Abstract
Previous studies indicated that fruit bats carry two betacoronaviruses, BatCoV HKU9 and BatCoV GCCDC1. To investigate the epidemiology and genetic diversity of these coronaviruses, we conducted a longitudinal surveillance in fruit bats in Yunnan province, China during 2009–2016. A total of 59 (10.63%) bat samples were positive for the two betacorona-viruses, 46 (8.29%) for HKU9 and 13 (2.34%) for GCCDC1, or closely related viruses. We identified a novel HKU9 strain, tentatively designated as BatCoV HKU9-2202, by sequencing the full-length genome. The BatCoV HKU9-2202 shared 83% nucleotide identity with other BatCoV HKU9 stains based on whole genome sequences. The most divergent region is in the spike protein, which only shares 68% amino acid identity with BatCoV HKU9. Quantitative PCR revealed that the intestine was the primary infection organ of BatCoV HKU9 and GCCDC1, but some HKU9 was also detected in the heart, kidney, and lung tissues of bats. This study highlights the importance of virus surveillance in natural reservoirs and emphasizes the need for preparedness against the potential spill-over of these viruses to local residents living near bat caves.
Electronic supplementary material
The online version of this article (10.1007/s12250-018-0017-2) contains supplementary material, which is available to authorized users.
Keywords: Betacoronavirus, Surveillance, Genetic diversity
Introduction
Coronaviruses are enveloped, single-stranded RNA viruses that belong to the subfamily Coronavirinae, family Coronaviridae, in the order Nidovirales. Based on the genetic distance and serological characterization, the family consists of four genera: alpha-, beta-, gamma-, and delta-coronaviruses (https://talk.ictvonline.org/ictv-reports/ictv_online_report/introduction/). Coronaviruses are important human pathogens that cause outbreaks of severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS) (de Groot et al. 2013; Drosten et al. 2003). Six human coronaviruses have been identified: human coronavirus 229E (HCoV-229E), HCoV-OC43, HCoV-HKU1, HCoV-NL63, SARS-CoV, and MERS-CoV (Hu et al. 2015). HCoV-229E, HCoV-OC43, HCoV-HKU1, and HCoV-NL63 are widespread in human populations and known to cause mild respiratory disease, while SARS-CoV and MERS-CoV had led to pandemics (Channappanavar and Perlman 2017). Stronger evidence showed that the direct ancestor of SARS-CoV, and likely MERS-CoV, originated in bats.
Bats are the only mammals capable of flight and represent approximately 20% species of all mammals (Hunter 2007). According to dietary differences, bats are distinguished as insectivores and frugivores (Stuckey et al. 2017). Frugivore bats are ideal bushmeat because of huge body and thick-flesh for local people in some districts in Africa and Southeast Asia (Mickleburgh et al. 2009). Meanwhile, frugivore bats in African or Pacific countries harbor diversity of virulent viruses, such as marburgvirus, hendra virus, and nipha virus (Shi 2013). In China, cross-reactive antibody or phylogenetically related viruses to henipaviruses, ebolaviruses and rabies virus have been detected in Chinese fruit bats (He et al. 2015; Jiang et al. 2010; Li et al. 2008; Yang et al. 2017; Yuan et al. 2012). In addition, genetically diverse reoviruses, adenoviruses, and coronaviruses have been detected or isolated from fruit bats (Du et al. 2010; Li et al. 2016; Tan et al. 2017).
Ro-BatCoV HKU9 and Ro-BatCoV GCCDC1 are two closely related but distinct betacoronavirus species found in Guangdong and Yunnan province, respectively. Both were found in the Chinese brown fruit bat Rousettus leschenaulti (Huang et al. 2016; Lau et al. 2010; Woo et al. 2007). HKU9 includes more variants and are genetically diverse, while GCCDC1 is less diverse. The greatest difference between these two viral species is the presence of p10 gene, which is thought to have been obtained from a reovirus, in the GCCDC1 genome (Huang et al. 2016; Lau et al. 2010). In Yunnan province, there are at least three fruit bat species, Eonycteris spelaea, R. leschenaultia, and an unclassified Rousettus species (He et al. 2015; Yang et al. 2017). These bats frequently cohabitate in the same cave and can only be distinguished by bat experts or molecular identification.
In this study, we conducted a longitudinal surveillance of the two betacoronaviruses in fruit bat samples collected during 2009–2016 in Yunnan province and reexamined the prevalence, genetic diversity, and host specificity of these viruses.
Materials and Methods
Sample Collection
Sampling was conducted as described previously (Li et al. 2005). Because of conservation concerns, for most captured bats, we collected fecal or anal samples and released the bats after sampling. Several bats were sacrificed for species identification and viral tissue tropism assays. Bat species were identified based on morphological characteristics and further confirmed by cytochrome b (Cytb) sequencing (Agnarsson et al. 2011). All samples were stored at − 80 °C until further analysis. All animal sampling processes were performed by veterinarians with approval from the Animal Ethics Committee of the Yunnan Institute of Endemic Diseases Control and Prevention.
Viral Detection
RNA was extracted from bat fecal or anal samples using the High Pure Viral RNA Kit (Roche, Basel, Switzerland). Partial RdRp was amplified using the SuperScript III One-Step RT-PCR and Platinum Taq Enzyme kit (Invitrogen, Carlsbad, CA, USA) by family-specific degenerate semi-nested PCR (Luna et al. 2007). Expected PCR products were gel-purified and subjected to sequencing using the Sanger ABI-PRISM platform (Applied Biosystems, Foster City, CA, USA). To exclude PCR contamination, the nucleotide sequences of the virus and bat Cytb of positive samples were evaluated by two independent PCRs by different experimenters. The partial RdRp sequences obtained in this study were submitted to GenBank under accession numbers MG762619–MG762664 for BatCoV HKU9 and MG762606–MG762618 for BatCoV GCCDC1.
Quantitative PCR (qPCR)
qPCR was used to investigate the tissue tropism of these viruses in various tissues. Total RNA was extracted from the hearts, livers, spleens, lungs, kidneys, brains, and intestines of six bats infected with bat coronaviruses HKU9 or GCCDC1 using the High Pure Viral RNA Kit. Partial RdRp representing HKU9 or GCCDC1 were cloned into the pGEM-T-easy Vector (Promega, Madison, WI, USA) and used as a positive control for quantitative analysis. Primers for the two different viruses were designed using IDT online software (https://sg.idtdna.com/site) (Supplementary Table S1). The assay was carried out in triplicate on a CFX connect Real-Time system (Bio-Rad, Hercules, CA, USA) with the One-Step RT-PCR SYBR Green kit (Vazyme, Nanjing, China). The PCR thermal cycling parameters were 50 °C for 5 min, 95 °C for 10 min, and 40 cycles of 95 °C for 5 min, and 60 °C for 30 s. An absolute quantitative method was used to determine the number of copies of the viruses referring to the standard control generated from the positive sets.
Amplification of Full-Length S, N, and P10 Gene
Primers targeting the S, N, and P10 gene were designed based on alignment of the reported HKU9 or GCCDC1 sequences (primer sequences provided upon request). The first round of PCR amplification was performed in a total volume of 25 μL using SuperScript III One-Step RT-PCR (Invitrogen) under the following parameters: 50 °C for 30 min, 94 °C for 5 min; 35 cycles of 94 °C for 30 s, 50 °C for 30 s, and 68 °C for 3 min; and a final extension at 68 °C for 10 min. The second round of PCR amplification was performed in a total volume of 50 μL using the Platinum Taq Enzyme kit (Invitrogen) under the following conditions: 94 °C for 5 min; 35 cycles of 94 °C for 30 s, 50 °C for 30 s, and 72 °C for 3 min; and a final extension at 72 °C for 10 min. Expected PCR products were gel-purified and sequenced directly using target primers. Weak bands were cloned into the pGEM T-easy vector and sequenced using the Sanger ABI-PRISM platform. Full-length N and p10 sequences were deposited into GenBank under the following accession numbers: MG762665–MG762673, MG762688–MG762692, and MG762675–MG762687.
Full-Length Genome Sequencing and Characterization
One positive sample (ID: 2202) was further sequenced using an Illumina platform at Novogene (Beijing, China). Briefly, the supernatant of homogenized intestine was centrifuged at 10,000×g for 10 min at 4 °C. The supernatant was filtered through a 0.45-μm polyvinylidene difluoride filter (Millipore, Billerica, MA, USA) to remove eukaryotic and bacterial-sized particles. The filtered samples were then centrifuged at 100,000×g for 2 h. The pellets were resuspended in 140 µL Hanks’ solution and RNA was extracted with the QIAamp viral RNA minikit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. Sequence-independent PCR amplification was conducted as previously described (Ge et al. 2012). PCR products greater than 500 base pairs were excised and extracted with a MinElute Gel Extraction Kit (Qiagen). The PCR products were adaptor-anchored, pooled, and sequenced on an Illumina platform.
The filtered sequence reads were aligned to sequences in the NCBI nonredundant nucleotide database (NT) and nonredundant protein database (NR) downloaded from the NCBI FTP server using BLASTn and BLASTx, respectively. All reads matched to coronavirus were extracted and assembled using megahit and trinity software. Based on the partial genome sequences of viruses, the remaining genome sequences were determined by inverse PCR, genome walking, and 5′- and 3′-rapid amplification of cDNA ends (RACE). Next, the nucleotide sequence of the full-genome (accession numbers: MG762674) and deduced amino acid sequences of the open reading frames (ORFs) were compared to those of related betacoronaviruses. For coronavirus species demarcation, seven independent replicase domains in the ORF1ab of the virus were selected for further analysis.
Phylogenetic Analysis
Partial RdRp sequences, full-length N gene sequences, and full-length genomic sequences obtained in this study were aligned with those of HUK9, GCCDC1, and related coronaviruses and representative betacoronaviruses using ClustalW. The phylogenetic tree was constructed by the neighbor-joining method with MEGA7.0 software with 1000 bootstrap replicates. According to the structure of the phylogenetic tree, the identities of all sequences from different lineages were calculated using ClustalW in MegAlign.
Virus Isolation
Vero E6 and primary intestine cell lines of E. spelaea and R. leschenaulti were used for virus isolation. Cells were cultured and inoculated with viral RNA-positive samples after tenfold dilution. The cells were incubated in culture medium containing 5% fetal bovine serum. After three blind passages, the cell culture supernatant was tested for the presence of live virus by nested RT-PCR.
Results
Prevalence of Betacoronavirus HKU9 and GCCDC1 and Related Viruses in Fruit Bats
A total of 555 fecal or anal samples from fruit bats were collected at four locations in Yunnan province, China in 2009–2016 (Fig. 1). By RT-PCR detection targeting partial RdRP, 46 (8.29%) samples were positive for HKU9 and 13 (2.34%) were positive for GCCDC1 or closely related viruses (Table 1). Different sampling times and sites showed different detection rates for HKU9. No positive results were detected in samples collected in Mengla, 2011 and Mojiang in 2013 (Table 1). HKU9 infection rates in Chuxiong, Mengla, and Jinghong were 18.59% (29/156), 5.32% (10/188), and 6.14% (7/114), respectively. GCCDC1 was not detected until 2015, with a positive rate of 5.26% in 2015 and significantly high positive rate in 2016 (18.86%) in Mengla.
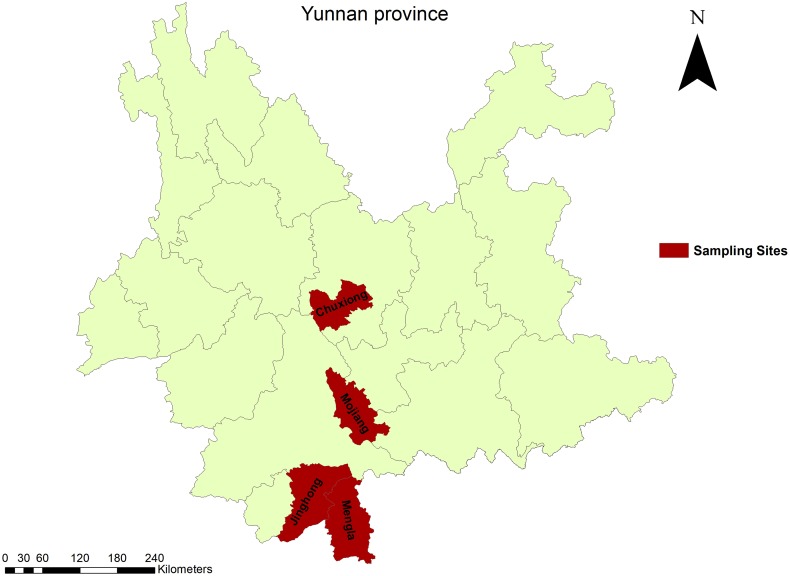
Fig. 1.
Map of sampling sites in Yunnan province of China. Red regions indicate the four districts where bat samples were collected.
Table 1.
Detection of BatCoV HKU9 and BatCoV GCCDC1 by RT-PCR in bat fecal or anal samples collected from four districts in the Yunnan province of China during 2009–2016.
| Year | Sampling sites | Virus | Total positives | |
|---|---|---|---|---|
| BatCoVHKU9 | BatCoV GCCDC1 | |||
| 2009 | Jinghong | 7/114 (6.14)a | 0/114 | 7/114 (6.14) |
| 2011 | Mengla | 0/28 | 0/28 | 0/28 |
| 2013 | Chuxiong | 28/42 (66.67) | 0/42 | 28/139 (2.01) |
| Mojiang | 0/97 | 0/97 | – | |
| 2014 | Mengla | 4/50 (8.00) | 0/50 | 5/164 (3.05) |
| Chuxiong | 1/114 (0.88) | 0/114 | – | |
| 2015 | Mengla | 5/57 (8.77) | 3/57 (5.26) | 8/57 (14.04) |
| 2016 | Mengla | 1/53 (1.89) | 10/53 (18.87) | 11/53 (2.08) |
| Total | – | 46/555 (8.29) | 13/555 (2.34) | 59/555 (10.63) |
aPositive samples/tested samples (%).
Phylogenetic Analysis
The amplified partial RdRp sequences in this study shared 74.4%–100% identity at the nucleotide (nt) level. A phylogenetic tree was conducted based on the alignment of partial RdRp sequences along with previously reported HKU9, GCCDC1, and related stains, as well as representative strains of other betacoronaviruses. The results revealed 59 sequences classified as two coronavirus species, HKU9 or GCCDC1 (Fig. 2A). All sequences from Rousettus bats were HKU9-related viruses and those from E. spelaea were GCCDC1-related viruses. In contrast to the GCCDC1 strains which are highly similar, the HKU9-related strains were highly diverse. Within the HKU9 species, the sequences in this study and previously reported sequences were divided into 5 lineages: Lineage 1 comprising 28 sequences and previously reported HKU9-10-2, HKU9-5-2, and HKU9-2 exclusively from R. leschenaulti; Lineage 2 comprising 5 sequences and previously reported HKU9-1 from R. leschenaulti; Lineage 3 comprising 10 sequences and previously reported HKU9-4 from unidentified Rousettus species R. sp.; Lineage 4 comprising the previously detected HKU9-3, 9-5, and 9-10 from R. leschenaulti; Lineage 5 comprising 3 sequences from Rousettus species. The other 13 sequences were exclusively from E. spelaea and grouped with previously reported BatCoV GCCDC1 (Huang et al. 2016).
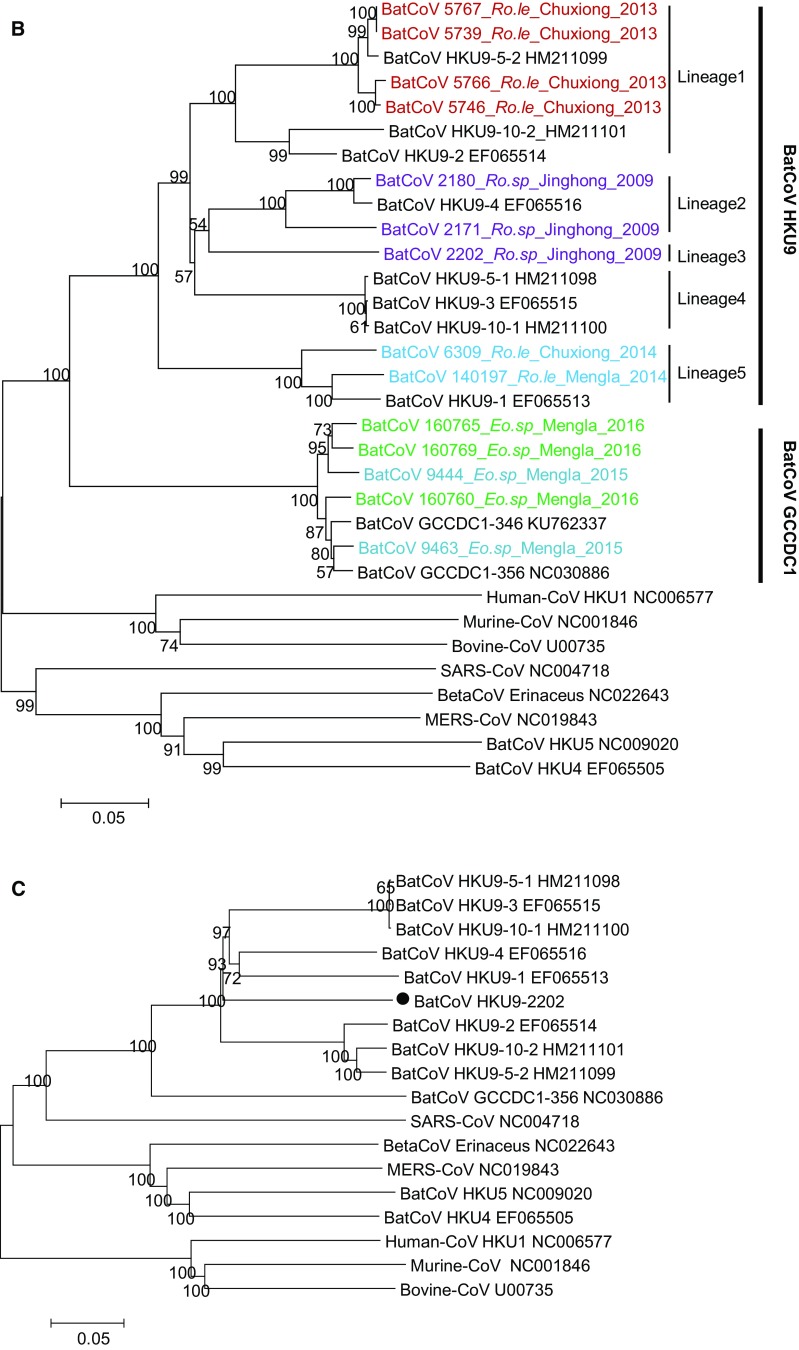
Fig. 2.
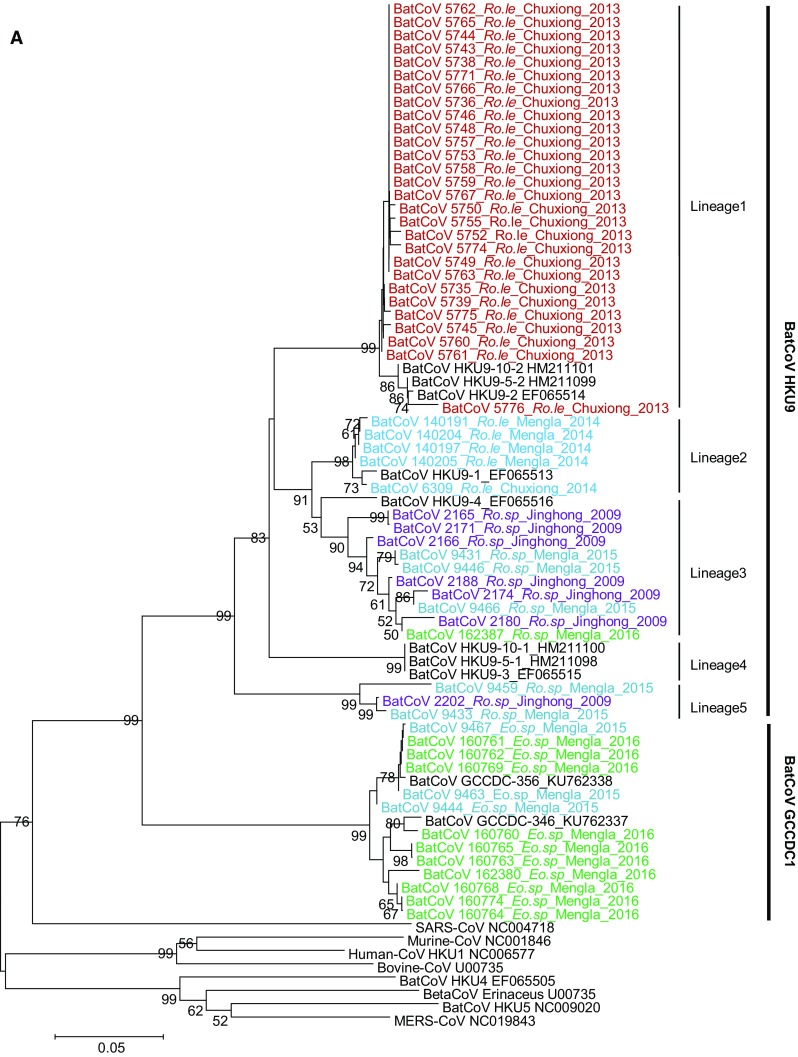
Phylogenetic analysis of the detected coronaviruses in this study. Partial RdRp sequences (A), complete nucleoprotein gene sequences (B), and full-length genomic sequence of BatCoV HKU9-2202 (C) were aligned with corresponding sequences of representative viral species in the genus Betacoronavirus. Phylogenetic trees were constructed using the neighbor-joining method implemented in MEGA7 and bootstrap values calculated from 1000 replicates. The sequence obtained in this study is labeled in color and named by the sample isolate identifier followed by bat species, location, and collection year.
To further characterize the relationships between the newly detected coronaviruses, we amplified the full-length sequences of S, N, and P10 gene from selected positive samples. We amplified N from 9 HKU9-related viruses and 5 GCCDC1-related viruses and P10 from 13 GCCDC1-related viruses. The amplifications of S failed for all positive samples. p10 amplified from this study shared 99%–100% similarity with previously reported sequences (Huang et al. 2016). The amplified N sequences of HKU9 and GCCDC1-related viruses showed 74.5%–100% and 95.2%–97.4% nt identity with each other, respectively. The phylogenetic tree constructed based on N showed a topology structure similar to that of RdRp (Fig. 2B).
Genomic Characterization of Novel Strains BatCoV HKU9-2202
The full-length genome sequence was obtained from one sample (BatCoV HKU9-2202) in lineage 5 by high-throughput sequencing and RACE. The genome of HKU9-2202 is 29,118 nt in length excluding the polyA tail, with a G/C content of 42%. The main ORFs of HKU9-2202 were predicted and deduced in the order: 5′-ORF1ab-Spike (S)-NS3-Envelope (E)-Membrane (M)-Nucleocapsid (N)-NS7a-NS7b-3′ (Table 2). The putative transcription regulatory sequences (TRSs) and their genomic localization were predicted based on the conserved core sequence (5′-ACGAAC-3′) of the TRSs of betacoronaviruses. Notably, in the putative TRS of E, there was a difference of one nucleotide with the consensus core sequences (Table 2).
Table 2.
Amino acid identity, TRS and sequence comparisons of BatCoV HKU9-2202 with BatCoV HKU9 and BatCoV GCCDCC1.
| ORFs | Nucleotide position (start to end) | Predict size (aa) of protein | Pairwise amino acid identity (%)a BatCoV HKU9-2202 vs |
Leader TRS and intergenic TRS | Distance from TRS and ATG | |
|---|---|---|---|---|---|---|
| HKU9-4b | GCCDC1c | |||||
| ORF1ab | 230–20991 | 6921 | 90.2 | 75.1 | CTTGAACGAACTTAA | 152 |
| S | 20951–24748 | 1266 | 63.8 | 61.2 | AGTGAACGAACTTGT | 42 |
| NS3 | 24745–25438 | 231 | 82.8 | 50.2 | AATAAACGAACAGCA | 3 |
| Ed | 25437–25667 | 77 | 96.1 | 67.1 | CAACGTCGAACTATA | 4 |
| M | 25672–26346 | 225 | 91.0 | 79.7 | CTTGAACGAACAAGA | 25 |
| N | 26410–27819 | 470 | 85.5 | 66.3 | TTTGAACGAACCTAT | 5 |
| NS7a | 27857–28450 | 198 | 23.2 | 32.3 | CTTGAACGAACATGA | 0 |
| NS7b | 28447–28890 | 148 | 30.5 | 27.4 | GGTTACGAACGTCT | 7 |
aCalculated with MegAlign using the Jotun Hein method.
bGenBank accession numbers of the referred HKU9-4: EF065516.
cGenBank accession numbers of the referred GCCDC1: NC030886.
dNucleotide site difference compared with the conserved TRS core sequence is underlined
Comparative genomic sequence analysis indicated that HKU9-2202 shared 83% nt identity with other previously reported BatCoV HKU9 strains. The most divergent regions were located in the S protein, which shared only 68% amino acid (aa) identity with those of other BatCoV HKU9. The aa identities of seven concatenated replicase domains, which were selected to define coronavirus species by the International Committee on Taxonomy of Viruses, shared 93% identity with other BatCoV HKU9, which was higher than the new species demarcation of 90%. Thus, the newly identified HKU9-2202 likely belongs to the BatCoV HKU9 species. To determine the evolutionary position of HKU9-2202, the full genome was subjected to phylogenetic analysis. HKU9-2202 formed a separate branch within the clade of BatCoV HKU9 species (Fig. 2C).
Tissue Tropism of batCoV HKU9 and GCCDC1-Related Virus
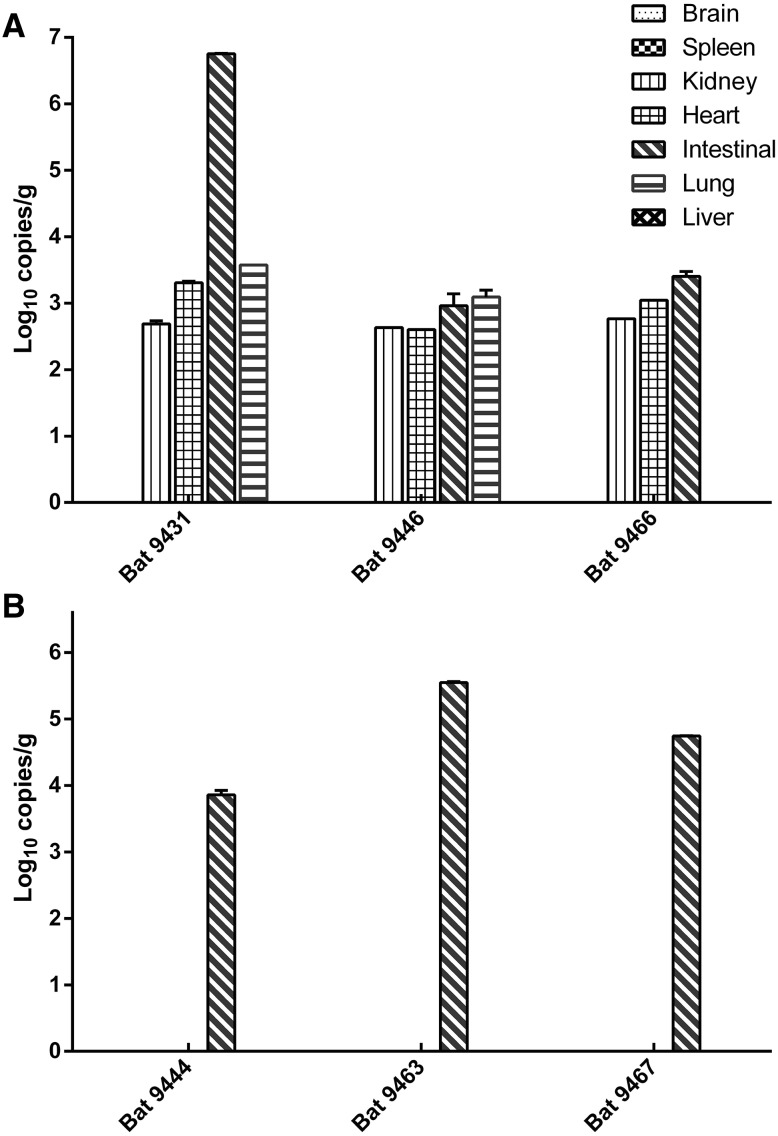
Tissues (heart, liver, spleen, lung, kidney, brain, intestine) from five bats positive for coronavirus were quantified by qPCR (Fig. 3). Higher virus genome copies were detected in all intestines and varied from 4.89 × 102 to 5.67 × 106 copies/g in different tissues. Three HKU9-positive bats (Bt9431, Bt9446 and Bt9466) showed wider tissue tropism, as demonstrated by the presence of viral RNA in the kidney, heart, and lung tissues (Fig. 3A). Three GCCDC1-positive bats (Bt9444, Bt9463, and Bt967) showed exclusive intestine tropism (Fig. 3B). The viral RNA was not detected in the brain, spleen, and liver tissues.
Fig. 3.
Tissue distribution of BatCoV HKU9 (A) and GCCDC1 (B) in positive bat samples.
Discussion
In this study, we conducted a longitudinal study of BatCoV HKU9 and BatCoV-GCCDC1 as well as related coronaviruses in fruit bats in 2009–2016. Highly diverse HKU9-related CoVs were found in Rousettus bats, while GCCDC1-related viruses found in E. spelaea showed high similarity. For HKU9-related CoVs, in addition to four previously reported lineages (Lau et al. 2010), a novel lineage was identified in this study. Previous studies reported that all group 2d coronaviruses within the betacoronavirus were from R. leschenaulti. In this study, we identified all bat species positive for coronavirus by sequencing the Cytb gene and found that HKU9 and GCCDC1 were from two different genera, Rousettus and Eonycteris, respectively. HKU9 consists 5 lineages. Lineage 1 and 2 are from R. leschenaulti and Lineages 3–5 are from an unidentified species Rousetta sp. These results suggest that the coronaviruses may undergo host restriction and have a long evolution history with their hosts.
We amplified multiple N genes and obtained the full-length genomic sequence of a novel HKU9 of linage 5 (BatCoV HKU9-2202). The most notable sequence difference between this novel HKU9 and previously identified BatCoV HKU9s is within the S gene. The S protein of HKU9-2202 shares 61%–68% aa identity to those of previously identified HKU9. The S protein plays a pivotal role in mediating coronavirus entry into host cells. Whether mutations in S are responsible for virulence and tissue tropism of HKU9-2202 requires further analysis.
Coronavirus is known to infect the host through the respiratory system and intestines (Masters and Perlman 2013). In this study, we found that intestine tissues are the major target of BatCoV HKU9 and GCCDC1. However, some HKU9 was also detected in the kidney and lung, suggesting that BatCoV HKU9 has wide tissue tropism and the potential to be transmitted by the oral-fecal route and respiratory routes to infect other animals.
There are at least five fruit bat species in China, all which are located in tropical regions. These fruit bats feed on fruits and flowers and have frequent contact with peoples and farms, thus increasing the risk of spillover of bat viruses to domestic animals and humans. In our previous study, we also found that these bats harbor novel genetically diverse filoviruses, some of which were found to co-infect with BatCoV HKU9 or GCCDC1 in the same individual (Huang et al. 2016; Yang et al. 2017). Our results improve the understanding of variable viruses carried by fruit bats in China. Further studies are needed to investigate the virome of these bat populations and understand the spillover potential of these bat viruses to other animals and humans.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Xing-Yi Ge, Shi-Yue Li, and Hui-Min Huang for assistance in bat sampling. This work was jointly supported by the China Natural Science Foundation (81290341 and 31621061 to ZLS), United States Agency for International Development Emerging Pandemic Threats PREDICT project (AID-OAA-A-14-00102), and National Institute of Allergy and Infectious Diseases of the National Institutes of Health (Award Number R01AI110964) to ZLS.
Author Contributions
ZLS and XLY designed and coordinated this study. RDJ, BJH, DSL, YZZ, and GJZ collected samples. YL, BL, XLY performed most of the experiments. ZLS, XLY, YL, BH, and HZL analyzed the data. YL, XLY, and ZLS drafted the manuscript. All authors read and approved the final manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Animal and Human Rights Statement
This study was approved by the Animal Ethics Committee of the Wuhan Institute of Virology. All institutional and national guidelines for the care and use of animals were followed.
References
- Agnarsson I, Zambrana-Torrelio CM, Flores-Saldana NP, May-Collado LJ. A time-calibrated species-level phylogeny of bats (Chiroptera, Mammalia) PLoS Curr. 2011;3:RRN1212. doi: 10.1371/currents.RRN1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R, Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin Immunopathol. 2017;39:529–539. doi: 10.1007/s00281-017-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot RJ, Baker SC, Baric RS, Brown CS, Drosten C, Enjuanes L, Fouchier RA, Galiano M, Gorbalenya AE, Memish ZA, Perlman S, Poon LL, Snijder EJ, Stephens GM, Woo PC, Zaki AM, Zambon M, Ziebuhr J. Middle east respiratory syndrome coronavirus (MERS-CoV): announcement of the coronavirus study group. J Virol. 2013;87:7790–7792. doi: 10.1128/JVI.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C, Gunther S, Preiser W, van der Werf S, Brodt HR, Becker S, Rabenau H, Panning M, Kolesnikova L, Fouchier RA, Berger A, Burguiere AM, Cinatl J, Eickmann M, Escriou N, Grywna K, Kramme S, Manuguerra JC, Muller S, Rickerts V, Sturmer M, Vieth S, Klenk HD, Osterhaus AD, Schmitz H, Doerr HW. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Du L, Lu Z, Fan Y, Meng K, Jiang Y, Zhu Y, Wang S, Gu W, Zou X, Tu C. Xi River virus, a new bat reovirus isolated in southern China. Arch Virol. 2010;155:1295–1299. doi: 10.1007/s00705-010-0690-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X, Li Y, Yang X, Zhang H, Zhou P, Zhang Y, Shi Z. Metagenomic analysis of viruses from bat fecal samples reveals many novel viruses in insectivorous bats in China. J Virol. 2012;86:4620–4630. doi: 10.1128/JVI.06671-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Feng Y, Zhang H, Xu L, Yang W, Zhang Y, Li X, Tu C. Filovirus RNA in fruit bats, China. Emerg Infect Dis. 2015;21:1675–1677. doi: 10.3201/eid2109.150260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Ge XY, Wang LF, Shi ZL. Bat origin of human coronaviruses. Virol J. 2015;12:221. doi: 10.1186/s12985-015-0422-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CP, Liu WJ, Xu W, Jin T, Zhao YZ, Song JD, Shi Y, Ji W, Jia H, Zhou YM, Wen HH, Zhao HL, Liu HX, Li H, Wang QH, Wu Y, Wang L, Liu D, Liu G, Yu HJ, Holmes EC, Lu L, Gao GF. A bat-derived putative cross-family recombinant coronavirus with a reovirus gene. PLoS Pathog. 2016;12:e1005883. doi: 10.1371/journal.ppat.1005883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter P. The nature of flight—the molecules and mechanics of flight in animals. EMBO Rep. 2007;8:811–813. doi: 10.1038/sj.embor.7401050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Wang L, Lu Z, Xuan H, Han X, Xia X, Zhao F, Tu C. Seroprevalence of rabies virus antibodies in bats from southern China. Vector Borne Zoonotic Dis. 2010;10:177–181. doi: 10.1089/vbz.2008.0212. [DOI] [PubMed] [Google Scholar]
- Lau SK, Poon RW, Wong BH, Wang M, Huang Y, Xu H, Guo R, Li KS, Gao K, Chan KH, Zheng BJ, Woo PC, Yuen KY. Coexistence of different genotypes in the same bat and serological characterization of Rousettus bat coronavirus HKU9 belonging to a novel Betacoronavirus subgroup. J Virol. 2010;84:11385–11394. doi: 10.1128/JVI.01121-10. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Li W, Shi Z, Yu M, Ren W, Smith C, Epstein JH, Wang H, Crameri G, Hu Z, Zhang H, Zhang J, McEachern J, Field H, Daszak P, Eaton BT, Zhang S, Wang LF. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Li Y, Wang J, Hickey AC, Zhang Y, Li Y, Wu Y, Zhang H, Yuan J, Han Z, McEachern J, Broder CC, Wang LF, Shi Z. Antibodies to Nipah or Nipah-like viruses in bats, China. Emerg Infect Dis. 2008;14:1974–1976. doi: 10.3201/eid1412.080359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Liu D, Ran X, Liu C, Guo D, Hu X, Tian J, Zhang X, Shao Y, Liu S, Qu L. Characterization and pathogenicity of a novel mammalian orthoreovirus from wild short-nosed fruit bats. Infect Genet Evol. 2016;43:347–353. doi: 10.1016/j.meegid.2016.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna LKD, Heiser V, Regamey N, Panning M, Drexler JF, Mulangu S, Poon L, Baumgarte S, Haijema BJ, Kaiser L, Drosten C. Generic detection of coronaviruses and differentiation at the prototype strain level by reverse transcription-PCR and nonfluorescent low-density microarray. J Clin Microbiol. 2007;45:1049–1052. doi: 10.1128/JCM.02426-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters PS, Perlman S. Coronaviridae. In: Knipe DM, Howley PM, editors. Fields virology. 6. Philadelphia: Lippincott Williams & Wilkins; 2013. pp. 825–858. [Google Scholar]
- Mickleburgh S, Waylen K, Racey P. Bats as bushmeat: a global review. Oryx. 2009;43:217. doi: 10.1017/S0030605308000938. [DOI] [Google Scholar]
- Shi Z. Emerging infectious diseases associated with bat viruses. Sci China Life Sci. 2013;56:678–682. doi: 10.1007/s11427-013-4517-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuckey MJ, Chomel BB, de Fleurieu EC, Aguilar-Setien A, Boulouis HJ, Chang CC. Bartonella, bats and bugs: a review. Comp Immunol Microbiol Infect Dis. 2017;55:20–29. doi: 10.1016/j.cimid.2017.09.001. [DOI] [PubMed] [Google Scholar]
- Tan B, Yang XL, Ge XY, Peng C, Liu HZ, Zhang YZ, Zhang LB, Shi ZL. Novel bat adenoviruses with low G+C content shed new light on the evolution of adenoviruses. J Gen Virol. 2017;98:739–748. doi: 10.1099/jgv.0.000739. [DOI] [PubMed] [Google Scholar]
- Woo PC, Wang M, Lau SK, Xu H, Poon RW, Guo R, Wong BH, Gao K, Tsoi HW, Huang Y, Li KS, Lam CS, Chan KH, Zheng BJ, Yuen KY. Comparative analysis of twelve genomes of three novel group 2c and group 2d coronaviruses reveals unique group and subgroup features. J Virol. 2007;81:1574–1585. doi: 10.1128/JVI.02182-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XL, Zhang YZ, Jiang RD, Guo H, Zhang W, Li B, Wang N, Wang L, Waruhiu C, Zhou JH, Li SY, Daszak P, Wang LF, Shi ZL. Genetically diverse filoviruses in Rousettus and Eonycteris spp. bats, China, 2009 and 2015. Emerg Infect Dis. 2017;23:482–486. doi: 10.3201/eid2303.161119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan JF, Zhang YJ, Li JL, Zhang YZ, Wang LF, Shi ZL. Serological evidence of ebolavirus infection in bats, China. Virol J. 2012;9:236. doi: 10.1186/1743-422X-9-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.