Dear Editor,
Zika virus (ZIKV), a pathogen within the genus Flavivirus, family Flaviviridae brought an international public health emergency due to its association with neonatal microcephaly case (Platt and Miner 2017). Currently the most reliable diagnostic test is PCR detection of ZIKV RNA from body fluid samples. Unfortunately, the short viremia window and asymptomatic/mild infections greatly reduce the success rate of PCRs (de Vasconcelos et al. 2018). Serological assays play a pivotal role in retrospective diagnosis of ZIKV infection, especially for pregnant women concerned about ZIKV infection during their early stages of pregnancy (de Vasconcelos et al. 2018). However, due to the high level of cross reactivity among related flaviviruses, all serology tests conducted in locations where flaviviruses co-circulate prove to be difficult for making a conclusive diagnosis, especially for diagnosis of ZIKV infection in dengue virus (DENV) pre-exposed individuals (David et al. 2017; L’Huillier et al. 2017; de Vasconcelos et al. 2018).
This difficulty is exacerbated by the lack of serum panels from individuals with an absolutely defined infection status, hence presenting a chicken-and-egg cycle in which any attempt to develop a specific assay has to rely on serum panels where infection status is not 100% defined. In this case, even the “gold standard” assay, the virus neutralization test (VNT), is inconclusive as ZIKV and DENV antibodies are known to cross-neutralize (Collins et al. 2017).
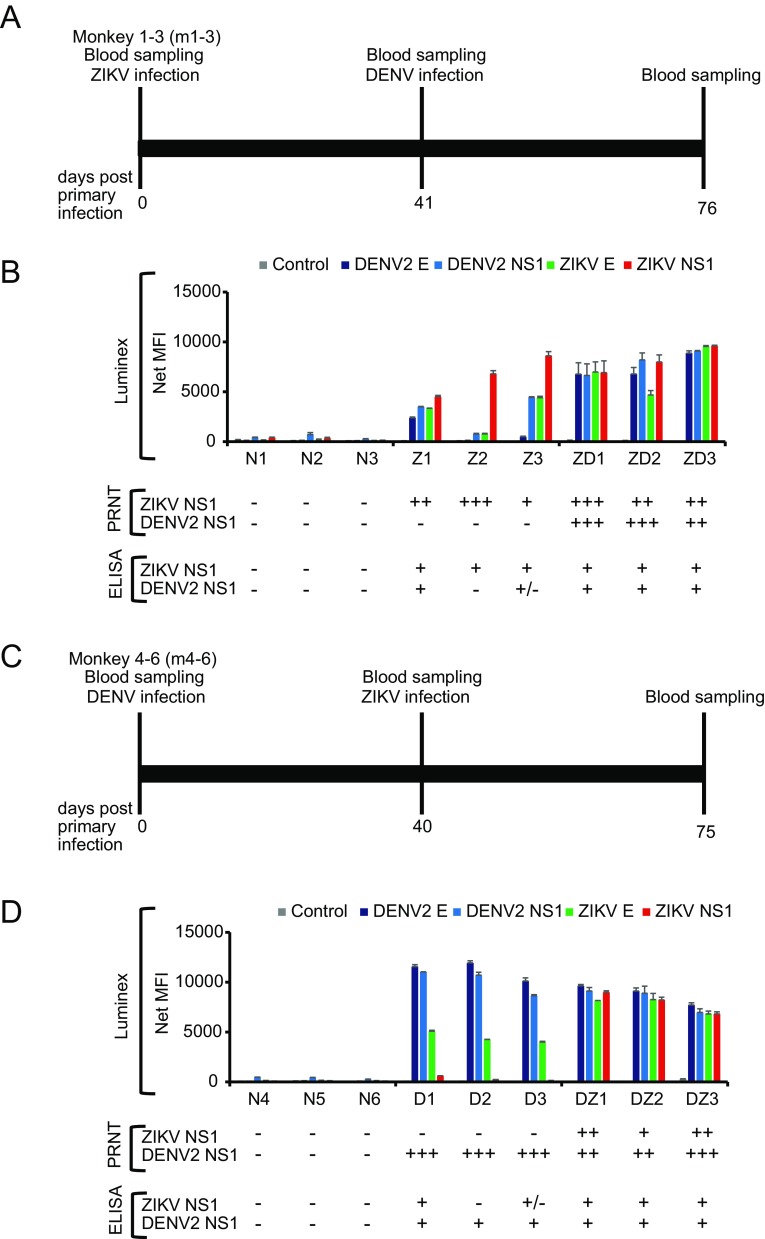
To address this challenge, we produced a panel of monkey sera covering four infection scenarios: (1) ZIKV alone (group Z); (2) DENV alone (D); (3) ZIKV followed by DENV (ZD); and (4) DENV followed by ZIKV (DZ). A dose of 105 pfu of ZIKV-Brazil (GenBank ID: KU365780.1) and/or DENV2 (GenBank ID: KU725663) was used for subcutaneous inoculation of cynomolgus macaque (Macaca fascicularis) with 3 animals per group. The infection study was approved by the Institutional Animal Care and Use Committee of the SingHealth Experimental Medicine Centre. Absence of ZIKV or DENV antibodies in these animals was confirmed using commercial ELISA kits for ZIKA (Anti-Zika Virus ELISA (IgG), Euroimmun, #EI2668-9601G) and DENV (Panbio Dengue IgG Indirect ELISA, Abbott, #01PE30), respectively. At 40 or 41 days post infection (dpi), seroconversion was evident for the D and Z group, respectively (data not shown). The animals were then challenged with the other virus using the same dose as the primary infection and bled 35 days post-secondary infection (Fig. 1A, Fig. 1C).
Fig. 1.
Antibody responses in four infection scenarios determined by Luminex, PRNT and ELISA. A Experimental timeline for the first group of three animals (m1, m2, m3). Monkeys were inoculated with 105 pfu of ZIKV by the subcutaneous route. On Day 41 post ZIKV infection, animals were infected with 105 pfu of DENV2. B Luminex, PRNT and ELISA analysis of serum samples collected after primary infection with ZIKV (41 dpi-ZIKV) are labelled Z and serum samples collected after secondary DENV infection are labelled as ZD (76 dpi-ZIKV/35 dpi-DENV2), respectively. C Experimental timeline for the second group of three animals (m4, m5, m6). Monkeys were inoculated with 105 pfu of DENV2 by the subcutaneous route. On Day 40 post DENV infection, animals were infected with 105 pfu of ZIKV. D Luminex, PRNT and ELISA analysis of the serum samples collected after primary infection with DENV (40 dpi-DENV2) are labelled D and serum samples collected after secondary ZIKV infection are labelled as DZ (75 dpi-DENV2/35 dip-ZIKV), respectively. Pre-bleed samples collected on Day 0 were labelled as N. A total of six animals (N1–N6) were used in this study with three animals in each infection group. Recombinant SARS-CoV N protein was used as control antigen at the same concentration as the other four antigens. For Luminex, all assays were done in duplicate and mean fluorescent intensity (MFI) was plotted for each sample. For ELISA, readings were defined as negative (−) for those with PanBio units below < 9 for DENV, relative units (RU) below < 16 for ZIKV; equivocal (±) for PanBio units 9–11 for DENV, RU 16–22 for ZIKV; positive (+) for those with PanBio above > 11 for DENV and RU above > 22 for ZIKV. For PRNT, samples with no neutralization activity was defined as negative (−); with neutralization activity at serum dilution of 1:10–1:20, 1:40–1:80 and 1:160–1:320 as (+), (++) and (+++), respectively.
As previously reported (David et al. 2017; L’Huillier et al. 2017), significant cross reactivity between the two viruses was observed using the commercial ELISA kits. To improve the specificity, we developed a multiplex Luminex assay similar as previously published (Wong et al. 2017). Recombinant E and NS1 proteins from ZIKV and DENV2, respectively, were obtained from The Native Antigen Company (UK) and 25 μg of each protein was coupled to the MagPlex beads (MagPlex-C Microspheres) at 5 μg/1 × 106 beads using the xMAP antibody coupling kit (#40-50016, both obtained from Luminex, USA), following manufacturer’s protocol.
As shown in Fig. 1B, while antibodies in group Z cross react with DENV, the ratio of Z/D is greater than 1 for either E or NS1 protein, which is true for all three animals in the group. Similarly, the D/Z ratio is greater than 1 for all three animals of group D (Fig. 1D). In another word, in the single virus infected groups (either D or Z), the ratio-based Luminex readouts were sufficiently specific to differentiate between Zika or dengue infection. See Supplementary Figure S1 and Table S1 for the original data, which showed that the Luminex readings with either E or NS1 antigens are statistically different between the primary infections by the two viruses. However, for groups ZD or DZ, the Z/D or D/Z the ratio is mostly indistinguishable and it is therefore impossible to deduce which virus was responsible for the most recent infection.
To further assess the cross reactivity of these serum panels, we conducted plaque reduction neutralization tests (PRNT) using previously published methods (Shan et al. 2017). The results (Fig. 1B, Fig. 1D) indicated that in all animals with only primary infection, neutralization was observed only with the homologous virus-serum pair. However, for the mixed infection groups, there was no clear “rule” observed. The ZD group seemed to have higher titer against the secondary infection virus whereas the DZ group is the opposite, with higher titer against the primary infection virus. For secondary flaviviral infections, it has been well documented that an anamnestic antibody response often occurs and the degree and specificity in relation to the two viruses can vary (Guzman et al. 2013; Priyamvada et al. 2016; Bardina et al. 2017).
While our findings are somewhat limited due to the small number of animals used in this study, the following important conclusions can be drawn from this study. (1) There is significant cross reactivity between anti-ZIKV and -DENV antibodies and any single virus/antigen-based assays won’t be reliable for conclusive sero-diagnosis; (2) although some samples showed that anti-ZIKV antibodies have more cross-reactivity with DENV than vice versa, the overall two-way cross-reactivity level is similar; (3) even with the small numbers used in this study, there is a clear individual-to individual variation in the context of cross reaction; (4) ratio-based readout from multiplex platforms, such as the E and NS1-based Luminex assay, seems to be sufficiently specific for diagnosis of primary infection with a single virus; and (5) no current test platform, even the “gold standard” PRNT, is able to determine the most recent secondary infection.
It might therefore be worthwhile to look into developing epitope-specific tests which could potentially detect ZIKV-specific antibodies in the presence of anti-DENV antibodies. Monoclonal antibody-based blocking ELISA (Balmaseda et al. 2017), peptide array (Jelsma et al. 2017) or peptide display phage library (Xu et al. 2015) are some of the approaches which can be deployed to address this challenging issue that needs an urgent solution. It should be cautioned that epitope-based serology is more prone to subtle immune response difference between individuals and a multiplex approach including multiple virus-specific epitopes may have to be deployed.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Edgar Macabe Pena and his team, especially Ryan Cabo Maniquiz and Redmond Belen Cheng, for assistance with monkey infection studies. We thank Veronika von Messling for providing expertise and assistance in planning of the animal studies, and Eng Eong Ooi and his lab members, especially Eugenia Ong and Esther Gan, for providing DENV2 viruses. The project was funded in part by NMRC Grant ZRRF16006 and CD-PHRG Grant CDPHRG/0006/2014.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Animal and Human Rights Statement
Cynomolgus macaques (Macaca fascicularis) were used for all studies. The experiments were approved by the SingHealth Institutional Animal Care and Use Committee of the SingHealth Experimental Medicine Centre (SEMC).
Contributor Information
Lin-Fa Wang, Phone: +65-65167256, Email: linfa.wang@duke-nus.edu.sg.
Danielle E. Anderson, Phone: +65-65767080, Email: danielle.anderson@duke-nus.edu.sg
References
- Balmaseda A, Stettler K, Medialdea-Carrera R, Collado D, Jin X, Zambrana JV, Jaconi S, Cameroni E, Saborio S, Rovida F, Percivalle E, Ijaz S, Dicks S, Ushiro-Lumb I, Barzon L, Siqueira P, Brown DWG, Baldanti F, Tedder R, Zambon M, de Filippis AMB, Harris E, Corti D. Antibody-based assay discriminates Zika virus infection from other flaviviruses. Proc Natl Acad Sci USA. 2017;114:8384–8389. doi: 10.1073/pnas.1704984114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardina SV, Bunduc P, Tripathi S, Duehr J, Frere JJ, Brown JA, Nachbagauer R, Foster GA, Krysztof D, Tortorella D, Stramer SL, Garcia-Sastre A, Krammer F, Lim JK. Enhancement of Zika virus pathogenesis by preexisting antiflavivirus immunity. Science. 2017;356:175–180. doi: 10.1126/science.aal4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins MH, McGowan E, Jadi R, Young E, Lopez CA, Baric RS, Lazear HM, de Silva AM. Lack of durable cross-neutralizing antibodies against Zika virus from dengue virus infection. Emerg Infect Dis. 2017;23:773–781. doi: 10.3201/eid2305.161630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David S, Angela S, Derek RS, Emelissa M, Nicole B, Charlene R, Leanne S, Kimberly H, Alyssia R, Maya T-A, Kai M, Kristina D, Elizabeth G, Joanne H, Rhonda M, Sharla B, Michael D. Evaluation of 5 commercially available Zika virus immunoassays. Emerg Infect Dis. 2017;23:1577. doi: 10.3201/eid2309.162043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vasconcelos ZFM, Azevedo RC, Thompson N, Gomes L, Guida L, Moreira MEL. Challenges for molecular and serological ZIKV infection confirmation. Childs Nerv Syst. 2018;34:79–84. doi: 10.1007/s00381-017-3641-5. [DOI] [PubMed] [Google Scholar]
- Guzman MG, Alvarez M, Halstead SB. Secondary infection as a risk factor for dengue hemorrhagic fever/dengue shock syndrome: an historical perspective and role of antibody-dependent enhancement of infection. Arch Virol. 2013;158:1445–1459. doi: 10.1007/s00705-013-1645-3. [DOI] [PubMed] [Google Scholar]
- Jelsma T, van der Wal FJ, Fijten H, Dailly N, van Dijk E, Loeffen WL. Pre-screening of crude peptides in a serological bead-based suspension array. J Virol Methods. 2017;247:114–118. doi: 10.1016/j.jviromet.2017.05.008. [DOI] [PubMed] [Google Scholar]
- L’Huillier AG, Hamid-Allie A, Kristjanson E, Papageorgiou L, Hung S, Wong CF, Stein DR, Olsha R, Goneau LW, Dimitrova K, Drebot M, Safronetz D, Gubbay JB. Evaluation of euroimmun anti-Zika virus IgM and IgG enzyme-linked immunosorbent assays for Zika virus serologic testing. J Clin Microbiol. 2017;55:2462–2471. doi: 10.1128/JCM.00442-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt DJ, Miner JJ. Consequences of congenital Zika virus infection. Curr Opin Virol. 2017;27:1–7. doi: 10.1016/j.coviro.2017.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priyamvada L, Quicke KM, Hudson WH, Onlamoon N, Sewatanon J, Edupuganti S, Pattanapanyasat K, Chokephaibulkit K, Mulligan MJ, Wilson PC, Ahmed R, Suthar MS, Wrammert J. Human antibody responses after dengue virus infection are highly cross-reactive to Zika virus. Proc Natl Acad Sci USA. 2016;113:7852–7857. doi: 10.1073/pnas.1607931113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan C, Xie X, Ren P, Loeffelholz MJ, Yang Y, Furuya A, Dupuis AP, II, Kramer LD, Wong SJ, Shi PY. A rapid Zika diagnostic assay to measure neutralizing antibodies in patients. EBioMedicine. 2017;17:157–162. doi: 10.1016/j.ebiom.2017.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong SJ, Furuya A, Zou J, Xie X, Dupuis AP, II, Kramer LD, Shi PY. A multiplex microsphere immunoassay for Zika virus diagnosis. EBioMedicine. 2017;16:136–140. doi: 10.1016/j.ebiom.2017.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu GJ, Kula T, Xu Q, Li MZ, Vernon SD, Ndung’u T, Ruxrungtham K, Sanchez J, Brander C, Chung RT, O’Connor KC, Walker B, Larman HB, Elledge SJ. Viral immunology. Comprehensive serological profiling of human populations using a synthetic human virome. Science. 2015;348:aaa0698. doi: 10.1126/science.aaa0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.