Abstract
Low molecular weight (LMW) thiols play an important role as thiol-cofactors for many enzymes and are crucial to maintain the reduced state of the cytoplasm. Most Gram-negative bacteria utilize glutathione (GSH) as major LMW thiol. However, in Gram-positive Actinomycetes and Firmicutes alternative LMW thiols, such as mycothiol (MSH) and bacillithiol (BSH) play related roles as GSH surrogates, respectively. Under conditions of hypochlorite stress, MSH and BSH are known to form mixed disulfides with protein thiols, termed as S-mycothiolation or S-bacillithiolation that function in thiol-protection and redox regulation. Protein S-thiolations are widespread redox-modifications discovered in different Gram-positive bacteria, such as Bacillus and Staphylococcus species, Mycobacterium smegmatis, Corynebacterium glutamicum and Corynebacterium diphtheriae. S-thiolated proteins are mainly involved in cellular metabolism, protein translation, redox regulation and antioxidant functions with some conserved targets across bacteria. The reduction of protein S-mycothiolations and S-bacillithiolations requires glutaredoxin-related mycoredoxin and bacilliredoxin pathways to regenerate protein functions.
In this review, we present an overview of the functions of mycothiol and bacillithiol and their physiological roles in protein S-bacillithiolations and S-mycothiolations in Gram-positive bacteria. Significant progress has been made to characterize the role of protein S-thiolation in redox-regulation and thiol protection of main metabolic and antioxidant enzymes. However, the physiological roles of the pathways for regeneration are only beginning to emerge as well as their interactions with other cellular redox systems. Future studies should be also directed to explore the roles of protein S-thiolations and their redox pathways in pathogenic bacteria under infection conditions to discover new drug targets and treatment options against multiple antibiotic resistant bacteria.
Abbreviations: Ac, acetyl; AcCys, acetyl cysteine; AhpE, membrane-associated peroxidase; AldA, aldehyde dehydrogenase A; Bca, BSH S-conjugate amidase; Brx, bacilliredoxin; BSH, bacillithiol; BshA, glycosyltransferase for GlcNAc-Mal biosynthesis; BshB, deacetylase producing GlcN-Mal; BshC, cysteine ligase for BSH biosynthesis; BSSB, oxidized bacillithiol disulfide; Bst, BSH-S-transferases; CA-MRSA, community acquired MRSA; CHP, cumene hydroperoxide; CoASH, coenzymeA; CoASSH, CoASH persulfide; Cys, cysteine; DHAP, dihydroxyacetone phosphate; DTT, dithiothreitol; EGT, ergothioneine; FA, formaldehyde; FeS, iron-sulfur; GapDH, glycolytic glyceraldehyde 3-phosphate dehydrogenase; GuaB, inosine-5-monophosphate dehydrogenases; GlcN, glucoseamine; GlcNAc, N-acetyl glucoseamine; GlxA/B, glyoxalases A and B; Grx, glutaredoxin; GSH, glutathione; GSSG, oxidized glutathione disulfide; Gst, GSH-S-transferases; H2O2, hydrogen peroxide; HED, hydroxyethyl disulfide; HTA, hemithioacetal; INH, isoniazid; Ins, myoinositol; LC-MS/MS, liquid chromatography tandem mass spectrometry; LMW, low molecular weight; Mal, malate; Mca, mycothiol-S-conjugate amidase; Met, methionine; MetE, methionine synthase; MetSO, methionine sulfoxide; MG, methylglyoxal; MgsA, methylglyoxal synthase; MRSA, methicillin-resistant Staphylococcus aureus; Mrx1, mycoredoxin1; MSH, mycothiol; MSONH2, MSH sulfinamide; MSNO, S-nitrosomycothiol; MsrA/B, methionine sulfoxide reductase A/B; MST, mycothiol-S-transferase; MT, metallothionein; Mtb, Mycobacterium tuberculosis; NADH, nicotinamide adenine dinucleotide; NADPH, nicotinamide adenine dinucleotide phosphate; HOCl, sodium hypochlorite; NFC, nitrofuranylcalanolide; OHP, organic hydroperoxide; OhrR, organic hydroperoxide repressor; PDB, Protein Data Bank; PpaC, inorganic pyrophosphatase; pKa, negative base-10 logarithm of the acid dissociation constant; PPP, pentose phosphate pathway; protein-SSB, BSH protein mixed disulfide; RES, reactive electrophilic species; RNS, reactive nitrogen species; RSS, reactive sulfur species; roGFP2, redox-sensitive green fluorescent protein; ROS, reactive oxygen species; SarZ, redox-sensing virulence regulator; STRING, Search Tool for the Retrieval of Interacting Genes/Proteins; Trx, thioredoxin; TrxR, thioredoxin reductase; YpdA, NADPH-dependent flavin oxidoreductase
Keywords: Protein S-thiolation, Bacillithiol, Mycothiol, Gram-positive bacteria
Graphical abstract
Highlights
-
•
Bacillithiol and mycothiol are major LMW thiols in many Gram-positive bacteria.
-
•
HOCl leads to widespread protein S-mycothiolation and S-bacillithiolation which function in thiol-protection and redox regulation.
-
•
Redox-sensitive metabolic and antioxidant enzymes are main targets for S-mycothiolation or S-bacillithiolation.
-
•
Mycoredoxin and bacilliredoxin pathways mediate reduction of S-thiolations.
1. Introduction
Low molecular weight (LMW) thiols play an important role in many cellular processes in all organisms. They are crucial to maintain the reduced state of the cytoplasm and function as thiol-cofactors of enzymes involved in detoxification of reactive oxygen, electrophilic, chlorine, nitrogen and sulfur species (ROS, RES, RCS, RNS, RSS), toxins and antibiotics, in metal storage, buffering and transport, and sulfide homeostasis [1], [2], [3]. The well-studied LMW thiol glutathione (GSH) is produced in most eukaryotes, Gram-negative bacteria, and some Gram-positive bacteria, such as Streptococci, Listeria, Lactobacilli and Clostridia [2], [4]. Most Gram-positive bacteria do not produce GSH and utilize instead alternative LMW thiols to cope with oxidative stress and redox regulation of metabolic enzymes. Actinomycetes utilize mycothiol (MSH) as their major LMW thiol (Fig. 1A) [5], [6]. In Firmicutes, such as Bacillus and Staphylococcus species, bacillithiol (BSH) plays a related role like GSH to control cellular redox homeostasis under oxidative stress and infection conditions (Fig. 1B) [3], [7]. There is also evidence that coenzyme A (CoASH) may substitute for the absence of BSH in some Firmicutes, such as S. aureus or Bacillus megaterium [2], [8]. In addition, S. aureus and Borrelia burgdorferi both encode a CoAS disulfide reductase, further indicating that CoASH plays a role to cope with oxidative stress [8], [9].
Fig. 1.
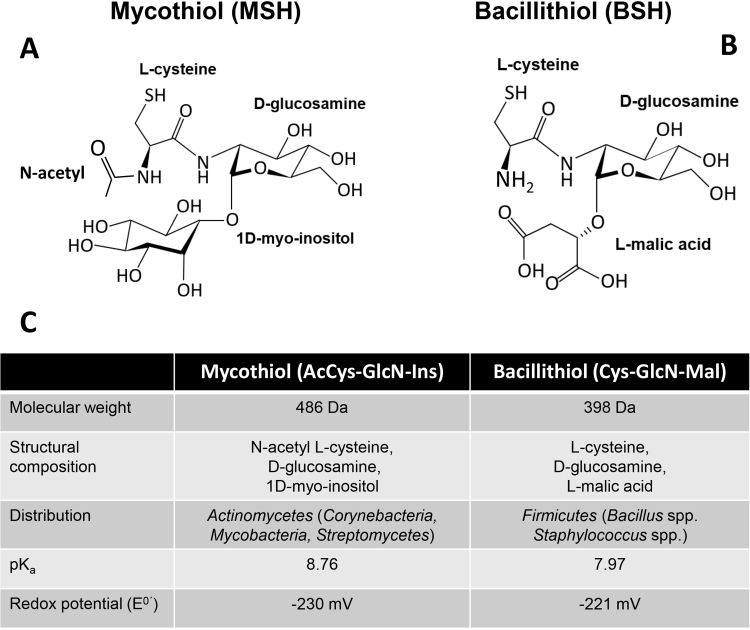
Structures, distributions and chemical properties of mycothiol and bacillithiol. (A, C) Mycothiol (MSH, AcCys-GlcN-Ins) is composed of acetyl cysteine, glucosamine and myoinositol and has a molecular weight (MW) of 486 Da. MSH has an standard thiol redox potential of −230 mV and the pKa of the mycothiolate anion was determined as 8.76. MSH is the major low molecular weight (LMW) thiol in high-GC Gram-positive Actinomycetes, including Corynebacteria, Mycobacteria and Streptomycetes. (B, C) Bacillithiol (BSH, Cys-GlcN-Ins) consists of cysteine, glucosamine and malate. BSH has an MW of 398 Da and a standard thiol redox potential of −221 mV. The pKa of the bacillithiolate anion was determined as 7.96. BSH is utilized as the major LMW thiol in low-GC content Gram-positive Firmicutes, such as Bacillus and Staphylococcus species.
This figure is adapted from [7].
The question arises why different bacteria utilize different LMW thiols? Phylogenetic analyses revealed that the γ-glutamylcysteine synthase GshA, which catalyzes the first step of GSH biosynthesis, likely evolved in cyanobacteria and was distributed then to other bacteria and eukaryotes by lateral gene transfer [10], [11]. Since the major function of GSH is the protection against oxygen toxicity, GSH biosynthesis was acquired with the evolution of oxygen by cyanobacteria. However, there was significant microbial diversity before the evolution of cyanobacteria, resulting in the evolution of alternative LMW thiols as protection mechanisms against increasing oxygen toxicity. Thus, it is not surprising that various prokaryotes utilize different LMW thiols [10].
LMW thiols are usually present in millimolar concentrations in the cytosol and are kept reduced by NADPH-dependent thiol-disulfide reductases [2], [12]. Under oxidative and hypochlorite stress, redox-sensitive protein thiols are susceptible to various forms of thiol-oxidations, including reversible S-thiolations with LMW thiols as well as intra- or intermolecular protein disulfides (Fig. 2). In the absence of adjacent thiols, protein thiols can be also overoxidized to irreversible Cys sulfinic or sulfonic acids. Protein S-thiolations can have protective functions and control the activity of metabolic enzymes or transcription factors (Fig. 2). This review summarizes the current knowledge about the role of protein S-thiolations in cellular physiology and under oxidative stress in Gram-positive bacteria, such as Bacillus subtilis, Staphylococcus aureus, Corynebacterium diphtheriae and Mycobacterium smegmatis. Molecular dynamics simulations have provided further insights into the structural protein changes upon S-thiolation, which are discussed for conserved S-bacillithiolated proteins at the molecular and mechanistic level.
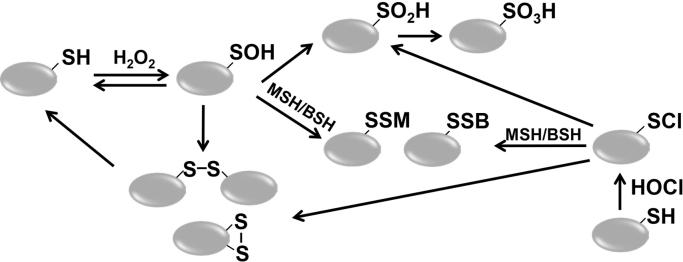
Fig. 2.
Thiol-chemistry in response to H2O2 and HOCl stress. Thiol-oxidation of Cys residues by H2O2 generates an unstable Cys sulfenic acid intermediate (Cys-SOH) that can rapidly react with LMW thiols, such as mycothiol (MSH) and bacillithiol (BSH), resulting in S-mycothiolations (Cys-SSM) and S-bacillithiolations (Cys-SSB) respectively. Alternatively, Cys oxidation can lead to the formation of intermolecular or intramolecular disulfides in proteins. In the absence of LMW thiols, protein thiols can be overoxidized to irreversible Cys sulfinic acid (Cys-SO2H) and sulfonic acid (Cys-SO3H). HOCl reacts with protein thiols via chlorination, generating an unstable sulfenylchloride intermediate (Cys-SCl) that reacts further with MSH, BSH or protein-thiols to reversible S-mycothiolations, S-bacillithiolations or protein disulfides. In the absence of adjacent thiols, sulfenylchloride rapidly leads to overoxidation generating Cys sulfinic and sulfonic acids.
2. Biosynthesis and functions of mycothiol and bacillithiol in Gram-positive bacteria
2.1. The biosynthetic pathway and functions of mycothiol in Actinomycetes
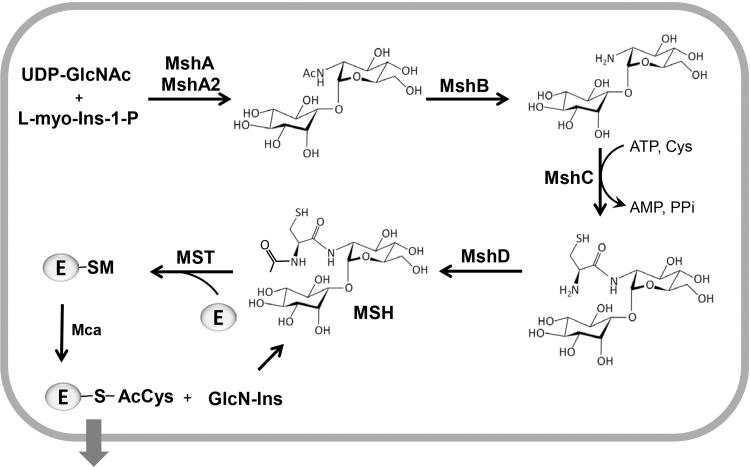
The cysteinyl pseudo-disaccharide mycothiol (MSH; AcCys-GlcN-Ins) is composed of acetyl cysteine (AcCys), glucosamine (GlcN) and myoinositol (Ins) and has a molecular weight of 486 Da. MSH is the major LMW thiol in all Actinomycetes, including Corynebacteria, Mycobacteria and Streptomycetes (Fig. 1AC) [6], [7], [13]. The biosynthesis of MSH is catalyzed in five enzymatic steps, which involve MshA, MshA2, MshB, MshC and MshD (Fig. 3). The first step of the MSH biosynthesis is catalyzed by the glycosyltransferase MshA, which conjugates myo-inositol-1-phosphate (Ins-P) to UDP-N-acetyl glucosamine (UDP-GlcNAc) leading to the formation of N-acetyl glucosamine myo-inositol-1-phosphate (GlcNAc-Ins-P). In the second step, the phosphatase MshA2 catalyzes dephosphorylation of GlcNAc-Ins-P to N-acetyl glucosamine myo-inositol (GlcNAc-Ins). The third step involves the metal-dependent deacetylase MshB for deacetylation of GlcNAc-Ins leading to GlcN-Ins [6], [14]. The MshB enzyme is a homolog of the MSH-conjugate amidase (Mca) that catalyzes the hydrolysis of MS-conjugates. In the fourth step of the MSH biosynthesis, the ATP-dependent ligase MshC ligates cysteine to GlcN-Ins leading to Cys-GlcN-Ins. The Cys ligase MshC is a homolog of the Cys-tRNA synthetase. The acetyltransferase MshD catalyzes the acetylation of the Cys amino group by acetyl-CoA as final step of MSH biosynthesis [6], [13], [15]. Under oxidative stress, MSH is oxidized to mycothiol disulfide (MSSM) which requires the NADPH-dependent mycothiol disulfide reductase (Mtr) for NADPH-dependent reduction of MSSM to maintain a high MSH: MSSM redox ratio [13], [16]. The levels of MSH vary strongly between different members of Actinomycetes with the highest levels of ~1–20 µmol/g raw dry weight (rdw) in Mycobacteria and much lower levels of 0.3 µmol/g rdw in C. diphtheriae [17], [18].
Fig. 3.
The biosynthesis pathway of MSH and detoxification of electrophiles in Actinomycetes. The MSH glycosyltransferase MshA conjugates myo-inositol-1-phosphate (Ins-P) to UDP-N-acetyl glucosamine (UDP-GlcNAc) leading to N-acetyl glucosamine myo-inositol-1-phosphate (GlcNAc-Ins-P). Dephosphorylation and deacetylation of GlcNAc-Ins-P is catalyzed by the phosphatase MshA2 and the deacetylase MshB, respectively. MshC ligates cysteine to GlcN-Ins in an ATP-dependent reaction. MshD acetylates the Cys moiety of Cys-GlcN-Ins to produce MSH. The mycothiol-S-transferase (MST) catalyzes the MSH-dependent detoxification of toxic electrophiles (E), xenobiotics and antibiotics. The mycothiol-S-conjugate amidase (Mca) cleaves the amide bond to produce mercapturic acids (AcCys-R) and GlcN-Ins. Mercapturic acids are exported and GlcN-Ins is recycled to MSH. This figure is adapted from [93].
The thiol pKa for MSH was determined as 8.76, which is only slightly more acidic (0.17 pH units) compared to the thiol pKa of GSH, and 0.4 pH units less acidic compared to the first microscopic thiol pKa value of free Cys (Fig. 1C) [19]. Since MSH is present at much higher levels compared to Cys in Actinomycetes, the mycothiolate anion is the most abundant LMW thiolate anion that reacts with oxidants and electrophiles in vivo. The thiol-redox potential of MSH is in the range of GSH and was calculated as E0′(MSSM/MSH) of −230 mV (Fig. 1C). However, the thiol-redox potential of MSH calculated with biophysical methods showed discrepancies compared to previously measured EMSH values determined as −300 mV using the Mrx1-roGFP2 biosensor [19], [20]. This indicates technical challenges to measure the exact MSH/MSSM ratio inside bacterial cells.
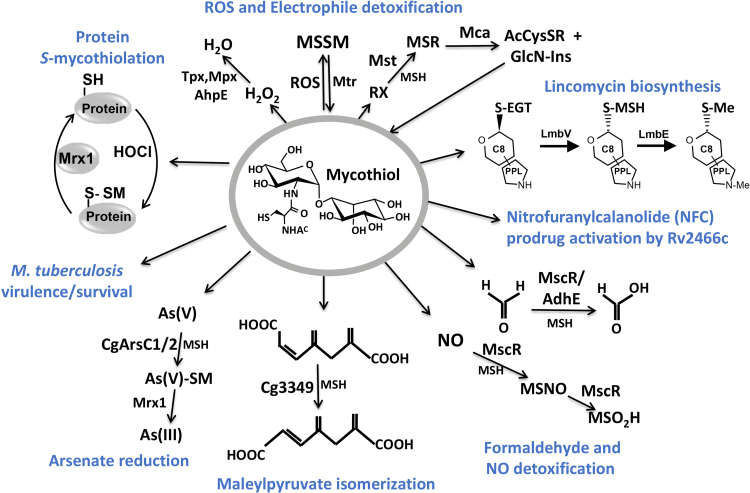
MSH plays an important role as thiol-cofactor for many enzymes that are involved in the detoxification of antibiotics, xenobiotics, ROS, RES, RNS, and other reactive species (Fig. 4) [2], [6], [7]. For a comprehensive overview of the detailed functions of MSH in different Actinomycetes the reader is referred to recent reviews which will be briefly outlined here and updated based on novel results [6], [7], [13].
Fig. 4.
The functions of mycothiol (MSH) in Actinomycetes. Mycothiol (MSH) is oxidized by ROS to mycothiol disulfide (MSSM) and regenerated by the mycothiol disulfide reductase (Mtr). The antioxidant enzymes Mpx, Tpx and AhpE were shown to function in H2O2 detoxification. Electrophiles (RX) are detoxified by the MSH S-transferase (MST) leading to MS-electrophiles (MSR) that are cleaved by the MSH S-conjugate amidase (Mca) to mercapturic acids (AcCyS-R) that are exported. MSH and ergothioneine (EGT) are involved in natural product biosynthesis of the lincosamide antibiotic lincomycin in Streptomyces lincolnensis. MSH functions as cofactor for the nitroreductase Rv2466c which activates the new mycobactericidal prodrug Nitrofuranylcalanolide (NFC) for reduction of the nitro group to amines. MSH is also a thiol-cofactor for detoxification of formaldehyde, S-nitrosomycothiol (MSNO) and arsenate. MscR is involved in MSNO detoxification generating MSH sulfinamide (MSONH2). MscR and AdhE both catalyze oxidation of formaldehyde to formate. In C. glutamicum, MSH is a cofactor for maleylpyruvate isomerase involved in isomerization of maleylpyruvate to fumaryl pyruvate. The arsenate reductases ArsC1/C2 catalyze conjugation of MSH to arsenate [Ars(V)] which is further reduced by Mrx1 to arsenite [Ars(III)]. MSH is also important for growth, survival and virulence in M. tuberculosis. Under HOCl stress, MSH functions in post-translational modification of proteins, termed as S-mycothiolations. This figure is adapted from [2].
MSH conjugates xenobiotics and antibiotics either spontaneously or enzyme-catalyzed by MSH S-transferases (MST), which belong to the DinB superfamily (Fig. 3, Fig. 4) [21]. These MSH-S-conjugates are hydrolyzed by the mycothiol-S-conjugate amidase (Mca), releasing mercapturic acid derivatives (AcCys-R) and GlcN-Ins. The GlcN-Ins is recycled to MSH and the toxic mercapturic acid derivatives are exported from the cell. Mca was shown to be involved in detoxification of MSH-S-conjugates with the antibiotics cerulenin and rifamycin in Mycobacteria [13]. MSH and ergothioneine (EGT) were also shown to function as sulfur donors through S-glycosylation reactions to mediate amino sugar transfer, activation and modification during the biosynthesis of the lincosamide antibiotic lincomycin in Streptomyces lincolnensis [22], [23]. This indicates a direct function of the LMW thiols MSH and EGT in the biosynthesis and molecular assembly of sulfur-containing natural products [22], [23].
MSH functions as thiol-cofactor for detoxification of formaldehyde, RNS, maleylpyruvate, methylglyoxal and arsenate and is required for activation of anti-mycobacterial prodrugs (Fig. 4). The MSH-dependent detoxification enzyme MscR is a dual function enzyme with S-nitrosomycothiol (MSNO) reductase and formaldehyde dehydrogenase activities [6], [13]. Formaldehyde is conjugated to MSH leading to S-hydroxymethyl-MSH, which is further oxidized to an S-formyl thioester and formate. MSNO is converted by MscR to MSH sulfinamide (MSONH2). The maleylpyruvate isomerase of C. glutamicum uses MSH as a cofactor for the enzymatic isomerization of maleylpyruvate to fumarylpyruvate [24]. MSH was shown as cofactor of the MSH-dependent arsenate reductases ArsC1/C2 in the detoxification of arsenate [25], [26].
MSH further contributes to antibiotic resistance in Mycobacterium tuberculosis (Mtb). MSH is required for activation of the pro-drugs isoniazid (INH) and nitrofuranylcalanolides (NFCs) which are potent anti-mycobacterial drugs [27], [28]. INH is activated by the catalase KatG and MSH, leading to NAD-INH adduct formation and inhibition of the enoyl-ACP reductase (InhA) of the mycolic acid biosynthesis pathway [29]. INH resistant Mtb isolates often carry spontaneous mutations in katG or mshA [14]. Novel antimycobacterial drugs, such as the nitrofurantoin derivatives NFC or the thienopyrimidine compound TP053 are activated by the MSH-dependent oxidoreductase Rv2466c, which was recently revealed as MSH-dependent nitroreductase to reduce nitro groups to amines [28], [30]. Importantly, NFC-resistant mutants were selected which had mutations in the gene encoding Rv2466c. This indicates that the MSH-dependent nitroreductase activity of Rv2466c is crucial for pro-drug activation [28]. Rv2466c has a DsbA-like structure and can also function as mycoredoxin-2 in demycothiolation with electrons from the MSH/Mtr pathway [30]. Rv2466c uses a monothiol-disulfide mechanism for reduction of S-mycothiolated proteins or intramolecular disulfides and a dithiol mechanism for prodrug activation (TP053) in vitro.
MSH further functions as reservoir of cysteine and is much less susceptible to auto-oxidation compared to cysteine [13]. In addition, MSH has important functions in the virulence and survival of the pathogen Mtb under infection conditions [31]. An essential function of MSH for growth and viability was found in Mtb since the mshC mutant could be only generated in the presence of a second copy of mshC [31].
2.2. Shotgun proteomics identified widespread protein S-mycothiolations in Actinomycetes
Under HOCl stress, MSH was shown to form mixed disulfides with protein thiols, termed as protein S-mycothiolations. Protein S-mycothiolations are widespread redox-modifications and were identified in many Actinomycetes, such as C. glutamicum, C. diphtheriae and M. smegmatis [18], [25], [32]. However, the extent of protein S-mycothiolation under HOCl stress differs with 58, 25 and 26 proteins identified in M. smegmatis, C. glutamicum and C. diphtheriae, respectively (Table S1). This higher level of S-mycothiolated proteins in Mycobacteria might be correlated with the 20-fold higher MSH level compared to Corynebacteria [17]. Overall, S-mycothiolated proteins were shown to be involved in many metabolic pathways, including glycolysis, gluconeogenesis, glycogen and maltodextrin degradation, as well as in the biosynthetic pathways for fatty acids, amino acids and nucleotides, cofactors, and protein translation. Several conserved S-mycothiolated proteins have antioxidant functions, such as peroxiredoxins (Tpx, Mpx, AhpE, AhpC) and the methionine sulfoxide reductase MsrA [18], [25], [32], [33].
In C. glutamicum, 25 proteins with S-mycothiolations were identified under HOCl stress and their level of oxidation was quantified using fluorescence-based thiol-redox proteomics [25] (Table S1). These S-mycothiolation targets function in glycolysis (Fba, Pta, XylB), glycogen and maltodextrin degradation (MalP), in the amino acid biosynthesis pathways for serine, cysteine, methionine (MetE, SerA, Hom), nucleotides and thiamine cofactors (GuaB, PurL, ThiD1, ThiD2), antioxidant functions (Tpx, Mpx), methionine sulfoxide reduction (MsrA), heme degradation (HmuO), and protein translation (RpsF, RpsC, RpsM, Tuf). Among these, Tuf, GuaB1, GuaB2, SerA and MetE are also conserved targets for S-thiolations across Gram-positive bacteria [25]. The most interesting S-mycothiolated metabolic enzyme in C. glutamicum was the maltodextrin phosporylase (MalP), which is involved in glycogen degradation during the stationary phase [34]. The malP mutant was very sensitive under HOCl stress indicating an essential function of MalP under oxidative stress [25]. Furthermore, the glycogen content was not decreased in C. glutamicum wild type under HOCl stress despite drastically reduced glucose uptake rates. Thus, S-mycothiolation of MalP is suggested to inhibit its function in glycogen degradation to save the source of energy under oxidative stress [25].
Many antioxidant enzymes were S-mycothiolated at their active site Cys residues and it was further investigated if S-mycothiolation functions in redox-regulation of peroxiredoxins (Tpx, Mpx, AhpE) and methionine sulfoxide reductases (MsrA, MsrB) [25], [33], [35], [36], [37], [38]. The thiol peroxidase (Tpx), an atypical 2-Cys peroxiredoxin was S-mycothiolated at its active and resolving Cys60 and Cys94 residues [25]. S-mycothiolation of Tpx in vitro inhibited its peroxidase activity. The MSH peroxidase (Mpx) was also S-mycothiolated at its peroxidatic Cys36 residue. The methionine sulfoxide reductase MsrA is involved in the repair of methionine sulfoxides and was S-mycothiolated at its conserved Cys91 residue. The cobalamin-independent methionine synthase (MetE) was S-mycothiolated at its Zn-binding active site Cys713 under HOCl stress and S-mycothiolation of MetE was shown to function in thiol-protection under acid stress [25], [39]. The inosine-5-monophosphate (IMP) dehydrogenases GuaB1 and GuaB2 are conserved S-thiolated proteins across bacteria. GuaB1 and GuaB2 were S-mycothiolated at their active site Cys302 and Cys317 respectively, forming the thioimidate intermediate [25].
In the pathogen C. diphtheriae, 26 S-mycothiolated proteins were identified under HOCl stress using shotgun liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis [18] (Table S1). These include five conserved targets for S-thiolations across Gram-positive bacteria, such as AhpC, the ribosomal proteins RplC and RpsM, the glycolytic enzyme glyceraldehyde-3-phosphate dehydrogenase (GapDH), and GuaB [18]. Other S-mycothiolated proteins are involved in energy metabolism, including the ribose 5-phosphate isomerase DIP1796 and the NADH dehydrogenases Ndh, GlpD and DIP1726. Further targets for S-mycothiolation function in the biosynthesis of amino acids (LeuB, DapA, GlnA), purine (PurA), iron sulfur-clusters (DIP1631) and cell wall metabolites (GlmS). The S-mycothiolated proteins contributed with 0.2–0.75% Cys abundance to the total Cys proteome of C. diphtheriae as revealed by shotgun proteomics [18]. Further biochemical studies of GapDH confirmed that protein S-mycothiolations function in redox-regulation and thiol-protection against overoxidation as referred in the following section.
In M. smegmatis, we identified 58 S-mycothiolated proteins under HOCl stress [32] (Table S1). The conserved peroxiredoxins Tpx, AhpC and OsmC are S-mycothiolated at their active and/or resolving Cys residues, which are involved in redox-regulation and detoxification processes in M. smegmatis. The global transcriptional regulator for iron uptake of the DtxR-family (IdeR) was S-mycothiolated at Cys102 in its primary iron-binding site [32]. Many abundant enzymes of the energy metabolism were further identified as S-mycothiolated in M. smegmatis. These are involved in the glycerol catabolism, glycolysis, the glyoxalate shunt and gluconeogenesis. The glycerol kinase GlpK3 and the glycerol dehydrogenase Adh2 are abundant S-mycothiolated proteins since glycerol is used as sole source of carbon and energy in M. smegmatis. The generation of dihydroxyacetone phosphate (DHAP) involves GlpK3 and Adh2. Thus, S-mycothiolation could prevent glycerol degradation under HOCl stress to save the carbon and energy source. The isocitrate lyase AceA and the myo-inositol-1-phosphate synthase Ino1 were further identified as abundant S-mycothiolated proteins in M. smegmatis. AceA is the key enzyme of the glyoxylate bypass of the TCA cycle in M. tuberculosis, which enables the use of carbon for biomass production via gluconeogenesis during growth on fatty acids as sole source of carbon and energy [40]. Thus, S-mycothiolation could inhibit AceA under oxidative stress to stop gluconeogenesis [32]. Furthermore, abundant enzymes involved in the biosynthesis of fatty acids as precursors for mycolic acids were S-mycothiolated in M. smegmatis, including acetyl-CoA carboxylases (AccD5 and AccD6), the enoyl-CoA hydratase (EchA6), the methoxy mycolic acid synthase (UmaA), the acyl-CoA dehydrogenase (MSMEG_0531), and the acyl-CoA-thioesterase (MSMEG_6208). Other S-mycothiolated proteins of M. smegmatis are involved in the nucleotide biosynthesis, including GuaB and GuaB2, and biosynthesis enzymes for thiamine (ThiG, MSMEG_4827), cobalamine (CobN), iron sulfur-cluster assembly (YfhF2) as well as ribosomal proteins (RplC, RpsM, RpsR2) and amino acyl tRNA synthetases (GatC, PheT) for protein translation [32]. Overall, many detailed future studies are required to elucidate the physiological role of the widespread protein S-mycothiolations in redox regulation and/or thiol-protection in Mycobacteria. In particularly, it would be interesting if mycothiolation functions in redox regulation of transcriptional regulators under infection conditions in M. tuberculosis. Recent biochemical studies revealed the formation of S-mycothiolated NsrR, a FeS-cluster-based Rrf2-family regulator and NO-sensor in many Gram-positive bacteria [41]. Using mass spectrometry, the formation of several iron nitrosyl species and S-thiolated forms of the Cys residues coordinating the FeS cluster have been reported for NsrR [41]. Thus, related S-nitrosylation and S-thiolation mechanisms could be relevant for redox-sensing of NsrR under in vivo conditions. In support of this notion, the NsrR-controlled hmp gene is also induced under HOCl stress in the RNAseq transcriptome of C. glutamicum and S. aureus [32], [42]. This might indicate that NsrR could sense HOCl stress via protein S-thiolation which remains to be investigated.
2.3. Protein S-mycothiolation is redox-controlled by the mycoredoxin-1 (Mrx1) and thioredoxin pathways
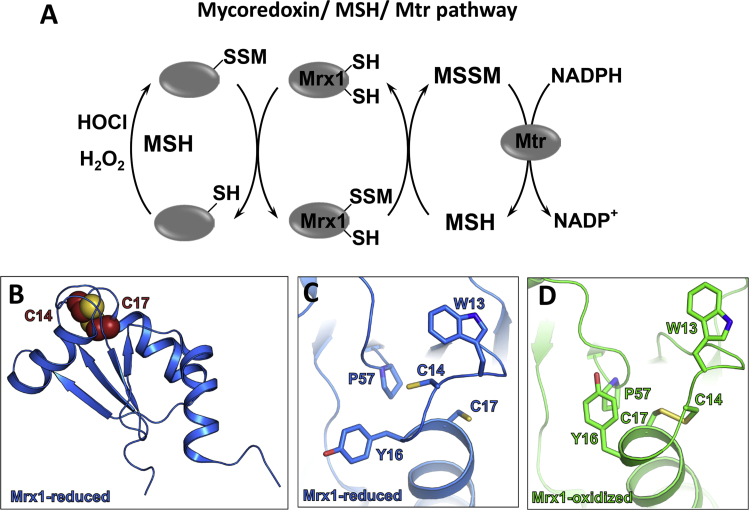
The redox regulation of protein S-mycothiolations requires specific thiol-disulfide reducing pathways to restore the protein activity. The Mrx1/MSH/Mtr system is specific for the de-mycothiolation of MSH-mixed protein disulfides (Fig. 5). Mycoredoxin-1 (Mrx1) was characterized as a glutaredoxin-homolog in Actinomycetes [43]. Mrx1 has a typical Trx-like fold with a CGYC catalytic active site, composed of a four-stranded antiparallel β-sheet and surrounded by three α-helices. The CGYC motif is located at the N-terminus of the first α-helix. The conserved proline residue (Pro57) is located in cis-conformation opposite to the active site. The active site Cys14 is solvent exposed in both oxidized and reduced forms of Mrx1, while the C-terminal resolving Cys17 is buried inside the protein [43]. Mrx1 has a negative redox potential of −218 mV that is in the same range as for glutaredoxins. Thus, Mrx1 can function as effective thiol disulfide reducing enzyme in Actinomycetes [44]. Mrx1 catalyzes the de-mycothiolation of MSH-mixed protein disulfides in a bimolecular nucleophilic substitution reaction analogous to the monothiol reaction mechanisms of glutaredoxins. During de-mycothiolation of the substrates, Mrx1 is S-mycothiolated at its active site Cys14. Oxidized Mrx1 is regenerated by MSH leading to formation of MSSM, which is reduced by Mtr to MSH with NADPH as electron donor. The electron transfer from the Mrx1/MSH/Mtr electron pathway to the MSH mixed disulfide substrate was shown in a hydroxyethyl disulfide (HED) assay [43].
Fig. 5.
Reversal of protein S-mycothiolation by the mycoredoxin-1 pathway. (A) In MSH-producing Actinomycetes, S-mycothiolated proteins are reduced by mycoredoxin-1 (Mrx1), resulting in an Mrx1-SSM intermediate that is reduced by MSH leading to MSSM. The NADPH-dependent mycothiol disulfide reductase Mtr recycles MSSM back to MSH. (B) The crystal structure of the reduced Mrx1 (PDB ID: 2LQO) and (C) its active site. (D) The oxidized Mrx1 active site in its intramolecular disulfide form (PDB ID: 2LQQ). The active and resolving Cys residues of Mrx1 are shown as spheres in (B) and as sticks in (C, D).
The peroxiredoxin Tpx of C. glutamicum was identified as first substrate for Mrx1 in vitro. S-mycothiolation of the peroxidatic Cys60 inhibited the peroxidase activity of Tpx, which could be restored by the Mrx1/MSH/Mtr electron pathway. Mass spectrometry confirmed the complete reduction of Tpx-SSM and MSH transfer to Mrx1, resulting in the formation of a transient Mrx1-SSM intermediate [25]. Another mycothiol peroxidase, Mpx of C. glutamicum controls intracellular ROS levels and was shown to be recycled by both, the Mrx1 and the thioredoxin/ thioredoxin reductase (Trx/TrxR) pathways [35], [38]. Reduction of the S-mycothiolated peroxidatic Cys36 of Mpx by Mrx1 proceeds via a monothiol mechanism as demonstrated by kinetic assays and MSH-specific Western blots [35]. Arsenate [Ars(V)] detoxification by the arsenate reductases ArsC1 and ArsC2 in C. glutamicum requires MSH and the Mrx1/MSH/Mtr pathway. ArsC1/C2 catalyze conjugation of MSH to As(V) resulting in an arseno-MSH conjugate (As(V)-SM). Mrx1 reduces the As(V)-SM conjugate to arsenite [As(III)] via a monothiol mechanism (Fig. 4) [26].
In C. diphtheriae, the glycolytic GapDH was the most abundant S-mycothiolated protein contributing with 0.75% to the total Cys proteome [18]. GapDH uses the active site Cys for the nucleophilic attack at the aldehyde group of glyceraldehyde-3-phosphate (G3P) to catalyze its phosphorylation to 1,3-bisphosphoglycerate and thereby generating NADH [45]. GapDH is S-mycothiolated at the conserved catalytic active site Cys153 resulting in reversible inhibition of GapDH activity under HOCl stress [18]. It was previously shown that H2O2-stress leads to an inactivation of GapDH upon S-thiolation, resulting in a metabolic re-configuration of central carbon metabolism [46], [47]. The lack of glycolysis is compensated by an increased glycolytic flux through the pentose-phosphate pathway (PPP) to provide increased levels of NAPDH as cofactor for thiol-disulfide oxidoreductases. The active site Cys of GapDH is highly reactive and susceptible for various post-translational thiol-modifications in response to ROS and RNS, including S-glutathionylation, S-nitrosylation and sulfenic acid formation in many eukaryotes [48], [49]. It was shown that the relatively high reactivity of the active site thiolate towards H2O2 depends on the stabilization of the transition state and a dedicated proton relay mechanism that promotes leaving group departure [50].
We demonstrated that S-mycothiolation protects the active site Cys of GapDH in C. diphtheriae against irreversible overoxidation under both H2O2 and HOCl treatments [18]. In our GapDH assays without MSH, increasing doses of H2O2 and HOCl resulted in an irreversible overoxidation of Cys153 to the sulfonic acid and partially Cys153-SS-Cys157 intramolecular disulfide bond formation [18]. In the presence of MSH, S-mycothiolation of the catalytic active site Cys153 could be verified by mass spectrometry and non-reducing MSH-specific Western blot analysis. Time course experiments indicated that inactivation of GapDH by S-mycothiolation was faster compared to its inactivation in the overoxidation pathway under HOCl and H2O2 stress [18]. In addition, intramolecular disulfides were detected under both, H2O2 and HOCl treatment in the presence and absence of MSH as an additional redox-regulatory mechanism of GapDH [18]. We also studied the redox-regulation of protein S-mycothiolation by the Mrx1 and Trx pathway. Reduction of S-mycothiolated GapDH occurred much faster by the Mrx1 pathway compared to Trx in vitro. Thus, Mrx1 plays probably the major role in demycothiolation of GapDH in vivo while Trx has only a minor role. These data are in agreement with the kinetics obtained for demycothiolation of Mpx by Mrx1, which was also faster compared to reduction by the Trx pathway [35].
MsrA and MsrB are further important redox enzymes which catalyze the reduction of two stereotypes of methionine sulfoxides (MetSO) to L-methionine [51]. While MsrA is specific for reduction of methionine-S-sulfoxide, MsrB shows activity for reduction of methionine-R-sulfoxides [52]. The Mrx1 and Trx pathway were shown to be involved in the recycling of the methionine sulfoxide reductase Cd-MsrA and Cg-MsrA during reduction of methionine sulfoxides in C. diphtheriae and C. glutamicum [36], [37], [53]. Two different redox relays operate in recycling of Cd-MsrA by the Trx and Mrx1 pathway upon MetSO reduction. Cd-MsrA has three Cys residues in positions 52, 206 and 215 with Cys52 as the active site. The disulfide relay which couples MetSO reduction by Cd-MrsA to the Trx pathway starts with formation of the Cys52-SOH upon attack of the MetSO substrate releasing reduced L-Met. Next, Cys52 is attacked by Cys206 or Cys216 resulting in intramolecular disulfides between Cys52 and either Cys206 or Cys215. This disulfide is reshuffled to a Cys206-Cys215 disulfide which is a substrate of Trx. In the MSH redox relay, Cys52-SOH forms an MSH mixed disulfide, followed by the subsequent transfer of MSH to Cys215 and Cys206. Cys206-SSM is finally the substrate that is recycled by the Mrx1 pathway. Thus, two different redox relays were discovered for Cd-MsrA upon MetSO reduction that require the Trx or Mrx1 pathway for regeneration [36]. Another study of Cg-MsrA’s catalytic mechanism in C. glutamicum revealed that Mrx1 reduces Cys56-SSM while Trx reduces the intramolecular disulfide between both C-terminal resolving Cys residues [53]. In addition, the Trx and Mrx1 pathways were required for reduction of Cg-MsrA under stress and the Trx pathway was found to be involved in Cg-MsrA regeneration under non-stress conditions. Apart from MsrA, the redox cycle of the methionine sulfoxide reductase MrsB of C. glutamicum was recently studied [37]. MsrB is specific for reduction of methionine-R-sulfoxides which requires the Trx pathway for recycling and was not important under oxidative stress in vivo [37].
The membrane-associated one-Cys peroxidase AhpE of M. tuberculosis was identified as S-mycothiolated at its active site Cys45 and was shown to be reduced by Mrx1 in vitro [33]. AhpE is S-mycothiolated during H2O2 detoxification. The reduction of AhpE-SSM was demonstrated in the presence of wild type Mrx1, indicating a dithiol mechanism. The peroxiredoxin AhpE comprises a molecular link between the peroxidase system and the Mrx1 redox pathway in Mycobacteria. Using NMR spectroscopy, the AhpE structures were resolved in the reduced, sulfenic acid, and sulfinic acid forms giving insights into the MSH-dependent reduction mechanism of AhpE [54].
In conclusion, recent studies on several identified S-mycothiolated antioxidant and metabolic enzymes (Tpx, Mpx, MsrA, and GapDH) revealed that both, the Mrx1 and Trx pathway are coupled to regeneration of their enzymatic activities under oxidative stress. However, Mrx1 was faster in demycothiolation of GapDH and Mpx compared to Trx indicating that Mx1 is the primary de-mycothiolating enzyme under oxidative stress. In concert with MSH and Mtr, Mrx1 can undergo mono- and dithiol mechanisms to reduce protein MSH-mixed disulfides. It remains to be further investigated whether mycoredoxin-2 (Rv2466c) plays a related or alternative role in reduction of S-mycothiolated proteins.
2.4. The biosynthetic pathway and functions of bacillithiol in Firmicutes
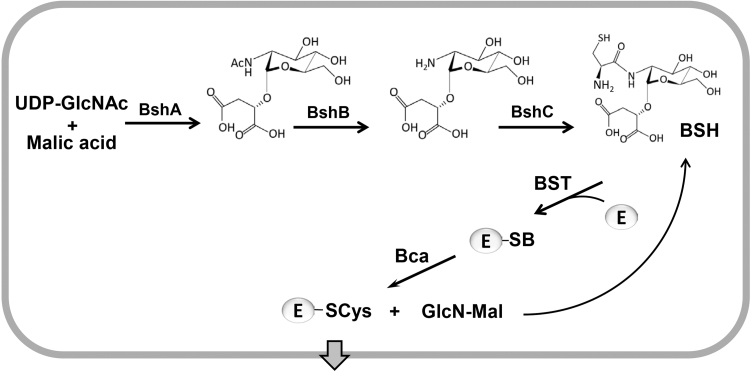
Bacillithiol (BSH, Cys-GlcN-malate) is the α-anomeric glycoside of L-cysteinyl-D-glucosamine with L-malic acid and has an MW of 398 Da (Fig. 1BC). BSH functions as the major LMW thiol in many Firmicutes bacteria, including Bacillus and Staphylococcus species [5]. The biosynthesis of BSH occurs in three steps by the enzymes BshA, BshB and BshC. The glycosyltransferase BshA catalyzes the conjugation of UDP-N-acetylglucosamine (UDP-GlcNAc) to L-malate leading to GlcNAc-Mal (Fig. 6). GlcNAc-Mal is further deacetylated by the deacetylase BshB to GlcN-Mal. The cysteine ligase BshC adds cysteine to GlcN-Mal as final step in the BSH biosynthesis [3], [55]. BSH is oxidized to BSSB under oxidative stress and high BSH/BSSB ratios were calculated as 100:1–400:1 in B. subtilis cells [56]. The putative BSSB reductase was suggested to function as NADPH-dependent flavin oxidoreductase YpdA which co-occurs with the BSH biosynthesis enzymes in BSH-producing bacteria [55]. However, experimental evidence is lacking to demonstrate a role of YpdA as BSSB reductase [3]. The standard thiol redox potential of BSH was calculated as E0′(BSSB/BSH) of −221 mV which is more positive compared to GSH (E0′(GSSG/GSH) is −240 mV) (Fig. 1C) [56]. However, microscopic pKa values of pKaSH= 7.97 and pKaSH= 9.55 were determined for the bacillithiolate anion (with protonated and deprotonated Cys amino group, respectively), suggesting a more acidic BSH thiolate anion with enhanced reactivity (Fig. 1C) [56].
Fig. 6.
The biosynthesis pathway of bacillithiol in Firmicutes. The glycosyltransferase BshA catalyzes the conjugation of UDP-N-acetylglucosamine (UDP-GlcNAc) to L-malate leading to N-acetylglucosamine malate (GlcNAc-Mal). GlcNAc-Mal is deacetylated by the deacetylase BshB to GlcN-Mal. The cysteine ligase BshC ligates cysteine to GlcN-Mal. The detoxification of toxic electrophiles (E) requires conjugation by the bacillithiol-S-transferase (BST) leading to BSH-S-conjugates which are cleaved by the BSH-S-conjugate amidase (Bca). Mercapturic acids are exported and GlcN-Mal is recycled to BSH. This figure is adapted from [93].
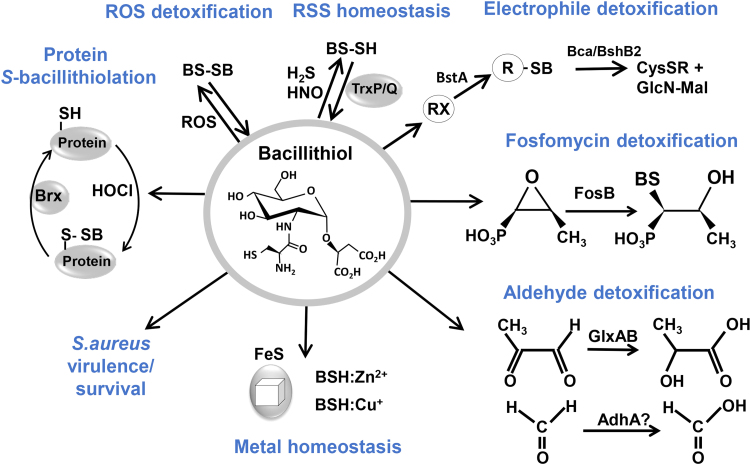
BSH functions in the detoxification of many thiol-reactive compounds, electrophiles, alkylating agents, toxic metals and antibiotics (Fig. 7) [3], [55]. BSH-deficient mutants in B. subtilis showed increased sensitivities toward hypochlorite, diamide, methylglyoxal, ROS, osmotic and acidic stress, alkylating agents, and fosfomycin. For a comprehensive overview of the many functions of BSH, the reader is referred to a recent review, which will be summarized and updated here only briefly [3].
Fig. 7.
The many functions of bacillithiol in Firmicutes. BSH is involved in detoxification of ROS, RES, HOCl, RSS and antibiotics (fosfomycin, rifampicin) in B. subtilis and S. aureus. ROS lead to oxidation of BSH producing bacillithiol disulfide (BSSB). Under sulfide stress, BSH persulfides (BSSH) were determined, which can be reduced by thioredoxins (TrxP and TrxQ). Electrophiles (RX) are conjugated to BSH by the BSH S-transferase BstA to form BS-electrophiles (BSR). The BSH S-conjugate amidases Bca or BshB2 cleave BSR into GlcNAc-Mal and mercapturic acids (CysSR) that are exported. BSH is used as cofactor for the epoxide hydrolase FosB in fosfomycin detoxification and functions as a cofactor for the glyoxalases GlxA and GlxB in B. subtilis in methylglyoxal detoxification. GlxA converts BSH-hemithioacetal to S-lactoyl-BSH that is a substrate for GlxB producing D-lactate. BSH-dependent detoxification of formaldehyde might be catalyzed by the formaldehyde dehydrogenase (AdhA) in B. subtilis. BSH functions in metal homeostasis as Zn2+ buffer and in FeS cluster assembly. In S. aureus, BSH is important for the virulence of S. aureus in macrophage infection assays. Under HOCl stress, proteins are S-bacillithiolated, which can be reversed by bacilliredoxins (Brx). This figure is adapted from [2].
The BSH-dependent thiol-S-transferase FosB is involved in the detoxification of the antibiotic fosfomycin and cleaves the ring structure of the BS-fosfomycin-conjugate [57]. The fosB and bsh mutants showed equal sensitivities towards fosfomycin treatment in B. subtilis and S. aureus indicating that FosB is a BSH-dependent S-transferase for fosfomycin detoxification. Co-crystallization of S. aureus FosB with BSH resulted in the formation of a BSH-mixed disulfide at the active site Cys9 of FosB of S. aureus in vitro [55], [58]. However, the electron density for the whole molecule of BSH was not complete in the FosB structure. The BSH S-transferase BstA catalyzes the conjugation of BSH to reactive electrophiles, such as chlorinated hydrocarbons and monobromobimane in vitro [21]. The resulting BS-electrophiles (BSR) are cleaved by the BSH S-conjugate amidases Bca or BshB2 into GlcNAc-Mal and mercapturic acids (CysSR). CysSR are exported from the cell by the potential efflux pumps encoded by the yfiS and yfiU genes [21].
BSH functions in detoxification of methylglyoxal as cofactor for the BSH-dependent glyoxalases GlxAB in B. subtilis [59]. Methylglyoxal reacts spontaneously with BSH to form BSH-hemithioacetal that is converted to S-lactoyl-BSH by GlxA. GlxB catalyzes the hydrolysis of S-lactoyl-BSH to lactate which is secreted. BSH is involved in detoxification of the toxic electrophile formaldehyde in the facultative methylotrophic bacterium Bacillus methanolicus which uses methanol as sole source of energy and carbon [60]. An unknown formaldehyde dehydrogenase is involved in oxidation of S-formyl-BSH in Bacillus methanolicus cells during growth on methanol, which is required for energy production [60].
BSH is further involved in detoxification of toxic heavy metals, such as tellurite and selenite and functions in metal homeostasis [61]. BSH can bind and store Zn2+ and functions as intracellular Zn2+ buffer in metal ion homeostasis [62]. The thiolate, amine, and carboxylate groups of BSH serve as ligands for metal coordination and can bind Zn2+ as (BSH)2:Zn2+ complex under Zn2+ stress. Treatment of the BSH-deficient mutant with Zn2+ resulted in a decreased accumulation of Zn2+ due to an increased expression of the CadA and CzcD efflux systems. BSH also protects against Zn2+ toxicity in cells lacking Zn2+ efflux pumps [62].
BSH is further implicated in Fe2+ homeostasis and is suggested to play a role in FeS cluster assembly and transport in S. aureus and B. subtilis [63], [64]. Mutants with defects in BSH biosynthesis showed a delayed growth upon depletion of Fe2+ or branched chain amino acids in the growth medium. The activities of several FeS-cluster enzymes (e.g. LeuCD, IlvD) were also decreased in the BSH-deficient mutant. These phenotypes suggest a potential role of BSH in FeS cluster biogenesis [64], [65]. Further growth analyses with nfu and sufA mutants indicate that BSH may participate in the biogenesis of FeS cluster proteins independently of the SufA and Nfu carriers [64], [65]. Finally, BSH has been shown to play a role in transport of copper to the Cu+ chaperone CopZ and Cu+ buffering to avoid Cu+ toxicity [66]. During Cu+ transport by BSH, CopZ was identified as S-bacillithiolated in vitro.
BSH was recently shown to control sulfide homeostasis under H2S and nitroxyl (HNO) stress in S. aureus [67], [68]. Strongly increased endogenous levels of BSH persulfide (BSSH), CoASH persulfide (CoASSH) and cysteine persulfide (CysSSH) were measured under Na2S and HNO treatment. Using a proteomics approach, several S-sulfhydrated proteins were identified by RSS stress including the glycolytic GapDH and the OhrR-type regulator MgrA. Biochemical experiments provided evidence that S-sulfhydration leads to inhibition of GapDH activity and functions in redox-regulation of the MgrA repressor to induce virulence factor expression. The thioredoxins (TrxP and TrxQ) enabled the specific reduction of protein S-sulfhydrations. The predicted CoASH disulfide reductases (Cdr) was suggested to function as CoASSH reductase [67].
BSH is also involved in virulence and survival of S. aureus under macrophage infection conditions as revealed by phenotype analysis of bshA mutants in clinical methicillin-resistant S. aureus (MRSA) isolates COL, USA300 and SH1000 [69], [70], [71]. S. aureus COL represents a hospital-acquired (HA) MRSA strain while USA300 was isolated as community-acquired (CA)-MRSA strain with strongly increased virulence due to many toxins encoded on prophages and other mobile genetic elements [72]. SH1000 is a natural bshC mutant belonging to the NCTC8325-4 lineage [73]. NCTC8325-4 is a laboratory model strain that was obtained from a sepsis isolate after removal of three resident prophages by UV. The survival of S. aureus COL and USA300 bshA mutants as well as of the natural bshC mutant of strain SH1000 were decreased in phagocytosis infections assays using murine macrophages and human whole-blood assays as compared to the wild type [69], [71]. These results suggest a role of BSH in the defense against the host-immune system and in the survival of S. aureus under infection conditions [69], [70], [71]. However, it remains to be shown if BSH is required for protection of S. aureus inside neutrophils against the antimicrobial activity of the myeloperoxidase (MPO) which is involved in HOCl production.
2.5. Physiological role of protein S-bacillithiolations in Firmicutes
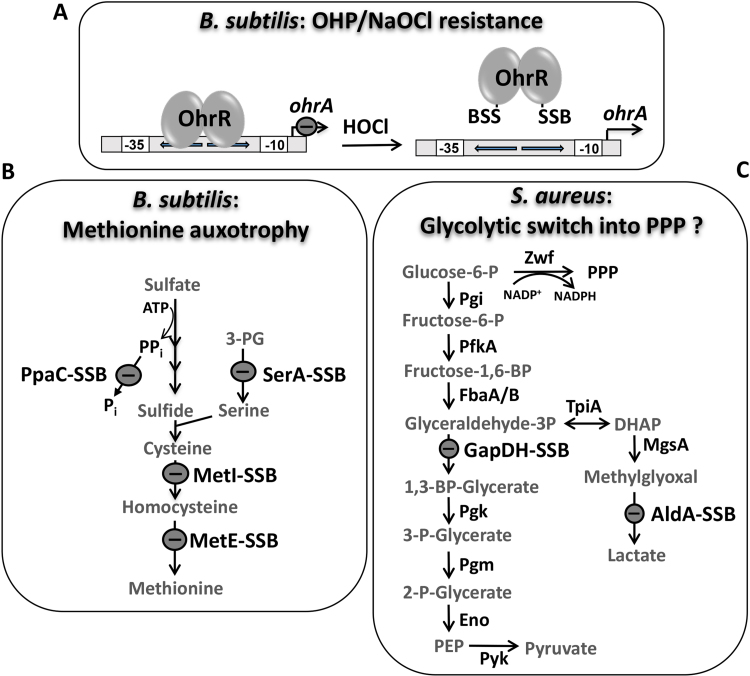
BSH plays an important role in post-translational modifications of proteins under oxidative stress in Gram-positive bacteria. In response to HOCl stress, protein thiols are oxidized to mixed disulfides with BSH, termed as protein S-bacillithiolation [74], [75], [76]. In total, we identified eight common and 29 unique S-bacillithiolated proteins using shotgun proteomics in B. subtilis, B. amyloliquefaciens, Bacillus pumilus, Bacillus megaterium and Staphylococcus carnosus (Table S2) [75], [76]. Many of the identified S-bacillithiolated proteins are involved in important cellular processes, such as the biosynthesis of amino acids, cofactors and nucleotides, protein translation, and redox-signalling of ROS. In B. subtilis, the MarR-type repressor OhrR was S-bacillithiolated under HOCl and organic hydroperoxide (OHP) stress at its redox-sensing Cys15. S-bacillithiolation of OhrR at its lone Cys residue leads to inactivation of its repressor function and derepression of transcription of the ohrA gene encoding the OhrA peroxiredoxin which confers resistance under HOCl and OHP stress (Fig. 8A) [74], [75]. MetE was identified as the most abundant S-bacillithiolated protein in B. subtilis, which was S-bacillithiolated at the active site Cys730 and at the non-conserved Cys719 [75]. S-bacillithiolation of MetE under HOCl stress resulted in a methionine auxotrophy phenotype perhaps to stop translation during recovery from oxidative stress (Fig. 8B). In addition, other enzymes involved in the Cys and Met biosynthesis pathways were identified as targets for S-bacillithiolation, including SerA, PpaC, MetI and YxjG. The translation elongation factor EF-Tu (TufA) and GuaB were also identified as abundant and essential S-bacillithiolated proteins under HOCl stress [25], [75], [76]. The bacilliredoxins BrxA, BrxB and BrxC were identified as S-bacillithiolated in B. subtilis and S. carnosus, which could be intermediates of the bacilliredoxin redox pathway [76].
Fig. 8.
Physiological roles of S-bacillithiolations of OhrR, MetE, GapDH and AldA under HOCl stress. (A, B) HOCl stress leads to S-bacillithiolation of OhrR and MetE in B. subtilis. (A) The repressor activity of the OhrR repressor is inhibited by S-bacillithiolation under HOCl and organic hydroperoxide (OHP) stress resulting in up-regulation of the OhrA peroxiredoxin that confers resistance. (B)S-bacillithiolation of the methionine synthase MetE and of other enzymes of the Cys and Met biosynthesis pathway (YxjG, PpaC, SerA, MetI) leads to methionine auxotrophy. (C) In S. aureus, the glycolytic GapDH and the aldehyde dehydrogenase AldA are the main targets for S-bacillithiolation under HOCl stress. S-bacillithiolation of their active site Cys residues leads to reversible inactivation. The physiological role of GapDH and AldA S-bacillithiolation under HOCl stress might be to redirect the glycolytic flux into the pentose phosphate pathway (PPP) for NADPH regeneration and perhaps to detoxify methylglyoxal by AldA and other glyoxalases. This figure is adapted from [3].
In S. aureus we determined the redox state of 228 Cys residues using OxICAT. We identified 58 HOCl-sensitive Cys residues that showed > 10% increased oxidation levels under HOCl stress. Among these HOCl-sensitive thiol-switches, 19 showed 20–30% increased oxidation that included the S-bacillithiolated proteins GapDH, AldA, GuaB and RpmJ. GapDH is S-bacillithiolated at the active site Cys151 and represents the most abundant S-bacillithiolated protein contributing with 4% to the total Cys proteome of S. aureus (Fig. 8C) [42]. In kinetic assays, GapDH of S. aureus was faster inactivated by H2O2 and HOCl stress in the presence of BSH due to S-bacillithiolation compared to overoxidation with the oxidants alone in vitro [42]. These experiments provide evidence that S-bacillithiolation is involved in redox regulation and thiol-protection of Cys151 of GapDH against overoxidation in S. aureus [42]. The oxidative inactivation of GapDH in S. aureus may induce metabolic re-configuration of carbon metabolism and redirection into the PPP as shown in eukaryotic organisms [47], [49].
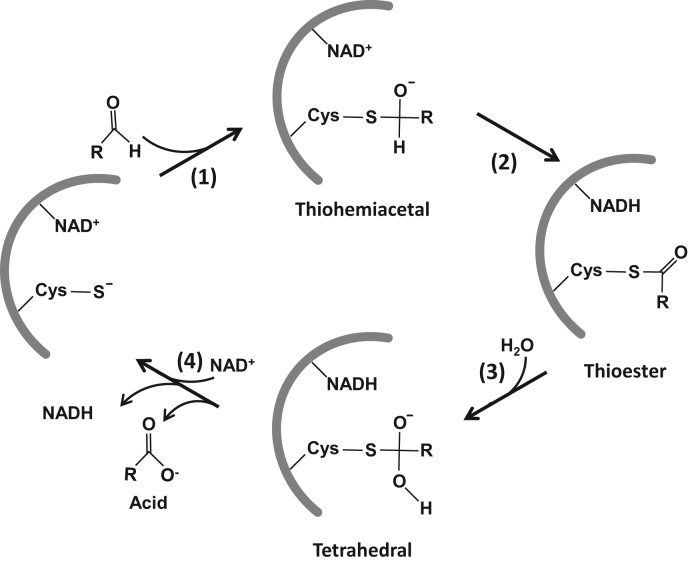
The aldehyde dehydrogenase AldA was identified as another target for S-bacillithiolation under HOCl stress in S. aureus (Fig. 8C) [77]. Aldehyde dehydrogenases (ALDHs) catalyze the NAD+-dependent oxidation of aldehydes to their carboxylic acids in four steps (Fig. 9) [78]. In the first step, a tetrahedral hemithioacetal-enzyme complex is formed. NAD+ binding repositions the catalytic cysteine, allowing the nucleophilic attack on the carbonyl carbon of an aldehyde substrate. The nucleophilicity of the catalytic cysteine is achieved by deprotonation through an adjacent conserved glutamate residue. In the second step, hybrid transfer from the hemithioacetal intermediate to NAD+ generates a covalent thioester intermediate. In the third step, NADH is released from the enzyme allowing hydrolysis of the thioester intermediate. Therefore, nucleophilic water attacks the thioester intermediate to generate a second tetrahedral intermediate. In the final step, hydrolysis of the thioacylenzyme complex releases a carboxylic acid product [78].
Fig. 9.
Catalytic mechanism of NAD+-dependent aldehyde dehydrogenases. NAD+ binds tightly to the enzymes active site, resulting in a conformational change and activation of the catalytic Cys thiol. Nucleophilic attack on the carbonyl carbon of the aldehyde substrate leads to the formation of a covalently bound tetrahedral thiohemiacetal intermediate (1). Hydride transfer from the tetrahedral thiohemiacetal intermediate to NAD+ generates a thioester intermediate (2). Deacylation of the thioester intermediate by nucleophilic attack of a water molecule produces a second tetrahedral intermediate (3). The acid product and NADH are released from the enzymes active site and a new NAD+ molecule binds (4). This figure is adapted from [94].
GapDH and AldA displayed both 29% increased oxidation under HOCl stress as revealed by the OxICAT approach in vivo [18], [77]. Treatment of AldA with H2O2 in the presence of BSH resulted in reversible S-bacillithiolation and loss of activity, which could be restored upon DTT reduction [77]. The purified AldA enzyme showed a broad substrate spectrum, catalyzing the oxidation of various aldehydes, including formaldehyde, methylglyoxal, acetaldehyde and glycolaldehyde. Thus, the question of the natural substrate for AldA remains unresolved. Transcriptional analysis further revealed an increased aldA transcription under formaldehyde, methylglyoxal and HOCl stress in a SigB-independent manner. Thus, our results point to an unknown redox-sensing regulator controlling expression of aldA under thiol-stress conditions. However, in survival phenotype assays, the aldA mutant was more sensitive to HOCl stress, but not to aldehydes. This suggests that unknown aldehyde substrates might be generated under HOCl stress as potential substrate for AldA [77].
2.6. Structural insights into the actives sites of targets for S-bacillithiolations
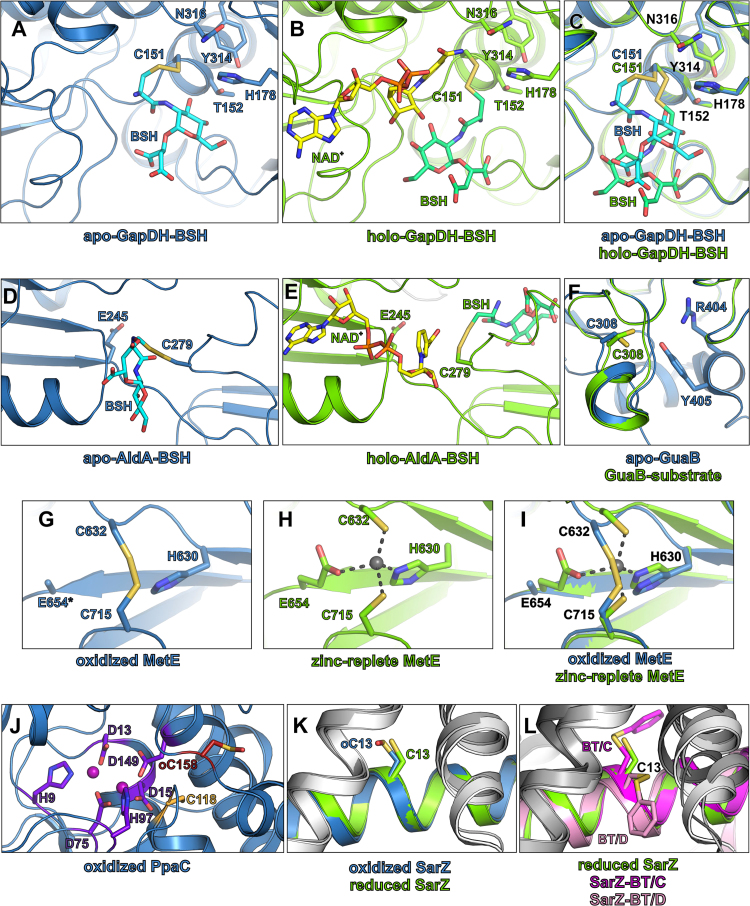
We were further interested in the structural changes of redox-sensitive proteins after S-bacillithiolation of their actives sites. Molecular docking and molecular dynamic simulations were used to model the S-bacillithiolated active sites C151 of GapDH (PDB ID: 3LC7, 3LVF) [79] and C279 of AldA (PDB ID: 3TY7) of S. aureus in the apo- and holo-enzyme structures, respectively (Fig. 10A-E) [42], [77]. Both Cys residues act as a nucleophile in catalysis, leading to the formation of a covalent intermediate [79], [80]. Interestingly, in aldehyde dehydrogenases, the Cys residue adopts a “resting” conformation in the apo-enzyme and an “attacking” conformation in the holo-enzyme [80]. The molecular docking of BSH into the AldA active site resulted in two different positions of the BSH depending on C279 activation state (Fig. 10D, E). While C279 is in “resting” position in the absence of the NAD+ cofactor, BSH partially occupies the cofactor-binding cavity. However, in the presence of NAD+, BSH is located closer to the substrate-binding pocket [77]. Although the conformation of C151 of GapDH is very similar in both apo- and holo-enzyme structures, molecular docking also resulted in two different positions of the BSH (Fig. 10A-C). The position of BSH in the apo- and holo-GapDH does not differ so significantly as in AldA. However, in the holo-GapDH BSH is still shifted towards the substrate-binding pocket, whereas in the apo-enzyme the BSH position partially overlaps with the NAD+-binding site [42]. However, in both models of S-bacillithiolated GapDH and AldA active sites, docking of the BSH did neither require major conformational changes in the overall structure of both proteins, nor in the conformation of the neighboring secondary structure elements.
Fig. 10.
Structural insights into conserved targets for S-bacillithiolation. (A, B) The S-bacillithiolated active site pocket of S. aureus GapDH in the apo- (A) and holo-enzyme (B). (C) The superposition of the GapDH active site of the apo- (blue) and holo-enzyme (green). The molecular docking of BSH was performed on the GapDH apo- (PDB ID: 3LC7) and holo-enzyme structures (PDB ID: 3LVF) (D, E) The S-bacillithiolated active site pocket of S. aureus AldA in the apo- (blue) (D) and holo-enzyme (green) (E). Molecular docking of BSH was performed on the AldA structure (PDB ID: 3TY7). (F) The active site of the B. anthracis GuaB in the apo-enzyme (blue) (PDB ID: 3TSB) and substrate-bound structure (green) (PDB ID: 3USB), the substrate is not shown. (G, H) The active site of the S. mutans MetE in the oxidized (PDB ID: 3L7R) (G) and in the zinc-replete structure (PDB ID: 3T0C) (H). In the oxidized structure, C632 and C715 form an intramolecular disulfide bond. The asterisk indicates a glutamate residue that was modelled as alanine due to the lack of the electron density at this position. (I) The superposition of the active site of the MetE oxidized (blue) and zinc-replete (green) structures. The zinc cation is shown as gray sphere. (J) The active site of the S. aureus PpaC (PDB ID: 4RPA). The active site residues and the neighboring cysteine residues are shown as sticks and coloured by atom type. The colour of the carbon atoms allows to distinguish between active site residues (purple) and neighboring cysteine residues that were found S-bacillithiolated (dark red) or were not S-bacillithiolated (light orange). The manganese cations are shown as pink spheres. (K, L) The redox-active cysteine residue of S. aureus SarZ. (K) Superposition of the reduced (blue) (PDB ID: 3HSE) and oxidized form (green) (PDB ID: 3HRM) of SarZ. (L) Superposition of the reduced (blue) and benzene thiol-bound SarZ in chain C (magenta) and in chain D (light pink) (PDB ID: 3HSR). One monomer of the SarZ dimer is shown in blue (reduced SarZ), green (oxidized SarZ), magenta (benzene thiol-bound SarZ chain C) or light pink (benzene thiol-bound SarZ chain D), whereas the other monomer is shown in shades of gray. All the important residues in all panels are shown as sticks and coloured by atom types (oxygen-red, nitrogen-blue, sulfur-yellow, phosphor-orange, carbon-variable) (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
We further compared these models of GapDH-SSB and AldA-SSB with other structures of conserved S-bacillithiolated proteins that were identified under HOCl stress previously. Among the targets for S-bacillithiolations, GuaB, MetE, the inorganic pyrophosphatase (PpaC), and the redox-sensing virulence regulator SarZ are conserved in Staphylococcus and Bacillus species [75], [76], [77]. Thus, we focused on the crystal structure analysis of S. aureus PpaC [81] and SarZ [82], GuaB of Bacillus anthracis [83], and MetE of Streptococcus mutans [84], which were available in the PDB database (Table 1).
Table 1.
The conserved targets for S-bacillithiolation and their available crystal structures.
| Protein | Organism | Related structures (PDB ID) | Cysteine residues | Function of the cysteine residue | Confirmed post-translational modificationa |
|---|---|---|---|---|---|
| AldA | Staphylococcus aureus | 3TY7 | C279 | active site residue | S-bacillithiolation |
| GapDH | Staphylococcus aureus | 3LC7 | C96 | - | - |
| 3LVF | C151 | active site residue | S-bacillithiolation, overoxidation | ||
| 5T73 | |||||
| GuaB | Staphylococcus aureus | - | C307 | active site | S-bacillithiolation |
| C326 | - | S-bacillithiolation | |||
| GuaB | Bacillus anthracis | 3TSB | C308 | active site | S-bacillithiolation |
| 3USB | C327 | - | S-bacillithiolation | ||
| 3TSD | |||||
| MetE | Staphylococcus aureus | - | C632 | zinc-binding site | - |
| C715 | zinc-binding site | S-bacillithiolation | |||
| MetE | Streptococcus mutans | 3L7R | C632 | zinc-binding site | - |
| 3T0C | C715 | zinc-binding site | S-bacillithiolation | ||
| PpaC | Staphylococcus aureus | 4RPA | C110 | - | overoxidation |
| C118 | - | - | |||
| C158 | - | S-bacillithiolation, overoxidation | |||
| SarZ | Staphylococcus aureus | 3HRM | C13 | redox-active | S-bacillithiolation |
| 3HSE | |||||
| 3HSR |
Also if confirmed in other species.
GuaB is another enzyme that uses NAD+ as a cofactor and contains C308 at the active site. The Cys acts as a nucleophile and forms a covalent adduct with the substrate [83], [85], similarly as discussed for GapDH and AldA. The conformation of the C308 in the substrate IMP-bound enzyme (PDB ID: 3USB) is different from the conformation adopted in the presence of the product (PDB ID: 3TSD) or in the apo-enzyme (PDB ID: 3TSB) (Fig. 10F). Although the two conformations were not termed “resting/attacking” as in AldA, it seems that the active site C308 of GuaB acts similarly to the C279 of AldA. Therefore, we would expect that GuaB might also bind BSH in two alternative positions depending on the C308 activation state. Interestingly, GuaB contains a flexible loop involving the residues 380–430, commonly termed the active site flap, that changes the conformation upon binding to a cofactor or substrate. The residues R404 and Y405 play an important role in the catalytic dyad that activates the water molecule through proton abstraction and allows the release of the product [83], [85]. The active site flap is well-resolved in the apo-enzyme structure, but in the IMP-GuaB the large part of the flap is missing due to the disorder of this part in the structure (Fig. 10F) [83]. We speculate that this structural element might also influence BSH binding and depending on its conformation, BSH could occupy different cavities in GuaB.
In contrast to GapDH, AldA and GuaB, the two active site Cys residues C632 and C715 of MetE play a different role in catalysis. The main function of these two Cys together with H630 and E654 is coordination of the zinc atom. Zinc acts as a Lewis acid and activates the thiol of a homocysteine which is one of the substrates of the MetE [84], [86]. In MetE of B. subtilis, only one active site C730 was S-bacillithiolated under HOCl stress (corresponding to C715 of S. aureus and S. mutans) [75]. The analysis of the crystal structures of S. mutans MetE (PDB ID: 3L7R, 3T0C) [84] suggests an explanation. The binding of the BSH to both Cys residues at the active site would not be possible due to the close location of C632 and C715 (Fig. 10G-I) and space limitation that do not allow for accommodation of two molecules of BSH.
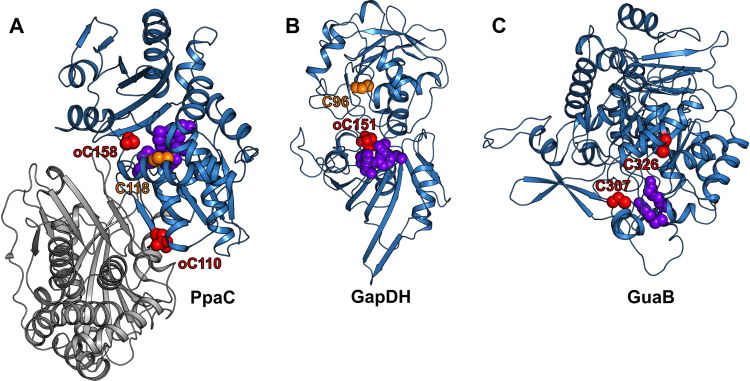
Another conserved target for S-bacillithiolation is the manganese-dependent inorganic pyrophosphatase PpaC which degrades pyrophosphate to inorganic phosphate. The active site of PpaC is composed of H9, D13, D15, D75, H97, D149 and two manganese atoms, but does not contain Cys (Fig. 10J) [81]. However, three conserved Cys residues C110, C118 and C158 are present in PpaC. C110 and C158 were identified as S-bacillithiolated in B. subtilis and S. aureus under HOCl stress [42], [75]. This observation is in a good agreement with the crystal structure of S. aureus PpaC (PDB ID: 4RPA)[81], where both, C110 and C158 are oxidized to sulfonic acids, indicating their susceptibilities to redox changes. C110 is buried in a cavity at the interface of the two monomers forming the PpaC dimer, whereas C158 is located at the protein surface (Fig. 11A). Thus, C110 and C158 are easily accessible for S-bacillithiolation or overoxidation. In contrast, C118 is buried in the N-terminal domain (Fig. 11A). Although C118 is close to the PpaC active site, it is difficult to access and was also not oxidized in the PpaC structure. Nevertheless, it remains intriguing whether S-bacillithiolation of the non-catalytic Cys residues in PpaC protects the enzyme against overoxidation or functions in redox-regulation which remains to be elucidated.
Fig. 11.
The distribution of the cysteine residues in S-bacillithiolation targets. All active site and cysteine residues of S. aureus PpaC (PDB ID: 4RPA) (A), S. aureus GapDH (PDB ID: 5T73) (B) and B. anthracis GuaB (PDB ID: 3TSB) (C) are shown as spheres. The cysteine residues for which the S-bacillithiolation was confirmed are highlighted in red, whereas the other cysteine residues are coloured orange. The active sites residues that are not cysteine are shown in purple. The overoxidized Cys residues are marked as oC. The chain B is shown in gray (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
Similar to PpaC, GapDH and GuaB also contain non-catalytic and non-conserved Cys residues, such as C96 of S. aureus GapDH that was neither S-bacillithiolated nor overoxidized and is buried in the structure like C118 in PpaC (Fig. 11B) [42]. In contrast, in GuaB of B. anthracis, the non-conserved C326 close to the active site was identified as S-bacillithiolated because it is accessible for BSH (Fig. 11C) [77].
The most interesting targets for S-bacillithiolation are redox-sensing regulators, such as the thiol-based OhrR repressor which was S-bacillithiolated at the conserved C15 under HOCl and CHP stress [74], [75]. In S. aureus, the OhrR-homolog SarZ was identified as redox-sensitive repressor and the structural changes were investigated during thiol-oxidation of the single C13, including S-thiolation with the synthetic benzene thiol [75], [82], [87], [88]. The crystal structures of SarZ in reduced (PDB ID: 3HSE), sulfenic acid (PDB ID: 3HRM), and mixed disulfide form (PDB ID: 3HSR)[82] provide the explanation for inhibition of SarZ repressor activity. Upon C13 oxidation, several α-helices of the DNA-binding helix-turn-helix motif change their conformations leading to dissociation of SarZ from the promoter DNA. Inhibition of SarZ repressor activity results in derepression of transcription of a large SarZ regulon, including the ohr peroxiredoxin gene and genes involved in cellular metabolism, virulence, and antibiotic resistance [87]. Interestingly, oxidation of SarZ to the C13 sulfenic acid does not lead to inactivation and structural changes of SarZ (Fig. 10K). Only further overoxidations of C13 to sulfonic acid or S-thiolation causes SarZ inactivation [82]. The structure of the S-thiolated C13-benzene thiol complex provides insights into possible binding sites for LMW thiols. Benzene thiol occupies different positions in different SarZ monomers present in the asymmetric unit of the S-thiolated SarZ structure. In monomer B and C, benzene thiol is oriented toward the surface of the protein, whereas in monomer D, it is buried in a hydrophobic cavity in the dimer interface (Fig. 10L). Benzene thiol might enter the cavity with C13 and the formation of the disulfide bond occurs at a position seen in monomer B, C. Afterwards, C13 changes the conformation yielding another orientation of benzene thiol, as in monomer D [82]. In the case of the BSH binding, the location of BSH in the hydrophobic pocket is less probable because BSH contains several charged groups.
In general, all discussed potential BSH-binding site are composed of positively or negatively charged residues that can interact with the amine, hydroxyl and carboxyl groups of the BSH. Our structural analysis shows that the S-bacillithiolated Cys residues in different proteins are easily accessible for BSH due to their location either in the active sites of proteins or at the protein surface. The Cys residues buried in protein structure, e.g. C96 of GapDH and C118 of GuaB, do not undergo S-bacillithiolation. In the majority of targets, S-bacillithiolation occurs at the catalytic active sites (C151 of GapDH, C279 of AldA, C308 of GuaB, C715 of MetE) or redox-sensing Cys residues of thiol-based redox sensors, such as C13 of SarZ or C15 of OhrR. S-thiolation of SarZ leads to conformational changes, which is the mechanism of derepression of SarZ controlled target genes [82]. The models of S-bacillithiolated GapDH and AldA do not support structural changes. This is in agreement with the catalytic reactions of substrate conversion in GapDH and AldA that are not accompanied by significant structural alterations. In contrast, the inactivation of SarZ requires structural changes in the DNA binding domains as redox-regulatory mechanism. Therefore, it is tempting to speculate that S-thiolation of enzymes in most cases does not lead to structural changes, although exceptions are possible.
2.7. Protein S-bacillithiolation is redox controlled by bacilliredoxins (Brx)
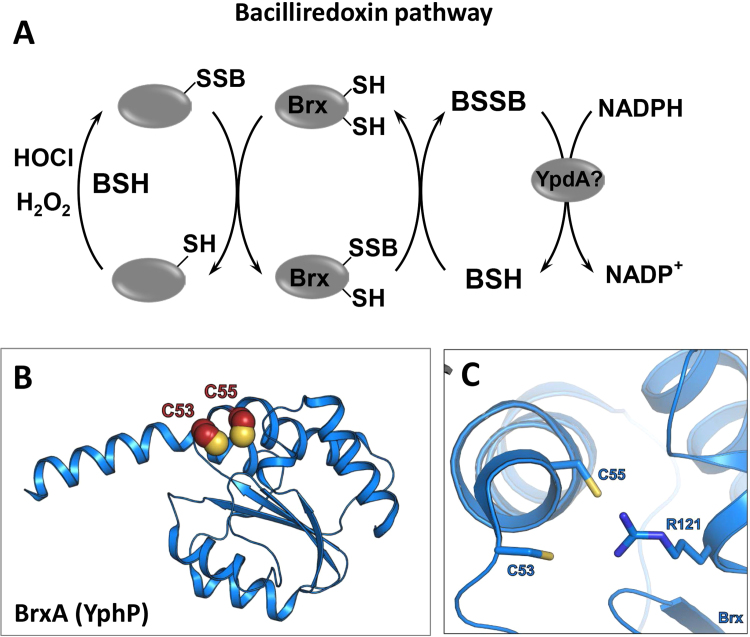
The reduction of S-bacillithiolated proteins in Firmicutes is catalyzed by the bacilliredoxin redox pathway (Fig. 12). The bacilliredoxins BrxA and BrxB were identified as paralogs of the DUF1094 family in B. subtilis, which shared 53% sequence identity [89], [90]. Phylogenomic profiling identified BrxA and BrxB as Trx-like proteins with unusual CGC active sites. The crystal structure of BrxA revealed overall structural similarities to thioredoxins. However, the redox potential was determined as −130 mV, which is much more positive compared to that of thioredoxin proteins [91]. Furthermore, BrxC was identified as candidate for a monothiol Brx which has a TCPIS active motif and is similar to monothiol Grx. However, the function of BrxC in de-bacillithiolation remains to be elucidated [92]. The flavin disulfide reductase YpdA was suggested as putative BSSB reductase in the STRING search due to its phylogenetic co-occurance together with the BSH biosynthesis enzymes [55]. However, the catalytic activity of YpdA in BSSB reduction could not be demonstrated yet.
Fig. 12.
Reversal of protein S-bacillithiolation by the bacilliredoxin pathway. (A) In BSH-producing Firmicutes, such as B. subtilis and S. aureus, a bacilliredoxin (Brx) redox pathway is proposed for reduction of S-bacillithiolated proteins. During the reduction of S-bacillithiolated proteins, a Brx-SSB intermediate is generated that could be reduced by BSH leading to oxidized BSSB. The reduction of BSSB may require the NADPH-dependent bacillithiol disulfide reductase YpdA which remains to be elucidated. (B) The crystal structure of BrxA (YphP) (PDB ID: 3FHK) and (C) its active site. The active and resolving Cys residues of Brx are displayed as spheres (B) and sticks (C).
This figure is adapted from [2].
Bacilliredoxins function analogous to glutaredoxins by the attack of the active site Cys on BSH-mixed protein disulfides, resulting in the transfer of BSH to the active site Cys of Brx (Fig. 12) [89]. In B. subtilis, the S-bacillithiolated OhrR repressor and MetE were identified as natural substrates for BrxA and BrxB in vitro. The DNA-binding activity of the OhrR repressor could be recovered after de-bacillithiolation by the BrxBC54A mutant protein. The de-bacillithiolation of MetE-SSB was demonstrated by BSH-specific Western blot analysis and mass spectrometry, but MetE reactivation could not be shown in vitro. While de-bacillithiolation of OhrR-SSB is catalyzed mainly by BrxB, reduction of MetE-SSB can be catalyzed by both BrxA and BrxB [89].
In S. aureus, the glycolytic GapDH was demonstrated as specific substrate for de-bacillithiolation by BrxA (SAUSA300_1321) during recovery from oxidative stress [42]. Using in vitro activity assays, the glycolytic GapDH activity could be restored after de-bacillithiolation by BrxA and the BrxCGA-resolving Cys mutant, but not by the BrxAGC-active site Cys mutant. BSH-specific Western blot analysis supported the reduction of GapDH-SSB in vitro [42]. However, in vivo evidence for the functions of Brx and YpdA in the oxidative stress response is still missing in Firmicutes.
3. Outlook for future research
In this review, we report an update in the research of the functions and properties of the LMW thiols mycothiol and bacillithiol, which are dominant scavengers of ROS and other reactive species in Actinomycetes and Firmicutes. MSH and BSH were also shown to contribute to the pathogenicity and antibiotic resistance mechanisms of major human pathogens, such as M. tuberculosis and S. aureus indicating the importance of LMW thiols in the defense against the host innate immune system during infections. Significant progress has been made in the structural and mechanistic characterization of many MSH and BSH-dependent detoxification enzymes, as well as MSH and BSH biosynthesis enzymes. However, the catalytic mechanism of the putative BshC ligase and the NADPH-dependent flavin disulfide reductase YpdA are still unknown. Similarly, other unknown MSH- or BSH-dependent formaldehyde dehydrogenases, quinone reductases, and peroxidases might play important roles in detoxification of reactive compounds. There is also significant progress for the role of BSH in FeS cluster assembly, Zn2+ and Cu+ homeostasis, but related roles of MSH have not been investigated and may be important to maintain metal homeostasis under infection conditions. Recent work also showed an involvement of BSH in sulfide homeostasis and sulfhydrations that are implicated in redox regulation of metabolic enzymes and virulence regulators. Finally, over the last years many targets for protein S-mycothiolation and S-bacillithiolation have been discovered through shotgun proteomics and quantitative thiol-redox proteomics. For some antioxidant enzymes (AhpE, Mpx, MsrA) and metabolic enzymes (GapDH), the roles of MSH and BSH in thiol-protection and redox-regulation under oxidative stress have been clearly shown. Molecular docking of BSH into the active sites of GapDH and AldA of S. aureus provided first insights that S-bacillithiolation does not require structural changes. Moreover, BSH can occupy two different positions in the active sites with the active site Cys in the attacking or resting state, depending on the presence of the NAD+ cofactor. Our structural comparison here revealed that these mechanisms could be relevant also for many other conserved targets for S-thiolations, such as GuaB. However, the physiological roles of these protein S-thiolations in cellular physiology in Actinomycetes and Firmicutes requires much more detailed future work. In addition, the functions of bacilliredoxins under oxidative stress remain to be elucidated. Thus, future research should be directed to elucidate the physiological roles of the LMW thiols BSH and MSH and the many targets for protein S-bacillithiolation and S-mycothiolation in bacterial physiology under stress and infection conditions.
Acknowledgements
This work was supported by an European Research Council (ERC) Consolidator grant (GA 615585) MYCOTHIOLOME (EU) and grants from the Deutsche Forschungsgemeinschaft, Germany (AN746/4-1 and AN746/4-2) within the SPP1710 on “Thiol-based Redox switches”, by the Research Training Group GRK1947 (project C01) and by the SFB973 project C08 to H.A. Protein crystal structure analysis was supported by an Alexander von Humboldt post-doc fellowship to A.P.-B.
Author disclosure statement
No competing financial interests exist.
Footnotes
Supplementary data associated with this article can be found in the online version at https://doi.org/10.1016/j.redox.2018.08.017.
Appendix A. Supplementary material
Supplementary material
References
- 1.Van Laer K., Hamilton C.J., Messens J. Low-molecular-weight thiols in thiol-disulfide exchange. Antioxid. Redox Signal. 2013;18(13):1642–1653. doi: 10.1089/ars.2012.4964. [DOI] [PubMed] [Google Scholar]
- 2.Loi V.V., Rossius M., Antelmann H. Redox regulation by reversible protein S-thiolation in bacteria. Front. Microbiol. 2015;6:187. doi: 10.3389/fmicb.2015.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chandrangsu P., Loi V.V., Antelmann H., Helmann J.D. The role of bacillithiol in Gram-positive Firmicutes. Antioxid. Redox Signal. 2018;28(6):445–462. doi: 10.1089/ars.2017.7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Potter A.J., Trappetti C., Paton J.C. Streptococcus pneumoniae uses glutathione to defend against oxidative stress and metal ion toxicity. J. Bacteriol. 2012;194(22):6248–6254. doi: 10.1128/JB.01393-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Newton G.L., Rawat M., La Clair J.J., Jothivasan V.K., Budiarto T., Hamilton C.J., Claiborne A., Helmann J.D., Fahey R.C. Bacillithiol is an antioxidant thiol produced in Bacilli. Nat. Chem. Biol. 2009;5(9):625–627. doi: 10.1038/nchembio.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Newton G.L., Buchmeier N., Fahey R.C. Biosynthesis and functions of mycothiol, the unique protective thiol of Actinobacteria. Microbiol. Mol. Biol. Rev. 2008;72(3):471–494. doi: 10.1128/MMBR.00008-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reyes A.M., Pedre B., De Armas M.I., Tossounian M.A., Radi R., Messens J., Trujillo M. Chemistry and redox biology of mycothiol. Antioxid. Redox Signal. 2018;28(6):487–504. doi: 10.1089/ars.2017.7074. [DOI] [PubMed] [Google Scholar]
- 8.elCardayre S.B. d., Davies J.E. Staphylococcus aureus coenzyme A disulfide reductase, a new subfamily of pyridine nucleotide-disulfide oxidoreductase. Sequence, expression, and analysis of cdr. J. Biol. Chem. 1998;273(10):5752–5757. doi: 10.1074/jbc.273.10.5752. [DOI] [PubMed] [Google Scholar]
- 9.Boylan J.A., Hummel C.S., Benoit S., Garcia-Lara J., Treglown-Downey J., Crane E.J., 3rd, Gherardini F.C. Borrelia burgdorferi bb0728 encodes a coenzyme A disulphide reductase whose function suggests a role in intracellular redox and the oxidative stress response. Mol. Microbiol. 2006;59(2):475–486. doi: 10.1111/j.1365-2958.2005.04963.x. [DOI] [PubMed] [Google Scholar]
- 10.Fahey R.C. Glutathione analogs in prokaryotes. Biochim. Biophys. Acta. 2013;1830(5):3182–3198. doi: 10.1016/j.bbagen.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 11.Copley S.D., Dhillon J.K. Lateral gene transfer and parallel evolution in the history of glutathione biosynthesis genes. Genome Biol. 2002;3(5) doi: 10.1186/gb-2002-3-5-research0025. (research0025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ritz D., Beckwith J. Roles of thiol-redox pathways in bacteria. Annu. Rev. Microbiol. 2001;55:21–48. doi: 10.1146/annurev.micro.55.1.21. [DOI] [PubMed] [Google Scholar]
- 13.Jothivasan V.K., Hamilton C.J. Mycothiol: synthesis, biosynthesis and biological functions of the major low molecular weight thiol in actinomycetes. Nat. Prod. Rep. 2008;25(6):1091–1117. doi: 10.1039/b616489g. [DOI] [PubMed] [Google Scholar]
- 14.Buchmeier N.A., Newton G.L., Koledin T., Fahey R.C. Association of mycothiol with protection of Mycobacterium tuberculosis from toxic oxidants and antibiotics. Mol. Microbiol. 2003;47(6):1723–1732. doi: 10.1046/j.1365-2958.2003.03416.x. [DOI] [PubMed] [Google Scholar]
- 15.Newton G.L., Av-Gay Y., Fahey R.C. A novel mycothiol-dependent detoxification pathway in mycobacteria involving mycothiol S-conjugate amidase. Biochemistry. 2000;39(35):10739–10746. doi: 10.1021/bi000356n. [DOI] [PubMed] [Google Scholar]
- 16.Kumar A., Nartey W., Shin J., Manimekalai M.S.S., Gruber G. Structural and mechanistic insights into mycothiol disulphide reductase and the mycoredoxin-1-alkylhydroperoxide reductase E assembly of Mycobacterium tuberculosis. Biochim. Biophys. Acta. 2017;1861(9):2354–2366. doi: 10.1016/j.bbagen.2017.05.007. [DOI] [PubMed] [Google Scholar]
- 17.Newton G.L., Arnold K., Price M.S., Sherrill C., Delcardayre S.B., Aharonowitz Y., Cohen G., Davies J., Fahey R.C., Davis C. Distribution of thiols in microorganisms: mycothiol is a major thiol in most actinomycetes. J. Bacteriol. 1996;178(7):1990–1995. doi: 10.1128/jb.178.7.1990-1995.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hillion M., Imber M., Pedre B., Bernhardt J., Saleh M., Loi V.V., Maass S., Becher D., Astolfi Rosado L., Adrian L., Weise C., Hell R., Wirtz M., Messens J., Antelmann H. The glyceraldehyde-3-phosphate dehydrogenase GapDH of Corynebacterium diphtheriae is redox-controlled by protein S-mycothiolation under oxidative stress. Sci. Rep. 2017;7(1):5020. doi: 10.1038/s41598-017-05206-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharma S.V., Van Laer K., Messens J., Hamilton C.J. Thiol redox and pKa properties of mycothiol, the predominant low-molecular-weight thiol cofactor in the actinomycetes. Chembiochem. 2016;17(18):1689–1692. doi: 10.1002/cbic.201600228. [DOI] [PubMed] [Google Scholar]
- 20.Bhaskar A., Chawla M., Mehta M., Parikh P., Chandra P., Bhave D., Kumar D., Carroll K.S., Singh A. Reengineering redox sensitive GFP to measure mycothiol redox potential of Mycobacterium tuberculosis during infection. PLoS Pathog. 2014;10(1):e1003902. doi: 10.1371/journal.ppat.1003902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Newton G.L., Leung S.S., Wakabayashi J.I., Rawat M., Fahey R.C. The DinB superfamily includes novel mycothiol, bacillithiol, and glutathione S-transferases. Biochemistry. 2011;50(49):10751–10760. doi: 10.1021/bi201460j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao Q., Wang M., Xu D., Zhang Q., Liu W. Metabolic coupling of two small-molecule thiols programs the biosynthesis of lincomycin A. Nature. 2015;518(7537):115–119. doi: 10.1038/nature14137. [DOI] [PubMed] [Google Scholar]
- 23.Zhang D., Tang Z., Liu W. Biosynthesis of lincosamide antibiotics: reactions associated with degradation and detoxification pathways play a constructive role. Acc. Chem. Res. 2018;51(6):1496–1506. doi: 10.1021/acs.accounts.8b00135. [DOI] [PubMed] [Google Scholar]
- 24.Feng J., Che Y., Milse J., Yin Y.J., Liu L., Ruckert C., Shen X.H., Qi S.W., Kalinowski J., Liu S.J. The gene ncgl2918 encodes a novel maleylpyruvate isomerase that needs mycothiol as cofactor and links mycothiol biosynthesis and gentisate assimilation in Corynebacterium glutamicum. J. Biol. Chem. 2006;281(16):10778–10785. doi: 10.1074/jbc.M513192200. [DOI] [PubMed] [Google Scholar]
- 25.Chi B.K., Busche T., Van Laer K., Bäsell K., Becher D., Clermont L., Seibold G.M., Persicke M., Kalinowski J., Messens J., Antelmann H. Protein S-mycothiolation functions as redox-switch and thiol protection mechanism in Corynebacterium glutamicum under hypochlorite stress. Antioxid. Redox Signal. 2014;20(4):589–605. doi: 10.1089/ars.2013.5423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ordonez E., Van Belle K., Roos G., De Galan S., Letek M., Gil J.A., Wyns L., Mateos L.M., Messens J. Arsenate reductase, mycothiol, and mycoredoxin concert thiol/disulfide exchange. J. Biol. Chem. 2009;284(22):15107–15116. doi: 10.1074/jbc.M900877200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hazbon M.H., Brimacombe M., Bobadilla del Valle M., Cavatore M., Guerrero M.I., Varma-Basil M., Billman-Jacobe H., Lavender C., Fyfe J., Garcia-Garcia L., Leon C.I., Bose M., Chaves F., Murray M., Eisenach K.D., Sifuentes-Osornio J., Cave M.D., Ponce de Leon A., Alland D. Population genetics study of isoniazid resistance mutations and evolution of multidrug-resistant Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2006;50(8):2640–2649. doi: 10.1128/AAC.00112-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Negri A., Javidnia P., Mu R., Zhang X., Vendome J., Gold B., Roberts J., Barman D., Ioerger T., Sacchettini J.C., Jiang X., Burns-Huang K., Warrier T., Ling Y., Warren J.D., Oren D.A., Beuming T., Wang H., Wu J., Li H., Rhee K.Y., Nathan C.F., Liu G., Somersan-Karakaya S. Identification of a mycothiol-dependent nitroreductase from Mycobacterium tuberculosis. ACS Infect. Dis. 2018;4(5):771–787. doi: 10.1021/acsinfecdis.7b00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Padiadpu J., Baloni P., Anand K., Munshi M., Thakur C., Mohan A., Singh A., Chandra N. Identifying and tackling emergent vulnerability in drug-resistant Mycobacteria. ACS Infect. Dis. 2016;2(9):592–607. doi: 10.1021/acsinfecdis.6b00004. [DOI] [PubMed] [Google Scholar]
- 30.Rosado L.A., Wahni K., Degiacomi G., Pedre B., Young D., de la Rubia A.G., Boldrin F., Martens E., Marcos-Pascual L., Sancho-Vaello E., Albesa-Jove D., Provvedi R., Martin C., Makarov V., Versees W., Verniest G., Guerin M.E., Mateos L.M., Manganelli R., Messens J. The antibacterial prodrug activator Rv2466c is a mycothiol-dependent reductase in the oxidative stress response of Mycobacterium tuberculosis. J. Biol. Chem. 2017;292(32):13097–13110. doi: 10.1074/jbc.M117.797837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sareen D., Newton G.L., Fahey R.C., Buchmeier N.A. Mycothiol is essential for growth of Mycobacterium tuberculosis Erdman. J. Bacteriol. 2003;185(22):6736–6740. doi: 10.1128/JB.185.22.6736-6740.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hillion M., Bernhardt J., Busche T., Rossius M., Maass S., Becher D., Rawat M., Wirtz M., Hell R., Ruckert C., Kalinowski J., Antelmann H. Monitoring global protein thiol-oxidation and protein S-mycothiolation in Mycobacterium smegmatis under hypochlorite stress. Sci. Rep. 2017;7(1):1195. doi: 10.1038/s41598-017-01179-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hugo M., Van Laer K., Reyes A.M., Vertommen D., Messens J., Radi R., Trujillo M. Mycothiol/mycoredoxin 1-dependent reduction of the peroxiredoxin AhpE from Mycobacterium tuberculosis. J. Biol. Chem. 2014;289(8):5228–5239. doi: 10.1074/jbc.M113.510248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seibold G.M., Eikmanns B.J. The glgX gene product of Corynebacterium glutamicum is required for glycogen degradation and for fast adaptation to hyperosmotic stress. Microbiology. 2007;153(Pt 7):2212–2220. doi: 10.1099/mic.0.2006/005181-0. [DOI] [PubMed] [Google Scholar]
- 35.Pedre B., Van Molle I., Villadangos A.F., Wahni K., Vertommen D., Turell L., Erdogan H., Mateos L.M., Messens J. The Corynebacterium glutamicum mycothiol peroxidase is a reactive oxygen species-scavenging enzyme that shows promiscuity in thiol redox control. Mol. Microbiol. 2015;96(6):1176–1191. doi: 10.1111/mmi.12998. [DOI] [PubMed] [Google Scholar]
- 36.Tossounian M.A., Pedre B., Wahni K., Erdogan H., Vertommen D., Van Molle I., Messens J. Corynebacterium diphtheriae methionine sulfoxide reductase a exploits a unique mycothiol redox relay mechanism. J. Biol. Chem. 2015;290(18):11365–11375. doi: 10.1074/jbc.M114.632596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Si M., Feng Y., Chen K., Kang Y., Chen C., Wang Y., Shen X. Functional comparison of methionine sulphoxide reductase A and B in Corynebacterium glutamicum. J. Gen. Appl. Microbiol. 2017;63(5):280–286. doi: 10.2323/jgam.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 38.Si M., Xu Y., Wang T., Long M., Ding W., Chen C., Guan X., Liu Y., Wang Y., Shen X., Liu S.J. Functional characterization of a mycothiol peroxidase in Corynebacterium glutamicum that uses both mycoredoxin and thioredoxin reducing systems in the response to oxidative stress. Biochem. J. 2015;469(1):45–57. doi: 10.1042/BJ20141080. [DOI] [PubMed] [Google Scholar]
- 39.Liu Y., Yang X., Yin Y., Lin J., Chen C., Pan J., Si M., Shen X. Mycothiol protects Corynebacterium glutamicum against acid stress via maintaining intracellular pH homeostasis, scavenging ROS, and S-mycothiolating MetE. J. Gen. Appl. Microbiol. 2016;62(3):144–153. doi: 10.2323/jgam.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 40.Munoz-Elias E.J., McKinney J.D. Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat. Med. 2005;11(6):638–644. doi: 10.1038/nm1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Crack J.C., Hamilton C.J., Brun N.E. Le. Mass spectrometric detection of iron nitrosyls, sulfide oxidation and mycothiolation during nitrosylation of the NO sensor [4Fe-4S] NsrR. Chem. Commun. 2018;54(47):5992–5995. doi: 10.1039/c8cc01339j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Imber M., Huyen N.T.T., Pietrzyk-Brzezinska A.J., Loi V.V., Hillion M., Bernhardt J., Thärichen L., Kolsek K., Saleh M., Hamilton C.J., Adrian L., Gräter F., Wahl M.C., Antelmann H. Protein S-bacillithiolation functions in thiol protection and redox regulation of the glyceraldehyde-3-phosphate dehydrogenase Gap in Staphylococcus aureus under hypochlorite stress. Antioxid. Redox Signal. 2018;28(6):410–430. doi: 10.1089/ars.2016.6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Laer K., Buts L., Foloppe N., Vertommen D., Van Belle K., Wahni K., Roos G., Nilsson L., Mateos L.M., Rawat M., van Nuland N.A., Messens J. Mycoredoxin-1 is one of the missing links in the oxidative stress defence mechanism of Mycobacteria. Mol. Microbiol. 2012;86(4):787–804. doi: 10.1111/mmi.12030. [DOI] [PubMed] [Google Scholar]
- 44.Aslund F., Berndt K.D., Holmgren A. Redox potentials of glutaredoxins and other thiol-disulfide oxidoreductases of the thioredoxin superfamily determined by direct protein-protein redox equilibria. J. Biol. Chem. 1997;272(49):30780–30786. doi: 10.1074/jbc.272.49.30780. [DOI] [PubMed] [Google Scholar]
- 45.Reis M., Alves C.N., Lameira J., Tunon I., Marti S., Moliner V. The catalytic mechanism of glyceraldehyde 3-phosphate dehydrogenase from Trypanosoma cruzi elucidated via the QM/MM approach. Phys. Chem. Chem. Phys. 2013;15(11):3772–3785. doi: 10.1039/c3cp43968b. [DOI] [PubMed] [Google Scholar]
- 46.Shenton D., Grant C.M. Protein S-thiolation targets glycolysis and protein synthesis in response to oxidative stress in the yeast Saccharomyces cerevisiae. Biochem. J. 2003;374(Pt 2):513–519. doi: 10.1042/BJ20030414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ralser M., Wamelink M.M., Kowald A., Gerisch B., Heeren G., Struys E.A., Klipp E., Jakobs C., Breitenbach M., Lehrach H., Krobitsch S. Dynamic rerouting of the carbohydrate flux is key to counteracting oxidative stress. J. Biol. 2007;6(4):10. doi: 10.1186/jbiol61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hildebrandt T., Knuesting J., Berndt C., Morgan B., Scheibe R. Cytosolic thiol switches regulating basic cellular functions: gapdh as an information hub? Biol. Chem. 2015;396(5):523–537. doi: 10.1515/hsz-2014-0295. [DOI] [PubMed] [Google Scholar]
- 49.Brandes N., Schmitt S., Jakob U. Thiol-based redox switches in eukaryotic proteins. Antioxid. Redox Signal. 2009;11(5):997–1014. doi: 10.1089/ars.2008.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peralta D., Bronowska A.K., Morgan B., Doka E., Van Laer K., Nagy P., Grater F., Dick T.P. A proton relay enhances H2O2 sensitivity of GAPDH to facilitate metabolic adaptation. Nat. Chem. Biol. 2015;11(2):156–163. doi: 10.1038/nchembio.1720. [DOI] [PubMed] [Google Scholar]
- 51.Drazic A., Winter J. The physiological role of reversible methionine oxidation. Biochim. Biophys. Acta. 2014;1844(8):1367–1382. doi: 10.1016/j.bbapap.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 52.Sharov V.S., Ferrington D.A., Squier T.C., Schoneich C. Diastereoselective reduction of protein-bound methionine sulfoxide by methionine sulfoxide reductase. FEBS Lett. 1999;455(3):247–250. doi: 10.1016/s0014-5793(99)00888-1. [DOI] [PubMed] [Google Scholar]
- 53.Si M., Zhang L., Chaudhry M.T., Ding W., Xu Y., Chen C., Akbar A., Shen X., Liu S.J. Corynebacterium glutamicum methionine sulfoxide reductase A uses both mycoredoxin and thioredoxin for regeneration and oxidative stress resistance. Appl. Environ. Microbiol. 2015;81(8):2781–2796. doi: 10.1128/AEM.04221-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kumar A., Balakrishna A.M., Nartey W., Manimekalai M.S.S., Gruber G. Redox chemistry of Mycobacterium tuberculosis alkylhydroperoxide reductase E (AhpE): structural and mechanistic insight into a mycoredoxin-1 independent reductive pathway of AhpE via mycothiol. Free Radic. Biol. Med. 2016;97:588–601. doi: 10.1016/j.freeradbiomed.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 55.Gaballa A., Newton G.L., Antelmann H., Parsonage D., Upton H., Rawat M., Claiborne A., Fahey R.C., Helmann J.D. Biosynthesis and functions of bacillithiol, a major low-molecular-weight thiol in Bacilli. Proc. Natl. Acad. Sci. USA. 2010;107(14):6482–6486. doi: 10.1073/pnas.1000928107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sharma S.V., Arbach M., Roberts A.A., Macdonald C.J., Groom M., Hamilton C.J. Biophysical features of bacillithiol, the glutathione surrogate of Bacillus subtilis and other firmicutes. Chembiochem. 2013;14(16):2160–2168. doi: 10.1002/cbic.201300404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roberts A.A., Sharma S.V., Strankman A.W., Duran S.R., Rawat M., Hamilton C.J. Mechanistic studies of FosB: a divalent-metal-dependent bacillithiol-S-transferase that mediates fosfomycin resistance in Staphylococcus aureus. Biochem. J. 2013;451(1):69–79. doi: 10.1042/BJ20121541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thompson M.K., Keithly M.E., Goodman M.C., Hammer N.D., Cook P.D., Jagessar K.L., Harp J., Skaar E.P., Armstrong R.N. Structure and function of the genomically encoded fosfomycin resistance enzyme, FosB, from Staphylococcus aureus. Biochemistry. 2014;53(4):755–765. doi: 10.1021/bi4015852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chandrangsu P., Dusi R., Hamilton C.J., Helmann J.D. Methylglyoxal resistance in Bacillus subtilis: contributions of bacillithiol-dependent and independent pathways. Mol. Microbiol. 2014;91(4):706–715. doi: 10.1111/mmi.12489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Müller J.E., Meyer F., Litsanov B., Kiefer P., Vorholt J.A. Core pathways operating during methylotrophy of Bacillus methanolicus MGA3 and induction of a bacillithiol-dependent detoxification pathway upon formaldehyde stress. Mol. Microbiol. 2015;98(6):1089–1100. doi: 10.1111/mmi.13200. [DOI] [PubMed] [Google Scholar]
- 61.Helmann J.D. Bacillithiol, a new player in bacterial redox homeostasis. Antioxid. Redox Signal. 2011;15(1):123–133. doi: 10.1089/ars.2010.3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ma Z., Chandrangsu P., Helmann T.C., Romsang A., Gaballa A., Helmann J.D. Bacillithiol is a major buffer of the labile zinc pool in Bacillus subtilis. Mol. Microbiol. 2014;94(4):756–770. doi: 10.1111/mmi.12794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fang Z., Dos Santos P.C. Protective role of bacillithiol in superoxide stress and Fe-S metabolism in Bacillus subtilis. Microbiol. Open. 2015;4(4):616–631. doi: 10.1002/mbo3.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rosario-Cruz Z., Chahal H.K., Mike L.A., Skaar E.P., Boyd J.M. Bacillithiol has a role in Fe-S cluster biogenesis in Staphylococcus aureus. Mol. Microbiol. 2015;98(2):218–242. doi: 10.1111/mmi.13115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rosario-Cruz Z., Boyd J.M. Physiological roles of bacillithiol in intracellular metal processing. Curr. Genet. 2016;62(1):59–65. doi: 10.1007/s00294-015-0511-0. [DOI] [PubMed] [Google Scholar]
- 66.Kay K.L., Hamilton C.J., Brun N.E. Le. Mass spectrometry of B. subtilis CopZ: Cu(i)-binding and interactions with bacillithiol. Metallomics. 2016;8(7):709–719. doi: 10.1039/c6mt00036c. [DOI] [PubMed] [Google Scholar]
- 67.Peng H., Zhang Y., Palmer L.D., Kehl-Fie T.E., Skaar E.P., Trinidad J.C., Giedroc D.P. Hydrogen sulfide and reactive sulfur species impact proteome S-sulfhydration and global virulence regulation in Staphylococcus aureus. ACS Infect. Dis. 2017;3(10):744–755. doi: 10.1021/acsinfecdis.7b00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Peng H., Shen J., Edmonds K.A., Luebke J.L., Hickey A.K., Palmer L.D., Chang F.J., Bruce K.A., Kehl-Fie T.E., Skaar E.P., Giedroc D.P. Sulfide homeostasis and nitroxyl intersect via formation of reactive sulfur species in Staphylococcus aureus. mSphere. 2017;2(3) doi: 10.1128/mSphere.00082-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pöther D.C., Gierok P., Harms M., Mostertz J., Hochgräfe F., Antelmann H., Hamilton C.J., Borovok I., Lalk M., Aharonowitz Y., Hecker M. Distribution and infection-related functions of bacillithiol in Staphylococcus aureus. Int. J. Med. Microbiol. 2013;303(3):114–123. doi: 10.1016/j.ijmm.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 70.Rajkarnikar A., Strankman A., Duran S., Vargas D., Roberts A.A., Barretto K., Upton H., Hamilton C.J., Rawat M. Analysis of mutants disrupted in bacillithiol metabolism in Staphylococcus aureus. Biochem. Biophys. Res. Commun. 2013;436(2):128–133. doi: 10.1016/j.bbrc.2013.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Posada A.C., Kolar S.L., Dusi R.G., Francois P., Roberts A.A., Hamilton C.J., Liu G.Y., Cheung A. Importance of bacillithiol in the oxidative stress response of Staphylococcus aureus. Infect. Immun. 2014;82(1):316–332. doi: 10.1128/IAI.01074-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Otto M. Community-associated MRSA: what makes them special? Int. J. Med. Microbiol. 2013;303(6–7):324–330. doi: 10.1016/j.ijmm.2013.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Novick R. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology. 1967;33(1):155–166. doi: 10.1016/0042-6822(67)90105-5. [DOI] [PubMed] [Google Scholar]
- 74.Lee J.W., Soonsanga S., Helmann J.D. A complex thiolate switch regulates the Bacillus subtilis organic peroxide sensor OhrR. Proc. Natl. Acad. Sci. USA. 2007;104(21):8743–8748. doi: 10.1073/pnas.0702081104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chi B.K., Gronau K., Mäder U., Hessling B., Becher D., Antelmann H. S-bacillithiolation protects against hypochlorite stress in Bacillus subtilis as revealed by transcriptomics and redox proteomics. Mol. Cell Proteom. 2011;10(11) doi: 10.1074/mcp.M111.009506. (M111 009506) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chi B.K., Roberts A.A., Huyen T.T., Bäsell K., Becher D., Albrecht D., Hamilton C.J., Antelmann H. S-bacillithiolation protects conserved and essential proteins against hypochlorite stress in firmicutes bacteria. Antioxid. Redox Signal. 2013;18(11):1273–1295. doi: 10.1089/ars.2012.4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Imber M., Loi V.V., Reznikov S., Fritsch V.N., Pietrzyk-Brzezinska A.J., Prehn J., Hamilton C., Wahl M.C., Bronowska A.K., Antelmann H. The aldehyde dehydrogenase AldA contributes to the hypochlorite defense and is redox-controlled by protein S-bacillithiolation in Staphylococcus aureus. Redox Biol. 2018;15:557–568. doi: 10.1016/j.redox.2018.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Halavaty A.S., Rich R.L., Chen C., Joo J.C., Minasov G., Dubrovska I., Winsor J.R., Myszka D.G., Duban M., Shuvalova L., Yakunin A.F., Anderson W.F. Structural and functional analysis of betaine aldehyde dehydrogenase from Staphylococcus aureus. Acta Crystallogr. D. Biol. Crystallogr. 2015;71(Pt 5):1159–1175. doi: 10.1107/S1399004715004228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mukherjee S., Dutta D., Saha B., Das A.K. Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 1 from methicillin-resistant Staphylococcus aureus MRSA252 provides novel insights into substrate binding and catalytic mechanism. J. Mol. Biol. 2010;401(5):949–968. doi: 10.1016/j.jmb.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 80.Huo L., Davis I., Liu F., Andi B., Esaki S., Iwaki H., Hasegawa Y., Orville A.M., Liu A. Crystallographic and spectroscopic snapshots reveal a dehydrogenase in action. Nat. Commun. 2015;6:5935. doi: 10.1038/ncomms6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gajadeera C.S., Zhang X., Wei Y., Tsodikov O.V. Structure of inorganic pyrophosphatase from Staphylococcus aureus reveals conformational flexibility of the active site. J. Struct. Biol. 2015;189(2):81–86. doi: 10.1016/j.jsb.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 82.Poor C.B., Chen P.R., Duguid E., Rice P.A., He C. Crystal structures of the reduced, sulfenic acid, and mixed disulfide forms of SarZ, a redox active global regulator in Staphylococcus aureus. J. Biol. Chem. 2009;284(35):23517–23524. doi: 10.1074/jbc.M109.015826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Makowska-Grzyska M., Kim Y., Wu R., Wilton R., Gollapalli D.R., Wang X.K., Zhang R., Jedrzejczak R., Mack J.C., Maltseva N., Mulligan R., Binkowski T.A., Gornicki P., Kuhn M.L., Anderson W.F., Hedstrom L., Joachimiak A. Bacillus anthracis inosine 5′-monophosphate dehydrogenase in action: the first bacterial series of structures of phosphate ion-, substrate-, and product-bound complexes. Biochemistry. 2012;51(31):6148–6163. doi: 10.1021/bi300511w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fu T.M., Almqvist J., Liang Y.H., Li L., Huang Y., Su X.D. Crystal structures of cobalamin-independent methionine synthase (MetE) from Streptococcus mutans: a dynamic zinc-inversion model. J. Mol. Biol. 2011;412(4):688–697. doi: 10.1016/j.jmb.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 85.Hedstrom L. IMP dehydrogenase: structure, mechanism, and inhibition. Chem. Rev. 2009;109(7):2903–2928. doi: 10.1021/cr900021w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Peariso K., Goulding C.W., Huang S., Matthews R.G., Penner-Hahn J.E. Characterization of the zinc binding site in methionine synthase enzymes of Escherichia coli: the role of zinc in the methylation of homocysteine. J. Am. Chem. Soc. 1998;120:8410–8416. [Google Scholar]
- 87.Chen P.R., Nishida S., Poor C.B., Cheng A., Bae T., Kuechenmeister L., Dunman P.M., Missiakas D., He C. A new oxidative sensing and regulation pathway mediated by the MgrA homologue SarZ in Staphylococcus aureus. Mol. Microbiol. 2009;71(1):198–211. doi: 10.1111/j.1365-2958.2008.06518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hillion M., Antelmann H. Thiol-based redox switches in prokaryotes. Biol. Chem. 2015;396(5):415–444. doi: 10.1515/hsz-2015-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gaballa A., Chi B.K., Roberts A.A., Becher D., Hamilton C.J., Antelmann H., Helmann J.D. Redox regulation in Bacillus subtilis: the bacilliredoxins BrxA(YphP) and BrxB(YqiW) function in de-bacillithiolation of S-bacillithiolated OhrR and MetE. Antioxid. Redox Signal. 2014;21(3):357–367. doi: 10.1089/ars.2013.5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hochgräfe F., Mostertz J., Albrecht D., Hecker M. Fluorescence thiol modification assay: oxidatively modified proteins in Bacillus subtilis. Mol. Microbiol. 2005;58(2):409–425. doi: 10.1111/j.1365-2958.2005.04845.x. [DOI] [PubMed] [Google Scholar]
- 91.Derewenda U., Boczek T., Gorres K.L., Yu M., Hung L.W., Cooper D., Joachimiak A., Raines R.T., Derewenda Z.S. Structure and function of Bacillus subtilis YphP, a prokaryotic disulfide isomerase with a CXC catalytic motif. Biochemistry. 2009;48(36):8664–8671. doi: 10.1021/bi900437z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Petersohn A., Brigulla M., Haas S., Hoheisel J.D., Völker U., Hecker M. Global analysis of the general stress response of Bacillus subtilis. J. Bacteriol. 2001;183(19):5617–5631. doi: 10.1128/JB.183.19.5617-5631.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Viars S., Valentine J., Hernick M. Structure and function of the LmbE-like superfamily. Biomolecules. 2014;4(2):527–545. doi: 10.3390/biom4020527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Munoz-Clares R.A., Gonzalez-Segura L., Diaz-Sanchez A.G. Crystallographic evidence for active-site dynamics in the hydrolytic aldehyde dehydrogenases. Implications for the deacylation step of the catalyzed reaction. Chem. Biol. Interact. 2011;191(1–3):137–146. doi: 10.1016/j.cbi.2010.12.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material