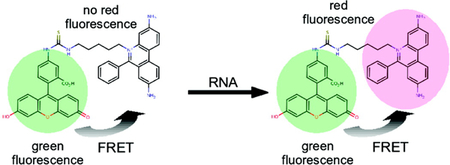
Graphical Abstract
Among the available techniques to detect and quantify the presence of duplex DNA or RNA, use of intercalating fluorophores has been widely employed due to the simplicity and versatility it affords.1–4 Though there are a number of intercalating fluorophores which are commercially available, phenanthridine derivatives such as ethidium bromide (EB) and propidium iodide (PI) are among the oldest and most often used as a result of the substantial increase in fluorescence intensity and lifetime that occurs upon intercalation.5–7 Moreover, the presence of an absorption band, albeit a weak one, in the visible region makes it possible to excite these fluorophores not only with UV light but also using the argon ion laser lines (488 nm, 514 nm).
The ability to use an argon ion laser for excitation is especially useful for cell imaging applications since it is often the laser source found in confocal imaging microscopy. The relatively weak absorption at the laser wavelength of phenanthridine derivatives, however, places such probes at a disadvantage since ultimately the fluorescent signal that is detected depends on the amount of light absorbed. An obvious way to improve the performance of these probes is to increase their ability to absorb visible light.
We report on the synthesis and spectral properties of a phenanthridine derivative containing a covalently linked fluorescein molecule that acts as an energy transfer donor (Figure 1). For simplicity, this molecule is abbreviated as FLEth to signify the simultaneous presence of both a fluorescein moiety and an ethidium bromide-like intercalator. Using a combination of spectroscopic and microscopic imaging techniques, we demonstrate that FLEth exhibits photophysical properties of both fluorophores and further-more is superior to propidium iodide in terms of signal intensity for the detection of RNA under visible light excitation. Also, since the FLEth molecule retains the fluorescent properties of the fluorescein donor, then it can simultaneously be used as both a whole cell marker and a probe for duplex RNA in cellular imaging.
Figure 1.
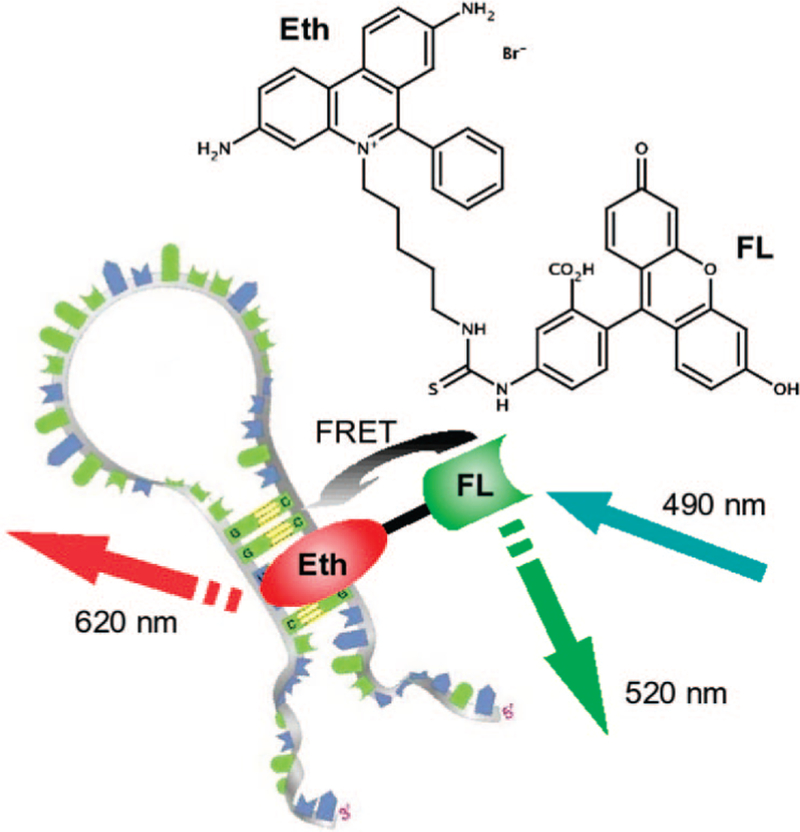
Chemical structure of FLEth and cartoon depicting the energy transfer process from fluorescein to the intercalated phenanthridine fluorophore.
The primary reasons for selecting fluorescein as the donor fluorophore are for its strong absorption in the visible, its high quantum yield of fluorescence (QY), and more importantly its established fluorescence resonance energy transfer (FRET) with PI:8 the energy transfer efficiency from the FL to Eth moiety is found to be about 77%. From the latter result, it is clear that the majority of photons absorbed by fluorescein are efficiently transferred to the intercalated fluorophore. Additionally, given that the transfer efficiency is not a 100%, FLEth remains measurably fluorescent even in the absence of duplex RNA.
The fluorescence signal of FLEth due to unquenched fluorescein is shown in Figure 2, which compares the spectra in the absence and presence of Turolla yeast RNA. The spectra were also compared in the absence and presence of an 18 base pair dsDNA. In the presence of dsDNA, there was only a slight difference in the shape and intensity of the fluorescence spectrum, suggesting a low binding affinity toward dsDNA (Supporting Information). On the other hand, the spectral change in the presence of yeast RNA is markedly different. In the absence of RNA, only the emission band of fluorescein is present, with a peak at 520 nm, while in the presence of yeast RNA, the fluorescein emission band remains, but now there is new band with a peak at 610 nm corresponding to the phenanthridine fluorophore. When the two spectra are compared, there is over a 9-fold increase in the fluorescence intensity at 610 nm, suggesting a stronger binding affinity of FLEth to duplex RNA than DNA. However, further studies need to be done to better understand this finding.
Figure 2.
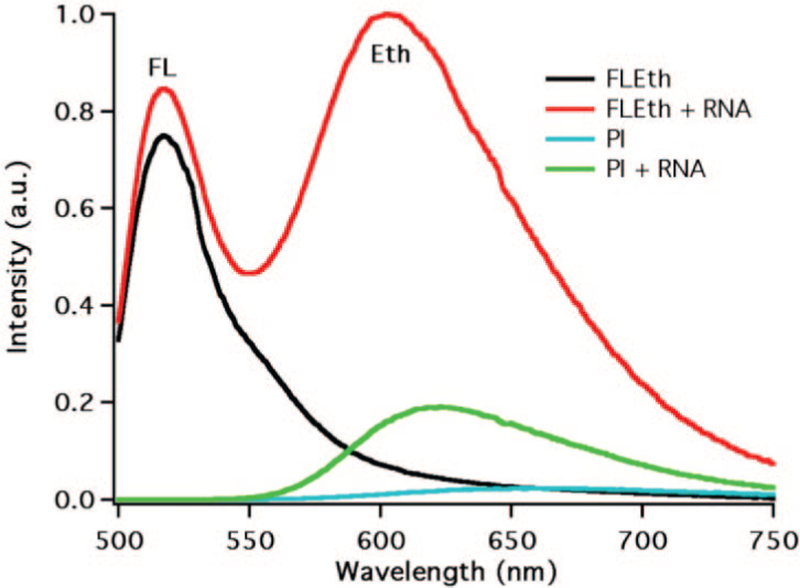
Fluorescence spectra comparison of the response of FLEth and PI to yeast RNA.
From Figure 2, the fluorescence intensity of FLEth can be compared to that of PI under the same experimental conditions. What is immediately apparent is just how much brighter FLEth is in the presence of RNA than is PI in the presence of RNA. The intensity of FLEth is approximately 5× greater at 610 nm, which effectively means that use of FLEth results in better signal to background (S/B) ratio. The fluorescence lifetime of the two probes was also compared. When RNA was bound, the lifetime of PI was measured to be 18 ± 0.1 ns while FLEth exhibited a two-component lifetime of 3 ± 0.1 ns (50%) and 20 ± 0.1 ns (50%) monitored at 620 nm. The shorter-lived species is from the residual emission of fluorescein at 620 nm, while the 20 ns is due to the bound phenanthridine fluorophore.
The relatively long-lived fluorescence lifetime of the FLEth probe also allows for time-resolved detection experiments to be performed. The ability to use this detection method is especially useful for monitoring probes in complex biological solutions, such as cell growth medium (CGM), which have native fluorescence that overlaps with that of the monitoring probe.9–12 In CGM, the steadystate S/B ratio of FLEth on addition of RNA is 7. Using the time-resolved method, which monitors the signal 15 ns after excitation, the S/B ratio is increased to 40 (see Figure 3). Such a dramatic increase in the S/B ratio occurs because, in the time window monitored, the emission signals from the CGM and fluorescein have essentially totally decayed.
Figure 3.
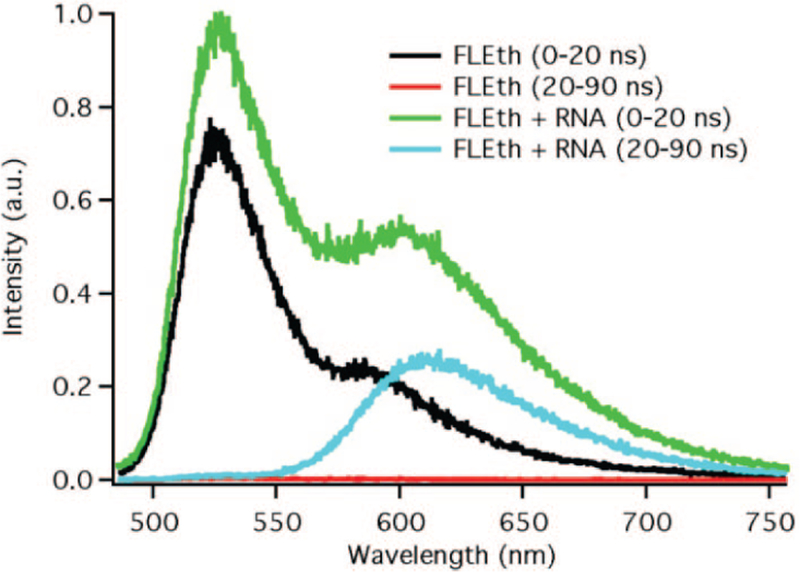
Time-resolved fluorescence spectra comparison of the response of FLEth to yeast RNA in cell growth medium at various time delays.
The combination of the dual color (green and red) emission and greater affinity for duplex RNA indicates that FLEth is a candidate as a probe for the in vivo imaging of RNA. To test this possibility, FLEth uptake into mammalian breast cancer cells was evaluated by employing a fluorescence microscope. Figure 4a shows an image obtained by detecting the emission of fluorescein, and it shows that the probe is distributed throughout the cells. In a sense, FLEth is serving as a whole cell marker. When the emission from the phenanthridine fluorophore is monitored for imaging, as done in Figure 4b, features which are not so apparent in Figure 4a become enhanced. Since FLEth appears to have a higher affinity for RNA, the regions that exhibit the greatest intensity are expected be those rich in RNA. The image shows that the probe’s intensity is less in the nucleus, which contains DNA, than it is in the surrounding cytoplasm. Furthermore, in the nucleolus, a dense region of RNA in the nucleus, the probe’s intensity is at a maximum.
Figure 4.
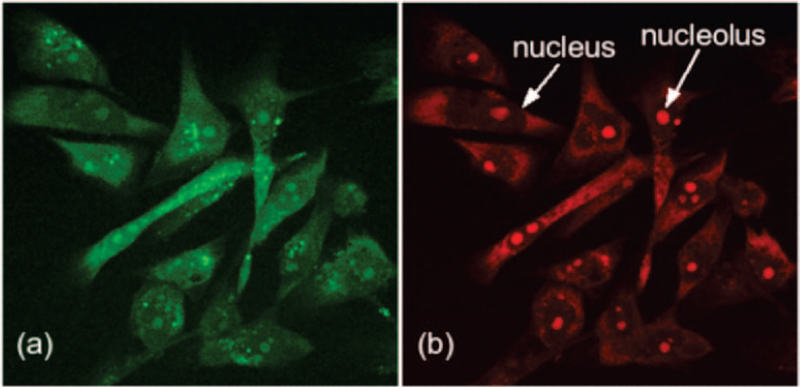
Fluorescence microscope images of FLEth uptake into mammalian cells. The dual color emission allows FLEth to be monitored at both the green channel (a) and red channel (b).
In conclusion, by simply linking fluorescein to a phenanthridine fluorophore to produce FLEth, we were able to create an RNA intercalator whose photophysical properties make it superior to other phenanthridine derivatives for the fluorescence detection of RNA using visible light excitation. It undergoes a 9-fold increase in signal intensity upon intercalation and is 5× brighter than PI as a result of the efficient energy transfer from fluorescein. Additionally, due the relatively long-lived fluorescence, the S/B ratio can be increased from 7 to 40 using the time-resolved detection technique. Cell imaging also confirms the ability of FLEth to be used simultaneously as a whole cell marker and as a probe for duplex RNA.
Acknowledgment
This work was supported by the Center of Excellence in Genomic Science Grant P50 HG002806 from the National Institutes of Health and NSF CHE-04–15516 and NSF CHE-07–17518.
Footnotes
Supporting Information Available: Experimental procedures and cell imaging information. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- (1).Briggs C; Jones M Acta Histochemica 2005, 107, 301–312. [DOI] [PubMed] [Google Scholar]
- 2).Eggleston AK; Rahim NA; Kowalczykowski SC Nucleic Acids Res 1996, 24, 1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3).Suzuki T; Fujikura K; Higashiyama T; Takata K J. Histochem. Cytochem 1997, 45, 49–53. [DOI] [PubMed] [Google Scholar]
- 4).Lauretti F; de Melo FL; Benati FJ; de Mello Volotao E; Santos N; Linhares REC; Nozawa C J. Virol. Methods 2003, 114, 29–35. [DOI] [PubMed] [Google Scholar]
- 5).Olmsted J; Kearns DR Biochemistry 1977, 16, 3647–365. [DOI] [PubMed] [Google Scholar]
- 6).Cui HC; Valdez JG; Steinkamp JA; Harry A; Crissman HA Cytometry Part A 2003, 52A, 46–55. [DOI] [PubMed] [Google Scholar]
- 7).Livache T; Fouque B; Teoulet R Anal. Biochem 2004, 217, 248–254. [DOI] [PubMed] [Google Scholar]
- 8).Johansson MC; Baldetorp B; Oredsson SM Anal. Cell. Pathol 1999, 19, 91–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9).Marti AA; Puckett CA; Dyer J; Stevens N; Jockusch S; Ju J; Barton JK; Turro NJ J. Am. Chem. Soc 2007, 129, 8680–8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10).Yuan J; Wang G; Majima K; Matsumoto K Anal. Chem 2001, 73, 1869–1876. [DOI] [PubMed] [Google Scholar]
- 11).Marriott G; Hekiecker M; Diamndis EP; Yan-Marritt Y Biophys. J 1994, 67, 957–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12).Seveus L; Vaisala M; Syrjanen S; Sandberg M; Kuusisto A; Harju R; Salo J; Hemmila I; Kojola H; Soini E Cytometry 1992, 13, 329–338. [DOI] [PubMed] [Google Scholar]


