Abstract
The PI3K-AKT-mTOR-signaling pathway is frequently activated in glioblastoma (GBM). Inhibition of phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta (PIK3CB)/p110β (a PI3K catalytic isoform) by RNAi substantially suppresses GBM growth with less toxicity to normal astrocytes. However, insufficient and non-specific small interfering RNA (siRNA) delivery may limit the efficacy of RNAi-based therapies against GBM. Here we prepared a novel methoxy-modified PIK3CB siRNA molecule (siPIK3CB) that was covalently conjugated to a [cyclo(Arg-Gly-Asp-D-Phe-Lys)-Ahx]2-Glu-PEG-MAL (biRGD) peptide, which selectively binds to integrin αvβ3 receptors. The αvβ3-positive U87MG cell line was selected as a representative for GBM. An orthotopic GBM xenograft model based on luciferase-expressing U87MG was established and validated in vivo to investigate bio-distribution and anti-tumor efficacy of biRGD-siPIK3CB. In vitro, biRGD-siPIK3CB specifically entered and silenced PIK3CB expression in GBM cells in an αvβ3 receptor-dependent manner, thus inhibiting cell cycle progression and migration and enhancing apoptosis. In vivo, intravenously injected biRGD-siPIK3CB substantially slowed GBM growth and prolonged survival by reducing tumor viability with silencing PIK3CB expression. Furthermore, biRGD-siPIK3CB led to mild tubulointerstitial injury in the treatment of GBM without obvious hepatotoxicity, whereas co-infusion of Gelofusine obviously alleviated this injury without compromising anti-tumor efficacy. These findings revealed a great translational potential of biRGD-siPIK3CB conjugate as a novel molecule for GBM therapy.
Keywords: siRNA delivery, gene silencing, orthotopic glioblastoma, tumor targeting
Introduction
RNAi is a post-transcriptional gene-regulatory process triggered by small interfering RNAs (siRNAs) that has the potential to eradicate diseases caused by mutation or gene overexpression.1, 2, 3 Since 2004, more than 20 RNAi therapies have been approved by the U.S. Food and Drug Administration (FDA) for clinical trials.4 Recently, Alnylam Pharmaceuticals has announced the completion of the rolling submission of a new drug application (NDA) to the FDA for Patisiran, an RNAi therapeutic targeting transthyretin (TTR) for the treatment of patients with hereditary transthyretin-mediated amyloidosis.5
Even though siRNA technology has shown remarkable results in both preclinical and clinical trials, systemic siRNA delivery has remained a major obstacle hindering the further development of RNAi in the clinic.6 siRNA is an ∼14-kDa macromolecule with a negative charge that has no bioavailability to cross cell membranes.3 As a consequence, unless it is incorporated into nanoparticles or conjugated to other molecules, intravenously injected siRNA is rapidly excreted by kidneys without reaching the target cells.7 Even though nanoparticles have enhanced delivery power, they come with substantial costs, such as elevated risk of toxicity with additional materials, inescapable poor diffusion coefficient, and poor pharmacokinetics.8 As an alternative to nanoparticles, vastly simplified smaller-molecular-weight siRNA conjugates, which directly link bioactive groups to siRNA thus allowing it to enter cells of interest, have already been examined in preclinical and clinical trials.9 For example, conjugation of N-acetylgalactosamine (GalNAc) to siRNA, which enables specific binding to asialoglycoprotein receptor on hepatocytes,10 has proven to be a successful therapeutic approach; currently there are six GalNAc-siRNA ongoing clinical trials for liver genes.11 Similarly, in this study we have conjugated siRNA to an optimized bivalent cyclic RGD peptide ([cyclo(Arg-Gly-Asp-D-Phe-Lys)-Ahx]2-Glu-PEG-MAL [biRGD]) that was different from conventional RGD dimers.12, 13
Glioblastoma (GBM) is the most common and among the deadliest types of primary brain malignancy. The 5-year overall survival for GBM patients is approximately 5%, with a prognosis of 14.6 months after diagnosis following total tumor resection, radiotherapy, chemotherapy, or a combination of these treatments.14, 15 It has been reported that the mutation and overactivation of the phosphatidylinositol 3-kinase (PI3K)/protein kinase B (Akt)/rapamycin-sensitive mTOR-complex (mTOR) pathway leads to the development of temozolomide resistance and GBM recurrence.16 Therefore, PI3K inhibitors have emerged as promising tools for treating GBM. Nevertheless, pan-PI3K inhibitors have shown modest benefits in the clinic for GBM because they are nonselective for PI3K isoforms, thus causing high toxicity in normal astrocytes.17 For this reason, it was necessary to develop a novel high-efficiency drug that could selectively inhibit specific PI3K catalytic isoforms, which presented a stronger association with the activities of PI3K signaling in GBM. Several studies have indicated that phosphatidylinositol-4,5-biphosphate 3-kinase catalytic subunit β (PIK3CB), which is a catalytic subunit p110 β of the PI3K gene family, is correlated with high risk and poor survival of GBM, while the inhibition of PIK3CB can remarkably suppress GBM cell growth in vitro and in vivo, with less cytotoxicity to astrocytes.17, 18, 19, 20 In the present study, we have constructed an siRNA conjugate that specifically silenced PIK3CB expression and inhibited GBM growth in vitro and in vivo.
When applying GBM treatment, intravenous-injected molecules must cross the blood-brain barrier (BBB) and the blood-brain tumor barrier (BBTB) to the tumor site.21 The BBB is the so-called neurovascular unit that is formed by the continuous brain endothelial cells interacting with surrounding cells like astrocytes, pericytes, and perivascular macrophages.22 With the progression of GBM, the original neurovascular endothelial cells in the BBB are integrated into newly formed blood vessels induced by tumors, then forming the BBTB.21 In GBM, both neovascular endothelial cells forming the BBTB and tumor cells highly express integrin αvβ3 receptors, which is a key molecule mediating tumor invasion and metastasis.23, 24 According to these characteristics, several RGD-based delivery systems have been designed to specifically bind to αvβ3 receptors, which mediate enhanced recognition and internalization of these systems to overcome the BBTB.25, 26, 27 In this study, we installed optimized biRGD peptide on siRNA conjugate in order to improve the penetration of the siRNA conjugate into GBM tissues via endocytosis and transportation by αvβ3 receptors.
In our previous studies, we have synthesized siRNA conjugates based on cyclo(Arg-Gly-Asp-D-Phe-Lys) (cRGD). Liu et al.28 have shown that cRGD-conjugated siRNA targeting Vascular Endothelial Growth Factor Receptor 2 (VEGFR2) mRNA could silence the expression of VEGFR2 in neovascular endothelial cells, thus inhibiting tumor angiogenesis. In addition, He and colleagues29 showed that cRGD-conjugated siRNA targeting epidermal growth factor receptor (EGFR) leads to an obvious reduction of EGFR expression and significant inhibition of tumor growth in a subcutaneous GBM model. However, the effect of cRGD-conjugated siRNA on orthotopic GBM has not yet been investigated. In the present study, we prepared a biRGD-siRNA conjugate by linking siRNA to a bivalent [cyclo(Arg-Gly-Asp-D-Phe-Lys)-Ahx]2-Glu-PEG-MAL peptide, which had a longer and more flexible linker between RGD motifs than conventional RGD dimers. Consequently, we established an intracranial orthotopic GBM model to observe whether this molecule could overcome multiple physiological barriers to exert anti-tumor effects through RNAi in vivo.
Results
Preparation and Serum Stability of biRGD-siRNA
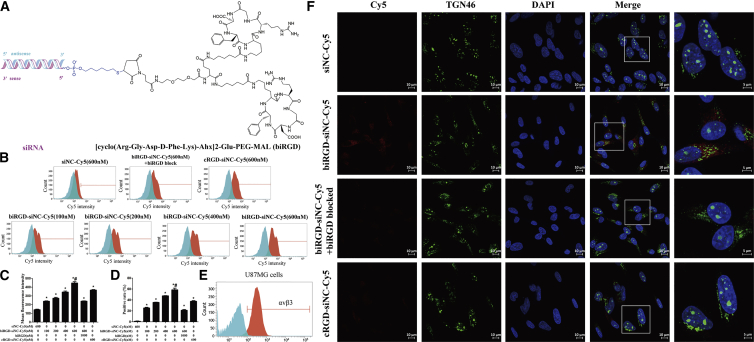
The diagram of biRGD-siRNA conjugate is shown in Figure 1A. To increase the stability and silencing potency while reducing the off-target effect and cytotoxicity, the backbone of siRNA was modified with 2′-O-methyl (2′-OMe) in the recommended nucleotides.30, 31, 32, 33 Thus, the double-stranded PIK3CB siRNA (siPIK3CB) (sense strand, 5′-AC(mC)(mU)G(mU)GGAUACUCAU(mU)A(mA)dTdT-3′; antisense strand, 5′-U(mU)AA(mU)GAGUAUCCA(mC)AG(mG)(mU)dTdT-3′) was selected as the representative siRNA drug. Here we linked the 5′-phosphate of the sense strand of siPIK3CB to a peptide through the thiol-maleimide linker, which contained optimized cyclic RGD dimers ([cyclo(Arg-Gly-Asp-D-Phe-Lys)-Ahx]2-Glu-PEG-MAL) that had a longer and more flexible linker (19 bonds excluding side arms of K-residues formed by two Ahx groups and Glu) between two cyclic RGD motifs than two cyclic RGD motifs in conventional E[c(RGDfK)]2. This may allow the optimized bivalent cyclic RGD to bind to integrin αvβ3 receptors with high affinity via achieving simultaneous receptor binding.13, 34, 35 High-performance liquid chromatography-mass spectrometry (HPLC-MS) results showed that the molecular mass of synthesized biRGD-siPIK3CB was the same as theoretical molecular mass. BiRGD-conjugated sense strand siPIK3CB was determined as 8,723.0 Da, which was acceptably close to the predicted 8,723.7 Da, as seen in Figure S1A. In addition, all siRNAs were HPLC-purified with a >90% purity, as seen in Figure S1B.
Figure 1.
The Schematic Depiction, Cellular Uptake, and Intracellular Distribution of biRGD-siRNA In Vitro
(A) The diagram of biRGD-siRNA conjugate. The [cyclo(Arg-Gly-Asp-D-Phe-Lys)-Ahx]2-Glu-PEG-MAL (biRGD) peptide was conjugated to the 5′-phosphate of sense strand of siRNA through the thiol-maleimide linker. The backbone of siRNA was modified with 2′-O-methyl (2′-OMe) ribose modification in the recommended nucleotides. (B) Cellular uptake levels of biRGD-siNC-Cy5, cRGD-siNC-Cy5, and siNC-Cy5 in U87MG cells incubated in Opti-MEM for 12 hr at 37°C were measured by flow cytometry. For additional specificity tests, before transfection with biRGD-siNC-Cy5, cells were also pre-treated with 1 μM unconjugated biRGD peptide (biRGD blocked). (C and D) Quantitative analysis of Cy5 mean fluorescence intensity (C) and positive rate (D). *p < 0.05 versus siNC-Cy5 group; #p < 0.05 versus biRGD-siNC-Cy5 with biRGD blocked group. (E) Analysis of expression of αvβ3 receptors on U87MG cells by flow cytometry. (F) Confocal fluorescence microscopy images of intracellular distribution of biRGD-siNC-Cy5, cRGD-siNC-Cy5, and siNC-Cy5. U87MG cells were transfected with biRGD-siNC-Cy5, cRGD-siNC-Cy5, or siNC-Cy5 incubated in Opti-MEM for 12 hr at 37°C. In addition, before transfection with biRGD-siNC-Cy5, cells were blocked with biRGD (1 μM). Cell nuclei were counterstained with DAPI (blue), siRNA was labeled with Cy5 (red), and TGN46 was labeled with Alexa Fluor 488 (green). Scale bar, 10 μm.
As seen in Figure S1C, incubation of 2′-OMe-modified biRGD-siPIK3CB conjugate in mouse serum revealed no detectable degradation up to 72 hr and low degradation at 96 hr. In similar conditions, 2′-OMe-modified siPIK3CB without conjugation to biRGD was degraded after 48 hr. These results indicated that the serum stability of siRNA modified with biRGD peptide was significantly improved.
Cellular Uptake and Intracellular Distribution of biRGD-siRNA In Vitro
For detecting cellular uptake and intracellular distribution, biRGD-negative control siRNA (siNC) conjugate, the representative biRGD-siRNA that had no homologous sequence in human and mouse transcriptomes, was labeled with fluorophore (indodicarbocyanine-5 [Cy5]). As shown in Figures 1B–1D, low uptake of non-conjugated siNC-Cy5 (600 nM) was observed in U87MG cells, which highly express integrin αvβ3 receptors (Figure 1E). In contrast, biRGD-siNC-Cy5 displayed enhanced cellular uptake that increased progressively, with concentrations ranging from 100 to 600 nM. No evidence of saturation of uptake was found in the tested concentration range. Moreover, the uptake of biRGD-siNC-Cy5 was approximately twice as much as cRGD-siNC-Cy5, which was prepared in our earlier report.28 Nevertheless, the uptake of biRGD-siNC-Cy5 was inhibited via blocking integrin αvβ3 receptors by pre-treating with excess biRGD peptide. In addition, no uptake of biRGD-siNC-Cy5 (600 nM) was observed in HeLa cells, which negatively express integrin αvβ3 receptors (Figures S2A and S2B).
For analyzing intracellular distribution of biRGD-siRNA, we examined the subcellular localization of the biRGD-siNC-Cy5 at 24 hr compared with an equal amount of siNC-Cy5, cRGD-siNC-Cy5, or biRGD-siNC-Cy5 blocking with excess biRGD peptide. As shown in Figure 1F, the non-conjugated siNC-Cy5 was not detectable, while the biRGD-siNC-Cy5 and cRGD-siNC-Cy5 displayed substantial uptake and wide distribution in the cell. Moreover, we chose TGN46 protein locating in trans-Golgi as the cytoplasmic perinuclear marker to further examine whether biRGD-siNC-Cy5 could reach trans-Golgi membrane and be released into the cytoplasm. The result of subcellular localization showed that part of biRGD-siNC-Cy5 uptaken by cells had a substantial overlap with TGN46 protein, suggesting that conjugates could be released into the cytoplasm as reported elsewhere.36, 37, 38 However, the intracellular distribution of biRGD-siNC-Cy5 was inhibited by pre-treating with excess biRGD peptide.
Gene-Silencing Efficiency of biRGD-siPIK3CB In Vitro
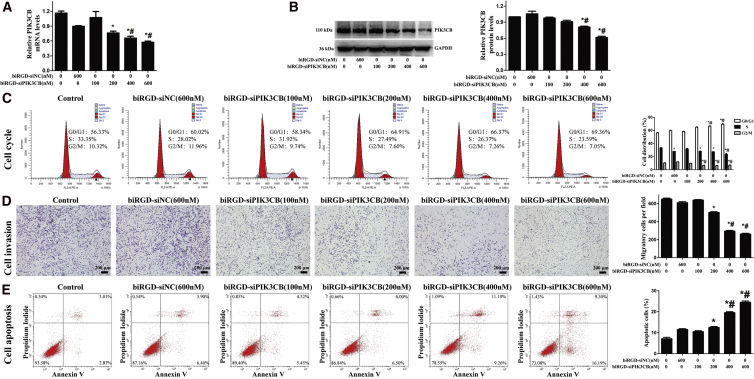
The biRGD-conjugated siRNA was tested in the absence of transfection agent. qRT-PCR analysis demonstrated that the PIK3CB mRNA level of U87MG cells transfected with 400 or 600 nM biRGD-siPIK3CB was significantly decreased (the remaining amount was about 60% and 50%, respectively; Figure 2A) after 48 hr compared with the control and biRGD-siNC groups. In addition, PIK3CB protein level was markedly reduced after 72-hr treatment with 400 and 600 nM biRGD-siPIK3CB (the remaining amount was about 70% and 60%, respectively) (Figure 2B), while almost no effect was observed in the control and biRGD-siNC groups. Taken together, these data demonstrated that biRGD-siRNA was able to efficiently induce targeted gene knockdown in U87MG cells.
Figure 2.
Analyses of Gene-Silencing Efficiency, Cell Cycle, Apoptosis, and Migration of biRGD-siPIK3CB In Vitro
Cells were incubated with different molecules for 24 hr in Opti-MEM without serum. Consequently, 10% FBS was added and cells were incubated for a certain amount of time (depending on the assay). (A) Quantitative analysis of PIK3CB mRNA level using qRT-PCR after 48-hr treatment of biRGD-siPIK3CB. The results were normalized with GAPDH mRNA level. (B) Quantitative analysis of PIK3CB protein level using western blot after 72-hr treatment of biRGD-siPIK3CB. The results were normalized with GAPDH protein level. (C) Cell cycle of U87MG cells was analyzed by flow cytometry after 48-hr treatment of biRGD-siPIK3CB. (D) Transwell migration of U87MG cells was analyzed under a microscope after 48-hr treatment of biRGD-siPIK3CB. (E) Apoptosis of U87MG cells was detected by flow cytometry after 72-hr treatment of biRGD-siPIK3CB. *p < 0.05 versus control group; #p < 0.05 versus biRGD-siNC group. Scale bar, 200 μm.
Influence of biRGD-siPIK3CB on Cell Cycle Progression, Invasion, and Apoptosis In Vitro
After 48-hr treatment, an increase in G0/G1 population and reduction in S-phase population were observed in cells treated with 400 and 600 nM biRGD-siPIK3CB compared to control and biRGD-siNC groups (Figure 2C). These data suggested that biRGD-siPIK3CB could inhibit cell cycle progression, thus affecting U87MG cell growth. Furthermore, transwell migration assay showed that biRGD-siPIK3CB could significantly inhibit U87MG cell invasion with increasing concentrations (Figure 2D). At 72 hr post-treatment, flow cytometry data showed that different concentrations of biRGD-siPIK3CB (400 and 600 nM) could significantly induce apoptosis of U87MG cells in vitro, with the apoptosis rates of 15.14% and 20.76%, respectively (Figure 2E), compared to other groups, which showed slight apoptotic effect. These results indicated that biRGD-siPIK3CB could promote U87MG cell apoptosis in vitro.
Bio-distribution and Intracranial Tumor Permeability of biRGD-siRNA In Vivo
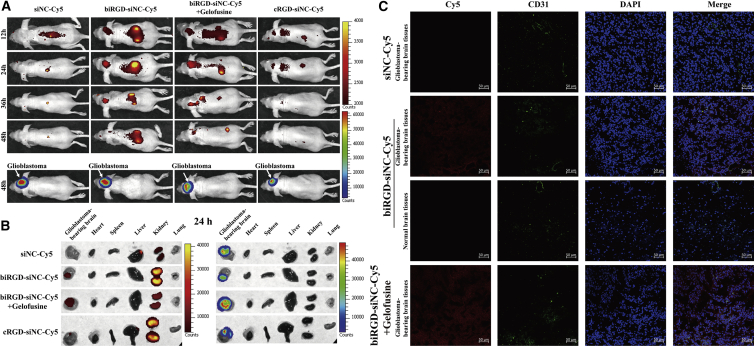
An intracranial orthotopic GBM (U87MG) xenograft expressing a luciferase reporter gene was established to investigate the bio-distribution and intracranial tumor permeability of the biRGD-siRNA in vivo. As shown in Figure 3A, biRGD-siNC-Cy5 could specifically target orthotopic GBM after intravenous injection (1 nmol/20 g). At 12, 24, and 36 hr, high red Cy5 fluorescent expressions of biRGD-siNC-Cy5 were detected within the tumor and kidneys, and the Cy5 fluorescence intensity of orthotopic GBM reached a peak at 24 hr after injection. However, when injected with siNC-Cy5, no red Cy5 fluorescent signal expression was found in the tumor location from 12 to 48 hr. In addition, biRGD-siNC-Cy5 mixed with Gelofusine (4 mg/20 g) injections did not affect tumor targeting of biRGD-siNC-Cy5, while it could reduce the fluorescence intensity of biRGD-siNC-Cy5 in the kidneys. After dissection, results demonstrated that biRGD-siNC-Cy5 could specifically target the GBM-bearing brain tissue and the co-injection of Gelofusine could efficiently reduce the renal retention of biRGD-siNC-Cy5 without affecting its tumor targeting (Figure 3B). Moreover, biRGD-siNC-Cy5 was not found in other main organs, i.e., heart, spleen, and lung.
Figure 3.
Distribution and Intracranial Tumor Permeability of biRGD-siRNA In Vivo
(A) In vivo imaging of mice bearing orthotopic U87MG-Luc glioblastoma with different treatments. Mice were injected intravenously with 1 nmol/20 g siNC-Cy5, biRGD-siNC-Cy5, biRGD-siNC-Cy5 mixed with Gelofusine (4 mg/20 g), or cRGD-siNC-Cy5 at single doses. The subsequent bio-distribution of different molecules labeled with Cy5 was detected at 12, 24, 36, and 48 hr using the IVIS (in red, Cy5 emission spectrum). Intracranial U87MG-Luc glioblastoma was detected at 48 hr using the IVIS (in luciferase emission spectrum). (B) Ex vivo fluorescence imaging of different major organs from part of orthotopic U87MG-Luc glioblastoma-bearing mice at 24 hr. Red Cy5 emission spectrum is indicated for siRNA molecules labled with Cy5 (left panel), and luciferase emission spectrum is indicated for intracranial U87MG-Luc glioblastoma (right panel). (C) Confocal microscopy images of normal brain tissues and the glioblastoma-bearing brain tissues collected from in vivo imaging assay. Cell nuclei were stained with DAPI (blue), blood vessel was marked with CD31 (green), and siRNA was labeled with Cy5 (red). Scale bar, 50 μm. All images were scaled to the same minimum and maximum color values.
To further verify whether the biRGD-siNC-Cy5 had intracranial tumor permeability, the GBM-bearing brains were frozen at –80°C, cut into sections, and stained with CD31. As shown in Figure 3C, no Cy5 fluorescent expression of siNC-Cy5 was found in GBM-bearing brain tissues, while little biRGD-siNC-Cy5 was observed in sections of normal brain tissues. Yet, significantly higher biRGD-siNC-Cy5 signal was found in GBM tissues, suggesting that biRGD-siRNA was taken up by tumor cells and co-injection of Gelofusine did not affect this phenomenon.
Influence of biRGD-siPIK3CB on Tumor Growth and Survival Time in Orthotopic GBM Mice and Its Evaluation of Immunogenicity and Toxicity
To test the potential of biRGD-siPIK3CB as an anti-cancer agent, we first investigated its effects on the orthotopic GBM development in a concentration-dependent manner. Briefly, nude mice bearing orthotopic U87MG-Luc GBM were injected intravenously 12 times over a 24-hr interval, and tumor size was monitored by the luminescence intensity via an in vivo imaging system (IVIS) on days 0, 7, and 12 after treatment. Consequently, mice were sacrificed after 12 days of treatment, and then GBM-bearing brain tissues were excised and photographed. In addition, tumors and kidneys from the mice were obtained for histopathologic analyses.
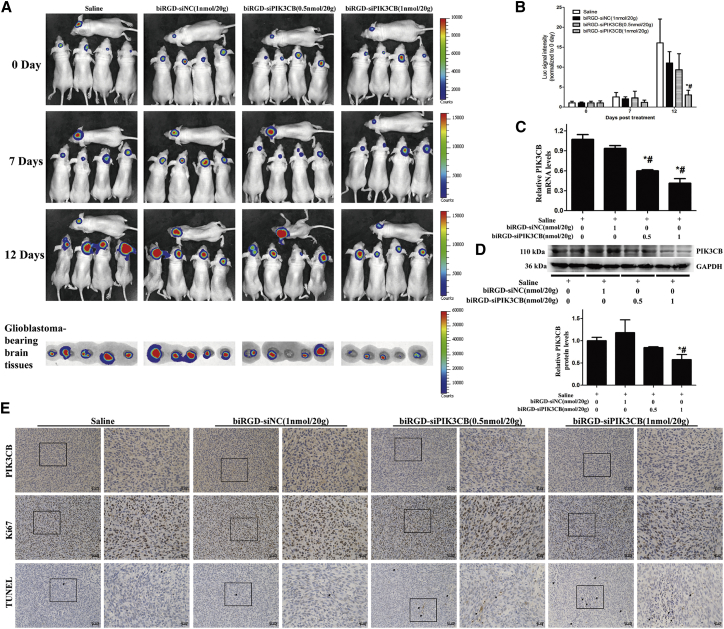
As shown in Figures 4A and 4B, no significant difference was found between saline and biRGD-siNC (1 nmol/20 g) groups, suggesting that biRGD-siNC did not have anti-tumor efficacy. After 12 days of treatment, significant tumor growth inhibition was observed in biRGD-siPIK3CB (1 nmol/20 g) groups compared with saline and biRGD-siNC (1 nmol/20 g) groups (p < 0.05), while no significance was found when treating mice with low-dose biRGD-siPIK3CB (0.5 nmol/20 g) (p > 0.05). These results suggested that biRGD-siPIK3CB had the potential as an anti-GBM agent and that doses higher than 1 nmol/20 g/day, administrated by tail intravenous injection, could effectively inhibit the proliferation of orthotopic GBM. Still, the results from pathological sections of kidneys from the biRGD-siPIK3CB (1 nmol/20 g) group showed mild pathological changes in kidneys, such as renal edema and interstitial hyperemia, compared with the saline group, indicating that long-term use of high-dose biRGD-siPIK3CB may cause tubulointerstitial injury because of renal reabsorption of biRGD-siPIK3CB (Figure S3A).39
Figure 4.
Anti-tumor Efficacy and Mechanism of biRGD-siPIK3CB in Orthotopic U87MG-Luc Glioblastoma Model
(A) IVIS luminescent imaging of glioblastoma-bearing mice and ex vivo glioblastoma-bearing brain tissues from each group. Conditions of the treatment were as follows: saline group, biRGD-siNC (1 nmol/20 g) group, biRGD-siPIK3CB (0.5 nmol/20 g) group, or biRGD-siPIK3CB (1 nmol/20 g) group. All animals were injected intravenously 12 times over a 24-hr interval. (B) The luminescent signal intensity of mice in all groups. (C and D) Silencing activity of different concentrations of biRGD-siPIK3CB in vivo. U87MG-Luc glioblastoma tissues from the anti-tumor assay were collected for qRT-PCR (C) or western blot (D) analysis; results were normalized with GAPDH level. (E) PIK3CB expression, cell proliferative activity, and the number of apoptotic cells in orthotopic U87MG-Luc glioblastoma tissues were detected by immunohistochemical staining. The relative expression level of PIK3CB protein was evaluated by mean optical density (IOD/area). Proliferative cells in tumor were stained by ki67 antibody, and the results were indicated with ki67-positive cells/total cells. The number of apoptotic cells in tumor was indicated by apoptotic index (TUNEL-positive cells/total cells) with TUNEL staining. Positive cells were showing brown. *p < 0.05 versus saline group; #p < 0.05 versus biRGD-siNC group. Scale bar, 50 or 20 μm.
qRT-PCR data showed an obviously low expression of PIK3CB mRNA in biRGD-siPIK3CB (0.5 nmol/20 g) and biRGD-siPIK3CB (1 nmol/20 g) groups compared with saline and biRGD-siNC (1 nmol/20 g) groups (Figure 4C). Furthermore, Figure 4D shows that PIK3CB protein level was significantly decreased in the biRGD-siPIK3CB (1 nmol/20 g) group. Similarly, immunohistochemical analysis using anti-PIK3CB antibody labeling displayed remarkably low PIK3CB protein levels in tumors from biRGD-siPIK3CB (0.5 nmol/20 g) and biRGD-siPIK3CB (1 nmol/20 g) groups (Figure 4E). In addition, in vitro results showed that PIK3CB reduction inhibited cell cycle progression and promoted cellular apoptosis. Therefore, we used ki67 staining and transferase-mediated deoxyuridine triphosphate-biotin nick end labeling (TUNEL) assay to evaluate proliferative activity and apoptosis of orthotopic GBM from in vivo assays, respectively. The proliferative cells in tumors from the biRGD-siPIK3CB (0.5 nmol/20 g) and biRGD-siPIK3CB (1 nmol/20 g) groups obviously decreased compared with those in the saline and biRGD-siNC (1 nmol/20 g) groups (Figure 4E). In addition, the results of the TUNEL assay demonstrated an obviously higher number of apoptotic tumor cells in the biRGD-siPIK3CB (0.5 nmol/20 g) and biRGD-siPIK3CB (1 nmol/20 g) groups compared to the saline and biRGD-siNC (1 nmol/20 g) groups, and the number of apoptotic tumor cells in the biRGD-siPIK3CB (1 nmol/20 g) group was about twice that in the biRGD-siPIK3CB (0.5 nmol/20 g) group (Figure 4E). Taken together, these results showed that biRGD-siPIK3CB could decrease proliferation and viability of GBM via silencing the expression of PIK3CB in vivo.
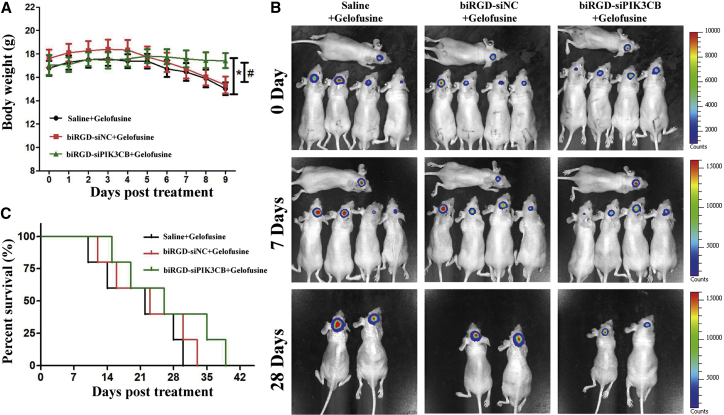
Clinical therapeutic benefits were mainly based on the quality of life and prolonged survival time in GBM patients.17 In addition, to alleviate renal injury from reabsorption of biRGD-siRNA, the overall survival in all treated groups was evaluated with co-injection of Gelofusine, which has been proven to reduce renal reabsorption of biRGD-siRNA (bio-distribution assays). As shown in Figures 5A and 5B, after 9 days of treatment, the body weight of mice in the biRGD-siPIK3CB group ranged between 17.1 ± 1.7 g to 17.3 ± 1.3 g, while it fluctuated between 16.8 ± 1.3 g to 15.9 ± 1.9 g and 17.6 ± 1.5 g to 16.2 ± 2.3 g in the saline and biRGD-siNC groups, respectively. No significant differences were observed between the saline and biRGD-siNC groups during the monitoring period. This indicated that the saline mixed with Gelofusine group and the biRGD-siNC mixed with Gelofusine group in the orthotopic GBM-bearing mice showed a rapid decrease in body weight, while the body weight of mice in the biRGD-siPIK3CB mixed with Gelofusine group was steady (p < 0.05). In addition, the median survival time of the biRGD-siPIK3CB group (26 days) was longer compared to the saline (22 days) and biRGD-siNC (23 days) groups (Figures 5B and 5C). Interestingly, co-injection of Gelofusine decreased renal edema and interstitial hyperemia induced by biRGD-siPIK3CB without affecting anti-tumor efficiency (Figure S3A). To sum up, biRGD-siPIK3CB could prolong the survival time of orthotopic GBM-bearing mice due to its combined efficacy in decreasing proliferation and promoting apoptosis, suggesting that biRGD-siRNA possessed excellent anti-tumor efficacy on GBM.
Figure 5.
Survival Monitoring of Orthotopic U87MG-Luc Glioblastoma-Bearing Mice Treated with biRGD-siPIK3CB
Animals were injected intravenously over a 48-hr interval with the following treatments, which were all mixed with Gelofusine (4 mg/20 g): saline group, biRGD-siNC (3 nmol/20 g) group, or biRGD-siPIK3CB (3 nmol/20 g) group. (A) Body weight of mice in all groups. Body weight of mice was measured daily until mice lost 40% of their primary body weight without glioblastoma, or animal deaths occurred in any group. (B) Tumor growth was monitored on days 0, 7, and 28 after treatment detected by IVIS luminescent imaging. (C) The survival curve of mice in all groups. *p < 0.05 versus saline group, #p < 0.05 versus biRGD-siNC group.
It has been reported that unmodified siRNAs could induce innate immune responses, which would confuse the anti-tumor effect.40 To alleviate such potential non-specific effects, we have introduced the chemical modification of 2′-O-methyl ribose modification inrecommended nucleotides into the siRNA backbone of biRGD-siPIK3CB.30, 31, 32, 33To test the possible immunostimulatory effects of biRGD-siPIK3CB conjugate, we examined the levels of interferon (IFN)-β and interleukin-6 (IL-6) in the serum collected from immunocompetent BALB/c mice 6hr after single higher-doses injection of saline, biRGD-siPIK3CB (5nmol/20 g), or biRGD-siPIK3CB (5nmol/20 g) mixed with Gelofusine. Yet, no significant difference was found in IL-6, IFN-β, and tumor necrosis factor alpha (TNF-α) levels of the serum among differenttreatment groups (FigureS3B), indicating that biRGD-siPIK3CBdid not induce a systemic immune response under such administration conditions. In addition, we detectedthe level of alanine transaminase (ALT) in the serum to evaluatethe hepatotoxicityof biRGD-siPIK3CB. Briefly, no statistically meaningful difference was detected among mice treated with saline,biRGD-siPIK3CB, or biRGD-siPIK3CB mixed with Gelofusine (FigureS3C), indicating that biRGD-siPIK3CB had no obvious hepatotoxicity.
Discussion
Due to its unmatched selectivity, high potency, and universality in treating diseases of all types, siRNA has attracted great interest as a potential therapeutic reagent.41 For this potential to be used, several issues, including poor delivery efficiency, off-target effects, immunogenicity, and instability of siRNA, have to be clearly addressed.3 The majority of currently available carrier systems in clinical trials are nanoparticles, including liposomes and polymer nanoparticles. Nevertheless, these nanoparticles have some disadvantages, such as complicated formulation processes, toxicity from their components, poor pharmacokinetics, and poor homogeneity and reproducibility, which directly limit the clinical transformation of siRNA nanoparticles.7, 8, 9, 41 As an alternative, many research groups has been focusing on siRNA conjugates, which are bioactive groups modified to obtain favorable physicochemical properties and biological benefits for clinical translation, and they are a current trend in the development of siRNA delivery systems.9, 10, 42, 43 In this study, we obtained an siRNA conjugate by linking to an optimized bivalent cyclic RGD peptide, which had relatively small size, good pharmacokinetics, enhanced delivery efficiency, and good reproducibility, all of which make it potentially suitable for clinical translation.
biRGD-siRNA Obtained Gene-Silencing Ability due to Its Enhanced Integrin αvβ3-Targeting Affinity via [cyclo (Arg-Gly-Asp-D-Phe-Lys)-Ahx]2-Glu-PEG-MAL Peptide and Improved Stability of siRNA
Our results showed that biRGD-siRNA was taken up by GBM cells via endocytosis of integrin αvβ3 receptors and then was delivered to the trans-Golgi, where it was eventually released into the cytoplasm. As a result, biRGD-siRNA could inhibit the expression of the target gene in GBM cells in vivo and in vitro. Several factors could account for this phenomenon. First, we constructed a bivalent [cyclo(Arg-Gly-Asp-D-Phe-Lys)-Ahx]2-Glu-PEG-MAL peptide (biRGD) with a longer and more flexible linker (19 bonds excluding side arms of K-residues formed by two Ahx groups and Glu) between two cyclic RGD motifs compared to the previous dimeric RGD moiety. It has been reported that the integrin αvβ3-targeting capability of cyclic RGD dimers could be further improved by increasing distance and flexibility of the linker between cyclic RGD motifs, which allowed cyclic RGD dimers to bind to two adjacent integrin αvβ3 receptors simultaneously.13, 34, 44 Second, extending the distance between the targeting [cyclo(Arg-Gly-Asp-D-Phe-Lys)-Ahx]2-Glu group and siRNA via 21 bonds could reduce the hindrance of the large biRGD group to the RNA-induced silencing complex (RISC) loading.9, 45 Third, in addition to Rab4-dependent short-loop pathway, some studies have indicated that RGD peptide is internalized into endosomal compartments by binding to αvβ3 receptors and then enters the long-loop pathway, relocating to Rab11-positive perinuclear recycling compartments (PNRCs) and trans-Golgi.37, 38, 46, 47 This required the siRNA conjugate to be sufficiently stable against nuclease degradation in the endosome, which provided the opportunity to reach the PNRC and trans-Golgi. Our results also showed that conjugating biRGD to the 5′-phosphate of sense strand of siRNA could substantially improve stability of siRNA, which may be related to increasing the steric hindrance of siRNA against 5′-exonuclease-mediated degradation in the endosome.12, 36, 48
biRGD-siRNA Had Substantial Accumulation in the Orthotopic Brain Tumor due to Its Effective Penetration through the BBTB and Prolonged Retention Time In Vivo
For GBM treatment using the intravenous administration route, siRNA conjugate must cross the BBB and the BBTB in order to be finally uptaken by GBM cells. As the GBM grows, the permeability of the BBB increases and the BBTB dominates between the tumor and its vessels due to angiogenesis-related structural change.49, 50 In our study, biRGD-siRNA was able to target and accumulate in the intracranial U87MG tumors and be uptaken by tumor cells, because biRGD-siRNA was powerfully recognized and internalized by integrin αvβ3 receptors expressed on neovascular endothelial cells forming the BBTB and GBM cells, which provided an effective measure through the tight junction of the BBTB into the orthotopic brain tumor.23, 24, 51, 52 In addition, increasing serum stability and molecular weight of biRGD-siRNA conjugate could further promote the accumulation in the orthotopic brain tumor by prolonging retention time in vivo. As a result, these characteristics of biRGD-siRNA could provide the premise for anti-GBM efficacy in vivo.
biRGD-siPIK3CB Could Substantially Slow GBM Growth and Prolong Survival of Animals via Inhibiting Cell Proliferation and Enhancing Apoptosis In Vitro and In Vivo
At present, the majority of PI3K inhibitors for treating GBM recruit pan-PI3K inhibitors, which are nonselective for PI3K isoforms, thus leading to high toxicity in normal astrocytes and bringing modest benefits in the clinic.17, 53 It has been found that PIK3CB or p110β present a stronger association with the activities of PI3K signaling than other PI3K isoforms in GBM, especially in recurrent GBM acquired radiotherapy and chemotherapy resistance, and that the inhibition of PIK3CB remarkably impeded GBM growth with less cytotoxicity to astrocytes.17, 20, 54, 55, 56 Here we prepared an efficient biRGD-siRNA conjugate for specifically silencing PIK3CB expression in GBM. Our data demonstrated that biRGD-siPIK3CB could substantially slow GBM growth in vitro and in vivo and prolong survival of the animals bearing intracranial orthotopic GBM via inhibiting cell proliferation and enhancing apoptosis with silencing PIK3CB expression, suggesting that biRGD-siPIK3CB conjugate can be used as a novel candidate RNAi drug for GBM therapy.
Gelofusine Acting as an Effective Renal Protective Agent Could Form a Compound Preparation with biRGD-siPIK3CB for GBM Therapy
Our previous reports suggested that cRGD-conjugated siRNA induced mild tubulointerstitial injury via activating the TLR3-signaling pathway due to being reabsorbed and trapped in the kidneys, whereas co-infusion of Gelofusine could attenuate this injury by reducing renal reabsorption of cRGD-siRNA.28, 29, 39 Similarly, our results also showed that biRGD-siPIK3CB had mild kidney injury, such as renal edema and interstitial hyperemia, in the treatment of GBM, whereas co-infusion of Gelofusine could substantially alleviate this injury without harming the anti-tumor efficacy of biRGD-siPIK3CB. Thus, Gelofusine acting as an effective renal protective agent could be used to form a compound preparation with biRGD-siRNA for future clinical applications.
Materials and Methods
Reagents and Materials
The U87MG cell line (human malignant GBM multiforme cell line, ATCC HTB-14) was purchased from ATCC. The U87MG-Luc cell line was generated in our laboratory through transfection with a reporter gene encoding firefly luciferase, as previously reported.57 The cells were cultured using DMEM supplemented with 10% fetal bovine serum (FBS) (Gibco, CA, USA), and they were grown in a humidified atmosphere of 5% CO2 at 37°C.
RNAiso Plus, SYBR Premix Ex Taq and PrimeScript RT reagent kit with gDNA Eraser were from TaKaRa (Japan). Opti-MEM, DMEM, and FBS were from Thermo Fisher Scientific (MA, USA). The 8-μm Transwell cell culture inserts were from Corning (NY, USA). Anti-Human integrin αvβ3 fluorescein isothiocyanate (FITC)-conjugated antibody (11-0519) was from eBioscience (CA, USA). D-Luciferin was from Bioworld Technology (MN, USA). ELISA kits were from Bio-Swamp (Shanghai, China). Anti-CD31 antibody (AF3628) was from R&D Systems (MN, USA). Anti-Ki67 (ARG53222) was from Arigo Biolaboratories (China). TUNEL in situ cell death detection kit-POD was from Roche (Switzerland). Anti-TGN46 (13573-1-AP), anti-PIK3CB (20584-1-AP), Alexa Fluor 488-conjugated goat anti-rabbit second antibody (SA00006-2), and anti-GAPDH (HRP-60004) were from Proteintech Group (China). Annexin V-FITC/propidium iodide (PI) apoptosis detection kit and cell cycle detection kit were obtained from KeyGEN Biotech (China). Gelofusine was from B. Braun Medical (Boulogne Billancourt, France).
Sequence and Modifications of siRNA
siRNA sequences and modifications for this study were as follows: human siPIK3CB sense strand 5′-AC(mC)(mU)G(mU)GGAUACUCAU(mU)A(mA)dTdT-3′, antisense strand 5′-U(mU)AA(mU)GAGUAUCCA(mC)AG(mG)(mU)dTdT-3′; and siNC sense strand 5′-(mC)(mG)(mU)GAUUGCGAGACUC(mU)(mG)(mA)dTdT-3′, antisense strand 5′-(mU)(mC)(mA)GAGUCUCGCAAUC(mA)(mC)(mG)dTdT-3′. mN represented 2′-O-methyl sugar-modified RNA nucleosides. siPIK3CB was the representative siRNA that effectively silenced the expression of Homo sapiens PIK3CB. siNC was the negative control siRNA that had no homologous sequence in human and mouse transcriptomes, and Cy5-labeled siNC (siNC-Cy5) was also prepared. All the siRNAs were synthesized by Guangzhou RiboBio.
Preparation of biRGD-siRNA Conjugate
The biRGD peptide, where Ahx stood for 6-Aminocaproic acid; PEG for 8-amino-3, 6-dioxaoctanoic acid; and MAL for β-maleimidopropionic acid, were synthesized by Guoping Pharmaceutical (China). To prepare biRGD-siRNA molecules, biRGD was covalently conjugated to the 5′ end of siRNA sense strand using thiol-maleamide linker, as previously described.28 The molecular weight of biRGD-sense strand siRNA was detected and characterized by the Oligo HTCS LC-MS system (Novatia). The purity of conjugated biRGD-sense strand siRNA (biRGD-ssRNA) and antisense strand siRNA were determined by HPLC. The analyses were performed employing an Agela C-8 column (25 cm × 4.6 mm) according to the following conditions: starting from 0.1 M triethylammonium acetate (pH 7.4), a linear gradient of 0%–50% MeCN was pumped at a flow rate of 1 mL/min for 0–30 min. To generate double-stranded siRNAs, antisense RNA molecules (50 μM) were mixed with biRGD-ssRNA conjugates (50 μM) in annealing buffer (10 mM Tris-HCl [pH 7.4], 50 mM NaCl, and 1 mM EDTA), heat denatured at 95°C for 3 min, and slowly cooled down to room temperature. biRGD-siPIK3CB, biRGD-siNC, and biRGD-siNC-Cy5 were connected by biRGD oligopeptide with siPIK3CB, siNC, and siNC-Cy5, respectively. cRGD-siNC and cRGD-siNC-Cy5 were also prepared as previously described.28 Desalting and filter purification of conjugated siRNA for in vivo applications were performed using Millipore centrifugation with a 0.22-μm sterile filtration membrane.
Serum Stability Assay
5 μL 20 μM biRGD-siPIK3CB or siPIK3CB was mixed with 5 μL mouse serum and incubated at 37°C for 0, 12, 24, 48, 72, and 96 hr. At each time point, aliquots were collected and subjected to electrophoresis in 1.2% non-denaturing agarose gels.
Cellular Uptake and Confocal Fluorescence Microscopy Assay
U87MG cells were transfected with different concentrations of biRGD-siNC-Cy5 (100, 200, 400, or 600 nM), cRGD-siNC-Cy5 (600 nM), and siNC-Cy5 (600 nM) in Opti-MEM for 12 hr at 37°C. To demonstrate the effect of biRGD binding to the αvβ3 receptor, U87MG cells were pre-treated with 1 μM unconjugated biRGD peptide (biRGD blocked) at 37°C for 1 hr prior to transfection with biRGD-siNC-Cy5, as previously reported.37 For the cellular uptake assay, the cells were washed with PBS three times to remove any extracellular molecules. Consequently, cells were digested with Tyrisin, washed twice with PBS, and resuspended in 0.1 mL PBS. For determining the expression level of integrin αvβ3 receptors of U87MG, 1 × 104 cell suspension was mixed with 5 μL anti-integrin αvβ3 FITC-conjugated antibody at 4°C for 30 min. Cells were washed twice with PBS and resuspended in 0.1 mL PBS. Fluorescence data were collected using a fluorescence-activated cell sorting (FACS) verse flow cytometer (BD Biosciences, USA) and analyzed using Cell Quest software.
For the confocal fluorescence microscopy assay, after transfection of different molecules at 600 nM, cells were washed with PBS and fixed immediately using 4% paraformaldehyde at room temperature for 15 min, then incubated with anti-TGN46 primary antibody followed by Alexa Fluor 488-conjugated goat anti-rabbit second antibody. Cells were then stained with DAPI (Roche, Switzerland) for 10 min at 37°C, and finally they were observed with confocal microscopy (Zeiss LSM 800, Germany; Cy5 excitation, 640 nm; emission, 680 nm).
Establishment of the In Vitro Assay
The cells were seeded into different types of plates (Corning, CA, USA) at a recommended density of cells per well and allowed to attach for 24 hr. The processing of different groups was as follows: control group, the processing condition was the same as those in other groups without any treatment; biRGD-siNC group, biRGD-siNC (600 nM) was added into different wells and incubated with cells for 24 hr in Opti-MEM without serum; and biRGD-siPIK3CB group, biRGD-siPIK3CB (100, 200, 400, or 600 nM) was separately added into different wells and incubated with cells for 24 hr in Opti-MEM without serum. Consequently, 10% serum was supplemented, and cells were incubated for 48 hr (for qRT-PCR, cell cycle analysis, and transwell migration assay) or 72 hr (for western blot and cell apoptosis analysis).
qRT-PCR and Western Blot Analysis
The qRT-PCR and western blot analysis were conducted as previously described.28 The primer sequences for qRT-PCR analysis used in this experiment were as follows: human PIK3CB forward primer 5′-TATTTGGACTTTGCGACAAGACT-3′, reverse primer 5′-TCGAACGTACTGGTCTGGATAG-3′; and human GAPDH forward primer 5′-CGGAGTCAACGGATTTGGTCGTAT-3′, reverse primer 5′-AGCCTTCTCCATGGTGGTGAAGAC-3′.
Cell Cycle and Cell Apoptosis Analyses
For cell cycle analysis, cells were harvested and suspended in cold 70% ethanol for fixation at 4°C overnight. The fixed cells were washed with PBS and successively incubated with RNase A (100 μg) and PI (50 μg/mL) for 15 min. The DNA content was then analyzed using a FACS verse flow cytometer, and the cell cycle profile was evaluated using the FACS suite software. For cell apoptosis analysis, cell apoptosis was measured using an Annexin V-FITC/PI apoptosis detection kit according to the manufacturer’s instructions. The cell apoptosis rates were analyzed using a FACS verse flow cytometer.
Transwell Migration Assay
The inserts were coated with 50 μL 2 mg/mL Matrigel matrix (Becton Dickinson, 356234), according to the manufacturer’s instruction. Following this, 5 × 104 cells suspended in serum-free DMEM were placed in the upper compartment of 8-μm-pore transwells, and 300 μL 10% FBS in DMEM was added to the lower compartment. The cells were allowed to migrate for 24 hr. For quantification, the cells in the lower compartment were stained with crystal violet and counted in five randomly chosen fields under a microscope.
Animal Handling
BALB/c nude mice (4–6 weeks old, ∼20 g) and BALB/c mice (4–6 weeks old, ∼20 g) were purchased from the Experimental Animal Center of Sun Yat-Sen University (Guangzhou, China). All the animals were housed in an environment with a temperature of 22°C ± 1°C, relative humidity of 50% ± 1%, and a light-dark cycle of 12-12 hr. In addition, all animal experiments (including the mice euthanasia procedure) were done in compliance with the regulations and guidelines of Sun Yat-Sen University institutional animal care and conducted according to the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) and Institutional Animal Care and Use Committee (IACUC) guidelines.
Intracranial Orthotopic GBM Model Establishment
An intracranial orthotopic GBM model was established to investigate bio-distribution and anti-tumor efficacy of biRGD-siRNA, according to a previously described method.58, 59 Briefly, U87MG-Luc cells were suspended in ice-cold PBS at 3 × 105 cells in 3 μL and kept on ice. Nude mice were anesthetized with chloral hydrate and attached to the base of a stereotaxic frame with ear bars. The skull was exposed through a 1-cm midline incision, and a burr hole was made 2 mm to the right of the bregma and 1 mm posterior to the coronal suture using a Dremel equipped with a 1-mm tip. Using a Hamilton syringe attached to the stereotaxic frame, 3 μL U87MG-Luc cells was then slowly injected over 2 min at a depth of 3 mm. After the injection, the syringe was kept in place for 1 min prior to withdrawal, and the incision was closed with Vetbond skin glue. The tumor growth was monitored using an IVIS (IVIS Lumina II, Caliper, USA).
In Vivo Distribution Assay
Mice bearing orthotopic GBM (n = 12) were injected intravenously with 1 nmol/20 g siNC-Cy5, biRGD-siNC-Cy5, biRGD-siNC-Cy5 mixed with Gelofusine (4 mg/20 g, Succinylated Gelatin Injectim that efficiently reduced the renal retention of cRGD-siRNA39), or cRGD-siNC-Cy5 at single doses, respectively. The subsequent bio-distribution was detected at 12, 24, 36, and 48 hr using the IVIS Spectrum at the appropriate wavelength (Cy5: λex = 640 nm, λem = 680 nm). At 24 hr, some mice were sacrificed, and tumors and major organs were excised and imaged.
Immunofluorescence Analysis
Tissues from the assessment of in vivo distribution were analyzed for subsequent frozen section fluorescence, as previously described.60 Tissues were observed by confocal microscopy (Zeiss LSM 800, Germany; Cy5 excitation, 640 nm; emission, 680 nm).
In Vivo Anti-tumor Effects and Survival Monitoring Assay
For evaluating anti-tumor effects of different concentration of biRGD-siPIK3CB, 20 nude mice bearing orthotopic U87MG-Luc GBM were randomly divided into 4 groups (2 males and 3 females in each group). Treatments were started when the tumors were detected by IVIS luminescent imaging. Conditions of the treatment were as follows: saline, biRGD-siNC (1 nmol/20 g), biRGD-siPIK3CB (0.5 nmol/20 g), or biRGD-siPIK3CB (1 nmol/20 g). All animals were injected intravenously 12 times over a 24-hr interval. To monitor in vivo anti-GBM efficacy, tumor size was monitored on days 0, 7, and 12 after treatment via fluorescence intensity detection by IVIS luminescent imaging. After being monitored on day 12, mice were sacrificed, and fluorescence intensity of the GBM-bearing brains was detected by IVIS luminescent imaging. Then tissues and serum were collected for qRT-PCR, western blot, immunohistochemistry, and toxicity studies.
For survival monitoring, 15 nude mice bearing orthotopic U87MG-Luc GBM were randomly divided into 3 groups (2 males and 3 females in each group). Animals were injected intravenously at a 48-hr interval with the following treatments: saline mixed with Gelofusine (4 mg/20 g), biRGD-siNC (3 nmol/20 g) mixed with Gelofusine (4 mg/20 g), or biRGD-siPIK3CB (3 nmol/20 g) mixed with Gelofusine (4 mg/20 g). The body weight of mice was measured daily until the animal’s death occurred; tumor size was monitored on days 0, 7, and 28 after treatment via fluorescence intensity detected by IVIS luminescent imaging. When mice lost 40% of their primary body weight without GBM, mice were sacrificed, and tissues and serum were collected for immunohistochemical analysis and toxicity studies.
Histopathological Evaluation
Whole tissues (tumors and kidneys) from in vivo assay were fixed immediately using 4% paraformaldehyde. Tissues were then embedded in paraffin wax, cut into 5-μm-thick sections, and stained with H&E for histological examination, TUNEL for labeling apoptotic cells or immunohistochemistry for analyzing Ki67-positive proliferative cells or PIK3CB expression with standard clinical laboratory protocols.
Immunogenicity Evaluation and Toxicity Assay
For the immune response studies, the serum was collected from immunocompetent BALB/c mice at 6 hr after the single injection of saline, biRGD-siPIK3CB (5 nmol/20 g), or biRGD-siPIK3CB (5 nmol/20 g) mixed with Gelofusine (4 mg/20 g), according to previous reports.40, 61 IL-6, IFN-β, and TNF-α levels in the serum were detected by an ELISA kit according to the manufacturer’s instructions. For analyzing hepatotoxicity, the serum was collected from in vivo anti-tumor assay, and the ALT (alanine transaminase) was measured by automated Aeroset Chemistry Analyzer (Abbott, USA), according to the manufacturer’s instructions. Images were viewed and captured using a light microscope (Zeiss, Germany). Percentages of positive staining area were quantified using the Image-Pro Plus software (Media Cybernetics, MD, USA).
Statistical Analysis
The results were expressed as the mean ± SE. Homogeneity-of-variance test was conducted first. The data were statistically analyzed through one-way ANOVA with a least significant difference or Dunnett T3 test for intergroup comparisons (SPSS software, version 19.0, SPSS, IL, USA). p < 0.05 was considered statistically significant.
Author Contributions
A.J. conceived the study, supervised experiments, and analyzed the data. B.C. designed the experiments, prepared the figures, and wrote the manuscript. Y.W., M.T., and S.H. performed and analyzed the in vitro experiments. Y.W., X.L., W.H., and J.L. conducted and analyzed the in vivo experiments. W.W., G.H., and X.B. provided siRNA samples and analyzed pathological results. Y.Y. and X.P. analyzed the data and revised the manuscript.
Conflicts of Interest
The authors have declared that no competing interests exist.
Acknowledgments
This work was supported by the Science and Technology Program of Guangzhou, China (201604020167), the Natural Science Foundation of Guangdong, China (2014A030310114), the Science and Technology Program of Guangdong Province, China (2013B091300014), Pearl River Nova Program of Guangzhou 2016 (201610010053), the National Nature Science Foundation of China (81773641, 81770173, and 81370449), and Guangzhou Key Medical Discipline Construction Project. This work was also supported by the President Foundation of Nanfang Hospital, Southern Medical University (2017B030). The authors thank the Guangdong Provincial Key Laboratory of Malignant Tumor Epigenetics and Gene Regulation for flow cytometry support.
Footnotes
Supplemental Information includes three figures and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.09.002.
Contributor Information
Yawei Yuan, Email: yuanyawei2015@outlook.com.
Xinghua Pan, Email: panvictor@smu.edu.cn.
Aimin Ji, Email: aiminji_007@163.com.
Supplemental Information
References
- 1.Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Elbashir S.M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 3.Wittrup A., Lieberman J. Knocking down disease: a progress report on siRNA therapeutics. Nat. Rev. Genet. 2015;16:543–552. doi: 10.1038/nrg3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaczmarek J.C., Kowalski P.S., Anderson D.G. Advances in the delivery of RNA therapeutics: from concept to clinical reality. Genome Med. 2017;9:60. doi: 10.1186/s13073-017-0450-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adams D., Suhr O.B., Dyck P.J., Litchy W.J., Leahy R.G., Chen J., Gollob J., Coelho T. Trial design and rationale for APOLLO, a Phase 3, placebo-controlled study of patisiran in patients with hereditary ATTR amyloidosis with polyneuropathy. BMC Neurol. 2017;17:181. doi: 10.1186/s12883-017-0948-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ozcan G., Ozpolat B., Coleman R.L., Sood A.K., Lopez-Berestein G. Preclinical and clinical development of siRNA-based therapeutics. Adv. Drug Deliv. Rev. 2015;87:108–119. doi: 10.1016/j.addr.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanasty R., Dorkin J.R., Vegas A., Anderson D. Delivery materials for siRNA therapeutics. Nat. Mater. 2013;12:967–977. doi: 10.1038/nmat3765. [DOI] [PubMed] [Google Scholar]
- 8.Pecot C.V., Calin G.A., Coleman R.L., Lopez-Berestein G., Sood A.K. RNA interference in the clinic: challenges and future directions. Nat. Rev. Cancer. 2011;11:59–67. doi: 10.1038/nrc2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee S.H., Kang Y.Y., Jang H.E., Mok H. Current preclinical small interfering RNA (siRNA)-based conjugate systems for RNA therapeutics. Adv. Drug Deliv. Rev. 2016;104:78–92. doi: 10.1016/j.addr.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 10.Zimmermann T.S., Karsten V., Chan A., Chiesa J., Boyce M., Bettencourt B.R., Hutabarat R., Nochur S., Vaishnaw A., Gollob J. Clinical Proof of Concept for a Novel Hepatocyte-Targeting GalNAc-siRNA Conjugate. Mol. Ther. 2017;25:71–78. doi: 10.1016/j.ymthe.2016.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alnylam (2018). Alnylam clinical development pipeline. http://www.alnylam.com/alnylam-rnai-pipeline/.
- 12.Arosio D., Casagrande C. Advancement in integrin facilitated drug delivery. Adv. Drug Deliv. Rev. 2016;97:111–143. doi: 10.1016/j.addr.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 13.Liu S. Radiolabeled cyclic RGD peptides as integrin alpha(v)beta(3)-targeted radiotracers: maximizing binding affinity via bivalency. Bioconjug. Chem. 2009;20:2199–2213. doi: 10.1021/bc900167c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ostrom Q.T., Gittleman H., Xu J., Kromer C., Wolinsky Y., Kruchko C., Barnholtz-Sloan J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2009-2013. Neuro-oncol. 2016;18(suppl_5):v1–v75. doi: 10.1093/neuonc/now207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furnari F.B., Fenton T., Bachoo R.M., Mukasa A., Stommel J.M., Stegh A., Hahn W.C., Ligon K.L., Louis D.N., Brennan C. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 16.Oike T., Suzuki Y., Sugawara K., Shirai K., Noda S.E., Tamaki T., Nagaishi M., Yokoo H., Nakazato Y., Nakano T. Radiotherapy plus concomitant adjuvant temozolomide for glioblastoma: Japanese mono-institutional results. PLoS ONE. 2013;8:e78943. doi: 10.1371/journal.pone.0078943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pridham K.J., Le L., Guo S., Varghese R.T., Algino S., Liang Y., Fajardin R., Rodgers C.M., Simonds G.R., Kelly D.F., Sheng Z. PIK3CB/p110β is a selective survival factor for glioblastoma. Neuro-oncol. 2018;20:494–505. doi: 10.1093/neuonc/nox181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wee S., Wiederschain D., Maira S.M., Loo A., Miller C., deBeaumont R., Stegmeier F., Yao Y.M., Lengauer C. PTEN-deficient cancers depend on PIK3CB. Proc. Natl. Acad. Sci. USA. 2008;105:13057–13062. doi: 10.1073/pnas.0802655105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Opel D., Westhoff M.A., Bender A., Braun V., Debatin K.M., Fulda S. Phosphatidylinositol 3-kinase inhibition broadly sensitizes glioblastoma cells to death receptor- and drug-induced apoptosis. Cancer Res. 2008;68:6271–6280. doi: 10.1158/0008-5472.CAN-07-6769. [DOI] [PubMed] [Google Scholar]
- 20.Pu P., Kang C., Zhang Z., Liu X., Jiang H. Downregulation of PIK3CB by siRNA suppresses malignant glioma cell growth in vitro and in vivo. Technol. Cancer Res. Treat. 2006;5:271–280. doi: 10.1177/153303460600500308. [DOI] [PubMed] [Google Scholar]
- 21.van Tellingen O., Yetkin-Arik B., de Gooijer M.C., Wesseling P., Wurdinger T., de Vries H.E. Overcoming the blood-brain tumor barrier for effective glioblastoma treatment. Drug Resist. Updat. 2015;19:1–12. doi: 10.1016/j.drup.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 22.Abbott N.J., Rönnbäck L., Hansson E. Astrocyte-endothelial interactions at the blood-brain barrier. Nat. Rev. Neurosci. 2006;7:41–53. doi: 10.1038/nrn1824. [DOI] [PubMed] [Google Scholar]
- 23.Mattern R.H., Read S.B., Pierschbacher M.D., Sze C.I., Eliceiri B.P., Kruse C.A. Glioma cell integrin expression and their interactions with integrin antagonists: Research Article. Cancer Ther. 2005;3A:325–340. [PMC free article] [PubMed] [Google Scholar]
- 24.Zitzmann S., Ehemann V., Schwab M. Arginine-glycine-aspartic acid (RGD)-peptide binds to both tumor and tumor-endothelial cells in vivo. Cancer Res. 2002;62:5139–5143. [PubMed] [Google Scholar]
- 25.Ruan H., Chen X., Xie C., Li B., Ying M., Liu Y., Zhang M., Zhang X., Zhan C., Lu W., Lu W. Stapled RGD Peptide Enables Glioma-Targeted Drug Delivery by Overcoming Multiple Barriers. ACS Appl. Mater. Interfaces. 2017;9:17745–17756. doi: 10.1021/acsami.7b03682. [DOI] [PubMed] [Google Scholar]
- 26.Chen C., Duan Z., Yuan Y., Li R., Pang L., Liang J., Xu X., Wang J. Peptide-22 and Cyclic RGD Functionalized Liposomes for Glioma Targeting Drug Delivery Overcoming BBB and BBTB. ACS Appl. Mater. Interfaces. 2017;9:5864–5873. doi: 10.1021/acsami.6b15831. [DOI] [PubMed] [Google Scholar]
- 27.Chen T., Song X., Gong T., Fu Y., Yang L., Zhang Z., Gong T. nRGD modified lycobetaine and octreotide combination delivery system to overcome multiple barriers and enhance anti-glioma efficacy. Colloids Surf. B Biointerfaces. 2017;156:330–339. doi: 10.1016/j.colsurfb.2017.05.038. [DOI] [PubMed] [Google Scholar]
- 28.Liu X., Wang W., Samarsky D., Liu L., Xu Q., Zhang W., Zhu G., Wu P., Zuo X., Deng H. Tumor-targeted in vivo gene silencing via systemic delivery of cRGD-conjugated siRNA. Nucleic Acids Res. 2014;42:11805–11817. doi: 10.1093/nar/gku831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He S., Cen B., Liao L., Wang Z., Qin Y., Wu Z., Liao W., Zhang Z., Ji A. A tumor-targeting cRGD-EGFR siRNA conjugate and its anti-tumor effect on glioblastoma in vitro and in vivo. Drug Deliv. 2017;24:471–481. doi: 10.1080/10717544.2016.1267821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bramsen J.B., Laursen M.B., Nielsen A.F., Hansen T.B., Bus C., Langkjaer N., Babu B.R., Højland T., Abramov M., Van Aerschot A. A large-scale chemical modification screen identifies design rules to generate siRNAs with high activity, high stability and low toxicity. Nucleic Acids Res. 2009;37:2867–2881. doi: 10.1093/nar/gkp106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Volkov A.A., Kruglova N.S., Meschaninova M.I., Venyaminova A.G., Zenkova M.A., Vlassov V.V., Chernolovskaya E.L. Selective protection of nuclease-sensitive sites in siRNA prolongs silencing effect. Oligonucleotides. 2009;19:191–202. doi: 10.1089/oli.2008.0162. [DOI] [PubMed] [Google Scholar]
- 32.Jackson A.L., Burchard J., Leake D., Reynolds A., Schelter J., Guo J., Johnson J.M., Lim L., Karpilow J., Nichols K. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA. 2006;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohrt T., Schwille P. siRNA modifications and sub-cellular localization: a question of intracellular transport? Curr. Pharm. Des. 2008;14:3674–3685. doi: 10.2174/138161208786898798. [DOI] [PubMed] [Google Scholar]
- 34.Wang L., Shi J., Kim Y.S., Zhai S., Jia B., Zhao H., Liu Z., Wang F., Chen X., Liu S. Improving tumor-targeting capability and pharmacokinetics of (99m)Tc-labeled cyclic RGD dimers with PEG(4) linkers. Mol. Pharm. 2009;6:231–245. doi: 10.1021/mp800150r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi J., Wang L., Kim Y.S., Zhai S., Liu Z., Chen X., Liu S. Improving tumor uptake and excretion kinetics of 99mTc-labeled cyclic arginine-glycine-aspartic (RGD) dimers with triglycine linkers. J. Med. Chem. 2008;51:7980–7990. doi: 10.1021/jm801134k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nair J.K., Attarwala H., Sehgal A., Wang Q., Aluri K., Zhang X., Gao M., Liu J., Indrakanti R., Schofield S. Impact of enhanced metabolic stability on pharmacokinetics and pharmacodynamics of GalNAc-siRNA conjugates. Nucleic Acids Res. 2017;45:10969–10977. doi: 10.1093/nar/gkx818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alam M.R., Ming X., Fisher M., Lackey J.G., Rajeev K.G., Manoharan M., Juliano R.L. Multivalent cyclic RGD conjugates for targeted delivery of small interfering RNA. Bioconjug. Chem. 2011;22:1673–1681. doi: 10.1021/bc200235q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alam M.R., Dixit V., Kang H., Li Z.B., Chen X., Trejo J., Fisher M., Juliano R.L. Intracellular delivery of an anionic antisense oligonucleotide via receptor-mediated endocytosis. Nucleic Acids Res. 2008;36:2764–2776. doi: 10.1093/nar/gkn115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cen B., Liao W., Wang Z., Gao L., Wei Y., Huang W., He S., Wang W., Liu X., Pan X., Ji A. Gelofusine Attenuates Tubulointerstitial Injury Induced by cRGD-Conjugated siRNA by Regulating the TLR3 Signaling Pathway. Mol. Ther. Nucleic Acids. 2018;11:300–311. doi: 10.1016/j.omtn.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robbins M., Judge A., MacLachlan I. siRNA and innate immunity. Oligonucleotides. 2009;19:89–102. doi: 10.1089/oli.2009.0180. [DOI] [PubMed] [Google Scholar]
- 41.Chakraborty C., Sharma A.R., Sharma G., Doss C.G.P., Lee S.S. Therapeutic miRNA and siRNA: Moving from Bench to Clinic as Next Generation Medicine. Mol. Ther. Nucleic Acids. 2017;8:132–143. doi: 10.1016/j.omtn.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nair J.K., Willoughby J.L., Chan A., Charisse K., Alam M.R., Wang Q., Hoekstra M., Kandasamy P., Kel’in A.V., Milstein S. Multivalent N-acetylgalactosamine-conjugated siRNA localizes in hepatocytes and elicits robust RNAi-mediated gene silencing. J. Am. Chem. Soc. 2014;136:16958–16961. doi: 10.1021/ja505986a. [DOI] [PubMed] [Google Scholar]
- 43.Zhou J., Lazar D., Li H., Xia X., Satheesan S., Charlins P., O’Mealy D., Akkina R., Saayman S., Weinberg M.S. Receptor-targeted aptamer-siRNA conjugate-directed transcriptional regulation of HIV-1. Theranostics. 2018;8:1575–1590. doi: 10.7150/thno.23085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ji S., Czerwinski A., Zhou Y., Shao G., Valenzuela F., Sowiński P., Chauhan S., Pennington M., Liu S. (99m)Tc-Galacto-RGD2: a novel 99mTc-labeled cyclic RGD peptide dimer useful for tumor imaging. Mol. Pharm. 2013;10:3304–3314. doi: 10.1021/mp400085d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matsuda S., Keiser K., Nair J.K., Charisse K., Manoharan R.M., Kretschmer P., Peng C.G., V Kel’in A., Kandasamy P., Willoughby J.L. siRNA conjugates carrying sequentially assembled trivalent N-acetylgalactosamine linked through nucleosides elicit robust gene silencing in vivo in hepatocytes. ACS Chem. Biol. 2015;10:1181–1187. doi: 10.1021/cb501028c. [DOI] [PubMed] [Google Scholar]
- 46.De Franceschi N., Hamidi H., Alanko J., Sahgal P., Ivaska J. Integrin traffic - the update. J. Cell Sci. 2015;128:839–852. doi: 10.1242/jcs.161653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aguzzi M.S., Fortugno P., Giampietri C., Ragone G., Capogrossi M.C., Facchiano A. Intracellular targets of RGDS peptide in melanoma cells. Mol. Cancer. 2010;9:84. doi: 10.1186/1476-4598-9-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Elkayam E., Parmar R., Brown C.R., Willoughby J.L., Theile C.S., Manoharan M., Joshua-Tor L. siRNA carrying an (E)-vinylphosphonate moiety at the 5′ end of the guide strand augments gene silencing by enhanced binding to human Argonaute-2. Nucleic Acids Res. 2017;45:5008. doi: 10.1093/nar/gkw1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jain R.K., di Tomaso E., Duda D.G., Loeffler J.S., Sorensen A.G., Batchelor T.T. Angiogenesis in brain tumours. Nat. Rev. Neurosci. 2007;8:610–622. doi: 10.1038/nrn2175. [DOI] [PubMed] [Google Scholar]
- 50.Gavard J., Gutkind J.S. VEGF controls endothelial-cell permeability by promoting the beta-arrestin-dependent endocytosis of VE-cadherin. Nat. Cell Biol. 2006;8:1223–1234. doi: 10.1038/ncb1486. [DOI] [PubMed] [Google Scholar]
- 51.Miura Y., Takenaka T., Toh K., Wu S., Nishihara H., Kano M.R., Ino Y., Nomoto T., Matsumoto Y., Koyama H. Cyclic RGD-linked polymeric micelles for targeted delivery of platinum anticancer drugs to glioblastoma through the blood-brain tumor barrier. ACS Nano. 2013;7:8583–8592. doi: 10.1021/nn402662d. [DOI] [PubMed] [Google Scholar]
- 52.Quader S., Liu X., Chen Y., Mi P., Chida T., Ishii T., Miura Y., Nishiyama N., Cabral H., Kataoka K. cRGD peptide-installed epirubicin-loaded polymeric micelles for effective targeted therapy against brain tumors. J. Control. Release. 2017;258:56–66. doi: 10.1016/j.jconrel.2017.04.033. [DOI] [PubMed] [Google Scholar]
- 53.Li X., Wu C., Chen N., Gu H., Yen A., Cao L., Wang E., Wang L. PI3K/Akt/mTOR signaling pathway and targeted therapy for glioblastoma. Oncotarget. 2016;7:33440–33450. doi: 10.18632/oncotarget.7961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wahl M., Chang S.M., Phillips J.J., Molinaro A.M., Costello J.F., Mazor T., Alexandrescu S., Lupo J.M., Nelson S.J., Berger M. Probing the phosphatidylinositol 3-kinase/mammalian target of rapamycin pathway in gliomas: A phase 2 study of everolimus for recurrent adult low-grade gliomas. Cancer. 2017;123:4631–4639. doi: 10.1002/cncr.30909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pitz M.W., Eisenhauer E.A., MacNeil M.V., Thiessen B., Easaw J.C., Macdonald D.R., Eisenstat D.D., Kakumanu A.S., Salim M., Chalchal H. Phase II study of PX-866 in recurrent glioblastoma. Neuro-oncol. 2015;17:1270–1274. doi: 10.1093/neuonc/nou365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Venkatesan S., Lamfers M.L., Dirven C.M., Leenstra S. Genetic biomarkers of drug response for small-molecule therapeutics targeting the RTK/Ras/PI3K, p53 or Rb pathway in glioblastoma. CNS Oncol. 2016;5:77–90. doi: 10.2217/cns-2015-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Y., Sun X., Huang M., Ke Y., Wang J., Liu X. A novel bispecific immunotoxin delivered by human bone marrow-derived mesenchymal stem cells to target blood vessels and vasculogenic mimicry of malignant gliomas. Drug Des. Devel. Ther. 2015;9:2947–2959. doi: 10.2147/DDDT.S79475. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 58.Ozawa T., James C.D. Establishing intracranial brain tumor xenografts with subsequent analysis of tumor growth and response to therapy using bioluminescence imaging. J. Vis. Exp. 2010;(41):1986. doi: 10.3791/1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu X., Wang X., Du W., Chen L., Wang G., Cui Y., Liu Y., Dou Z., Wang H., Zhang P. Suppressor of fused (Sufu) represses Gli1 transcription and nuclear accumulation, inhibits glioma cell proliferation, invasion and vasculogenic mimicry, improving glioma chemo-sensitivity and prognosis. Oncotarget. 2014;5:11681–11694. doi: 10.18632/oncotarget.2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen J., Shetty S., Zhang P., Gao R., Hu Y., Wang S., Li Z., Fu J. Aspirin-triggered resolvin D1 down-regulates inflammatory responses and protects against endotoxin-induced acute kidney injury. Toxicol. Appl. Pharmacol. 2014;277:118–123. doi: 10.1016/j.taap.2014.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zamanian-Daryoush M., Marques J.T., Gantier M.P., Behlke M.A., John M., Rayman P., Finke J., Williams B.R. Determinants of cytokine induction by small interfering RNA in human peripheral blood mononuclear cells. J. Interferon Cytokine Res. 2008;28:221–233. doi: 10.1089/jir.2007.0090. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.