Dear Editor,
Aquatic birds are thought to be the main reservoir for avian influenza viruses (AIVs) (Olsen et al. 2006). Pandemic emergences caused by these viruses threaten the international poultry industry giving rise to enormous economic losses. In addition, more and more reported cases of bird-to-human transmission alert us the importance of AIVs surveillance in natural foci for identifying, monitoring, and predicting the emergence of pathogens that have a potential threat (Poovorawan et al. 2013; Subbarao et al. 1998). Influenza A viruses are enveloped viruses with eight distinct negative-strand RNA segments which belong to the Orthomyxoviridae family. The RNA segments 4 and 6 encode for hemagglutinin (HA) and neuraminidase (NA) glycoproteins respectively. HAs are viral surface proteins that bind to sialic acid residues at the host cell surface and ensure further virus entry. NAs are considered to be responsible for release of progeny virions. Based on the antigenic properties of the HA and NA glycoproteins, influenza A viruses are categorized into different subtypes. Currently, 16 HA and 9 NA antigenic subtypes have been detected in wild birds and poultry all over the world. So far, influenza A viruses of the H1N1, H2N2, and H3N2 have been observed to cause pandemics (Webster et al. 1992). Occasionally, AIVs that circulate in poultry (e.g., those of subtypes H5, H6, H7, H9 and H10) cross the species barrier and infect humans. Especially, H5, H7, and H9 AIVs represent the top three subtypes that are candidates to cause the next human influenza pandemic. Conventional methods for AIV detection include haemagglutination (HA) test, hemagglutination-inhibition (HI) test, neutralization (NT) and ELISA test. These methods required strict condition of sample collection, relatively amount of virus particles and virus propagation and isolation from cell culture or embryonated chicken eggs which tend to be costly and time consuming. Molecular detection methods based on polymerase chain reaction (PCR) or real-time PCR show superior sensitivity and are now becoming acceptable as the new gold standard. However, the limited multiplexing capacity of PCR or real-time PCR made AIVs surveillance of natural foci insufficient. Furthermore, AIVs surveillance of natural background are costly and labor intensive, so these require multiplexing technologies that would provide influenza virus typing and subtyping in a single reaction to increase throughput capacity and reduce the cost of testing. In this study, we aimed to develop a multi-analyte suspension assay (MASA) combing one-tube multiplex reverse transcription-PCR (RT-PCR) with bead hybridization and detection for high-throughput AIVs subtyping based on Luminex xMAP technology.
To design the subtype-specific amplified primers and capture probes, a total of 27,549 Asian AIVs subtypes sequences were downloaded from the NCBI Genbank database and Global Initiative on Sharing All Influenza Data (GISAID, http://www.gisaid.org/) database (Supplementary Table S1). These AIVs sequences were reported to be isolated from 32 countries in Asia, and 57% of which was isolated from China, 12% of which was from Vietnam and the sequence of Korea and Indonesia was 5% each (data not shown). These sequences covered 12 subtypes of AIVs, including H3 (n = 276), H4 (n = 280), H5 (n = 7107), H6 (n = 1581), H7 (n = 745), H9 (n = 4711), H10 (n = 96), N1 (n = 4986), N2 (n = 4799), N6 (n = 1605), N8 (n = 647), and N9 (n = 716). Next, multiple sequence alignments (MSA) were performed for each subtypes using Clustal Omega (http://www.clustal.org/omega/) to identify conserved nucleotide sequences within the HA or NA gene.
The coverage of a primer can be improved by introducing degeneracies, and a higher degeneracy facilitates higher coverage and also leads to unspecific amplification (Linhart and Shamir 2005). Based on the alignment results of each AIVs subtypes, we use DegePrime to design broad-range primers that target each conserved region of HA or NA gene in Asia. DegePrime is a computer program that finds a degenerate oligomer of as high coverage as possible for each position of a multiple sequences alignment (Hugerth et al. 2014). We placed constraints on the primers in term of minimum and maximum length (20 bp ≤ L ≤ 25 bp), maximum allowed differences in melting temperatures (Tm) (54 °C ≤ Tm ≤ 64 °C), GC % range allowed (30%–60%), length of amplicon (L ≤ 600 bp), maximum degeneracy allowed per primer (d ≤ 4), and maximum length of homopolymer (h ≤ 4). A set of primers and probes with different length and degeneracies was designed.
To pick out the subtype-specific primers and probes with high specificity, we used thermodynamics-based PCR primer specificity-checking program, MFEprimer-2.0, to evaluate these primers and probes candidates (Qu et al. 2012). This program can evaluate the binding stability of the entire primer sequences, run a virtual PCR amplification and filter out the predicted amplicons by size and other parameters. A total of 19 pairs of primers and 12 probes were picked out (Table 1). These primers were checked by using the above mentioned 12 subtypes of AIVs sequences as evaluation database, and each primer was specific and had no cross-reactivity (Supplementary Table S2). The percentages of predicted amplicons represent the coverage (sensitivity) of the subtype-specific primer pairs. The coverage of each primer pairs ranged from 82% to 97% (Supplementary Figure S1).
Table 1.
Subtype-specific primers and capture probes used for avian influenza virus subtyping.
| Subtypes | Functiona | Sequences (5′–3′)b | Location (bp) |
|---|---|---|---|
| H3 | Forward-1 | CATACGGRGCATGTCCCAARTA | 968–989 |
| Reverse-1* | TGGTATATCTTGAARCARCCAT | 1457–1478 | |
| Forward-2 | TGCATHACTCCAAATGGAAGCA | 907–928 | |
| Reverse-2* | CATTRCCCATGTCYTCAGCATT | 1438–1459 | |
| Probe† | TAGARAATGGATGGGAAGGRATGRTAGATGGTTGGTATGG | 1082–1121 | |
| H4 | Forward-1 | ACACRGGRAACCCTGTGATATG | 56–77 |
| Reverse-1* | CCATTCARATGRTCACATCCTG | 257–278 | |
| Reverse-2* | TCCCATTGTTSGCRAGTATGCT | 364–385 | |
| Probe† | ACYGATGATCAAGTRGAAGTGGTCACTGCACARGAACTGG | 124–163 | |
| H5 | Forward | TCTAGYATGCCATTCCACAAYA | 907–928 |
| Reverse* | CATCTAGRAAYCCGTCTTCCAT | 1249–1270 | |
| Probe† | TAGARGGAGGATGGCARGGRATGGTAGATGGTTGGTATGG | 1288–1309 | |
| H6 | Forward-1 | TACCAATCGARAACTGYGATGC | 860–881 |
| Reverse-1* | TCAGCRTTGTATGTCCAAACRT | 1301–1322 | |
| Forward-2 | ATMGCYCCTTGGTATGCATACA | 793–814 | |
| Reverse-2* | TGTCCAAACRTCCAGAAAYCCA | 1290–1311 | |
| Probe† | GAAGGAGGATGGACTGGRATGATWGATGGRTGGTATGGCT | 1063–1102 | |
| H7 | Forward | TYCAGAGTGGAGTACAGGTTGA | 827–848 |
| Reverse* | TCTGARTCAGCCAGATCAATTG | 1337–1358 | |
| Probe† | GATTACAAAAGCACTCAATCGGCAATTGATCAAATAACAG | 1126–1165 | |
| H9 | Forward-1 | GCTCCATGGTATGGACACRTTCT | 782–803 |
| Reverse-1* | TCWACCTCRCTGAATTCATGATC | 1214–1235 | |
| Forward-2 | GATAGGRCCAAGGCCYCTTG | 672–691 | |
| Reverse-2* | CCTTGRTCATTTGAATGCTGRA | 1085–1106 | |
| Probe† | GCAGTYGGTCTGAGGAATGTGCCTKCTAGATCWAGTAGAG | 976–1015 | |
| H10 | Forward | GGAGAGGTCTATTYGGTGCAAT | 1016–1037 |
| Reverse* | TGTTTCTTATGCTCTCCATGCA | 1462–1483 | |
| Probe† | AAACCAAYACTGAGTTCGAGTCAATAGAATCTGARTTCAG | 1193–1232 | |
| N1 | Forward | AGAGAGCCRTTCATMTCATGCT | 292–313 |
| Reverse* | GATAACAGGAGCAYTCCTCRTA | 766–787 | |
| Probe† | GAGTCTGTTGCTTGGTCRGCAAGTGCTTGYCATGATGGCA | 463–502 | |
| N2 | Forward-1 | TGCATAGCATGGTCYAGYTCAA | 514–535 |
| Reverse-1* | CCAACAAGYCCTGAGCAYACAT | 938–959 | |
| Reverse-2* | ATGCRCTTCCATCAGTCATYAC | 709–730 | |
| Probe† | AGAACYCAGGAGTCAGAATGYGTYTGCATCAATGGAACTT | 661–700 | |
| N6 | Forward-1 | AGGAYGCTCATATACTRGTCACAA | 296–319 |
| Reverse-1* | AYGTGCTTGACCAYCCTATGCA | 493–514 | |
| Forward-2 | GACCAAGCCSCTATGTGARGT | 228–248 | |
| Reverse-2* | TYCTTGATATGCCATCRTGGCA | 517–538 | |
| Probe† | TGCAGGATGTTTGCTCTGAGYCARGGMACAACACTCAGAG | 352–391 | |
| N8 | Forward-1 | CAGGCAAGRTTTGARGCAGTAG | 582–603 |
| Reverse-1* | GGRTAGCATGARCATTCTTCTA | 1164–1185 | |
| Forward-2 | TTTGAAGCTGTRGCRTGGTCA | 514–534 | |
| Reverse-2* | CCCATRGGRCTAGTGCATGA | 1000–1020 | |
| Probe† | TGCATTCAAGGTGARTGTTWTTGGGTAATGACRGATGGAC | 691–730 | |
| N9 | Forward | CTGACTGGAGTGGYTACAGTGG | 1187–1208 |
| Reverse* | CCAGTTCCATTGTCCMAGGAAT | 1344–1365 | |
| Probe† | TRTGGTGGACCAGYAATAGYATAGTATCGATGTGTTCCAG | 1298–1337 |
a*, Primer labeled with 5′ biotin; †, probe modified with 5′ amino-C12 linker.
bDegenerate base: R, G, M, Y, W, and S.
The MASA based on Luminex xMAP technology which can detect up to 100 DNA targets in one single reaction. In multiplex PCR step of the MASA, each primer pair of subtypes identify their target template sequences for amplification, and 5′ terminus-biotinylated primers labeled the target strand of the amplicons. After PCR amplification, mixed capture probes were added for hybridization. The capture probes modified with amino-C12 linker at the 5′ terminus were coupled to Luminex carboxylated microspheres with specific spectral addresses by a carbodiimide-based procedure. Each capture probe was complementary in sequence to the labeled strands of the PCR products. For detection, streptavidin–phycoerythrin (SA–PE) was added into the hybridization product to quantify the biomolecular interaction that has occurred at the microsphere surface. The signals produced for each microsphere were detected as they pass by two separate lasers in the Luminex 100 analyzer. Previously, we developed a MASA of Influenza Panel which targeted important subtypes circulating or emerging in China. The specificity of Influenza Panel was evaluated with 4 type influenza viruses H1N1, H3N2, H5N1, and H7N9 which were kindly provided by Microorganisms and Viruses Culture Collection Center, Wuhan Institute of Virology, CAS. As shown in Fig. 1A, the MASA was able to detect all target type or subtypes of influenza virus, whereas no signal corresponding to other subtypes was detected. We also assessed the specificity of H5, N1 and H7 with other influenza viruses (Fig. 1B–D). All the H5, N1 and H7 subtypes were detected accurately, and the median fluorescence intensity of other subtypes was below 50.
Fig. 1.
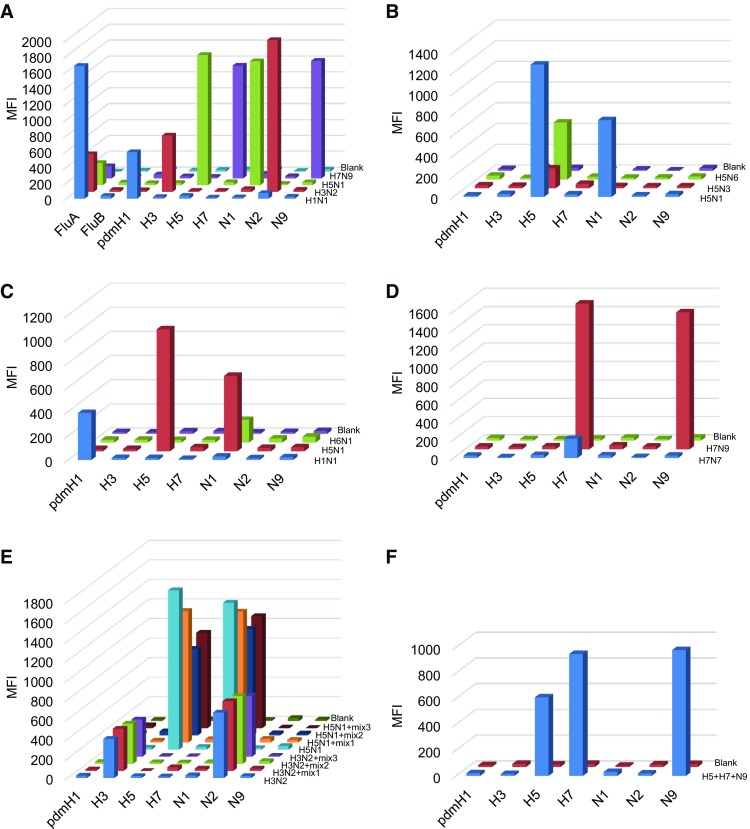
A The specificity of Influenza Panel was carried out with H1N1, H3N2, H5N1, and H7N9. B–D The specificity of H5, N1, and H7 was evaluated with H5N1, H5N3, H5N6, H1N1, H6N1, H7N7 and H7N9. E The mixed-sample detection capacity of H3, H5, N1 and N2 was carried out using three mixed samples respectively. Mix 1 included HSV-1, HSV-2, adenovirus. Mix 2 included SFTSV, JEV, EV71 and sendai virus. Mix 3 included HSV-1, HSV-2, adenovirus, SFTSV, JEV, EV71 and sendai virus. F The analysis of multi-analyte detection. H5, H7 and N9 subtypes were mixed and subjected to MASA. DEPC-H2O was used as a blank control. Biotin-labeled PCR products were separated by probe-coupled microspheres and are presented in terms of dye signal median fluorescence intensity (MFI) on the y axis. Each peak was identified by microspheres coupled with specific capture probes and is indicated on the x axis.
Because of the infections with multiple pathogens occurring naturally, the capability for mixed-sample was also investigated using a set of combinations of pathogens (HSV-1, HSV-2, adenovirus, SFTSV, JEV, EV71 and sendai virus). As shown in Fig. 1E, MASA could accurately detect and differentiate each subtype in the mixtures. We also mixed H5, H7 and N9 subtypes together and assessed the ability of multi-analyte detection. As shown in Fig. 1F, MASA detected each subtype specifically in multiple-target sample. Consistently, another influenza Panel based on xMAP technology showed that the analytical sensitivity was 4–10 copies vRNA/μL for seasonal H1N1, H3N2, and pandemic H1N1 (Xiaoyan et al. 2014). Munro et al. (2013) reported that the limits of detection of seasonal H1 and 2009 H1 were 10-fold higher for multiplex Luminex assay compared to real-time PCR.
In summary, we used xMAP technology to develop a multiplexed suspension assay for simultaneous AIVs subtyping in a single reaction, including H3, H4, H5, H6, H7, H9, H10, N1, N2, N6, N8, and N9. The evaluation data from MFEprimer-2.0 showed that these subtype-specific amplified primers had high specificity and coverage for Asian AIVs. Our studies provide a rapid, specific, high-throughput and economical method for AIVs surveillance of natural foci and further evaluations with kinds of collected samples are needed.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Accession numbers of 27,549 Asian AIVs subtypes sequences downloaded from NCBI and GISAID database (XLSX 582 kb)
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (No. 81501430) and the Basic Work Program of the Ministry of Science and Technology of China (2013FY113500).
Conflict of interest
The authors declare that they have no conflicts of interest.
Animal and Human Rights Statement
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- Hugerth LW, Wefer HA, Lundin S, Jakobsson HE, Lindberg M, Rodin S, Engstrand L, Andersson AF. DegePrime, a program for degenerate primer design for broad-taxonomic-range PCR in microbial ecology studies. Appl Environ Microbiol. 2014;80:5116–5123. doi: 10.1128/AEM.01403-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linhart C, Shamir R. The degenerate primer design problem: theory and applications. J Comput Biol. 2005;12:431–456. doi: 10.1089/cmb.2005.12.431. [DOI] [PubMed] [Google Scholar]
- Munro SB, Kuypers J, Jerome KR. Comparison of a multiplex real-time PCR assay with a multiplex Luminex assay for influenza virus detection. J Clin Microbiol. 2013;51:1124–1129. doi: 10.1128/JCM.03113-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen B, Munster VJ, Wallensten A, Waldenstrom J, Osterhaus AD, Fouchier RA. Global patterns of influenza a virus in wild birds. Science. 2006;312:384–388. doi: 10.1126/science.1122438. [DOI] [PubMed] [Google Scholar]
- Poovorawan Y, Pyungporn S, Prachayangprecha S, Makkoch J. Global alert to avian influenza virus infection: from H5N1 to H7N9. Pathog Glob Health. 2013;107:217–223. doi: 10.1179/2047773213Y.0000000103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu W, Zhou Y, Zhang Y, Lu Y, Wang X, Zhao D, Yang Y, Zhang C. MFEprimer-2.0: a fast thermodynamics-based program for checking PCR primer specificity. Nucleic Acids Res. 2012;40:W205–W208. doi: 10.1093/nar/gks552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbarao K, Klimov A, Katz J, Regnery H, Lim W, Hall H, Perdue M, Swayne D, Bender C, Huang J, Hemphill M, Rowe T, Shaw M, Xu X, Fukuda K, Cox N. Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science. 1998;279:393–396. doi: 10.1126/science.279.5349.393. [DOI] [PubMed] [Google Scholar]
- Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiaoyan L, Jinying C, Mei K, Xu S, Ming Z, Liru G. Development of high-throughput liquid chips for respiratory virus detection. Clin Lab. 2014;60:347–361. doi: 10.7754/Clin.Lab.2013.130334. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Accession numbers of 27,549 Asian AIVs subtypes sequences downloaded from NCBI and GISAID database (XLSX 582 kb)


