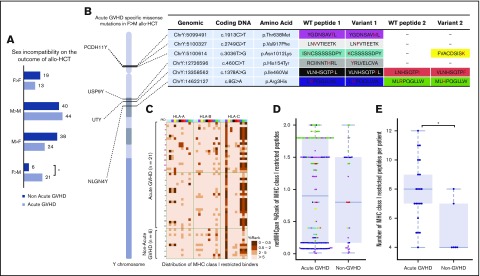
Figure 3.
Y-chromosome variants associate with acute GVHD. (A) Acute GVHD in sex-matched and sex-mismatched donor–recipient pairs, including a statistically significant association (star) in female-to-male (F>M) allogeneic stem cell transplant. Variants were identified in 4 genes (PCDH11Y, USP9Y, UTY, and NLGN4Y), which are displayed with approximate locations on the Y chromosome (B). Precise genomic coordinates and nucleotide and amino acid positions are tabulated, with variant residues shown in red. In some cases, alternative proteasomal cleavage prediction resulted in multiple peptides. (C-E) HLA-restricted affinity prediction for each color-coded peptide is shown for acute GVHD and non-GVHD patients (C) and summarized (D), with application of the recommended threshold for strong binders per male recipient (E). WT, wild-type.
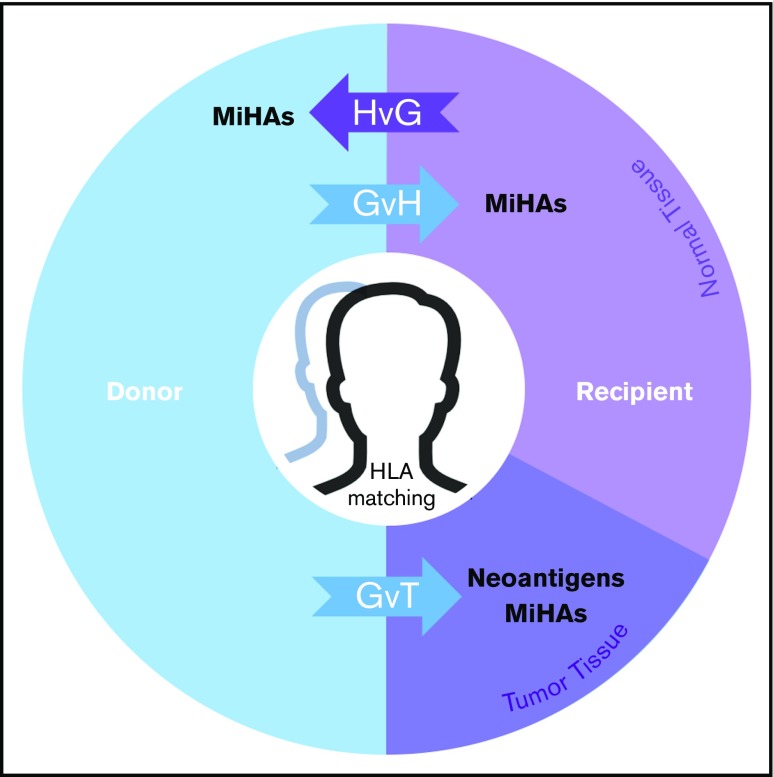
Figure 5.
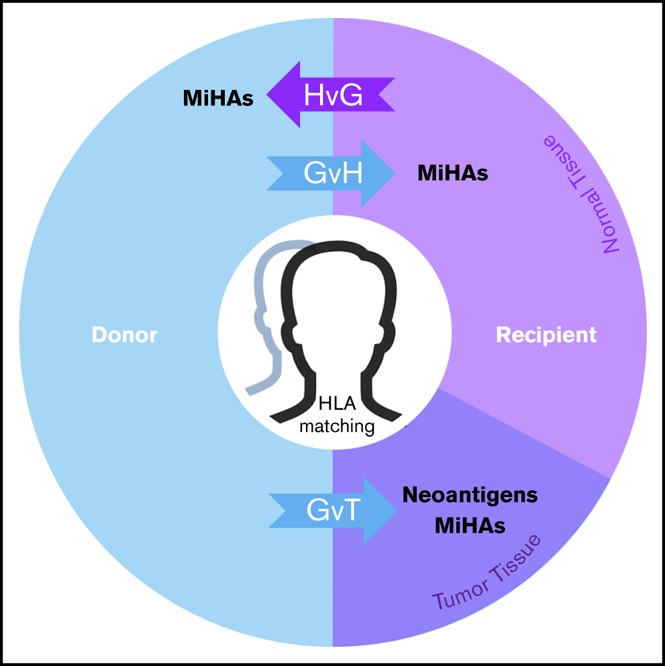
MiHAs contribute to the therapeutic benefits and adverse effects of allo-HCT. In the donor-to-recipient direction, germline-encoded variant peptides (some of which may be presented by recipient HLA molecules) are expressed on both normal and tumor tissue and thus may contribute to GVT or graft-versus-host (GvH) effects. Tumor-specific somatic mutations that encode immunoreactive neoantigens contribute to GVT. In the recipient-to-donor direction, MiHAs may have host-versus-graft (HvG) effects in various clinical contexts leading to rejection in HCT and solid organ transplant (SOT) or miscarriage in pregnancy. Matching of HLAs reduces alloreactive responses from donor or host immune systems in transplantation settings. With cord blood HCT, HLA matching is usually performed at fewer (6) loci. This model does not fully illustrate the genomic and immunological complexities of graft predominance with multiple unit infusion. With haploidentical pairs, GvH effects in the recipient are controlled nongenetically with prophylaxis. Predictable patterns of germline inheritance determine match rates at HLA (4/8) and MiHAs (50%) with consequent effects on GVT. The presence and therapeutic benefit of neoantigens (because they are not heritable) are predicted to be independent of graft source.
Visualization of X-Y paralogous regions (Figure 4A) were performed with manual curation using the BLAST-like alignment tool.51
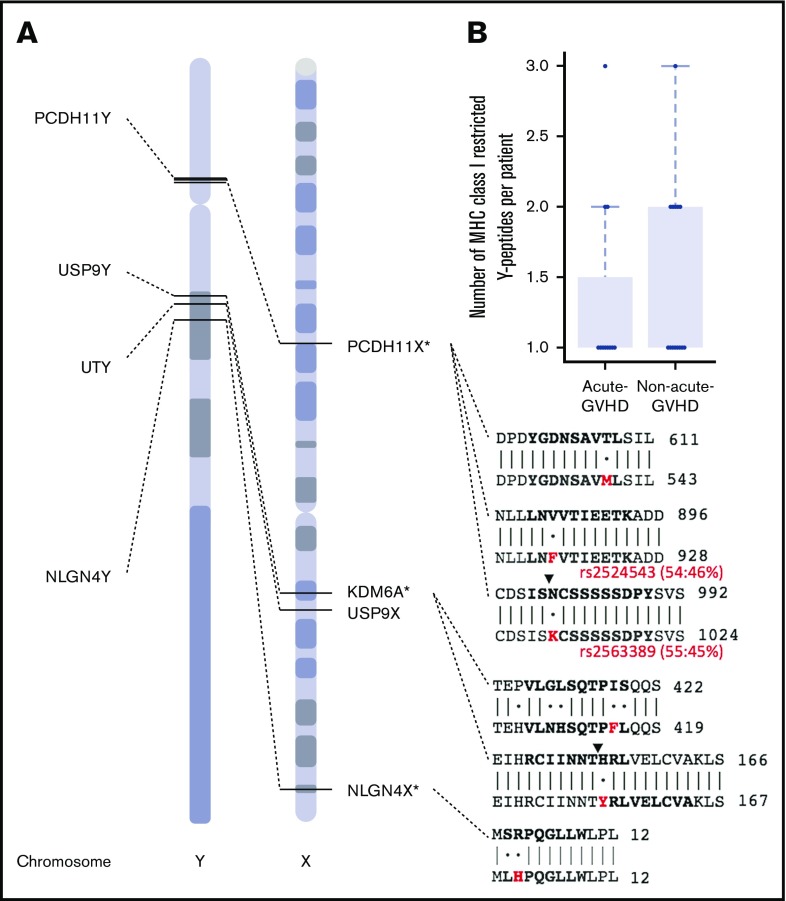
Figure 4.
Paralogous X-Y mismatching explains acute GVHD risk in male recipients with female donors. (A) Six missense variants on the Y chromosome are exclusive to sex-mismatched male patients with acute GVHD. These variants correspond to 9 variant peptides, which are restricted to 4 genes. Genomic positions for PCDH11Y, USP9Y, UTY, and NLGN4Y are shown with dotted lines to paralogous genes on the X chromosome. Protein coding sequence alignments indicating identity matches (bars) and mismatches (dots) are shown for regions that contain individual variant residues (red) and peptides with high-affinity prediction (bold). Ending amino acid coordinates are given to the right of each sequence alignment. Two male-specific variants are named biallelic polymorphisms rs2524543 and rs2563389 with minor allele frequencies 46% and 45%, respectively (red). Note the predicted cleavage sites (black triangles) created by coding variants in PCDH11Y and USP9Y. *For clarity, other alternative cleavage sites are not shown (see Figure 4 for details), and chromosome X is shown in reverse (3′) orientation. (B) The number of predicted high-affinity binding peptides per patient in male-to-male allogeneic HCT recipients with and without acute GVHD.
Statistics
P values were calculated using the χ2 test for Table 1 and the Wilcoxon rank sum test with continuity correction for Figures 2, 4A-C, 5A,D-E, and 6A,D-E. All other P values were calculated using hypergeometric tests with sample and population counts limited to patients with specified MHC restriction. Benjamini-Hochberg false discovery was applied to correct for multiple hypothesis testing in supplemental Table 1. All tests were performed in R with default parameters.
Figure 2.
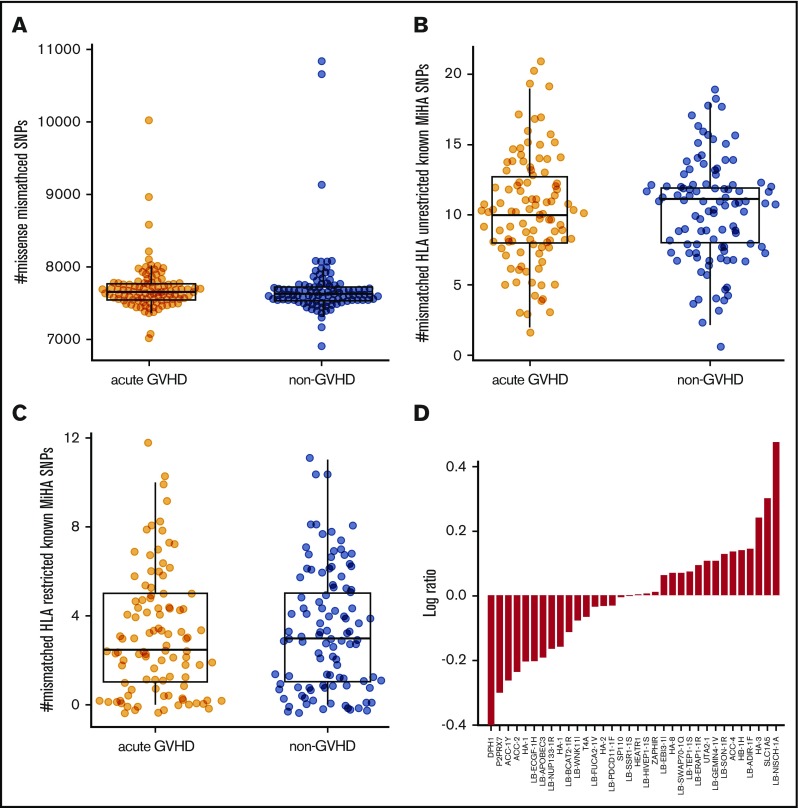
Autosomal variants do not associate with GVHD. The number of patient-specific missense variants (A) as well as known unrestricted (B) and HLA-restricted (C) MiHAs is comparable in the acute GVHD and non-GVHD groups. (D) Known, HLA-restricted MiHAs ordered by log-odds ratio (acute GVHD to non-GVHD). SNP, single-nucleotide polymorphism.
Results
Donor-recipient matching extends beyond five HLA loci
There is strong evidence that HLA-DPB1 T-cell epitope (TCE) matching correlates with allo-HCT outcome.52-58 Generally speaking, mismatched alleles between donor and recipient may be benign (permissible) or alloreactive (nonpermissible) in either direction (graft versus host or host versus graft), with clinical consequences that include GVHD or rejection, respectively. Several methods are available to determine the direction and permissibility of HLA-DPB1 mismatching. Although pairs in this cohort were not explicitly matched at this locus at the time of transplant, a retrospective analysis revealed 16%, 60%, 68%, and 76% of donor–recipient pairs were matched by HLA-DPB1 allele, TCE permissibility,52 expression,59 or functional distance,60 respectively (supplemental Figure 1). This is consistent with baseline likelihoods of finding HLA-DPB1 matches with productive 10/10 searches.61 We found that HLA-DPB1 allele mismatching did not associate with acute GVHD (P < .92), whereas TCE mismatching did associate as expected (P < .038; 1-sided Fisher’s exact test), leaving 56 out of 102 acute GVHD cases (55%) unaccounted for by mismatching at 6 HLA loci.
We hypothesized that HLA-matched unrelated donor–recipient pairs share genetic material outside the MHC. We used IBD inference40 to measure broad genomic similarity (see Methods), which revealed matching at the MHC regions as expected (Figure 1A). Overall, high rates of IBD were observed at the MHC, indicated by many outliers in randomized pairs, which can be attributed to very strong and recent natural selection acting upon these loci in the human population.62 Genetic similarities extended further, albeit to a lesser degree, across chromosome 6 (Figure 1B) and genome-wide (P < 2.2e-16; Figure 1C). Unexpectedly, there was a single outlier in control experiments where donors and patients were randomly paired. This simulated pair shared 50% of their DNA, likely representing a parent–child or full siblings. To protect confidentiality, we did not analyze the relationship further.
Autosomal MiHAs do not associate with acute GVHD
To investigate patient-specific variation further, we developed an integrative bioinformatics workflow adapted from neoantigen discovery to perform comparative analysis of all HLA-matched donor–recipient pairs regardless of TCE permissibility (supplemental Figure 2).
The acute GVHD and non-GVHD groups displayed comparable numbers of missense variants (P = .32; Figure 2A) and known MiHAs (P = .80; Figure 2B) restricted with patient HLA (P = .76; Figure 2C). Ordering MHC-restricted MiHAs by log ratio (Figure 2D) revealed DPH1 (rs35394823) and LB-NISCH-1A (rs887515) as the lowest and highest ranking; however, no associations achieved statistical significance. Thus, we expanded our study to include preexisting single-nuclear polymorphism microarray data from nonoverlapping patient samples. With the addition of 988 HLA-matched donor–recipient pairs (456 acute GVHD, 532 non-GVHD), no statistically significant associations were identified for 17 known MiHAs represented in both data sets (supplemental Table 1).
Y-chromosome–encoded variants associate with acute GVHD
There were 89 sex-mismatched cases in our cohort (Figure 3A). Male recipients with female donors (F>M) were more likely to develop acute GVHD (78%) than male recipients with male donors (52%, P < .02) Sequence analysis of the entire Y chromosome of F>M pairs identified only 6 missense variants (relative to the reference genome hg38) encoding a total of 9 variant peptides in 10 out of 21 recipients (48%) with acute GVHD. By contrast, the Y chromosomes of all 6 non-GVHD males matched the reference (Figure 3B). The variant peptides were confined to 4 genes: PCDH11Y, USP9Y, and UTY, which have reactive minor histocompatibility epitopes determined in vitro,63,64 and NLGN4Y, a neuroligin with unknown HCT significance. Except PCDH11Y, which is specific to the brain and heart, all genes have broad tissue expression (supplemental Figure 3) and thus make qualified candidates for MiHA presentation in GVHD-affected tissues. Filtering by class I MHC restriction (Figure 3C) revealed several variant and reference peptides with strong affinity for their respective HLA allele in both the acute GVHD and non-GVHD groups (Figure 3D); however, there were significantly more predicted binders per GVHD male (P < .015; Figure 3E), suggesting a possible compound effect of multiple Y-linked MiHAs. HLA-DPB1 alone did not explain the association, as 12 out of 21 F>M patients with acute GVHD (57%) were permissibly matched compared with 3 out of 5 without GVHD (60%) (P < .26; 1 patient was not typed at HLA-DBP1.)
Paralogous X-Y mismatching explains acute GVHD risk in male recipients with female donors
Risk of chronic GVHD from allo-HCT is higher in male patients with female donors because of B-cell alloreactivity,7,65-67 which is detectable by antibody response that occurs after,68 but not before, transplant.69 Results presented here extend risk in this patient segment to the acute form of GVHD by implicating male-specific variants in four genes. Although definitive clinical recommendations require confirmatory analysis, it is possible to investigate the genetic basis for risk in this cohort.
PCDH11Y, USP9Y, UTY, and NLGN4Y have paralogs on the X chromosome70 with 72%, 91%, 86%, and 24% amino acid identity, respectively. We mapped Y-encoded variant peptides from each male patient to paralogous sites on their female donor’s X chromosomes. All the variant peptides observed in males with acute GVHD mismatched corresponding sites (Figure 4A). By contrast, the 6 males without GVHD were X-Y matched at these sites, suggesting their donor-female immune systems were educated, and consequently nonalloreactive, to same-as-self peptides encoded at these positions.
The only other category with increased (albeit statistically insignificant) risk of acute GVHD were male recipients with male donors (Figure 3A). Y-Y mismatching was explored as a possible explanation; however, the number of predicted high-affinity binders per patient (maximum 3) was comparable between recipients with and without acute GVHD in the M>M direction (P = .52; Figure 4B) and considerably lower than acute GVHD recipients in the F>M direction (maximum 12; Figure 3E). Furthermore, all female donors, regardless of recipient sex, lacked variants representing high-affinity binding peptides. These findings associate acute GVHD risk, with explanatory genetic factors, specifically in male patients with female donors, at least in this cohort.
Discussion
Allo-HCT is a curative option for many disorders, yet side effects limit its widespread application. GVHD remains a principal barrier to more effective treatment and improved quality of life, but immune responses that contribute to therapeutic benefit and adverse events are physiologically coupled (Figure 5). In malignant conditions, tumor and normal cells are genetically distinct and analytically separable. In the context of allo-HCT, immunoreactive peptides resulting from tumor-specific somatic mutations (neoantigens) may contribute specifically to GVL. On the other hand, the tissue-specific expression and immunogenicity of germline polymorphisms (MiHAs) determine their relative contributions to GVL or GVHD. As treatment options advance, it is important to precisely define genetic factors that affect (or do not affect) clinical outcomes.
Despite research associating several autosomal MiHAs with clinical outcomes, none are routinely matched in allo-HCT. Target tissue expression partially determines the predominance of GVT or GVHD. For example, HA-2 (rs61739531) is expressed in cells of hematopoietic origin71 where there is evidence for GVL in AML with low risk of GVHD.72 However, expression patterns are not wholly determinant. For example, ZAPHIR (rs2074071) associates with GVT, but not GVHD, in renal cell carcinoma patients receiving nonmyleoablative allo-HCT.73 Similarly, other ubiquitously expressed MiHAs are associated with GVL in chronic myelogenous leukemia without evidence of GVHD, suggesting complex alloreactivities from antigen processing, presentation, and costimulation.74 This is consistent with studies of cancer vaccines where therapeutic benefit results from the synergistic effects of multiple cancer-specific neoepitopes combined with immune checkpoint blockade.75,76 In all cases, MHC restriction is an important qualifier, but filtering patients by HLA reduces the number of samples available for retrospective analysis.
Here, we analyzed common autosomal MiHAs with characterized HLA restriction in separate cohorts of 205 and 988 matched samples. We extended the capabilities of commonly used bioinformatics tools to aid comparative genomic analysis of donor–recipient pairs, incorporating MHC matching and antigen restriction as well as HLA-predicted binding affinity and tissue expression into a common workflow. This study was designed specifically to interrogate acute GVHD in AML patients who were in remission at the time of transplant. Consequently, leukemic cell counts were relatively low, and whole-genome sequences represented primarily germline polymorphism. Thus, bioinformatics analysis focused on MiHAs with broad tissue expression patterns. Future studies will apply these methods to patients with active disease, analyzing somatic variants (possible neoantigens) expressed in cells of hematopoietic origin within larger cohorts that are balanced for GVL-related outcomes including relapse.
Our analysis of autosomes revealed no statistically significant associations with acute GVHD among individual MiHAs. These results confirm a recent genome-wide association study of unrelated allo-HCT where MHC mismatching outweighed other genetic factors as contributors to GVHD risk.77 As with neoantigens, it seems plausible that multiple recipient-specific variants contribute to GVHD; however, unlike clonal expansion of somatic mutations in cancer, population-genetic mechanisms account for the co-occurrence of germline-encoded MiHAs. Indeed, there is evidence that arbitrary HLA-matched donor–recipient pairs may present thousands of MiHAs,78 which have a cumulative effect on T-cell responses.79 Although MiHAs are individually common (with minor allele frequencies in our cohort ranging from 19% to 61%; supplemental Table 1), alloreactive combinations may be rare, making it difficult to power case–control studies. Indeed, segmenting patients into subsets sharing ≥2 MiHAs lacked statistical power even when HLA restriction was limited to common alleles. Larger unrelated cohorts are necessary. Additionally, we plan comparable studies in related and haploidentical HCT pairs where shared donor–recipient haplotypes should reduce the number of MiHA combinations under consideration. These studies will also assess whether results reported here are relevant for patients receiving non–calcineurin-based GVHD prophylaxis.
Our comprehensive analysis of sex-linked variation revealed multiple MiHAs encoded on the Y chromosome that associate with acute GVHD specifically in F>M allo-HCT patients. Relative to other chromosomes, the Y is better suited to case–control MiHA association studies, because it lacks population-scale genetic variability due to extremely low rates of diversifying recombination.80 Furthermore, since genetic and phenotypic sex are tightly coupled, it is easy to presegment genomic analysis into clinically weighted categories such as sex match or mismatch. Our limited cohort of primarily white patients suggests the majority of Y haplotypes in this population increase risk of acute GVHD for males with female donors. This is consistent with previous observations of increased chronic GVHD and lower relapse in F>M allo-HCT,81-83 adding a genetic basis for choosing HLA-matched male donors over nonparous females.84,85 However, in cases where a female donor is otherwise the best option for male patients, results reported here may help select a more suitable match.
Supplementary Material
The full-text version of this article contains a data supplement.
Acknowledgments
The authors thank Darryl Carter, Bronwen Shaw, Mary Horowitz, Amir Toor, and Yung-Tsi Bolon for insightful critical review of the manuscript.
Funding for this work was provided by HLI, Inc., as well as the US Office of Naval Research (ONRN00014-17-1-2388).
Authorship
Contribution: W.W., H.H., M. Halagan, M. Heuer, M. Haagenson, N.M.P., J.U., M.M., and C.J.K. developed the bioinformatics workflow and analyzed data; M. Heuer and J.E.B. developed and administered compute infrastructure and information technology; C.V.-G. and S.S. collected and curated data; R.H.S., A.T., and W.B. performed sample preparation, WGS, and primary data analysis; and C.J.K. wrote the manuscript.
Conflict-of-interest disclosure: W.B. is an employee of Human Longevity, Inc. N.M.P. is founder of Root Deep Insight, Inc., a personal immunogenomics company. The remaining authors declare no competing financial interests.
Correspondence: Caleb J. Kennedy, Center for International Blood and Marrow Transplant Research, Minneapolis, MN 55401; e-mail: caleb.kennedy@nmdp.org.
REFERENCES
- 1.Copelan EA. Hematopoietic stem-cell transplantation. N Engl J Med. 2006;354(17):1813-1826. [DOI] [PubMed] [Google Scholar]
- 2.Horowitz MM, Gale RP, Sondel PM, et al. . Graft-versus-leukemia reactions after bone marrow transplantation. Blood. 1990;75(3):555-562. [PubMed] [Google Scholar]
- 3.Miller JS, Warren EH, van den Brink MR, et al. . NCI First International Workshop on The Biology, Prevention, and Treatment of Relapse After Allogeneic Hematopoietic Stem Cell Transplantation: report from the Committee on the Biology Underlying Recurrence of Malignant Disease following Allogeneic HSCT: graft-versus-tumor/leukemia reaction. Biol Blood Marrow Transplant. 2010;16(5):565-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferrara JL, Levine JE, Reddy P, Holler E. Graft-versus-host disease. Lancet. 2009;373(9674):1550-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gooley TA, Chien JW, Pergam SA, et al. . Reduced mortality after allogeneic hematopoietic-cell transplantation. N Engl J Med. 2010;363(22):2091-2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oostvogels R, Lokhorst HM, Mutis T. Minor histocompatibility Ags: identification strategies, clinical results and translational perspectives. Bone Marrow Transplant. 2016;51(2):163-171. [DOI] [PubMed] [Google Scholar]
- 7.Popli R, Sahaf B, Nakasone H, Lee JY, Miklos DB. Clinical impact of H-Y alloimmunity. Immunol Res. 2014;58(2-3):249-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim SJ, Gill JS. H-Y incompatibility predicts short-term outcomes for kidney transplant recipients. J Am Soc Nephrol. 2009;20(9):2025-2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wagner S. H-Y antigen in kidney transplant: does gender matter? Gend Med. 2012;9(5):387-388. [DOI] [PubMed] [Google Scholar]
- 10.Pfeffer PF, Thorsby E. HLA-restricted cytotoxicity against male-specific (H-Y) antigen after acute rejection of an HLA-identical sibling kidney: clonal distribution of the cytotoxic cells. Transplantation. 1982;33(1):52-56. [DOI] [PubMed] [Google Scholar]
- 11.Christiansen OB, Steffensen R, Nielsen HS. Anti-HY responses in pregnancy disorders. Am J Reprod Immunol. 2011;66(Suppl 1):93-100. [DOI] [PubMed] [Google Scholar]
- 12.Nielsen HS, Steffensen R, Lund M, et al. . Frequency and impact of obstetric complications prior and subsequent to unexplained secondary recurrent miscarriage. Hum Reprod. 2010;25(6):1543-1552. [DOI] [PubMed] [Google Scholar]
- 13.Molldrem J, Dermime S, Parker K, et al. . Targeted T-cell therapy for human leukemia: cytotoxic T lymphocytes specific for a peptide derived from proteinase 3 preferentially lyse human myeloid leukemia cells. Blood. 1996;88(7):2450-2457. [PubMed] [Google Scholar]
- 14.Molldrem JJ, Lee PP, Wang C, Champlin RE, Davis MMA. A PR1-human leukocyte antigen-A2 tetramer can be used to isolate low-frequency cytotoxic T lymphocytes from healthy donors that selectively lyse chronic myelogenous leukemia. Cancer Res. 1999;59(11):2675-2681. [PubMed] [Google Scholar]
- 15.Gao L, Bellantuono I, Elsässer A, et al. . Selective elimination of leukemic CD34(+) progenitor cells by cytotoxic T lymphocytes specific for WT1. Blood. 2000;95(7):2198-2203. [PubMed] [Google Scholar]
- 16.Cai A, Keskin DB, DeLuca DS, et al. . Mutated BCR-ABL generates immunogenic T-cell epitopes in CML patients. Clin Cancer Res. 2012;18(20):5761-5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pont MJ, van der Lee DI, van der Meijden ED, et al. . Integrated whole genome and transcriptome analysis identified a therapeutic minor histocompatibility antigen in a splice variant of ITGB2. Clin Cancer Res. 2016;22(16):4185-4196. [DOI] [PubMed] [Google Scholar]
- 18.Lindsley RC, Saber W, Mar BG, et al. . Prognostic mutations in myelodysplastic syndrome after stem-cell transplantation. N Engl J Med. 2017;376(6):536-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lazarian G, Guièze R, Wu CJ. Clinical implications of novel genomic discoveries in chronic lymphocytic leukemia. J Clin Oncol. 2017;35(9):984-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alexandrov LB, Nik-Zainal S, Wedge DC, et al. ; ICGC PedBrain. Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Falchook GS, Leidner R, Stankevich E, et al. . Responses of metastatic basal cell and cutaneous squamous cell carcinomas to anti-PD1 monoclonal antibody REGN2810. J Immunother Cancer. 2016;4(1):70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang AC, Postow MA, Orlowski RJ, et al. . T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature. 2017;545(7652):60-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Budczies J, Bockmayr M, Klauschen F, et al. . Mutation patterns in genes encoding interferon signaling and antigen presentation: a pan-cancer survey with implications for the use of immune checkpoint inhibitors. Genes Chromosomes Cancer. 2017;56(8):651-659. [DOI] [PubMed] [Google Scholar]
- 24.Strønen E, Toebes M, Kelderman S, et al. . Targeting of cancer neoantigens with donor-derived T cell receptor repertoires. Science. 2016;352(6291):1337-1341. [DOI] [PubMed] [Google Scholar]
- 25.Rajasagi M, Shukla SA, Fritsch EF, et al. . Systematic identification of personal tumor-specific neoantigens in chronic lymphocytic leukemia. Blood. 2014;124(3):453-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tran E, Ahmadzadeh M, Lu YC, et al. . Immunogenicity of somatic mutations in human gastrointestinal cancers. Science. 2015;350(6266):1387-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burkhardt UE, Wu CJ. Boosting leukemia-specific T cell responses in patients following stem cell transplantation. OncoImmunology. 2013;2(11):e26587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu YC, Robbins PF. Targeting neoantigens for cancer immunotherapy. Int Immunol. 2016;28(7):365-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Srinivasan M, Sedmak D, Jewell S. Effect of fixatives and tissue processing on the content and integrity of nucleic acids. Am J Pathol. 2002;161(6):1961-1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones S, Anagnostou V, Lytle K, et al. . Personalized genomic analyses for cancer mutation discovery and interpretation. Sci Transl Med. 2015;7(283):283ra53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garofalo A, Sholl L, Reardon B, et al. . The impact of tumor profiling approaches and genomic data strategies for cancer precision medicine. Genome Med. 2016;8(1):79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sampson JK, Sheth NU, Koparde VN, et al. . Whole exome sequencing to estimate alloreactivity potential between donors and recipients in stem cell transplantation. Br J Haematol. 2014;166(4):566-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deeg HJ, Antin JH. The clinical spectrum of acute graft-versus-host disease. Semin Hematol. 2006;43(1):24-31. [DOI] [PubMed] [Google Scholar]
- 34.Spellman S, Setterholm M, Maiers M, et al. . Advances in the selection of HLA-compatible donors: refinements in HLA typing and matching over the first 20 years of the National Marrow Donor Program Registry. Biol Blood Marrow Transplant. 2008;14(9 suppl):37-44. [DOI] [PubMed] [Google Scholar]
- 35.Dehn J, Setterholm M, Buck K, et al. . HapLogic: a predictive human leukocyte antigen-matching algorithm to enhance rapid identification of the optimal unrelated hematopoietic stem cell sources for transplantation. Biol Blood Marrow Transplant. 2016;22(11):2038-2046. [DOI] [PubMed] [Google Scholar]
- 36.Raczy C, Petrovski R, Saunders CT, et al. . Isaac: ultra-fast whole-genome secondary analysis on Illumina sequencing platforms. Bioinformatics. 2013;29(16):2041-2043. [DOI] [PubMed] [Google Scholar]
- 37.Li H, Handsaker B, Wysoker A, et al. ; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Danecek P, Auton A, Abecasis G, et al. ; 1000 Genomes Project Analysis Group. The variant call format and VCFtools. Bioinformatics. 2011;27(15):2156-2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Madbouly A, Wang T, Haagenson M, et al. . Investigating the association of genetic admixture and donor/recipient genetic disparity with transplant outcomes. Biol Blood Marrow Transplant. 2017;23(6):1029-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Browning BL, Browning SR. Detecting identity by descent and estimating genotype error rates in sequence data. Am J Hum Genet. 2013;93(5):840-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cleary JG, Braithwaite R, Gaastra K, et al. . Joint variant and de novo mutation identification on pedigrees from high-throughput sequencing data. J Comput Biol. 2014;21(6):405-419. [DOI] [PubMed] [Google Scholar]
- 42.Cingolani P, Platts A, Wang L, et al. . A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 2012;6(2):80-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Keşmir C, Nussbaum AK, Schild H, Detours V, Brunak S. Prediction of proteasome cleavage motifs by neural networks. Protein Eng. 2002;15(4):287-296. [DOI] [PubMed] [Google Scholar]
- 44.Nielsen M, Andreatta M. NetMHCpan-3.0; improved prediction of binding to MHC class I molecules integrating information from multiple receptor and peptide length datasets. Genome Med. 2016;8(1):33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coordinators NR; NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2016;44(D1):D7-D19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jacobsohn DA, Vogelsang GB. Acute graft versus host disease. Orphanet J Rare Dis. 2007;2(1):35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Consortium GT; GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348(6235):648-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Consortium GT; GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45(6):580-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Keen JC, Moore HM. The Genotype-Tissue Expression (GTEx) project: linking clinical data with molecular analysis to advance personalized medicine. J Pers Med. 2015;5(1):22-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kryuchkova-Mostacci N, Robinson-Rechavi M. A benchmark of gene expression tissue-specificity metrics. Brief Bioinform. 2017;18(2):205-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12(4):656-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zino E, Frumento G, Marktel S, et al. . A T-cell epitope encoded by a subset of HLA-DPB1 alleles determines nonpermissive mismatches for hematologic stem cell transplantation. Blood. 2004;103(4):1417-1424. [DOI] [PubMed] [Google Scholar]
- 53.Zino E, Vago L, Di Terlizzi S, et al. . Frequency and targeted detection of HLA-DPB1 T cell epitope disparities relevant in unrelated hematopoietic stem cell transplantation. Biol Blood Marrow Transplant. 2007;13(9):1031-1040. [DOI] [PubMed] [Google Scholar]
- 54.Crocchiolo R, Zino E, Vago L, et al. ; Italian Bone Marrow Donor Registry. Nonpermissive HLA-DPB1 disparity is a significant independent risk factor for mortality after unrelated hematopoietic stem cell transplantation. Blood. 2009;114(7):1437-1444. [DOI] [PubMed] [Google Scholar]
- 55.Fleischhauer K, Shaw BE, Gooley T, et al. ; International Histocompatibility Working Group in Hematopoietic Cell Transplantation. Effect of T-cell-epitope matching at HLA-DPB1 in recipients of unrelated-donor haemopoietic-cell transplantation: a retrospective study. Lancet Oncol. 2012;13(4):366-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shaw BE, Robinson J, Fleischhauer K, Madrigal JA, Marsh SG. Translating the HLA-DPB1 T-cell epitope-matching algorithm into clinical practice. Bone Marrow Transplant. 2013;48(12):1510-1512. [DOI] [PubMed] [Google Scholar]
- 57.Fleischhauer K, Fernandez-Viña MA, Wang T, et al. . Risk associations between HLA-DPB1 T-cell epitope matching and outcome of unrelated hematopoietic cell transplantation are independent of HLA-DPA1. Bone Marrow Transplant. 2014;49(9):1176-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pidala J, Lee SJ, Ahn KW, et al. . Nonpermissive HLA-DPB1 mismatch increases mortality after myeloablative unrelated allogeneic hematopoietic cell transplantation. Blood. 2014;124(16):2596-2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goyal RK, Lee SJ, Wang T, et al. . Novel HLA-DP region susceptibility loci associated with severe acute GvHD. Bone Marrow Transplant. 2017;52(1):95-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Crivello P, Zito L, Sizzano F, et al. . The impact of amino acid variability on alloreactivity defines a functional distance predictive of permissive HLA-DPB1 mismatches in hematopoietic stem cell transplantation. Biol Blood Marrow Transplant. 2015;21(2):233-241. [DOI] [PubMed] [Google Scholar]
- 61.Tram K, Stritesky G, Wadsworth K, Ng J, Anasetti C, Dehn J. Identification of DPB1 permissive unrelated donors is highly likely. Biol Blood Marrow Transplant. 2017;23(1):81-86. [DOI] [PubMed] [Google Scholar]
- 62.Albrechtsen A, Moltke I, Nielsen R. Natural selection and the distribution of identity-by-descent in the human genome. Genetics. 2010;186(1):295-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ofran Y, Kim HT, Brusic V, et al. . Diverse patterns of T-cell response against multiple newly identified human Y chromosome-encoded minor histocompatibility epitopes. Clin Cancer Res. 2010;16(5):1642-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vogt MH, Goulmy E, Kloosterboer FM, et al. . UTY gene codes for an HLA-B60-restricted human male-specific minor histocompatibility antigen involved in stem cell graft rejection: characterization of the critical polymorphic amino acid residues for T-cell recognition. Blood. 2000;96(9):3126-3132. [PubMed] [Google Scholar]
- 65.Nakasone H, Sahaf B, Miklos DB. Therapeutic benefits targeting B-cells in chronic graft-versus-host disease. Int J Hematol. 2015;101(5):438-451. [DOI] [PubMed] [Google Scholar]
- 66.Miklos DB, Kim HT, Miller KH, et al. . Antibody responses to H-Y minor histocompatibility antigens correlate with chronic graft-versus-host disease and disease remission. Blood. 2005;105(7):2973-2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sahaf B, Yang Y, Arai S, Herzenberg LA, Herzenberg LA, Miklos DB. H-Y antigen-binding B cells develop in male recipients of female hematopoietic cells and associate with chronic graft vs. host disease. Proc Natl Acad Sci USA. 2013;110(8):3005-3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nakasone H, Tian L, Sahaf B, et al. . Allogeneic HY antibodies detected 3 months after female-to-male HCT predict chronic GVHD and nonrelapse mortality in humans. Blood. 2015;125(20):3193-3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nakasone H, Sahaf B, Tian L, et al. ; Center for International Blood and Marrow Transplantation. Presensitization to HY antigens in female donors prior to transplant is not associated with male recipient post-transplant HY antibody development nor with clinical outcomes. Haematologica. 2016;101(1):e30-e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lahn BT, Pearson NM, Jegalian K. The human Y chromosome, in the light of evolution. Nat Rev Genet. 2001;2(3):207-216. [DOI] [PubMed] [Google Scholar]
- 71.Sellami MH, Ben Ahmed A, Kaabi H, Jridi A, Dridi A, Hmida S. HA-1 and HA-2 minor histocompatibility antigens in Tunisians. Tissue Antigens. 2010;75(6):720-723. [DOI] [PubMed] [Google Scholar]
- 72.Mutis T, Verdijk R, Schrama E, Esendam B, Brand A, Goulmy E. Feasibility of immunotherapy of relapsed leukemia with ex vivo-generated cytotoxic T lymphocytes specific for hematopoietic system-restricted minor histocompatibility antigens. Blood. 1999;93(7):2336-2341. [PubMed] [Google Scholar]
- 73.Broen K, Levenga H, Vos J, et al. . A polymorphism in the splice donor site of ZNF419 results in the novel renal cell carcinoma-associated minor histocompatibility antigen ZAPHIR. PLoS One. 2011;6(6):e21699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Griffioen M, Honders MW, van der Meijden ED, et al. . Identification of 4 novel HLA-B*40:01 restricted minor histocompatibility antigens and their potential as targets for graft-versus-leukemia reactivity. Haematologica. 2012;97(8):1196-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ott PA, Hu Z, Keskin DB, et al. . An immunogenic personal neoantigen vaccine for patients with melanoma. Nature. 2017;547(7662):217-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sahin U, Derhovanessian E, Miller M, et al. . Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature. 2017;547(7662):222-226. [DOI] [PubMed] [Google Scholar]
- 77.Martin PJ, Levine DM, Storer BE, et al. . Genome-wide minor histocompatibility matching as related to the risk of graft-versus-host disease. Blood. 2017;129(6):791-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jameson-Lee M, Koparde V, Griffith P, et al. . In silico derivation of HLA-specific alloreactivity potential from whole exome sequencing of stem-cell transplant donors and recipients: understanding the quantitative immunobiology of allogeneic transplantation. Front Immunol. 2014;5:529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Abdul Razzaq B, Scalora A, Koparde VN, et al. . Dynamical system modeling to simulate donor T cell response to whole exome sequencing-derived recipient peptides demonstrates different alloreactivity potential in HLA-matched and -mismatched donor-recipient pairs. Biol Blood Marrow Transplant. 2016;22(5):850-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wilson Sayres MA, Lohmueller KE, Nielsen R. Natural selection reduced diversity on human y chromosomes. PLoS Genet. 2014;10(1):e1004064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stern M, Brand R, de Witte T, et al. . Female-versus-male alloreactivity as a model for minor histocompatibility antigens in hematopoietic stem cell transplantation. Am J Transplant. 2008;8(10):2149-2157. [DOI] [PubMed] [Google Scholar]
- 82.Gratwohl A, Stern M, Brand R, et al. ; European Group for Blood and Marrow Transplantation and the European Leukemia Net. Risk score for outcome after allogeneic hematopoietic stem cell transplantation: a retrospective analysis. Cancer. 2009;115(20):4715-4726. [DOI] [PubMed] [Google Scholar]
- 83.Gratwohl A. The EBMT risk score. Bone Marrow Transplant. 2012;47(6):749-756. [DOI] [PubMed] [Google Scholar]
- 84.Loren AW, Bunin GR, Boudreau C, et al. . Impact of donor and recipient sex and parity on outcomes of HLA-identical sibling allogeneic hematopoietic stem cell transplantation. Biol Blood Marrow Transplant. 2006;12(7):758-769. [DOI] [PubMed] [Google Scholar]
- 85.Kollman C, Spellman SR, Zhang MJ, et al. . The effect of donor characteristics on survival after unrelated donor transplantation for hematologic malignancy. Blood. 2016;127(2):260-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.