Abstract
Porcine circoviruses (PCV) include PCV1, PCV2, and the new-emerging PCV3. PCV2 is pathogenic to pigs, but the pathogenicity of PCV3 in pigs is debatable. Recently, there have been frequent reports of PCV2 and PCV3 co-infections in clinical samples. Thus, it would be practical to develop a duplex PCR method to detect PCV2 and PCV3 simultaneously. In this study, specific primers and probes were designed to target PCV2 cap and PCV3 rep genes. A duplex real-time PCR method was then developed to detect the two viruses. The assay was found to be highly specific, sensitive, and reproducible for PCV2/3 without cross-reactions with other swine pathogens. The sensitivity of this assay was 2.9 copies for the PCV2 plasmid and 22.5 copies for the PCV3 plasmid. The established assay was then used to detect PCV2/3 infection in 340 clinical samples collected in the first half of 2017. The results showed that the co-infection rate of PCV2/3 in the samples was 27.6%. Our study provides an important tool that can be used to perform urgently needed surveys for the two porcine circoviruses to evaluate their impact on the swine industry.
Electronic supplementary material
The online version of this article (10.1007/s12250-018-0025-2) contains supplementary material, which is available to authorized users.
Keywords: Porcine circovirus 2 (PCV2), Porcine circovirus 3 (PCV3), Co-infection, Real-time PCR
Introduction
Porcine circoviruses (PCV), including PCV1, PCV2, and PCV3, are small, non-enveloped, single-stranded circular DNA viruses that belong to the genus Circovirus in the family Circoviridae (Zhai et al. 2014). PCV1 is a contaminant of a porcine kidney cell line and is non-pathogenic to pigs. In contrast, PCV2 is considered the primary etiological agent of porcine circovirus-associated diseases and has caused great economic losses to the swine industry worldwide (Zhai et al. 2014). The new-emerging PCV3 is associated with porcine dermatitis, nephropathy syndrome and reproductive failure, and cardiac and multisystemic inflammation, and it has been recently reported in the United States, Poland, South Korea, and China (Kwon et al. 2017; Palinski et al. 2017; Phan et al. 2016; Shen et al. 2017; Stadejek et al. 2017). Since its first report in December 2016, the co-infection of PCV3 with PCV2 has been frequently reported in clinical samples of diseased pigs (Ku et al. 2017). However, PCV2 and PCV3 were tested separately using different PCR methods in the above reports. It is more practical to detect PCV2 and PCV3 in clinical samples simultaneously. Therefore, in this study, we developed a duplex PCR method targeting the cap and rep gene for identifying PCV2 and PCV3 simultaneously.
Materials and Methods
Construction of PCV2 and PCV3 Plasmids
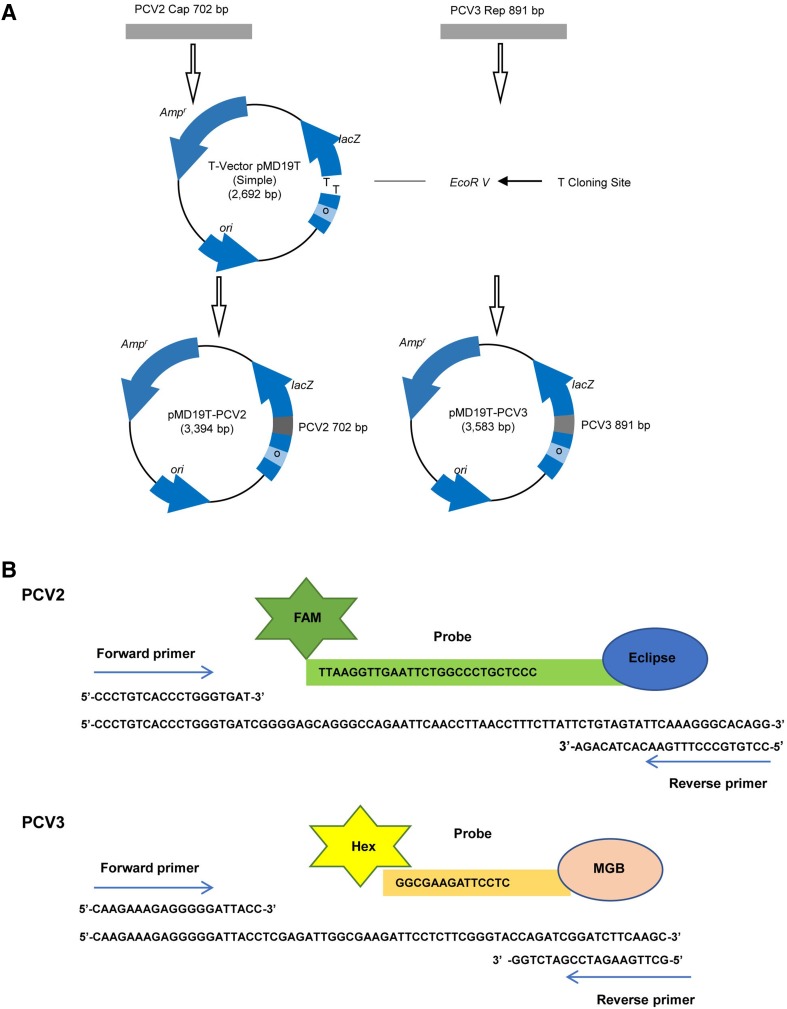
The cap gene sequence of PCV2 (GenBank Access No. AY556475.1) and rep gene sequence of PCV3 (GenBank Access No. MF769805) were synthesized and cloned into the pMD19 vector (Fig. 1). The constructed plasmids pMD19-PCV2 and pMD19-PCV3 were transformed into TOP10 competent cells (Thermo Fisher Scientific, USA). The constructed plasmids were then extracted using QIAGEN Plasmid Midi Kits and used as templates for real-time PCR optimization. The plasmids were quantified using a ND-2000c spectrophotometer (NanoDrop, Wilmington, USA), and the copy numbers of the plasmids were calculated as previously described (Yun et al. 2006). To test the sensitivity of the established real-time PCR, tenfold continuous dilutions of the constructed plasmids with TE buffer (10 mmol/L Tris–HCl, 1 mmol/L EDTA) were used as templates.
Fig. 1.
A Schematic presentation of the PCV2 and PCV3 plasmid construction. B Target fragment and primers.
Viruses
PCV1 (NVC-PCV101), porcine reproductive and respiratory syndrome virus (JXA1), porcine pseudorabies virus (HN1201), porcine parvovirus (NVC-PPV01), classical swine fever virus (NVC-CSFV01), and porcine epidemic diarrhea virus (NVC-PEDV01) were stored at the National Research Center for Veterinary Medicine. Viral DNA and RNA were extracted using TIANamp virus genomic DNA/RNA kits (Tiangen, Beijing China) according to the manufacturer’s instructions before being used as the templates for the specificity test in this study.
Primers and Probes for PCV2/3 Real-Time PCR
DNASTAR software (DNASTAR Inc., Madison, WI, USA) was used to confirm the highly conserved regions within the cap and rep gene in the PCV2 and PCV3 genomes. Primer Express 3.0 software (Applied Biosystems, Foster City, CA, USA) was then used to design the primers and probes. For PCV2, the forward primer was PCV2-F: 5′-CCCTGTCACCCTGGGTGAT-3′, the reverse primer was PCV2-F: 5′-CCTGTGCCCTTTGAATACTACAGA-3′, and the TaqMan probe was PCV2-P: FAM-5′-TAAGGTTGAATTCTGGCCCTGCTCCC3′-Eclipse. For PCV3, the forward primer was PCV3-F: 5′-CAAGAAAGAGGGGGATTACC-3′, the reverse primer was PCV3-R: 5′-GCTTGAAGATCCGATCTGG-3′, and the TaqMan probe was PCV3-P: hex-5′-GGCGAAGATTCCTC-3′-MGB. The lengths of the PCV2 and PCV3 amplicons were 82 and 69 base pairs, respectively.
Real-Time PCR
Real-time PCR was performed using an ABI 7500 instrument (Applied Biosystems, Foster City, USA). The real-time PCR mixture contained 12.5 µL Premix Ex Taq (Takara, Dalian, China), 1 µL template DNA, 1 µL primers (1 µmol/L), 1 µL probe (0.5 µmol/L), and sterile water to bring the final volume to 25 µL. The amplification parameters were set as 95 °C for 30 s, followed by 40 cycles of 95 °C for 10 s, and 60 °C for 30 s. The threshold was established as the default of the 7500 system SDS software before further raw data processing.
Sensitivity and Specificity of PCV2/3 Duplex Real-Time PCR
The sensitivity of the established duplex real-time PCR was determined using serial tenfold dilutions (103–1010 dilutions for pMD19-PCV2 and 101–109 dilutions for pMD19-PCV3) of plasmids as templates. The standard curves for PCV2 and PCV3 were generated, and the threshold cycle (Ct) of these standard dilutions was plotted against the log value of the copy number of the corresponding standard plasmid.
The specificity of the established duplex real-time PCR was tested using the DNA or cDNA of above-mentioned porcine viruses as templates, which was subjected to real-time PCR to detect PCV2 and PCV3.
Reproducibility of the PCV2/3 Duplex Real-Time PCR
To assess the intra- and inter-assay reproducibility of the real-time PCR, 107, 105, and 103 copies of plasmids were tested five different times. The intra- and inter-assay coefficients of variation (CV) for the Ct values were calculated.
Application of the PCV2/3 Duplex Real-Time PCR to Clinical Samples
Three hundred forty clinical samples, including lungs, lymph nodes, and tonsils, were collected from diseases pigs on 15 farms in Henan province in the first half of 2017. Viral DNA was extracted as described above and used as templates to detect PCV2 and PCV3 using the established duplex real-time PCR. This study was performed in strict accordance with the regulations in the guide for the care and use of laboratory animals of the National Research Center for Veterinary Medicine.
Results
Optimization of the PCV2/3 Duplex Real-Time PCR
Sixteen PCV3 strains were retrieved from GenBank and aligned to design the primers and probes (Supplementary Table S1). The primers and probes were optimized to contain the strongest fluorescence and the lowest threshold cycle with different concentrations. The final optimized concentrations of the primers and probes were 1 µL primers (1 µmol/L) and 1 µL probe (0.5 µmol/L) for both PCV2 and PCV3.
Specificity of the PCV2/3 Duplex Real-Time PCR
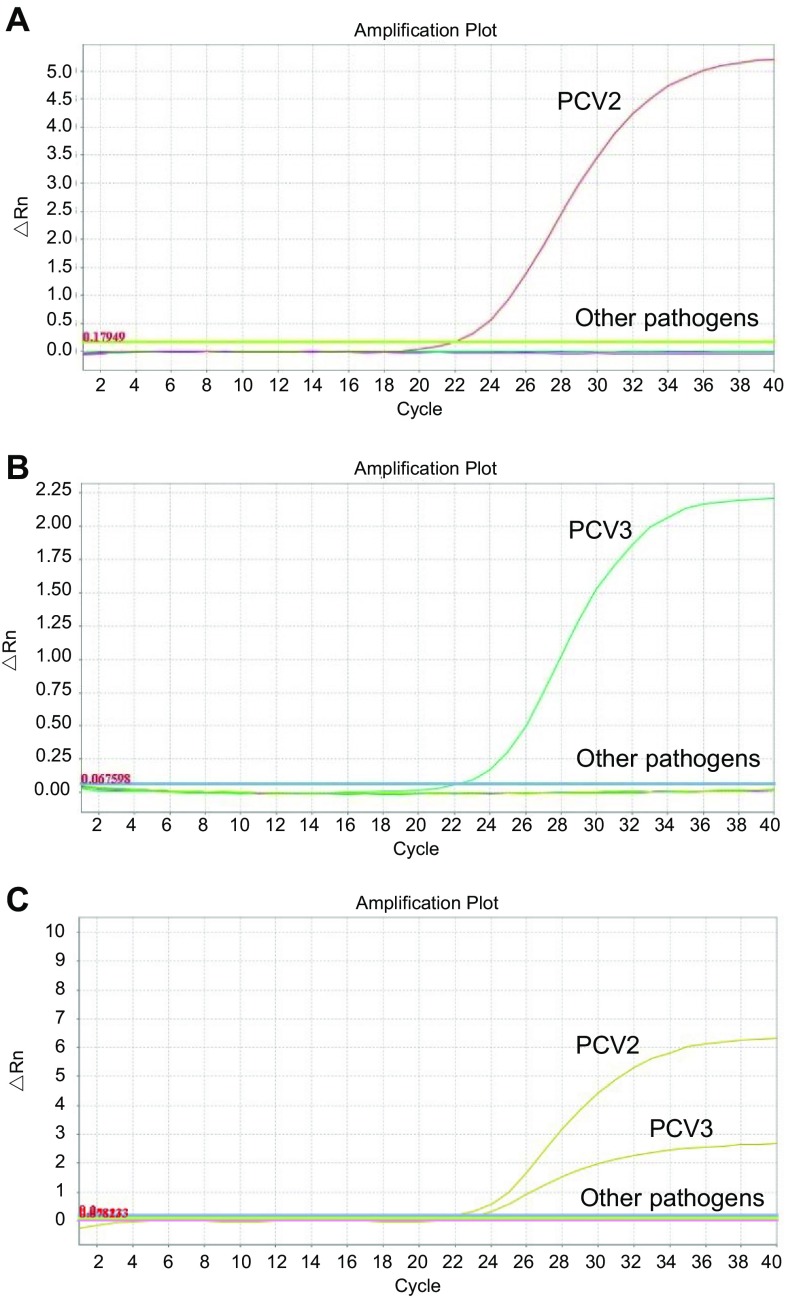
To assess the specificity of the duplex real-time PCR, the DNAs or cDNAs from PCV1, porcine reproductive and respiratory syndrome virus, porcine pseudorabies virus, porcine parvovirus, classical swine fever virus, and porcine epidemic diarrhea virus were used as templates in the established PCV2/3 duplex real-time PCR. As shown in Fig. 2, the duplex real-time PCR assay was able to detect and differentiate between PCV2 and PCV3 independently and simultaneously. In contrast, the other swine pathogens were not detected, demonstrating the high specificity of the established PCV2/3 duplex real-time PCR.
Fig. 2.
Specificity of the PCV2/3 duplex real-time PCR. A Specificity of the duplex real-time PCR for PCV2. B Specificity of the duplex real-time PCR for PCV3. C Specificity of the duplex real-time PCR for PCV2 and PCV3. The DNAs or cDNAs of swine viruses were used as templates. Other pathogens included PCV1, porcine reproductive and respiratory syndrome virus, porcine pseudorabies virus, porcine parvovirus, classical swine fever virus, and porcine epidemic diarrhea virus.
Sensitivity and Reproducibility of the PCV2/3 Duplex Real-Time PCR
The sensitivity of the PCV2/3 duplex real-time PCR was tested using serial tenfold dilutions of the constructed pMD19-PCV2 and pMD19-PCV3 plasmids. As shown in Fig. 3, the detection limits of the duplex real-time PCR were 2.9 copies for PCV2 and 22.5 copies for PCV3. The reproducibility of the assay was determined by testing three concentrations of plasmids five different times. The intra-assay CVs of 107, 105, and 103 copy numbers of PCV2 were 0.31%, 0.44%, and 0.41%, and the respective inter-assay CVs were 1.33%, 1.52%, and 1.14%. For PCV3, the intra-assay CVs of 107, 105, and 103 copy numbers were 0.53%, 0.48%, and 0.67%, and the respective inter-assay CVs were 1.69%, 1.47%, and 1.54%.
Fig. 3.
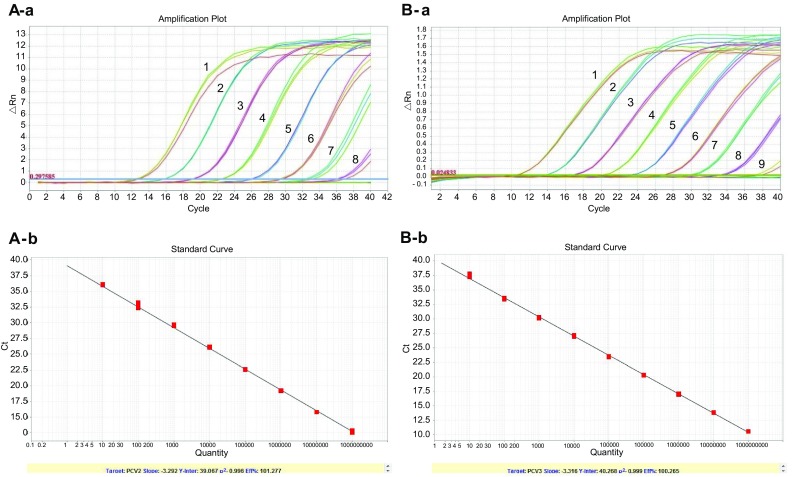
Sensitivity of the PCV2/3 duplex real-time PCR. A Sensitivity of the duplex real-time PCR for PCV2. (1–8): 103–1010 dilutions of pMD19-PCV2 plasmid were used as the templates for real-time PCR. B Sensitivity of the duplex real-time PCR for PCV3. (1–9): 101–109 dilutions of pMD19-PCV3 plasmid were used as the templates for real-time PCR.
Application of the PCV2/3 Duplex Real-Time PCR to Clinical Samples
The established PCV2/3 duplex real-time PCR was applied to detect PCV2 and PCV3 DNAs in 340 clinical samples. The results showed that 68.2% of clinical samples (232/340) were PCV2 PCR positive, 44.4% of clinical samples (151/340) were PCV3 PCR positive, and 27.6% of clinical samples (94/340) were both PCV2 and PCV3 PCR positive (Table 1). The positive PCR products were confirmed by gene sequencing (Genewiz, Suzhou, China). Among the different types of tissues, the highest frequency of PCV2-positive clinical tissues was found in tonsil samples, followed by lung and lymph node samples. In contrast, the highest frequency of PCV3-positive clinical tissues was found in lung samples, followed by lymph node and tonsil samples (Table 1).
Table 1.
Summary of PCV2/3 detection in 340 clinical samples using the established real-time PCR.
| Samples | PCV2+ | PCV3+ | PCV2+ PCV3+ |
|---|---|---|---|
| Lung | 75/98 (76.5%) | 66/98 (67.3%) | 34/98 (34.7%) |
| Lymph node | 93/162 (57.4%) | 75/162 (46.3%) | 51/162 (31.5%) |
| Tonsil | 64/80 (80.0%) | 10/80 (12.5%) | 9/80 (11.3%) |
| Total | 232/340 (68.2%) | 151/340 (44.4%) | 94/340 (27.6%) |
Discussion
PCV3 has been reported to be prevalent in the United States, Poland, South Korea, and China since 2016 (Ku et al. 2017; Kwon et al. 2017; Palinski et al. 2017; Stadejek et al. 2017). Clinically, porcine dermatitis, nephropathy syndrome and reproductive failure, and cardiac and multisystemic inflammation have been reported to be associated with PCV3 infection (Ku et al. 2017; Phan et al. 2016). Therefore, it is imperative to conduct PCV3 epidemiological surveys to evaluate its impact on the swine industry. PCV2 has been frequently found to be co-infected with PCV3 in clinical samples (Ku et al. 2017). In previous reports, PCV2 and PCV3 were detected separately using individual conventional PCR or real-time PCR (Wang et al. 2017). It is more practical to develop a duplex real-time PCR method to detect and differentiate between PCV2 and PCV3 DNAs in clinical samples simultaneously. Therefore, we, for the first time, developed such a duplex PCR and applied it to clinical samples.
In the present study, the established PCV2/3 duplex real-time PCR showed high specificity and sensitivity but low intra- and inter-assay CVs, which indicated that the assay can provide accurate and producible detection. Furthermore, the duplex real-time PCR can be utilized as a single PCV2 or PCV3 assay or combined into a duplex assay without impacting the quality of the results, making it more adaptable to field samples. Similar to PCV2, PCV3 was reported to mainly infect macrophages in lymphoid tissues (Palinski et al. 2017). Therefore, 340 clinical samples were then used to detect PCV2 and PCV3 DNAs using the established duplex real-time PCR. Consistent with previous reports, the co-infection rate of PCV2 and PCV3 in these clinical samples reached 27.6%, which indicated that PCV3 may play a role in PCV2-associated diseases (Wang et al. 2017). The above results were consistent with a previous report in which 22.3% of 76 clinical PCV3-positive samples were found to be co-infected with PCV2 (Fu et al. 2018). To date, all efforts to isolate PCV3 from primary cells and cell lines have failed. Therefore, successful virus isolation from cells will be the top priority for future studies on PCV3.
To conclude, a duplex real-time PCR method for the detection of PCV2 and PCV3 was established in this study. The assay was proved to be sensitive, specific, reliable, and feasible for clinical samples. This newly developed assay represents a useful tool for the clinical diagnosis of PCV2 and PCV3 infections.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by Grants from the National Key Research and Development Program (2016YFD0500703), Major Science and Technology Projects in Henan Province (171100110200), and Luoyang HeLuo Talent Plan (Dr. Kegong Tian).
Author Contributions
MS and KGT designed the study. XDL and MMQ performed the experiments and wrote the manuscript.
Conflict of interest
The authors declare that they do not have any conflict of interest.
Animal and Human Rights Statement
All animal trials were performed in strict accordance with the regulations in the guide for the care and use of laboratory animals of the National Research Center for Veterinary Medicine.
Contributor Information
Ming Sun, Phone: +86-10-82898320, Email: sunming@anheal.com.
Kegong Tian, Phone: +86-379-60687971, Email: tiankg@263.net.
References
- Fu X, Fang B, Ma J, Liu Y, Bu D, Zhou P, Wang H, Jia K, Zhang G. Insights into the epidemic characteristics and evolutionary history of the novel porcine circovirus type 3 in southern China. Transbound Emerg Dis. 2018;65:e296–e303. doi: 10.1111/tbed.12752. [DOI] [PubMed] [Google Scholar]
- Ku X, Chen F, Li P, Wang Y, Yu X, Fan S, Qian P, Wu M, He Q. Identification and genetic characterization of porcine circovirus type 3 in China. Transbound Emerg Dis. 2017;64:703–708. doi: 10.1111/tbed.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon T, Yoo SJ, Park CK, Lyoo YS. Prevalence of novel porcine circovirus 3 in Korean pig populations. Vet Microbiol. 2017;207:178–180. doi: 10.1016/j.vetmic.2017.06.013. [DOI] [PubMed] [Google Scholar]
- Palinski R, Pineyro P, Shang P, Yuan F, Guo R, Fang Y, Byers E, Hause BM. A novel porcine circovirus distantly related to known circoviruses is associated with porcine dermatitis and nephropathy syndrome and reproductive failure. J Virol. 2017;91:e01879-16. doi: 10.1128/JVI.01879-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan TG, Giannitti F, Rossow S, Marthaler D, Knutson TP, Li L, Deng X, Resende T, Vannucci F, Delwart E. Detection of a novel circovirus PCV3 in pigs with cardiac and multi-systemic inflammation. Virol J. 2016;13:184. doi: 10.1186/s12985-016-0642-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Liu X, Zhang P, Wang L, Liu Y, Zhang L, Liang P, Song C. Genome characterization of a porcine circovirus type 3 in South China. Transbound Emerg Dis. 2017;65:264–266. doi: 10.1111/tbed.12639. [DOI] [PubMed] [Google Scholar]
- Stadejek T, Wozniak A, Milek D, Biernacka K. First detection of porcine circovirus type 3 on commercial pig farms in Poland. Transbound Emerg Dis. 2017;64:1350–1353. doi: 10.1111/tbed.12672. [DOI] [PubMed] [Google Scholar]
- Wang J, Zhang Y, Liu L, Pang X, Yuan W. Development of a TaqMan-based real-time PCR assay for the specific detection of porcine circovirus 3. J Virol Methods. 2017;248:177–180. doi: 10.1016/j.jviromet.2017.07.007. [DOI] [PubMed] [Google Scholar]
- Yun JJ, Heisler LE, Hwang II, Wilkins O, Lau SK, Hyrcza M, Jayabalasingham B, Jin J, McLaurin J, Tsao MS, Der SD. Genomic DNA functions as a universal external standard in quantitative real-time PCR. Nucleic Acids Res. 2006;34:e85. doi: 10.1093/nar/gkl400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai SL, Chen SN, Xu ZH, Tang MH, Wang FG, Li XJ, Sun BB, Deng SF, Hu J, Lv DH, Wen XH, Yuan J, Luo ML, Wei WK. Porcine circovirus type 2 in China: an update on and insights to its prevalence and control. Virol J. 2014;11:88. doi: 10.1186/1743-422X-11-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.