Summary
Centrioles account for centrosomes and cilia formation. Recently, a link between centrosomal components and human developmental disorders has been established. However, the exact mechanisms how centrosome abnormalities influence embryogenesis and cell fate are not understood. PLK4-STIL module represents a key element of centrosome duplication cycle. We analyzed consequences of inactivation of the module for early events of embryogenesis in human embryonic stem cells (hESCs) and human induced pluripotent stem cells (hiPSCs). We demonstrate that blocking of PLK4 or STIL functions leads to centrosome loss followed by both p53-dependent and -independent defects, including prolonged cell divisions, upregulation of p53, chromosome instability, and, importantly, reduction of pluripotency markers and induction of differentiation. We show that the observed loss of key stem cells properties is connected to alterations in mitotic timing and protein turnover. In sum, our data define a link between centrosome, its regulators, and the control of pluripotency and differentiation in PSCs.
Keywords: centrosome, centriole, stem cell, differentiation, self-renewal, cell cycle, pluripotency, acentrosomal
Graphical Abstract
Highlights
-
•
Blocking of PLK4-STIL module in hESCs/hiPSCs leads to:
-
•
Centrosome loss, prolonged and error-prone mitosis;
-
•
p53-dependent differentiation;
-
•
Reduction of pluripotency linked to altered protein turnover
Recently a link has been established between centrosome and developmental disorders, yet the mechanisms connecting centrosome and cell fate are not understood. Cajanek and colleagues analyzed consequences of centrosome loss using hESCs/hiPSCs. They demonstrated that PLK4/STIL inhibition-mediated centrosome removal leads to loss of key stem cell properties connected to alterations in protein turnover and mitotic timing, which triggers p53-dependent differentiation.
Introduction
The centrosome, an organelle named by Theodor Boveri at the end of the 19th century, has been studied for a long time, but its functions and mechanisms of regulation are still incompletely understood. The centrosome typically acts as a microtubule organizing center (MTOC), taking part in cell division, cell shape organization, and cell motility (Conduit et al., 2015, Khodjakov and Rieder, 2001, Piel et al., 2001). Its core consists of two centrioles, microtubule-based structures with nine-fold radial symmetry, embedded in a protein matrix termed pericentriolar material (Bornens and Gönczy, 2014, Nigg and Stearns, 2011).
The centrosome duplicates once per cell cycle. As a cell divides, each daughter cell inherits one centrosome, so its number in the cells remains stable, similar to DNA content (Bornens and Gönczy, 2014, Nigg and Stearns, 2011). To date, hundreds of centrosomal proteins participating in centrosome biogenesis have been identified (Andersen et al., 2003, Gupta et al., 2015), with PLK4-STIL module having a pivotal role in the orchestration of centriole duplication (Arquint and Nigg, 2016, Bettencourt-Dias et al., 2005, Habedanck et al., 2005, Tang et al., 2011).
Overexpression of essential centrosome regulators, including PLK4, leads to centrosome amplification, whereas their depletion causes loss of centrosomes (Bazzi and Anderson, 2014, Bettencourt-Dias et al., 2005, Habedanck et al., 2005, Leidel et al., 2005, Strnad et al., 2007, Tang et al., 2011). Deregulation of the centrosome duplication cycle is implicated in the etiology of various disorders such as ciliopathies, microcephaly, primordial dwarfism, and cancer (Chavali et al., 2014, Gambarotto and Basto, 2016, Gönczy, 2015, Nigg et al., 2014). However, the consequences of centrosome abnormalities for cell fate have started to be revealed only recently. Inhibition of PLK4 depletes centrioles in various human somatic cell lines, leading to p53-dependent G1 arrest (Lambrus et al., 2015, Wong et al., 2015). In contrast, in vivo study using Drosophila demonstrated that centrosomes are not required for a substantial part of fly embryogenesis (Basto et al., 2006). The requirement for correct embryo development has been further addressed in mice. Mouse embryos without centrosomes die during gestation (Bazzi and Anderson, 2014, Hudson et al., 2001, Izraeli et al., 1999), and amplification of centrosomes after PLK4 overexpression in developing mouse brain leads to microcephaly-like phenotype (Marthiens et al., 2013). That being said, it is becoming clear that cellular outcomes of centrosome abnormalities differ between different models and perhaps even specific cell types (Basto et al., 2008, Levine et al., 2017, Marthiens et al., 2013, Vitre et al., 2015).
Human pluripotent stem cells (PSCs) encompassing both human embryonic stem cells (hESCs) and human induced pluripotent stem cells (hiPSCs) are able to self-renew and to differentiate into all cell types in the human body (Takahashi et al., 2007, Thomson et al., 1998). Pluripotency, governed by a network of transcription factors including OCT-4, SOX-2, and NANOG (Jaenisch and Young, 2008, Kashyap et al., 2009), is tightly connected to cell-cycle regulation (Becker et al., 2006, Pauklin and Vallier, 2013). Importantly, hESCs/hiPSCs hold great promise to model both physiological and pathophysiological aspects of human embryogenesis (Lancaster et al., 2013, Park et al., 2008, Shahbazi et al., 2016). Noteworthy, early passages of human PSCs seem prone to centrosome abnormalities (Brevini et al., 2009, Holubcová et al., 2011). Given these unique properties, we elected to investigate the consequences of halted centrosome duplication cycle in early embryonic events using hESCs and hiPSCs.
Here, we present our analyses of molecular and functional consequences of the inactivation of PLK4-STIL module and centrosome loss for human PSCs. We show that upon centrosome loss, the cells are in principle still able to undergo cell division. Such acentrosomal mitosis is twice as long and leads to mitotic errors and p53 stabilization, which is reflected by gradual loss of self-renewal potential. Interestingly, the observed p53 increase does not lead to significant apoptosis, but to loss of pluripotency and induction of differentiation. Finally, our data demonstrate that the loss of pluripotency regulators after PLK4 inhibition is p53-independent and linked to altered protein turnover.
Results
Blocking of PLK4 or STIL Leads to Centrosome Loss Followed by Decreased Proliferation of Stem Cells
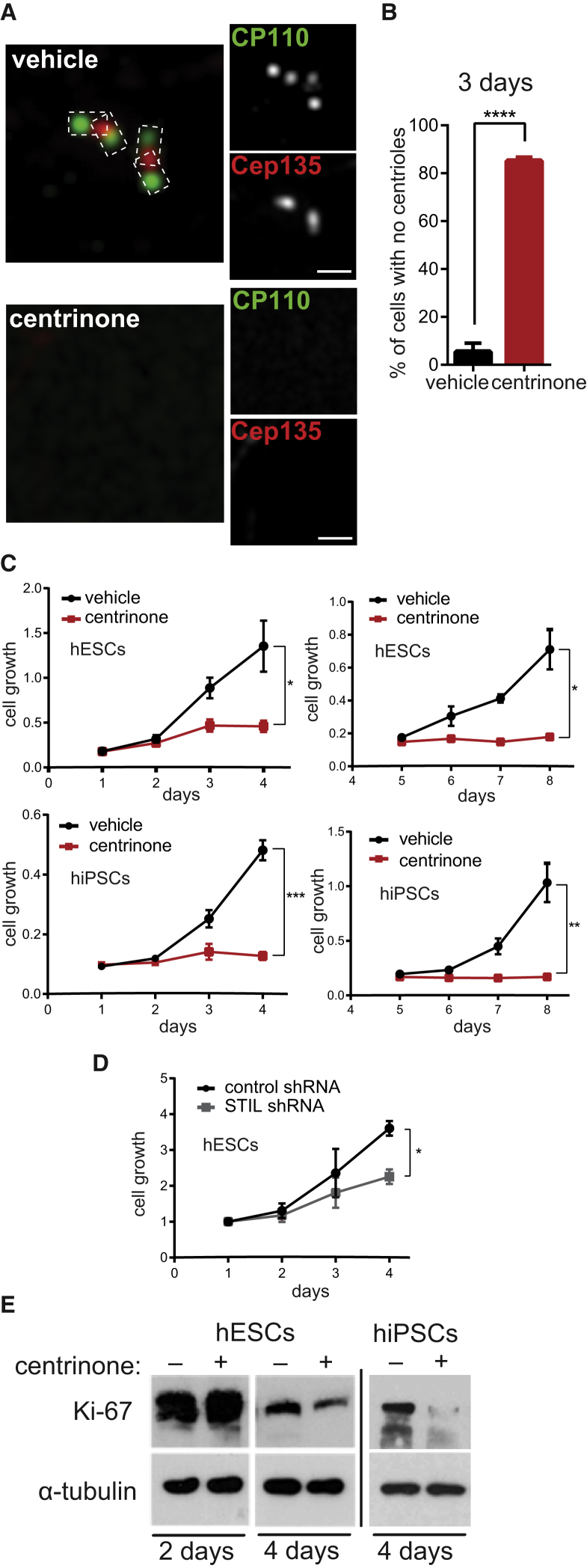
To assess the role of centrosomes in PSCs we used a PLK4 inhibitor, centrinone (Wong et al., 2015). First, we examined the efficacy of centrosome depletion in hESCs following treatment with centrinone. Using immunofluorescence staining for proximal centriolar marker Cep135 (Kleylein-Sohn et al., 2007) and distal centriolar marker CP110 (Chen et al., 2002), we detected the loss of centrosomes in about 40% of hESCs after 2 days (Figures S1A and S1B), and after 3 days the centrosome was depleted in almost 85% of hESCs (Figures 1A and 1B). We were also able to deplete centrosomes in hESCs using PLK4 or STIL short hairpin RNA (shRNA) (Figures S1C and S1D).
Figure 1.
Blocking of PLK4 or STIL Leads to Centrosome Loss Followed by Decreased Proliferation of Stem Cells
(A and B) Immunofluorescence (A) of 3-day vehicle- and centrinone-treated hESCs: centrosomes were visualized by antibody staining of distal marker CP110 (green) and proximal marker Cep135 (red). Scale bars, 1 μm. (B) Quantification of centrosome depletion, N > 150.
(C and D) Growth curves: cell number was measured at indicated time points by crystal violet assay, in vehicle- and centrinone-treated cells (C) or after STIL shRNA transfection (D).
(E) Western blot analyses of Ki-67 expression in 4-day vehicle- and centrinone-treated cells, with α-tubulin as a loading control.
Data are presented as mean ± SEM (∗p < 0.05, ∗∗p < 0.005, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001). See also Figure S1.
It has been recently demonstrated that the loss of centrosomes is detrimental for proliferation of non-transformed human somatic cells, but has little effect on cancer cells (Fong et al., 2016, Lambrus et al., 2015, Meitinger et al., 2016, Mikule et al., 2007, Wong et al., 2015). Given reported similarities in cycle control between embryonic stem cells and cancer cells (Kim et al., 2010), we examined consequences of centrosome depletion for PSC proliferation. Intriguingly, centrinone-treated hESCs/hiPSCs showed impaired proliferation from day 2 and virtually halted their growth past day 5 (Figure 1C). In addition, we also observed a negative effect on proliferation of hESCs following STIL knockdown (Figure 1D). Noteworthy, the negative effect of centrosome loss on proliferation was even more pronounced in the case of hESC-derived neural stem cells (NSCs) (Figure S1E). On the other hand, centrinone treatment showed only a minor effect on proliferation of U2OS cells (Figure S1F), in agreement with the previous report (Wong et al., 2015), even though the efficiency of centrosome depletion was comparable with that of hESCs (Figure S1G).
To corroborate this result, we examined the expression of Ki-67, a marker of proliferating cells. As shown in Figure 1E, centrinone treatment reduced expression levels of Ki-67. In addition, a decrease in the number of Ki-67+ cells was detected in the centrinone condition also by immunofluorescence (Figure S1H, quantified in Figure S1I).
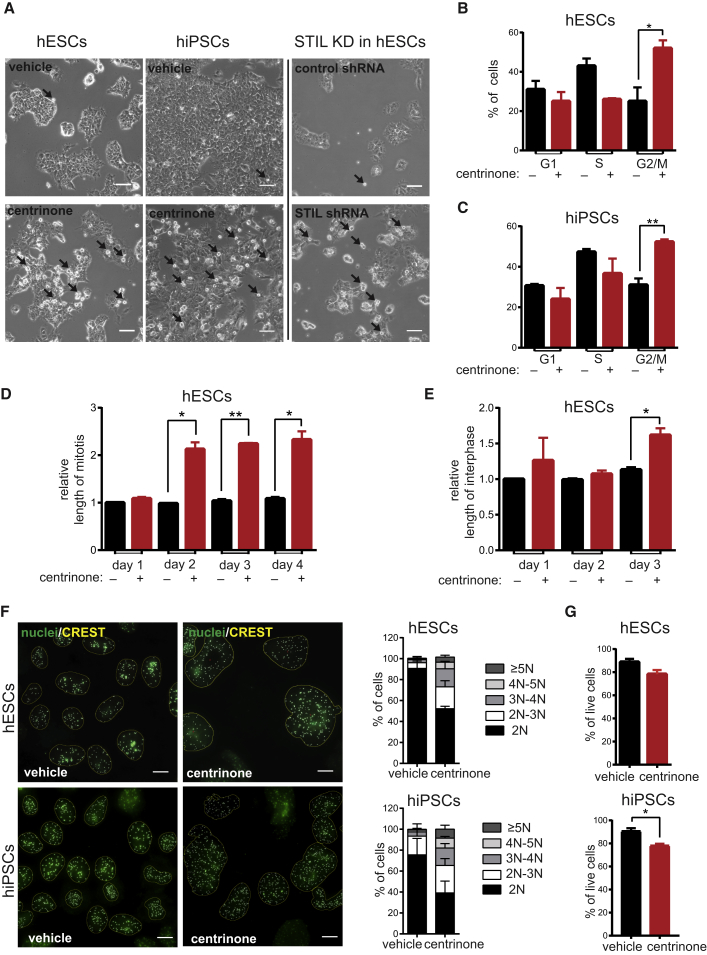
Centrosome Depletion Following PLK4 or STIL Blocking Leads to Prolonged Mitosis and Mitotic Defects
Centrosome loss has been reported to cause various mitotic defects in somatic cell lines (Sir et al., 2013, Wong et al., 2015). Indeed, we noted accumulation of rounded cells in the centrinone-treated cultures and following STIL knockdown (Figure 2A). Furthermore, our subsequent fluorescence-activated cell sorting (FACS) analysis proved that centrinone treatment leads to accumulation of hESCs/hiPSCs in G2/M phase (Figures 2B and 2C).
Figure 2.
Centrosome Depletion Following PLK4 or STIL Blocking Leads to Prolonged Mitosis and Mitotic Defects
(A) Phase-contrast images of 2-day vehicle- and centrinone-treated hESCs and hiPSCs or 2 days after STIL shRNA transfection. Arrows indicate mitotic cells. Scale bars, 50 μm.
(B–E) Cell-cycle distribution of 3-day vehicle- and centrinone-treated hESCs (B) or hiPSCs (C) analyzed by FACS. Measurement of relative length of mitosis (D) or interphase (E) by live imaging of H2A-GFP hESCs after indicated time of treatment. Data are normalized to the vehicle treatment condition on day 1 (n = 2, N > 40).
(F) Immunofluorescence analyses of centromere number in 4-day vehicle- or centrinone-treated hESCs and hiPSCs. Centromeres were visualized by CREST staining (yellow), nuclei were counterstained by Hoechst (green). Scale bars, 10 μm. Panels on the right show centromere quantification and corresponding intervals of chromosome numbers (n = 2, N > 90).
(G) Quantification of viability measurement by annexin V/PI staining in 2-day vehicle- and centrinone-treated hESCs and hiPSCs.
Data are presented as mean ± SEM (∗p < 0.05, ∗∗p < 0.005). See also Figure S2.
Next, we analyzed the length of mitosis by live imaging of the reporter H2A-GFP line derived from the same paternal hESC line. As shown in Figure 2D, completion of mitosis between days 2 and 4 took for the treated cells approximately twice as long as controls. In addition, centrinone-treated hESCs showed 1.5-fold prolonged interphase on day 3 compared with control (Figure 2E). All these data indicated an intriguing possibility that centrosome-less hESCs are viable and able to divide, even though for a limited time for the latter. In agreement with this hypothesis we found bipolar mitotic spindles even in acentrosomal cells (Figure S2A). In addition, we quantified the number of cells successfully finishing mitosis in our live imaging experiments. We focused on mitoses past the third day of centrinone treatment, when the majority of treated cells already lacks centrosomes (Figures 1A and 1B). Interestingly, we found 68.1% ± 1.9% of cells able to successfully go through mitosis within the 30-hr period we examined. This observation suggested that acentrosomal mitoses seem possible, but also confirmed our earlier observation (Figure 1C) that proliferation after centrosome loss is inefficient (note 1.5 times longer interphase of centrinone-treated cells; Figure 2E). In addition, we observed cytokinesis failure in approximately 15% of divisions (Figure S2D). To fully prove that acentrosomal hESCs can divide, we performed live imaging experiments with γ-tubulin-GFP hESCs following centrinone treatment (Figure S2G). To conclude, these data argue that centrosome-depleted hESCs are in principle able to successfully finish mitotic division and give rise to two daughter cells, albeit only for a limited time.
In the course of our experiments we noted that nuclei of centrinone-treated cells became bigger and acquired morphology different from control. In agreement with this, FACS analysis detected a modest increase of aneuploid cells after 3 days of centrinone treatment (Figures S2B and S2C). Since it is not possible to distinguish diploid cells residing in G2/M phase from tetraploid cells residing in G1 phase (Figures 2B and 2C) using this approach, it prompted us to quantify the chromosome number. The analysis was done at day 4, when the changes in cell morphology observed during live imaging were most pronounced. Previous work indicated that while centrosome loss during mouse embryogenesis does not lead to notable aneuploidy (Bazzi and Anderson, 2014), somatic cell lines show an increase in chromosomal abnormalities after the centrosome loss (Sir et al., 2013, Wong et al., 2015). Interestingly, our analyses revealed that centrinone treatment of hESCs/hiPSCs led to changes in chromosome number (Figure 2F), arguing that centrosome loss promotes genome instability in PSCs.
Next, to elucidate the survival potential of centrinone-treated cells, we assessed the number of early and late apoptotic cells by annexin V and propidium iodide (PI) staining. We found a modest difference in the number of viable (annexin V/PI negative) cells between centrinone condition and control (Figure 2G). Intriguingly, the proportion of apoptotic cells was notably elevated in hESC-derived NSCs following the centrinone treatment, in contrast to similarly treated cultures of hESCs/hiPSCs (Figure S2E). In addition, we compared the effects of centrinone with those of etoposide, a commonly used DNA-damage-inducing agent. Interestingly, while etoposide triggered a pronounced increase of apoptotic cells in hESC/hiPSC cultures, the percentage of apoptotic cells in centrinone-treated NSCs was similar to the NSC etoposide condition (Figure S2F).
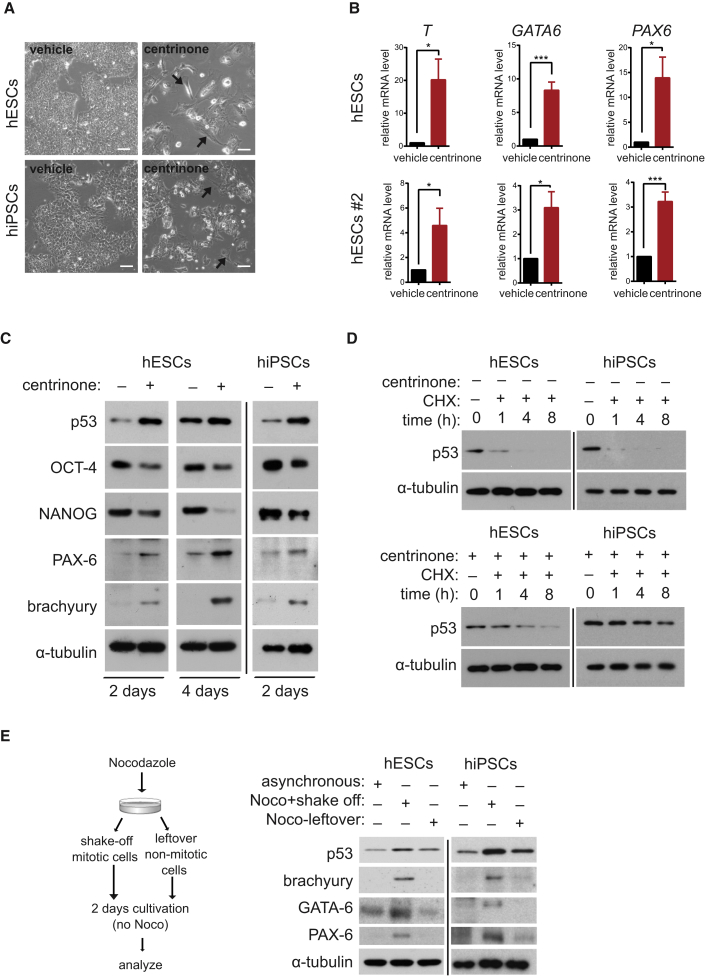
Blocking PLK4 or STIL Promotes Stem Cell Differentiation
Key aspects of PSC biology, the ability to self-renew and to differentiate, are intimately connected to cell-cycle regulation (Becker et al., 2006, Pauklin and Vallier, 2013). Given the phenotypes we found, we examined the impact of centrosome depletion after blocking PLK4 or STIL on those two features.
First, we observed that centrinone-treated cells lost typical stem cell morphology (Figure 3A), suggesting that centrosome loss affects stem cell differentiation. In agreement with this observation, we found a defect in polymerization of microtubules in centrosome-depleted hESCs (Figure S3A). Next, we examined expression of differentiation makers: ectodermal marker PAX-6, endodermal marker GATA-6, and mesodermal marker brachyury. Indeed, mRNA levels of all examined markers were upregulated after centrinone treatment (Figure 3B). Similar effects were confirmed also on the protein level (Figures 3C and S3B). Importantly, our analyses further revealed that protein levels of pluripotency markers OCT-4 and NANOG were decreased in centrinone-treated cells (Figures 3C and S3B). In addition, we detected higher protein levels of p53 in the centrinone conditions, thus confirming and extending previous observations on centrosome loss in somatic cells and mouse embryos (Bazzi and Anderson, 2014, Insolera et al., 2014, Lambrus et al., 2015, Mikule et al., 2007, Wong et al., 2015). Similar effects were also observed after PLK4/STIL shRNA (Figures S3C–S3E). Of note, we did not find a correlation between levels of aneuploidy and brachyury expression (Figures S3F and S3G).
Figure 3.
Blocking PLK4 or STIL Promotes Stem Cell Differentiation
(A) Phase-contrast images of hESCs or hiPSCs after 8 or 4 days of treatment, respectively. Arrows point to observed morphological changes. Scale bars, 50 μm.
(B) Analyses of mRNA levels of T, GATA6, and PAX6 in hESCs and hESCs #2 after 4 days of treatment. Data are presented as relative fold change over control.
(C–E) Western blot analyses of hESCs and hiPSCs after indicated time of treatment, with α-tubulin as a loading control. (C) Analyses of effects on pluripotency and differentiation by the indicated antibodies. (D) Analyses of effects of treatment (2 days) on protein turnover of p53 after indicated time (hours) of inhibition of translation by cycloheximide (CHX). (E) Analyses of the effect of temporal mitotic arrest by 6 hr of nocodazole treatment. Left panel shows scheme of the experiment. Controls (asynchronous cells) and treated samples (Noco+shake off, Noco-leftover) were probed for protein levels of p53, brachyury, GATA-6, and PAX-6 2 days after nocodazole washout. Noco-leftover condition represents non-mitotic nocodazole-treated cells.
Data are presented as mean ± SEM (∗p < 0.05, ∗∗∗p < 0.001). See also Figure S3.
Next, we examined centrinone effects in relation to those of retinoic acid (RA), a commonly used differentiation agent. Intriguingly, upregulation of p53 after centrosome loss was even higher than the effect of RA (Figure S3I). Furthermore, combination of centrinone and RA treatments enhanced PAX-6 protein levels, if compared with either RA treatment alone or untreated condition (Figure S3H).
In the course of our experiments we noted a temporal increase of Ser139 phosphorylated H2AX, a hallmark of DNA-damage response (DDR) (Rogakou et al., 1998) (Figure S3J). Given that DDR typically acts upstream of p53 activation (Brooks and Gu, 2010), we examined a possible role for DDR kinases in the observed increase of p53 levels. However, inhibition of ATM, ATR, and DNA-PK did not prevent the increase (Figure S3K). These data suggest that accumulation of p53 after PLK4 or STIL blocking-induced centrosome loss in hESCs is independent of DDR signaling.
Given this result, we considered alternative routes of p53 upregulation. First, we found that knockdown of dicer-1, a key regulator of microRNA biogenesis, does not prevent p53 upregulation (Figure S3L), indicating that microRNA machinery is not involved in the p53 activation. Next, using cycloheximide (CHX) to block translation, we found that p53 protein moiety is stabilized following centrinone treatment (Figure 3D). Furthermore, we examined effect of prolonged mitosis on p53 stabilization. To mimic the effect of centrosome depletion on the mitotic length, we treated hESCs for 2 hr (Figure S3M) or hESCs/hiPSCs for 6 hr by nocodazole to arrest them in mitosis, isolated mitotic cells by shake-off, then released them by washout and analyzed them 2 days following release. Remarkably, we found upregulated p53, brachyury, and GATA-6 in cells that experienced temporal mitotic arrest (Figure 3E). Of note, we occasionally observed an increase also in PAX-6 levels (Figure 3E), perhaps reflecting requirement of concomitant downregulation of OCT4/NANOG for the efficient PAX-6 induction (see Figure S4B). Importantly, cells treated with nocodazole, but not passing through mitotic arrest (“Noco-leftover”), showed expression of examined markers at levels comparable with that of control. Thus, the data demonstrate that prolonged mitosis is sufficient to trigger p53 upregulation and cell differentiation.
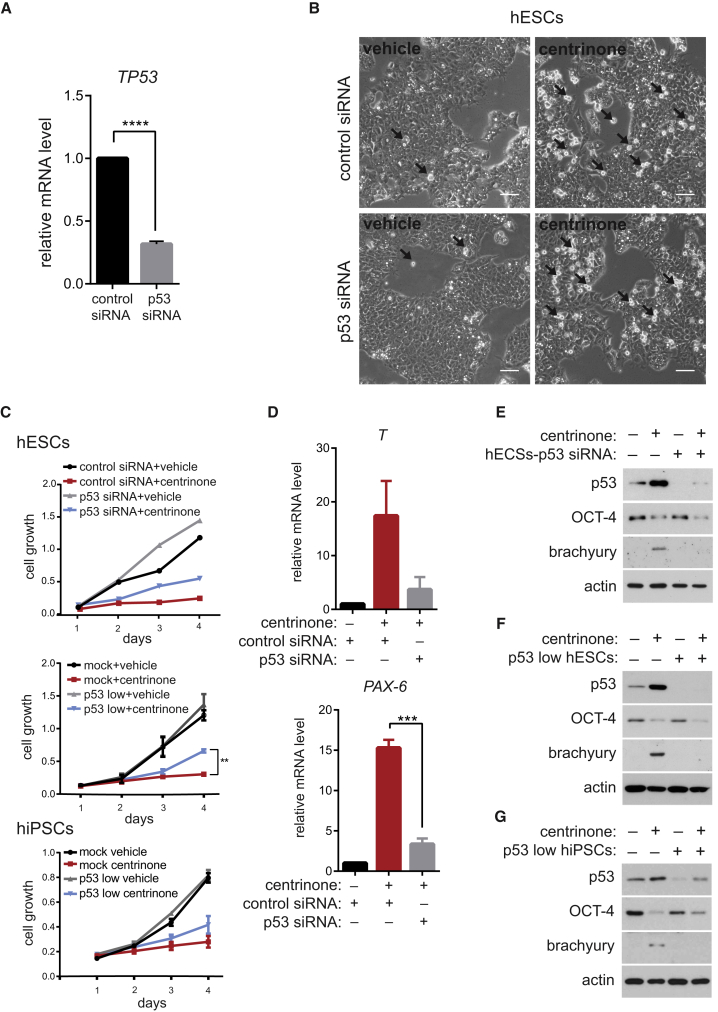
Differentiation Induced by Blocking of PLK4 or STIL Is p53 Dependent
Previous studies linked p53 to induction of differentiation in PSCs (Jain et al., 2012, Lin et al., 2005, Qin et al., 2007, Zhang et al., 2014). On the other hand, there are contradictory reports about the expression and/or activity of p53 in hESCs/hiPSCs (Aladjem et al., 1998, Maimets et al., 2008, Momcilovic et al., 2009, Qin et al., 2007, Wang et al., 2016, Zhang et al., 2014). To examine the role of p53 in the differentiation observed in our experiments, we first downregulated it using small interfering RNA (siRNA) (Figure 4A). We found that the knockdown of p53 has no apparent effect on mitotic cell accumulation following centrinone treatment (Figure 4B). Importantly, downregulation of p53 expression by either siRNA in hESC cultures or CRISPR/Cas9 system (p53 low hESCs/hiPSCs) only partially rescued the proliferation defect seen after centrosome loss (Figure 4C). Thus, these results not only confirmed and extended previous observations from somatic cells (Fong et al., 2016, Lambrus et al., 2016, Meitinger et al., 2016, Wong et al., 2015), but also suggested the interesting possibility that some of the phenotypes caused by centrinone treatment in hESCs/hiPSCs might be p53 independent. To this end, we examined the requirement of p53 for centrosome loss-induced differentiation of PSCs. We found that depletion of p53 prevented the upregulation of T and PAX6 mRNA in hESCs (Figure 4D). Furthermore, centrinone treatment led to higher protein levels of brachyury in control condition, but not in cells with depleted p53 by siRNA (Figure 4E). Intriguingly, expression of OCT-4 was not rescued by p53 depletion. To corroborate this finding, we performed a similar experiment using p53 low hESCs/hiPSCs (Figures 4F and 4G). As expected, we found full dependency of the induction of brachyury expression on the presence of p53. Importantly, however, OCT-4 protein was downregulated even in conditions without detectable levels of p53 (Figure 4F). Thus, these data demonstrated that the induction of differentiation markers after centrinone-mediated centrosome loss requires p53, while the loss of pluripotency markers is p53 independent.
Figure 4.
Differentiation Induced by Blocking of PLK4 or STIL Is p53 Dependent
Cells were transfected with either control or p53 siRNA, or the expression of p53 was permanently downregulated by CRISPR/Cas9 (p53 low cells) and subsequently treated as indicated.
(A) Analyses of mRNA levels of TP53 after siRNA transfection in hESCs, showing the efficiency of p53 knockdown. Data are presented as relative fold change over control.
(B) Phase-contrast images of hESCs following siRNA transfection and 2 days of treatment; black arrows indicate mitotic cells. Scale bars, 50 μm.
(C) Number of cells in described conditions was measured at indicated time points by crystal violet assay and plotted as growth curves. First panel shows siRNA data (n = 1), second panel shows analyses of p53 low hESCs and their respective controls (n = 3), and third panel shows p53 low hiPSCs (n = 3).
(D) Expression of differentiation markers (T and PAX6) after siRNA transfection and 4 days of treatment in hESCs, analyzed by qPCR. Data are presented as relative fold change over control (first column).
(E–G) Western blot analyses of rescue of the centrinone treatment-induced effects by p53 downregulation either by siRNA (2 days of treatment, E) or CRISPR/Cas9 (p53 low hESCs/hiPSCs; 3 days of treatment, F and G). Samples were probed with indicated antibodies, with actin as a loading control.
Data are presented as mean ± SEM (∗∗p < 0.005, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001).
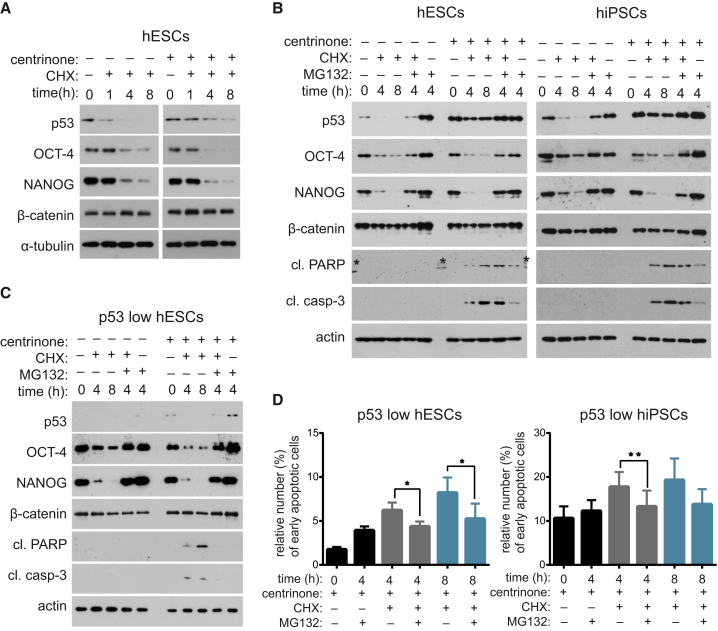
Loss of Pluripotency after PLK4 Inhibition and Centrosome Depletion Is Linked to Altered Protein Turnover
To elucidate the mechanism responsible for the downregulation of regulators of pluripotency after centrinone treatment, we first analyzed its effects on POU5F1 and NANOG mRNA levels. Surprisingly, we found no difference in either POU5F1 or NANOG mRNA levels (Figure S4A). This result indicated post-transcriptional regulation and prompted us to examine protein stability of OCT-4/NANOG. Interestingly, we found increased turnover of OCT-4/NANOG in the centrinone-treated hESCs (Figure 5A). Given that levels of p53 and β-catenin, included as controls in our experiments, did not follow the same trend as OCT-4/NANOG, these data confirmed the specificity of the effect. Next, we examined whether prolonged mitosis, induced by nocodazole, is sufficient to alter turnover of OCT-4/NANOG. However, in contrast to the effects seen on upregulation of p53 and other differentiation markers (Figure 3E), prolonged mitosis did not show an effect on OCT-4/NANOG turnover (Figure S4B).
Figure 5.
Loss of Pluripotency after PLK4 Inhibition and Centrosome Depletion Is Linked to Altered Protein Turnover
hESCs (mock: A and B, or p53 low: C and D) and hiPSCs (mock: B, or p53 low: D) were treated with centrinone (2 days) and indicated chemicals, and analyzed by western blot for protein expression (A–C) or by annexin V/PI staining for apoptosis (D).
(A) Western blot analyses of centrinone treatment effect on protein turnover after block of protein synthesis for indicated time by cycloheximide (CHX). Note the increased turnover of OCT-4 and NANOG, and the decreased turnover of p53 in centrinone conditions. β-Catenin was included in all depicted experiments as additional control for specificity; α-tubulin/actin served as loading controls.
(B and C) Analysis of rescue effects of inhibition of proteasome (MG132) on altered protein turnover following centrinone treatment (B). Where indicated, CHX was added together with MG132 for indicated time to analyze turnover rate of p53, OCT-4, NANOG, and β-catenin. Cleaved PARP and cleaved caspase-3 were used to probe for apoptosis (asterisks show non-specific antibody binding to marker). (C) Analysis of p53 low hESCs.
(D) Viability measurement by annexin V/PI staining of p53 low hESCs/hiPSCs in the indicated conditions (hESCs, n = 4; hiPSCs, n = 3).
Data are presented as mean ± SEM (∗p < 0.05, ∗∗p < 0.005). See also Figure S4.
To identify the degradation pathway responsible for turnover of OCT-4/NANOG, we used MG132 and chloroquine, inhibitors of proteasome and lysosome, respectively. While the treatment with chloroquine showed no effect (Figure S4C), addition of MG132 to CHX-treated cells showed a rescue effect on the drop in protein levels of OCT-4 and NANOG, indicating that these transcription factors are subjected to proteasomal degradation in both control and centrinone-treated cells (Figures 5B and S4C). As PLK4 was shown to regulate protein turnover of SAS-6 (Puklowski et al., 2011), we examined its possible direct role in turnover of OCT-4. First we tested that washing out centrinone is sufficient for reactivation of PLK4 in hESCs (Figures S4D–S4G), in agreement with previous reports on somatic cells (Lambrus et al., 2015, Wong et al., 2015). Importantly, however, restoring PLK4 activity in centrosome-depleted cells did not show any rescue effect on OCT-4 (Figure S4H).
We observed that the centrinone-treated cells were somewhat more sensitive, as they showed increased levels of cleaved poly(ADP ribose) polymerase (PARP) and cleaved caspase-3 (Figure 5B). Importantly, this phenomenon was triggered by the CHX treatment, as hESCs/hiPSCs treated only with centrinone showed no upregulation of apoptotic markers. Given these results, we hypothesized that the observed stress response of centrinone-treated cells to CHX treatment might reflect the accelerated loss of specific proteins in these cells. To this end, we aimed to test the causality between the observed priming of centrinone-treated stem cells to enter the apoptotic pathway and altered protein turnover. However, MG132 treatment, in agreement with its typical use in anti-tumor therapy (Goldberg, 2012), led to an increase of p53 and cell-death markers in hESCs/hiPSCs, indicated by the appearance of cleaved PARP and caspase-3 (Figure 5B). To bypass these undesired effects of MG132, we turned to p53 low hESCs. Interestingly, co-treatment of centrosome-depleted and CHX-treated hESCs with MG132 showed full rescue of the drop in OCT-4/NANOG levels (Figure 5C). Moreover, CHX treatment caused modest effects on cleaved PARP and caspase-3, respectively. Interestingly, those were fully rescued by the MG132 treatment. To corroborate our hypothesis, we tested the ability of MG132 to rescue the activation of the apoptotic pathway after centrosome loss in p53 low hESCs/hiPSCs. As expected, centrinone treatment alone led to only a negligible fraction of early apoptotic cells, while co-treatment with CHX increased this fraction (Figure 5D). Remarkably, addition of MG132 was able to notably decrease the proportion of early apoptotic cells, specifically in centrinone + CHX conditions. Together, these results demonstrated that p53 is not necessary for the increased protein turnover of OCT-4/NANOG in centrinone-treated cells, and that block of excessive protein degradation in centrosome-depleted cells is sufficient to lower the stress response and priming of these cells to apoptosis.
Discussion
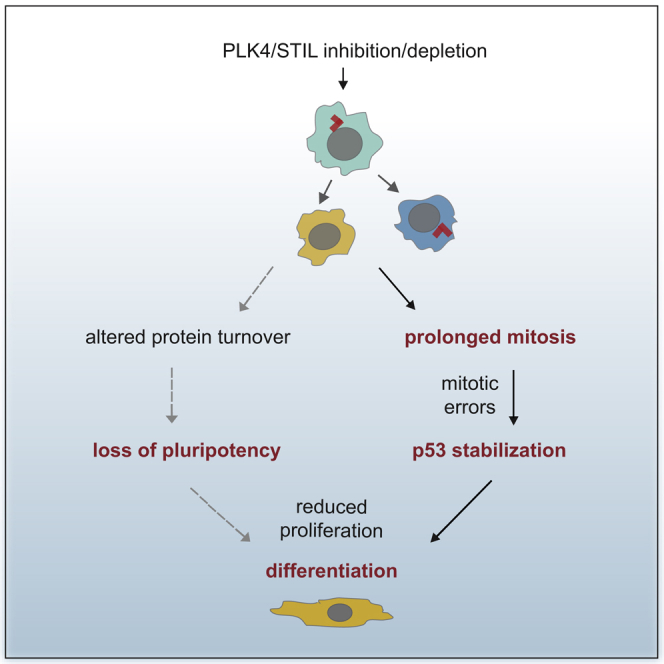
Centrosome abnormalities are related to detrimental developmental defects. Here we have explored the link between the loss of centrosome and the cell fate in hESCs/hiPSCs and have shown that depletion/inhibition of PLK4 or depletion of STIL lead to centrosome depletion, and in turn to prolonged mitosis, which consequently leads to p53 upregulation and subsequent differentiation. We further established the PLK4 inhibition-mediated and/or centrosome depletion-mediated loss of pluripotency independent of p53 and linked to altered protein turnover.
Our data indicate that a large portion of acentrosomal hESCs/hiPSCs is able to divide. Importantly, our experiments further showed that centrosome loss promotes aneuploidy in hESCs/hiPSCs, a phenomenon usually seen in somatic/cancer cell lines but not in vivo in mouse embryos (Bazzi and Anderson, 2014, Insolera et al., 2014, Sir et al., 2013, Wong et al., 2015). It remains to be determined whether this “mouse embryo versus human cells” difference reflects specific aspects of cell culture or is linked to an acentrosomal period of early embryogenesis in mouse (Szollosi et al., 1972). Either way, our data establish that the loss of centrosome in hESCs/hiPSCs contributes to genome instability.
The proliferation rate of centrosome-less hESCs/hiPSCs was impaired, consistent with reports on human somatic cells (Lambrus et al., 2015, Wong et al., 2015) or mouse embryo (Bazzi and Anderson, 2014). Studies on somatic cells also proposed that the proliferation defect seen after centrosome loss is fully dependent on p53 (Lambrus et al., 2015, Wong et al., 2015). Interestingly, however, depletion of p53 by RNAi or CRISPR/Cas9 showed only moderate rescue of proliferation defect after centrosome loss, while it completely prevented the induction of differentiation markers. Even though we cannot formally exclude the effects of different experimental designs, we conclude that self-renewal defect in centrosome-depleted hESCs/hiPSCs following PLK4 or STIL blocking is dependent on p53 only partially.
Previous studies linked p53 to induction of differentiation in PSCs (Jain et al., 2012, Lin et al., 2005, Qin et al., 2007, Zhang et al., 2014). However, the ability of activated p53 to directly repress transcription of any gene has been recently challenged (Allen et al., 2014), and the possible role of p53 in direct repression of pluripotency factors is rather controversial (Aladjem et al., 1998, Maimets et al., 2008, Momcilovic et al., 2009, Qin et al., 2007, Wang et al., 2016, Zhang et al., 2014). In addition, a recent report by Gogendeau et al. (2015) postulated that aneuploidy-induced differentiation of NSCs in Drosophila is largely p53-independent. With all that said, our data clearly point out the requirement of p53 for the induction of differentiation markers after inactivation of PLK4-STIL module and centrosome loss. Importantly, however, loss of pluripotency markers upon centrinone treatment was not rescued by p53 depletion, suggesting that the loss of pluripotency is p53-independent. This is in agreement with recent reports on Nanog expression during differentiation of p53 null mouse ESCs (Shigeta et al., 2013, Wang et al., 2016) and work on p53-deficient mice reporting no developmental defects (Donehower et al., 1992). Furthermore, this model predicts that the control of pluripotency and the induction of differentiation following centrosome depletion are interconnected but autonomously regulated phenomena, with p53 playing an instructive role in the latter. That being said, we speculate that the involvement of p53 in centrosome loss-driven differentiation explains why we and others (Amps et al., 2011, Ben-David et al., 2014, Taapken et al., 2011, Zhang et al., 2016) find no evidence for aneuploidy being the main driving force of upregulation of differentiation markers, as reported for p53-independent differentiation of Drosophila NSCs (Gogendeau et al., 2015).
Activation of p53 is a typical response to DDR. However, observations from us (this study) and others (Lambrus et al., 2015, Wong et al., 2015) demonstrate that the activation of p53 after centrosome loss is DDR-independent. Recent studies provided hints about events upstream of p53 activation in centrosome-less somatic cells by pointing out the requirement for TP53BP1 and USP28 (Fong et al., 2016, Lambrus et al., 2016, Meitinger et al., 2016). It is possible that this module operates also in PSCs. However, the exact nature of the putative stress signal activating p53 remains elusive. Our data indicate that the prolonged mitosis, one of the earliest consequences of centrosome loss, is sufficient to trigger p53 upregulation and differentiation in hESCs/hiPSCs. This finding has two pertinent consequences. First, any experiments with PSCs involving mitotic drugs need to be interpreted with caution, as such treatment may directly interfere with their undifferentiated status. Furthermore, it raises a question about the role of prolonged mitoses following centrosome loss. As already mentioned, prolonged mitosis is sufficient to trigger differentiation via induction of p53. However, removal of p53 is not able to sustain self-renewal of centrosome-depleted hESCs/hiPSCs. In addition, we were not able to mimic the effect of PLK4 or STIL blocking on downregulation of OCT-4/NANOG by the prolongation of mitosis. Thus, it seems plausible that defects observed in hESCs/hiPSCs following inactivation of the PLK4-STIL module reflect impairment of both mitotic and non-mitotic function of the centrosome.
We found that the decrease of OCT-4/NANOG after centrinone treatment is caused by faster turnover of these proteins, independently of p53. The fact that active PLK4 is not able to rescue downregulation of OCT-4 in centrosome-depleted cells supports the conclusion that the enhanced turnover is a consequence of centrosome loss rather than inhibition of PLK4. We speculate that altered proteasomal activity, in combination with elevated stress response, might contribute to this phenomenon (Bryja et al., 2017, Gerdes et al., 2007, Vora and Phillips, 2015). In addition, the increased degradation of NANOG/OCT-4 could be a consequence of more complex metabolic changes. This prediction is supported by the rescue effect of MG132 treatment on induction of apoptosis in our experiments, possibly due to prevention of a loss of pro-survival factors upon block of protein synthesis (Portt et al., 2011).
In sum, our study defines a novel role for PLK4-STIL module and the centrosome in the regulation of key stem cell properties. It identifies both p53-dependent and -independent consequences of inactivation of the module in PSCs and connects them to alterations in mitotic timing and protein metabolism. Future studies on the links between centrosome, proteasome regulation, and apoptotic response could contribute to a better understanding of the pathology of centrosome-related diseases.
Experimental Procedures
Cell Lines
hESCs (line CCTL14, https://hpscreg.eu/cell-line/MUNIe007-A), hESCs 2 (CCTL12, https://hpscreg.eu/cell-line/MUNIe005-A; “hESCs #2”) (Adewumi et al., 2007, Bohaciakova et al., 2017), and hiPSCs (derived as described previously; Barta et al., 2016) were cultured, treated, and analyzed as described in detail in Supplemental Experimental Procedures.
Statistical Analysis
All statistical analyses were done using Student's t test and graphically visualized in GraphPad Prism Software v. 6.0 (GraphPad Software, La Jolla, CA; www.graphpad.com). All data are presented as mean ± SEM from three independent experiments, unless otherwise stated, and p values <005 were considered significant (∗p < 0.05, ∗∗p < 0.005, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 in figures).
Western Blots
Detailed protocol is described in Supplemental Experimental Procedures; western blot quantifications and all independent repeats are shown in Supplemental Western Blot Data.
Additional experimental procedures are provided in Supplemental Information. A list of used antibodies, primers, and shRNA constructs is provided in Table S1.
Authors Contributions
T.R. designed and performed experiments, and analyzed and interpreted data; D.B. performed experiments, and analyzed and interpreted data; M.E. and V.P. performed experiments and analyzed data; T.B. and A.H. provided critical reagents; L.C. conceived and supervised the study, analyzed and interpreted data, and together with T.R. wrote the manuscript. All authors approved the final version of the manuscript.
Acknowledgments
We thank Erich Nigg, Andrew Shiau, Borivoj Vojtesek, and Anthony Hyman for sharing reagents, Lumir Krejci for access to DV Elite, Pavlina Janovska for help with FACS analyses, Klara Koudelkova and Karolina Hanzakova for assistance, Vitezslav Bryja, Zdenek Andrysik, and members of the L.C. lab for critical comments and suggestions. We acknowledge the core facility CELLIM of CEITEC, supported by the MEYS CR (LM2015062 Czech-BioImaging). This work was supported by grants from the SoMoPro II Program (project 4SGA8574), cofinanced by European Union and the South-Moravian Region; Czech Science Foundation (16-03269Y); Swiss National Science Foundation (IZ11Z0_166533); follow-up research fund from Federation of Biochemical and Biophysical Societies (FEBS); Grant Agency of Masaryk University (GAMU grant category E), and funds from Medical Faculty MU to Junior researcher (ROZV/24/LF2016) to L.C. D.B. and T.B. were supported by the Czech Science Foundation (15-18316Y, 16-24004Y, and 18-25429Y), and funds from Medical Faculty MU to Junior researcher (ROZV/25/LF/2017, ROZV/24/LF/2018 and MUNI/G/1131/2017). A.H. was supported by Czech Science Foundation (15-11707S) and by the project no. LQ1605 (MYES CR, NPU II). The content of this publication reflects only the authors' views, and the European Union is not liable for any use that may be made of the information contained therein.
Published: September 6, 2018
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, four figures, and one table and can be found with this article online at https://doi.org/10.1016/j.stemcr.2018.08.008.
Supplemental Information
References
- Adewumi O., Aflatoonian B., Ahrlund-Richter L., Amit M., Andrews P.W., Beighton G., Bello P.A., Benvenisty N., Berry L.S., Bevan S. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat. Biotechnol. 2007;25:803–816. doi: 10.1038/nbt1318. [DOI] [PubMed] [Google Scholar]
- Aladjem M.I., Spike B.T., Rodewald L.W., Hope T.J., Klemm M., Jaenisch R., Wahl G.M. ES cells do not activate p53-dependent stress responses and undergo p53- independent apoptosis in response to DNA damage. Curr. Biol. 1998;8:145–155. doi: 10.1016/s0960-9822(98)70061-2. [DOI] [PubMed] [Google Scholar]
- Allen M.A., Andrysik Z., Dengler V.L., Mellert H.S., Guarnieri A., Freeman J.A., Sullivan K.D., Galbraith M.D., Luo X., Kraus W.L. Global analysis of p53-regulated transcription identifies its direct targets and unexpected regulatory mechanisms. Elife. 2014;3 doi: 10.7554/eLife.02200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amps K., Andrews P.W., Anyfantis G., Armstrong L., Avery S., Baharvand H., Baker J., Baker D., Munoz M.B., Beil S. Screening ethnically diverse human embryonic stem cells identifies a chromosome 20 minimal amplicon conferring growth advantage. Nat. Biotechnol. 2011;29:1132–1146. doi: 10.1038/nbt.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen J.S., Wilkinson C.J., Mayor T., Mortensen P., Nigg E.A., Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- Arquint C., Nigg E.A. The PLK4-STIL-SAS-6 module at the core of centriole duplication. Biochem. Soc. Trans. 2016;44:1253–1263. doi: 10.1042/BST20160116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barta T., Peskova L., Collin J., Montaner D., Neganova I., Armstrong L., Lako M. Inhibition of miR-145 enhances reprogramming of human dermal fibroblasts to induced pluripotent stem cells. Stem Cells. 2016;34:246–251. doi: 10.1002/stem.2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basto R., Lau J., Vinogradova T., Gardiol A., Woods C.G., Khodjakov A., Raff J.W. Flies without centrioles. Cell. 2006;125:1375–1386. doi: 10.1016/j.cell.2006.05.025. [DOI] [PubMed] [Google Scholar]
- Basto R., Brunk K., Vinadogrova T., Peel N., Franz A., Khodjakov A., Raff J.W. Centrosome amplification can initiate tumorigenesis in flies. Cell. 2008;133:1032–1042. doi: 10.1016/j.cell.2008.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzi H., Anderson K.V. Acentriolar mitosis activates a p53-dependent apoptosis pathway in the mouse embryo. Proc. Natl. Acad. Sci. USA. 2014;111:E1491–E1500. doi: 10.1073/pnas.1400568111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker K.A., Ghule P.N., Therrien J.A., Lian J.B., Stein J.L., Van Wijnen A.J., Stein G.S. Self-renewal of human embryonic stem cells is supported by a shortened G1 cell cycle phase. J. Cell. Physiol. 2006;209:883–893. doi: 10.1002/jcp.20776. [DOI] [PubMed] [Google Scholar]
- Ben-David U., Arad G., Weissbein U., Mandefro B., Maimon A., Golan-lev T., Narwani K., Clark A.T., Andrews P.W., Benvenisty N. Aneuploidy induces profound changes in gene expression, proliferation and tumorigenicity of human pluripotent stem cells. Nat. Commun. 2014;5:4825. doi: 10.1038/ncomms5825. [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias M., Rodrigues-Martins A., Carpenter L., Riparbelli M., Lehmann L., Gatt M.K., Carmo N., Balloux F., Callaini G., Glover D.M. SAK/PLK4 is required for centriole duplication and flagella development. Curr. Biol. 2005;15:2199–2207. doi: 10.1016/j.cub.2005.11.042. [DOI] [PubMed] [Google Scholar]
- Bohaciakova D., Renzova T., Fedorova V., Barak M., Kunova-Bosakova M., Hampl A., Cajanek L. An efficient method for generation of knockout human. Stem Cells Dev. 2017;26:1521–1527. doi: 10.1089/scd.2017.0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornens M., Gönczy P. Centrosomes back in the limelight. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014;369 doi: 10.1098/rstb.2013.0452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brevini T.A.L., Pennarossa G., Antonini S., Paffoni A., Rebulla P., Scanziani E., De Eguileor M., Benvenisty N. Cell lines derived from human parthenogenetic embryos can display aberrant centriole distribution and altered expression levels of mitotic spindle check-point transcripts. Stem Cell Rev. Rep. 2009;5:340–352. doi: 10.1007/s12015-009-9086-9. [DOI] [PubMed] [Google Scholar]
- Brooks C.L., Gu W. New insights into p53 activation. Cell Res. 2010;20:614–621. doi: 10.1038/cr.2010.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryja V., Červenka I., Čajánek L. The connections of Wnt pathway components with cell cycle and centrosome: side effects or a hidden logic ? Crit. Rev. Biochem. Mol. Biol. 2017;52:614–637. doi: 10.1080/10409238.2017.1350135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavali P.L., Pütz M., Gergely F. Small organelle, big responsibility: the role of centrosomes in development and disease. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014;369 doi: 10.1098/rstb.2013.0468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Indjeian V.B., McManus M., Wang L., Dynlacht B.D. CP110, a cell cycle-dependent CDK substrate, regulates centrosome duplication in human cells. Dev. Cell. 2002;3:339–350. doi: 10.1016/s1534-5807(02)00258-7. [DOI] [PubMed] [Google Scholar]
- Conduit P.T., Wainman A., Raff J.W. Centrosome function and assembly in animal cells. Nat. Rev. Mol. Cell Biol. 2015;16:611–624. doi: 10.1038/nrm4062. [DOI] [PubMed] [Google Scholar]
- Donehower L.A., Harvey M., Slagle B.L., McArthur M.J., Montgomery C.A., Butel J.S., Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- Fong C.S., Mazo G., Das T., Goodman J., Kim M., O’Rourke B.P., Izquierdo D., Tsou M.F.B. 53BP1 and USP28 mediate p53-dependent cell cycle arrest in response to centrosome loss and prolonged mitosis. Elife. 2016;5 doi: 10.7554/eLife.16270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambarotto D., Basto R. Consequences of numerical centrosome defects in development and disease. In: Lüders J., editor. The Microtubule Cytoskeleton: Organisation, Function and Role in Disease. Springer Vienna; 2016. pp. 117–149. [Google Scholar]
- Gerdes J.M., Liu Y., Zaghloul N.A., Leitch C.C., Lawson S.S., Kato M., Beachy P.A., Beales P.L., DeMartino G.N., Fisher S. Disruption of the basal body compromises proteasomal function and perturbs intracellular Wnt response. Nat. Genet. 2007;39:1350–1360. doi: 10.1038/ng.2007.12. [DOI] [PubMed] [Google Scholar]
- Gogendeau D., Siudeja K., Gambarotto D., Pennetier C., Bardin A.J., Basto R. Aneuploidy causes premature differentiation of neural and intestinal stem cells. Nat. Commun. 2015;6:8894. doi: 10.1038/ncomms9894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg A.L. Development of proteasome inhibitors as research tools and cancer drugs. J. Cell Biol. 2012;199:583–588. doi: 10.1083/jcb.201210077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönczy P. Centrosomes and cancer: revisiting a long-standing relationship. Nat. Rev. Cancer. 2015;15:639–652. doi: 10.1038/nrc3995. [DOI] [PubMed] [Google Scholar]
- Gupta G.D., Coyaud É., Gonçalves J., Mojarad B.A., Liu Y., Wu Q., Gheiratmand L., Comartin D., Tkach J.M., Sally W.T. A dynamic protein interaction landscape of the human centrosome-cilium interface. Cell. 2015;163:1484–1499. doi: 10.1016/j.cell.2015.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habedanck R., Stierhof Y.-D., Wilkinson C.J., Nigg E.A. The Polo kinase Plk4 functions in centriole duplication. Nat. Cell Biol. 2005;7:1140–1146. doi: 10.1038/ncb1320. [DOI] [PubMed] [Google Scholar]
- Holubcová Z., Matula P., Sedláčková M., Vinarský V., Doležalová D., Bárta T., Dvořák P., Hampl A. Human embryonic stem cells suffer from centrosomal amplification. Stem Cells. 2011;29:46–56. doi: 10.1002/stem.549. [DOI] [PubMed] [Google Scholar]
- Hudson J.W., Kozarova A., Cheung P., Macmillan J.C., Swallow C.J., Cross J.C., Dennis J.W. Late mitotic failure in mice lacking Sak, a polo-like kinase. Curr. Biol. 2001;11:441–446. doi: 10.1016/s0960-9822(01)00117-8. [DOI] [PubMed] [Google Scholar]
- Insolera R., Bazzi H., Shao W., Anderson K.V., Shi S.-H. Cortical neurogenesis in the absence of centrioles. Nat. Neurosci. 2014;17:1528–1535. doi: 10.1038/nn.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izraeli S., Lowe L.A., Bertness V.L., Good D.J., Dorward D.W., Kirsch I.R., Kuehn M.R. The SIL gene is required for mouse embryonic axial development and left – right specification. Nature. 1999;399:691–694. doi: 10.1038/21429. [DOI] [PubMed] [Google Scholar]
- Jaenisch R., Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132:567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A.K., Allton K., Iacovino M., Mahen E., Milczarek R.J., Zwaka T.P., Kyba M., Barton M.C. p53 regulates cell cycle and microRNAs to promote differentiation of human embryonic stem cells. PLoS Biol. 2012;10 doi: 10.1371/journal.pbio.1001268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashyap V., Rezende N.C., Scotland K.B., Shaffer S.M., Persson J.L., Gudas L.J., Mongan N.P. Regulation of stem cell pluripotency and differentiation involves a mutual regulatory circuit of the NANOG, OCT4, and SOX2 pluripotency transcription factors with polycomb repressive complexes and stem cell microRNAs. Stem Cells Dev. 2009;18:1093–1108. doi: 10.1089/scd.2009.0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A., Rieder C.L. Centrosomes enhance the fidelity of cytokinesis in vertebrates and are required for cell cycle progression. J. Cell Biol. 2001;153:237–242. doi: 10.1083/jcb.153.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Woo A.J., Chu J., Snow J.W., Fujiwara Y., Kim C.G., Cantor A.B., Orkin S.H. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143:313–324. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleylein-Sohn J., Westendorf J., Le Clech M., Habedanck R., Stierhof Y.-D., Nigg E.A. Plk4-induced centriole biogenesis in human cells. Dev. Cell. 2007;13:190–202. doi: 10.1016/j.devcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Lambrus B.G., Uetake Y., Clutario K.M., Daggubati V., Snyder M., Sluder G., Holland A.J. P53 protects against genome instability following centriole duplication failure. J. Cell Biol. 2015;210:63–77. doi: 10.1083/jcb.201502089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrus B.G., Daggubati V., Uetake Y., Scott P.M., Clutario K.M., Sluder G., Holland A.J. A USP28-53BP1-p53-p21 signaling axis arrests growth after centrosome loss or prolonged mitosis. J. Cell Biol. 2016;214:143–153. doi: 10.1083/jcb.201604054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster M.A., Renner M., Martin C.-A., Wenzel D., Bicknell L.S., Hurles M.E., Homfray T., Penninger J.M., Jackson A., Knoblich J.A. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidel S., Delattre M., Cerutti L., Baumer K., Gönczy P. SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat. Cell Biol. 2005;7:115–125. doi: 10.1038/ncb1220. [DOI] [PubMed] [Google Scholar]
- Levine M.S., Bakker B., Boeckx B., Moyett J., Lu J., Vitre B., Spierings D.C., Lansdorp P.M., Cleveland D.W., Lambrechts D. Centrosome amplification is sufficient to promote spontaneous tumorigenesis in mammals. Dev. Cell. 2017;40:313–322. doi: 10.1016/j.devcel.2016.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T., Chao C., Saito S., Mazur S.J., Murphy M.E., Appella E., Xu Y. p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat. Cell Biol. 2005;7:165–171. doi: 10.1038/ncb1211. [DOI] [PubMed] [Google Scholar]
- Maimets T., Neganova I., Armstrong L., Lako M. Activation of p53 by nutlin leads to rapid differentiation of human embryonic stem cells. Oncogene. 2008;27:5277–5287. doi: 10.1038/onc.2008.166. [DOI] [PubMed] [Google Scholar]
- Marthiens V., Rujano M.A., Pennetier C., Tessier S., Paul-Gilloteaux P., Basto R. Centrosome amplification causes microcephaly. Nat. Cell Biol. 2013;15:731–740. doi: 10.1038/ncb2746. [DOI] [PubMed] [Google Scholar]
- Meitinger F., Anzola J.V., Kaulich M., Richardson A., Stender J.D., Benner C., Glass C.K., Dowdy S.F., Desai A., Shiau A.K. 53BP1 and USP28 mediate p53 activation and G1 arrest after centrosome loss or extended mitotic duration. J. Cell Biol. 2016;214:155–166. doi: 10.1083/jcb.201604081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikule K., Delaval B., Kaldis P., Jurcyzk A., Hergert P., Doxsey S. Loss of centrosome integrity induces p38-p53-p21-dependent G1-S arrest. Nat. Cell Biol. 2007;9:160–170. doi: 10.1038/ncb1529. [DOI] [PubMed] [Google Scholar]
- Momcilovic O., Choi S., Varum S., Bakkenist C., Schatten G., Navara C. Ionizing radiation induces ATM dependent checkpoint signaling and G2 but not G1 cell cycle arrest in pluripotent human embryonic stem cells. Stem Cells. 2009;27:1822–1835. doi: 10.1002/stem.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg E.A., Stearns T. The centrosome cycle: centriole biogenesis, duplication and inherent asymmetries. Nat. Cell Biol. 2011;13:1154–1160. doi: 10.1038/ncb2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg E.A., Čajánek L., Arquint C. The centrosome duplication cycle in health and disease. FEBS Lett. 2014;588:2366–2372. doi: 10.1016/j.febslet.2014.06.030. [DOI] [PubMed] [Google Scholar]
- Park I.-H., Arora N., Huo H., Maherali N., Ahfeldt T., Shimamura A., Lensch M.W., Cowan C., Hochedlinger K., Daley G.Q. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauklin S., Vallier L. The cell cycle state of stem cells determines cell fate propensity. Cell. 2013;155:135–147. doi: 10.1016/j.cell.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piel M., Nordberg J., Euteneuer U., Bornens M. Centrosome-dependent exit of cytokinesis in animal cells. Science. 2001;291:1550–1554. doi: 10.1126/science.1057330. [DOI] [PubMed] [Google Scholar]
- Portt L., Norman G., Clapp C., Greenwood M., Greenwood M.T. Anti-apoptosis and cell survival: a review. Biochim. Biophys. Acta. 2011;1813:238–259. doi: 10.1016/j.bbamcr.2010.10.010. [DOI] [PubMed] [Google Scholar]
- Puklowski A., Homsi Y., Keller D., May M., Chauhan S., Kossatz U., Grünwald V., Kubicka S., Pich A., Manns M.P. The SCF-FBXW5 E3-ubiquitin ligase is regulated by PLK4 and targets HsSAS-6 to control centrosome duplication. Nat. Cell Biol. 2011;13:1004–1009. doi: 10.1038/ncb2282. [DOI] [PubMed] [Google Scholar]
- Qin H., Yu T., Qing T., Liu Y., Zhao Y., Cai J., Li J., Song Z., Qu X., Zhou P. Regulation of apoptosis and differentiation by p53 in human embryonic stem cells. J. Biol. Chem. 2007;282:5842–5852. doi: 10.1074/jbc.M610464200. [DOI] [PubMed] [Google Scholar]
- Rogakou E.P., Pilch D.R., Orr A.H., Ivanova V.S., Bonner W.M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139∗. J. Biol. Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- Shahbazi M.N., Jedrusik A., Vuoristo S., Recher G., Hupalowska A., Bolton V., Fogarty N.M.E., Campbell A., Devito L.G., Ilic D. Self-organization of the human embryo in the absence of maternal tissues. Nat. Cell Biol. 2016;18:700–708. doi: 10.1038/ncb3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigeta M., Ohtsuka S., Nishikawa-Torikai S., Yamane M., Fujii S., Murakami K., Niwa H. Maintenance of pluripotency in mouse ES cells without Trp53. Sci. Rep. 2013;3:2944. doi: 10.1038/srep02944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sir J.H., Pütz M., Daly O., Morrison C.G., Dunning M., Kilmartin J.V., Gergely F. Loss of centrioles causes chromosomal instability in vertebrate somatic cells. J. Cell Biol. 2013;203:747–756. doi: 10.1083/jcb.201309038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strnad P., Leidel S., Vinogradova T., Euteneuer U., Gönczy P. Regulated HsSAS-6 levels ensure formation of a single procentriole per centriole during the centrosome duplication cycle. Dev. Cell. 2007;13:203–213. doi: 10.1016/j.devcel.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szollosi D., Calarco P., Donahue R.P. Absence of centrioles in the first and second meiotic spindles of mouse oocytes. J. Cell Sci. 1972;11:521–541. doi: 10.1242/jcs.11.2.521. [DOI] [PubMed] [Google Scholar]
- Taapken S.M., Nisler B.S., Newton M.A., Sampsell-Barron T.L., Leonhard K.A., McIntire E.M., Montgomery K.D. Karyotypic abnormalities in human induced pluripotent stem cells and embryonic stem cells. Nat. Biotechnol. 2011;29:313–314. doi: 10.1038/nbt.1835. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Tang C.-J.C., Lin S.-Y., Hsu W.-B., Lin Y.-N., Wu C.-T., Lin Y.-C., Chang C.-W., Wu K.-S., Tang T.K. The human microcephaly protein STIL interacts with CPAP and is required for procentriole formation. EMBO J. 2011;30:4790–4804. doi: 10.1038/emboj.2011.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S., Jones J.M. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Vitre B., Holland A.J., Kulukian A., Shoshani O., Hirai M., Wang Y., Maldonado M., Cho T., Boubaker J., Swing D.A. Chronic centrosome amplification without tumorigenesis. Proc. Natl. Acad. Sci. USA. 2015;112:E6321–E6330. doi: 10.1073/pnas.1519388112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vora S., Phillips B.T. Centrosome-associated degradation limits β-catenin inheritance by daughter cells after asymmetric division. Curr. Biol. 2015;25:1005–1016. doi: 10.1016/j.cub.2015.02.020. [DOI] [PubMed] [Google Scholar]
- Wang Q., Zou Y., Nowotschin S., Kim S.Y., Li Q.V., Soh C.-L., Su J., Zhang C., Shu W., Xi Q. The p53 family coordinates Wnt and nodal inputs in mesendodermal differentiation of embryonic stem cells. Cell Stem Cell. 2016;20:70–86. doi: 10.1016/j.stem.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong Y.L., Anzola J.V., Davis R.L., Yoon M., Motamedi A., Kroll A., Seo C.P., Hsia J.E., Kim S.K., Mitchell J.W. Reversible centriole depletion with an inhibitor of Polo-like kinase 4. Science. 2015;348:1155–1160. doi: 10.1126/science.aaa5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Cheng L., Jia Y., Liu G., Li C., Song S., Bradley A. Aneuploid embryonic stem cells exhibit impaired differentiation and increased neoplastic potential. EMBO J. 2016;35:2285–2300. doi: 10.15252/embj.201593103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.N., Chung S.K., Xu Z., Xu Y. Oct4 maintains the pluripotency of human embryonic stem cells by inactivating p53 through sirt1-mediated deacetylation. Stem Cells. 2014;32:157–165. doi: 10.1002/stem.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.