Abstract
Autosomal dominant mutations in leucine-rich repeat kinase 2 (LRRK2) are associated with Parkinson’s disease (PD). Most pathogenic LRRK2 mutations result in amino acid substitutions in the central ROC (Ras of complex proteins)–C-terminus of ROC–kinase triple domain and affect enzymatic functions of the protein. However, there are several variants in LRRK2, including the risk factor G2385R, that affect PD pathogenesis by unknown mechanisms. Previously, we have shown that G2385R LRRK2 has decreased kinase activity in vitro and altered affinity to LRRK2 interactors. Specifically, we found an increased binding to the chaperone Hsp90 (heat shock protein 90 kDa) that is known to stabilize LRRK2, suggesting that G2385R may have structural effects on LRRK2. In the present study, we further explored the effects of G2385R on LRRK2 in cells. We found that G2385R LRRK2 has lower steady-state intracellular protein levels compared with wild-type LRRK2 due to increased protein turnover of the mutant protein. Mechanistically, this is a consequence of a higher affinity of G2385R compared with the wild-type protein for two proteins involved in proteasomal degradation, Hsc70 and carboxyl-terminus of Hsc70-interacting protein (CHIP). Overexpression of CHIP decreased intracellular protein levels of both G2385R mutant and wild-type LRRK2, while short interfering RNA CHIP knockdown had the opposite effect. We suggest that the G2385R substitution tilts the equilibrium between refolding and proteasomal degradation toward intracellular degradation. The observation of lower steady-state protein levels may explain why G2385R is a risk factor rather than a penetrant variant for inherited PD.
Introduction
There is growing evidence that genetic factors play an important role in the development of familial and sporadic Parkinson’s disease (PD). Mutations in the leucine-rich repeat kinase 2 (LRRK2) gene are one, relatively common, genetic cause of PD [1]. Pathogenic mutations are clustered in the central catalytic core [ROC (Ras of complex proteins)–COR (C-terminus of ROC)–kinase tridomain] of the LRRK2 protein and affect enzymatic functions of the protein [2]. Specifically, the G2019S substitution located in the kinase domain augments phosphorylation activity of LRRK2 [3–5], while the R1441C/G and Y1699C mutations found in the ROC–COR bidomain disrupt GTPase function of the protein [6–8]. These observations suggest that inherited PD may result from altered LRRK2 function and have led to the proposal that kinase inhibitors might be useful therapeutically in LRRK2 cases and, perhaps, also in other forms of PD [9].
In addition to pathogenic mutations, several risk factors in the LRRK2 gene have been reported. Among these, the G2385R variant, found in some Asian populations [10,11], is located on the outer surface of the C-terminal WD40 domain. Substitution of this residue with arginine could potentially interfere with intra- and/or intermolecular interactions of LRRK2 [12]. Previously, we demonstrated that G2385R LRRK2 has an altered pattern of interaction with known LRRK2 partner proteins including a diminished affinity for 14–3-3 proteins [13], probably mediated by a decrease in phosphorylation of two residues, S910 and S935 [14]. At the same time, the G2385R has stronger binding to Hsp90 (heat shock protein 90 kDa) [13], an important LRRK2 chaperone [15]. The diminished phosphorylation activity of G2385R LRRK2 led us to suggest that this variant is a partial loss-of-function mutation [13].
These observations suggested that, despite being a relatively modest substitution at the C-terminus of a large protein, G2385R has structural effects on LRRK2 that affect both enzymatic activity and protein interactions. This led us to ask whether there are other effects of this mutation on LRRK2 that might be a consequence of altered protein folding. Here, we report that the G2385R is less stable than the wild-type variant. Mechanistically, stability depends on the E3 protein–ubiquitin ligase CHIP (carboxyl-terminus of Hsc70-interacting protein), which has enhanced binding to the mutant protein compared with wild-type LRRK2.
Materials and methods
Plasmids
HaloTag LRRK2 constructs in pHT2 vector and 3xFLAG-HD-LRRK2 constructs in Gateway-modified p3xFlag-CMV™−7.1 N-terminal Met-3xFlag vector (Sigma) were described previously [15,16]. The 3xFLAG-tagged construct of LRRK2 in pCHMWS plasmid was generously provided by Dr J.-M. Taymans (Jean-Pierre Aubert Research Center, Lille, France). 2xMyc-CHIP constructs were obtained from Dr Leonard Petrucelli (Mayo Clinic, Jacksonville, FL, U.S.A.). Point mutations were introduced by using the QuikChange II XL Site-Directed Mutagenesis Kit (Stratagene). All plasmids were fully sequenced.
Chemicals and antibodies
Chemicals and reagents used in the present study were GTP (Sigma, Cat# G8877), protease inhibitor cocktail (Roche, Cat# 04693159001), HALT phosphatase inhibitor cocktail (Thermo Scientific, Cat# 78427), Geldanamycin (Sigma, Cat# G3381), GTP beads (Jena Bioscience, Cat# NU-412–10), EZview Red Protein G Affinity Gel (Sigma, Cat# E3403), EZview Red ANTI-FLAG M2 Affinity Gel (Sigma, Cat# F2426), HaloTag® Biotin Ligand (Promega, Cat# G8281), ON-TARGETplus Non-targeting Pool (GE, Cat# D-001810–10-20), ON-TARGETplus Human STUB1 siRNA (short interfering RNA) SMARTpool: ON-TARGETplus STUB1 siRNA (GE, Cat# L007201–00-0020), and ON-TARGETplus Mouse STUB1 siRNA SMARTpool (GE, Cat# L-063143–01-0005). Odyssey® Blocking Buffer (PBS) (Li-COR, Cat# 927–40000), QuikChange II XL Site-Directed Mutagenesis Kit (Stratagene, Cat# 200521), Streptavidin MagneSphere® Paramagnetic Particles (Promega, Cat# Z5481), 10× TBS (KD Medical, Cat# RGF-3385), 4× SDS sample buffer (Invitrogen, Cat# NP0008), TRIzol Reagent (Life Technologies, Cat# 15596018), SuperScript III First-Strand Synthesis System for RT-PCR (Life Technologies, Cat# 18080–051), Power SYBR Green PCR Master Mix (Life Technologies, Cat# 4367659), Pierce Silver Stain Kit (Thermo, Cat#24612), papain (Worthington, Cat# LK003176), Basal Medium Eagle (Sigma, Cat# B1522), B27 supplement (Thermo Scientific, Cat# 17504–044), N2 supplement (Thermo Scientific, Cat# 17502–048), glutaMAX-I (Thermo Scientific, Cat# 35050–061), pre-coated coverslips (Corning, Cat# 354087), 0.45% glucose (Sigma, Cat# G8769), and cytosine arabinoside (Sigma, Cat# PHR1787).
Antibodies used were anti-CHIP (Cell Signaling, Cat# C3B6), anti-Cyclophilin B (Abcam, Cat# ab16045), anti-c-myc (Roche, Cat# 11667149001), anti-HaloTag® (Promega, Cat# G9211), anti-GAPDH (glyceraldehyde 3-phosphate dehydrogenase; Sigma, Cat# 68795), anti-LRRK2 (Epitomics, Cat# 3514–1), anti-LRRK2 (Millipore, Cat# MABN40), anti-Hsc70 (Abcam, Cat# ab19136), anti-Hsp90 (Cell Signaling, Cat# 4877), anti-FLAG (Sigma, Cat# F1804), anti-ubiquitin VU-1 (LifeSensors, Cat# VU101), HRP-conjugated secondary antibodies (Jackson Immunoresearch, Cat# 711–036-152, 715–036-151), and IRDye® fluorescent-conjugated secondary antibodies (LiCor Biosciences, Cat# 926–68020, 926–68021, 926–32210, 926–32211).
RNA isolation, reverse transcription, and real-time qPCR
RNA was extracted from HEK293FT cells transiently transfected with 3xFLAG-HD-tagged LRRK2 constructs using Trizol (Invitrogen) and stored at −20°C until use. RNA concentration was determined using a Nanodrop spectrophotometer (Thermo Scientific). One microgram of extracted RNA was transformed into cDNA using the SuperScript III First-Strand Synthesis SuperMix for qRT-PCR (Invitrogen). Quantitative PCR was performed using the power SYBR-green dye mix (Applied Biosystems) and primer pairs that amplified LRRK2 and cyclophilin B (PPIB) as a reference gene. Each sample was run in quadruplicate on a 7900HT Fast Real-time PCR system (Applied Biosystems) and relative gene expression was calculated as previously described [17].
Primer sequences used were: human LRRK2 forward 5׳-TGATGATGAGGGGGAAGAAG-3׳, human LRRK2 reverse 5׳-TCCCTATGAGCTGGGAAATG-3׳, mouse LRRK2 forward 5׳-AAAGCTGTGCCGACTGAGTT-3׳, mouse LRRK2 reverse 5׳- TACAAAGCCACTTGGGTTCC-3׳, 3xFLAG-HD forward 5׳-GTTTTCCCAGTCAC GACGTT-3׳, 3xFLAG-HD reverse 5׳-ATGGCGGTCATATTGGACAT-3׳, human PPIB forward 5׳-GCACAGG AGGAAAGAGCATC-3׳, and human PPIB reverse 5׳-AGCCAGGCTGTCTTGACTGT-3׳.
Co-immunoprecipitation
HEK293FT cells were transfected with 3xFLAG-HD-LRRK2 using Lipofectamine 2000 (Life Technologies) for 48 h followed by lysis in buffer containing 20 mM Tris–HCl (pH 7.5), 150 mM NaCl, 3 mM KCl, 10% (v/v) glycerol, 1 mM EDTA, 0.3% (v/v) Triton X-100 supplemented with protease inhibitor cocktail (Roche) and HALT phosphatase inhibitor cocktail (Thermo Scientific). Lysates were spun down at 20 000 g for 10 min, and supernatants were pre-cleared with EZview Red Protein G Affinity Gel for 1 h followed by immunoprecipitation with EZview Red ANTI-FLAG M2 Affinity Gel (both Sigma) for 2 h and gentle washing (six times) with buffer containing 20 mM Tris–HCl (pH 7.5), 150 mM NaCl, 3 mM KCl, and 0.1% Triton. LRRK2 was eluted with Gentle Ag/Ab Elution Buffer (pH 6.6) (Thermo Scientific) with 0.01% (v/v) Triton and 150 μg/ml 3xFLAG® peptide for 30 min followed by desalting of the eluate with Zeba Spin Desalting Columns (Thermo Scientific) having a 7K molecular mass cutoff. Transfection with siRNA was made using the DharmaFECT1 transfection reagent (GE Healthcare) according to the manufacturer’s instructions. The amount of immunoprecipitated binding partner was normalized to that of the LRRK2 construct eluted from the beads, estimated by densitometry using Image J (https://imagej.nih.gov/ij/).
Pull-down assay
HEK293FT cells were separately transfected with 3xFLAG-HD-LRRK2-wt, 3xFLAG-HD-G2385R, empty vector, or Myc-CHIP using Lipofectamine 2000. Transfected cells were collected after 24 h and lysed with buffer containing 20 mM Tris–HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1% (v/v) Triton X-100, and 10% (v/v) glycerol, supplemented with protease inhibitor cocktail (Thermo Scientific), pre-cleared with EZview Red Protein G Affinity Gel, and incubated with ANTI-FLAG M2 Affinity Gel (for all FLAG-plasmid-transfected cells). After 1 h of incubation, the pre-cleared Myc-CHIP lysates were equally added to all FLAG lysates and incubated for an additional 4 h, followed by six washes with buffer containing 20 mM Tris–HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 0.1% (v/v) Triton X-100, and 10% (v/v) glycerol. Lysates were eluted for 30 min using 150 μg/ml 3xFLAG peptide in kinase buffer and 400 mM NaCl and loaded for Western blot analysis. LRRK2 purity was assessed by loading a 4–18% PAGE gel followed by the Silver Stain according to the manufacture’s protocol (Thermo Scientific).
Pulse chase
HaloTag interchangeable labeling technology (Promega) was used to analyze the stability of the LRRK2 protein as previously described [15]. Briefly, HEK-293 cells were transiently transfected with HaloTag WT or G2385R mutant LRRK2 36 h prior to pulse-labeling with 5 μM HaloTag TMR Biotin ligand (Promega) in DMEM supplemented with 10% FBS for 3 h. After labeling, cells were washed three times with Opti-MEM (Invitrogen) supplemented with 10% FBS (Invitrogen) and chased in the presence or absence of 1 μM Geldanamycin (GA) (Sigma). Cells were collected at appropriate time points and lysed with buffer containing 20 mM Tris–HCl (Ph 7.5), 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3VO4, 1 mg/ml leupeptin (Cell Signaling), and protease inhibitor cocktail (Roche). The biotin-labeled LRRK2 was precipitated by Streptavidin Magnesphere® Paramagnetic particles (Promega) for 2 h at 4°C followed by three washes with PBS supplemented with 1% Triton X-100 and one wash with PBS only. Biotinylated LRRK2 was eluted in NuPAGE sample buffer (Invitrogen) for 5 min at 95°C and subjected to Western blot analysis using Halo antibody (Promega).
Primary cortical neuron preparation, transfection, and counting
Animal studies were performed in accordance with an animal study protocol approved by the Institutional Animal Care and Use Committee of the National Institute on Aging, NIH. Primary cortical neurons were prepared from C57BL6/J newborn (P0) pups by dissociation with papain (Worthington) and plated onto poly-D-lysine/laminin–pre-coated 24-well plates at 0.35 × 106 cells/well in Basal Medium Eagle (Sigma) supplemented with B27, N2, glutaMAX-I (Thermo Scientific), and 0.45% glucose (Sigma). Cytosine arabinoside (3 mM) (Sigma) was added at 2 DIV (days in vitro) to inhibit glial cell growth. Neurons were transfected at 5 DIV (days in vitro) with pooled ON-TARGETplus siRNA for CHIP (GE), or non-targeting control (NTC) siRNA (GE), at a final concentration of 25 nM. Cells were fixed and stained for the neuronal marker MAP2 and Hoechst for immunocytochemistry. Twelve images of cortical neurons per sample were acquired with a Zeiss confocal microscope. Each image was quantified for the percent of cells with intact nuclei by a researcher blinded to the treatment group.
Statistical analysis
One-way ANOVA with Tukey’s honest significance post hoc tests was used for individual comparisons of multiple LRRK2 constructs. Two-way ANOVA with Bonferroni post hoc tests was applied in experiments where different LRRK2 constructs were used in the absence and presence of Geldanamycin with mutants and treatment as factors. The relationship between CHIP binding and LRRK2 levels was evaluated using Pearson’s correlation. Cell death in primary cortical neurons was evaluated using unpaired t-tests with Welch’s correction for unequal variance. Data are shown as mean ± SEM from independent experiments and statistical inference shown using: *P < 0.05; **P < 0.01; ***P < 0.001; ns, non-significant.
Results
The G2385R mutation destabilizes LRRK2
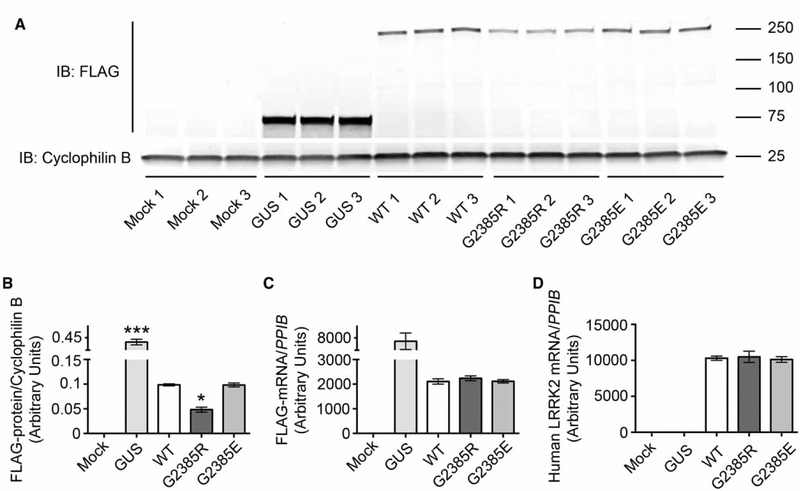
To test if there are general effects of the G2385R mutation on LRRK2, we first analyzed steady-state levels of 3xFLAG-LRRK2 expression in HEK293FT cells after transient transfection (Figure 1A). We noted that the G2385R LRRK2 variant was expressed at lower levels than wild-type protein (Figure 1B). Interestingly, substitution of the neutral glycine 2385 with a negatively charged glutamate did not affect the expression level of LRRK2, suggesting a specific effect of the G2385R mutation. Mechanistically, lower steady-state levels of the mutant protein could be either due to instability of mRNA and/or to an enhanced degradation rate of the protein product. To dissect these possibilities, we first estimated the mRNA level of expression for the same constructs, but did not find significant differences between WT and G2385R LRRK2 using two sets of primers (Figure 1C,D).
Figure 1.
Steady-state mRNA and protein expression levels for different LRRK2 mutants.
(A) Steady-state levels of protein expression of WT, G2385R, and G2385E LRRK2 in HEK293FT cells transiently transfected with an equal amount of 3xFLAG-HD-LRRK2 plasmid DNA. Quantification of protein expression levels (B), FLAG-tagged mRNA (C), and human LRRK2 mRNA (D) normalized to cyclophilin B protein and mRNA expression, respectively (n = 3 independent experiments).
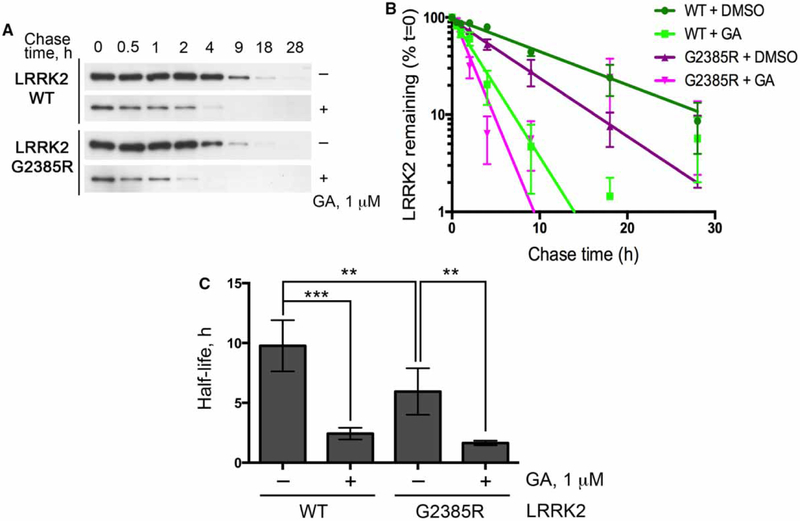
These results nominate protein degradation as a possible mechanism for the lower steady-state levels of G2385R LRRK2. To test this hypothesis, we performed pulse-chase experiments with Halo-tagged LRRK2 where biotinylation can be controlled in a transient manner (Figure 2A,B). The half-life of WT LRRK2 estimated using this technique, t1/2-WT = 9.7 ± 2.1 h, was within the range reported in previous studies using this protein [15,18]. Notably, LRRK2 with the G2385R substitution had a significantly shorter half-life than wild-type protein (t1/2-G2385R = 5.9 ± 1.9 h; Figure 2C). Furthermore, inhibition of Hsp90 with 1 μM GA decreased the half-life of both WT and G2385R mutant LRRK2 (t1/2-WT-GA = 2.4 ± 0.5 vs. t1/2-G2385R-GA = 1.6 ± 0.2 h), supporting prior observations that this co-chaperone controls the overall turnover rate of LRRK2. These results establish that the G2835R mutant LRRK2 has a higher turnover rate compared with wild-type protein.
Figure 2.
The G2385R substitution destabilizes LRRK2.
(A) HEK293FT cells transiently transfected with WT or G2385R LRRK2 were pulse-labeled with biotinylated Halo ligand, treated with DMSO or 1 mM GA, and harvested at indicated time points. (B) Time courses of the decay and (C) estimated half-life of biotinylated proteins (n = 6 independent experiments).
The G2385R LRRK2 mutant has enhanced proteasomal degradation rate
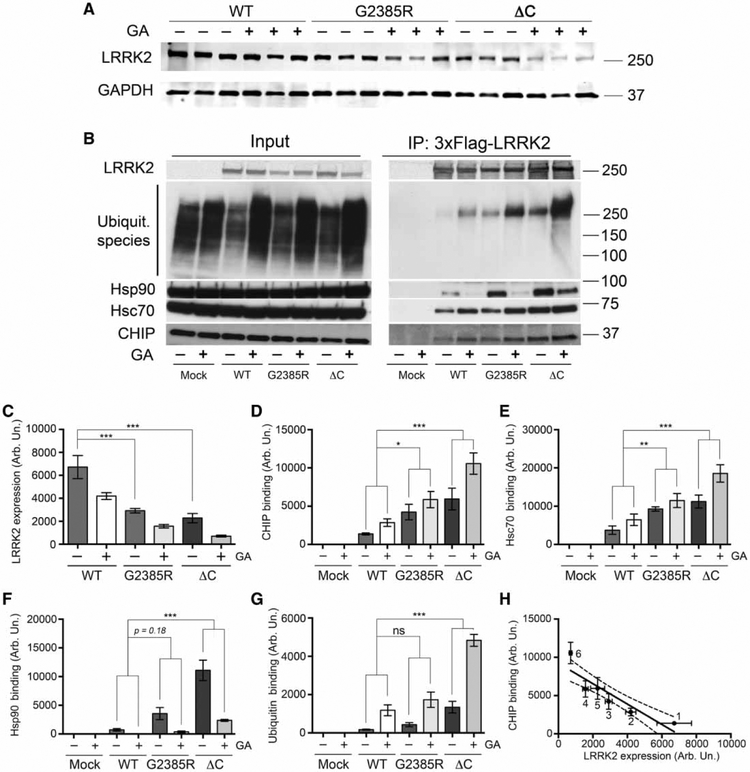
We next sought to understand the mechanism(s) by which G2385R has a higher turnover rate than wild-type LRRK2. It has been suggested that the intracellular degradation of LRRK2 is at least partially executed by the proteasomal degradation pathway [19,20], specifically by the co-chaperone CHIP [21]. CHIP possesses an E3 protein–ubiquitin ligase activity and can ubiquitylate interacting partners [22] including LRRK2 [19]. We therefore considered CHIP as a potential candidate E3 ligase for the effects of G2385R on LRRK2 stability. The G2385R and ΔC LRRK2 [C-terminus-deleted LRRK2 (1–2498)] had lower expression levels but higher affinity to CHIP compared with wild-type protein (Figure 3A–F). Inhibition of Hsp90 with GA further decreased LRRK2 steady-state levels, consistent with the protein turnover data in Figure 2, and promoted binding of CHIP to LRRK2. Under the same conditions, G2385R had higher levels of ubiquitin addition than wild-type protein (Figure 3B,G). There was an overall negative correlation between LRRK2 steady-state levels and CHIP binding (Figure 3H). These data demonstrate that both G2385R and ΔC LRRK2, which may be misfolded, are more prone to CHIP-mediated proteasomal degradation than wild-type protein.
Figure 3.
Levels of WT, G2385R, and ΔC LRRK2 variant expression are inversely related to CHIP binding.
(A) Steady-state levels of protein expression of WT, G2385R, and ΔC LRRK2 (C-terminus-deleted LRRK2, containing amino acids 1–2498) in HEK293FT cells were transiently transfected with an equal amounts of plasmid DNA and pretreated for 3 h with DMSO or 1 μM GA prior to cell lysis. (B) CHIP-binding ability of different LRRK2 mutants in HEK293FT cells transfected with an adjusted amount of plasmid DNA in the absence (DMSO) or presence of 1 μM GA for 3 h. Quantifications of protein expression levels normalized to GAPDH (n = 3) (C) and CHIP, Hsc70, Hsp90, and ubiquitin binding (n = 6) (D–G) normalized to GAPDH. (E) Correlation analysis of steady-state protein expression levels and CHIP binding by different LRRK2 mutants.
CHIP binds multiple LRRK2 domains including the WD40 domain
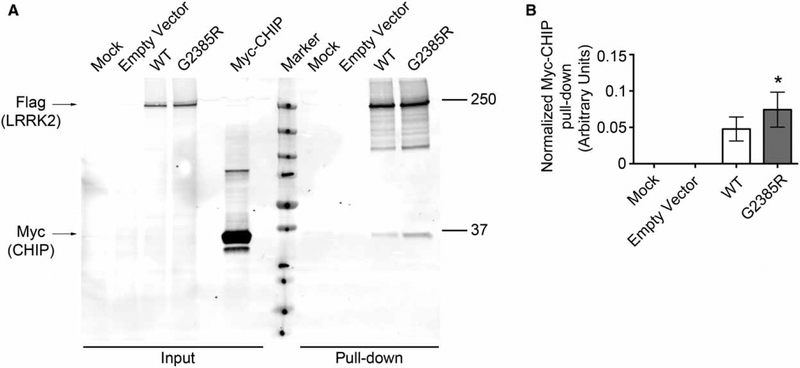
The enhanced CHIP binding and associated ubiquitylation could feasibly be either due to a direct recognition of the mutant protein by CHIP or an indirect event due to other changes in the cell. We therefore performed two sets of experiments to try to distinguish these possibilities. First, we found that we could recapitulate the enhanced binding of CHIP to G2385R LRRK2 using an in vitro pull-down assay (Figure 4). Examination of the purified LRRK2 (Supplementary Figure S1) demonstrated that there is some contamination with the chaperone Hsp70. However, this was found in both WT and G2385R LRRK2, suggesting that CHIP may work with Hsp70 to control overall levels of LRRK2 stability.
Figure 4.
CHIP pull-down with wild-type and G2385R LRRK2.
HEK293FT cells were separately transfected with 2xMyc-CHIP and 3xFLAG-LRRK2 constructs which were then purified and incubated together. (A) Expression of wild-type and G2385R LRRK2 and pull-down of CHIP. (B) Quantifications of CHIP pull-down (n = 3 independent experiments). Purity of the material used in this pull-down assay was assessed by Silver staining (see Supplementary Figure S1).
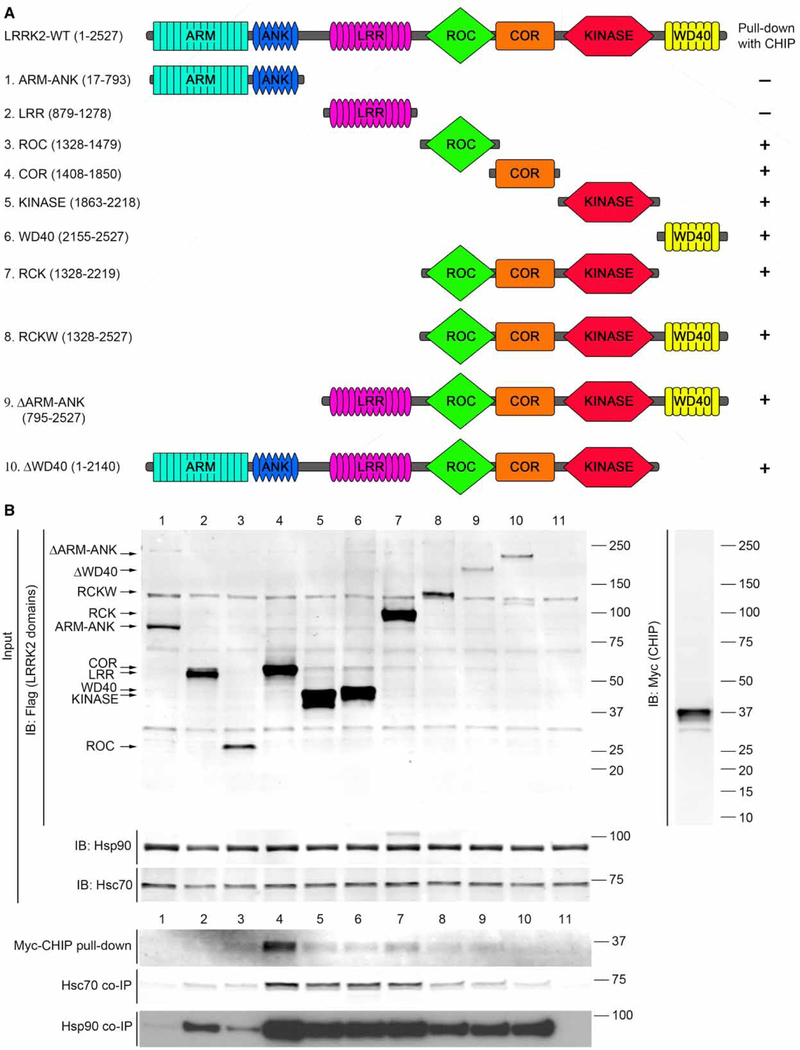
Second, given the enhanced binding of CHIP/Hsp70 to G2385R LRRK2, where the mutation is in the C-terminal WD40 domain, we revisited the mapping of interactions between LRRK2 and CHIP using a series of deletion constructs (Figure 5A). Previous reports have mapped the binding domains of CHIP to LRRK2 but with inconsistent results. Specifically, while Ko et al. [19] nominated the ROC domain of LRRK2 as being specifically responsible for CHIP recruitment, Ding and Golderg [20] found that CHIP could bind to both the N-and C-terminal portions of LRRK2. For the isolated domains, we found an apparently strong interaction of Hsc70 or Hsp90 with the COR domain and also detectable interaction with other single domains including the isolated WD40 domain (Figure 5B). Therefore, we conclude that chaperone/co-chaperone complexes can bind to multiple regions of LRRK2, including the WD40 domain, where G2385R is located. Collectively, these results are consistent with CHIP directly recognizing different regions of LRRK2, probably via affinity to hydro-phobic surfaces. These data also support the hypothesis that G2385R destabilizes the structure of LRRK2, increasing the number of exposed hydrophobic residues, in turn leading to enhanced turnover of the mutant protein by CHIP and other chaperones.
Figure 5.
CHIP pull-down with various LRRK2 domains.
(A) Schematic diagrams of tested LRRK2 constructs. (B) Expression of domains, pull-down of CHIP, and co-immunoprecipitation of Hsc70 and Hsp90 after separate transient transfection of 2xMyc-CHIP and 3xFLAG-LRRK2 constructs in HEK293FT cells.
CHIP knockdown enhances LRRK2 expression, while CHIP overexpression diminishes LRRK2 levels
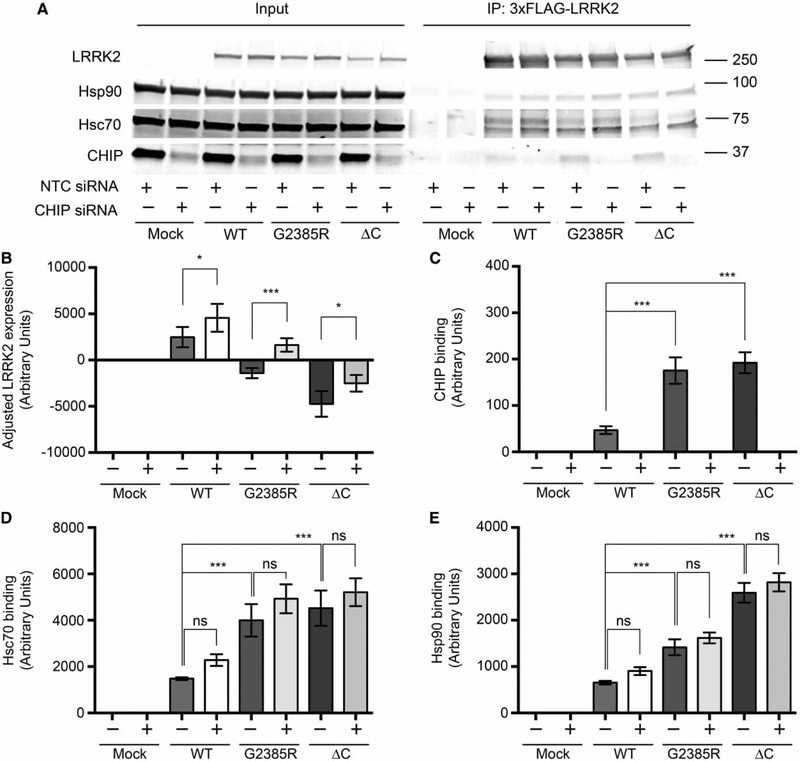
These results suggest that CHIP preferentially binds G2385R LRRK2 compared with the wild-type protein. To test whether this binding is functionally important for intracellular degradation of LRRK2, we transiently knocked down CHIP in HEK293FT cells. Consistent with the idea that CHIP controls LRRK2 levels, protein expression was higher for all LRRK2 constructs in HEK293FT cells after CHIP siRNA treatment (Figure 6A,B). The apparent binding of LRRK2 to Hsc70 or Hsp90 was not affected, suggesting that these two chaperones bind independently of CHIP. Reciprocally, overexpression of CHIP resulted in lower steady-state LRRK2 levels (Figure 7).
Figure 6.
CHIP knockdown increases protein expression levels of WT, G2385R, and ΔC LRRK2 variants.
HEK293FT cells were transiently transfected with smart pool NTC or CHIP siRNA (Thermo Scientific) 24 h before transfection with an equal amount of 3xFLAG-HD-LRRK2 plasmid DNA. After 48 h from transfection with LRRK2 constructs, cells were treated for 3 h with DMSO or 1 μM GA prior to lysis. (A) Co-immunoprecipitation of Hsp90, Hsc70, and CHIP with 3xFLAG-HD-LRRK2 constructs. (B) Adjusted steady-state levels of WT, G2385R, and ΔC LRRK2 protein expression in the presence of NTC (−) or CHIP (+) siRNA in input. (C–E) CHIP, Hsc70, and Hsp90-binding ability of different LRRK2 mutants normalized to the amount of immunoprecipitated LRRK2 (n = 6).
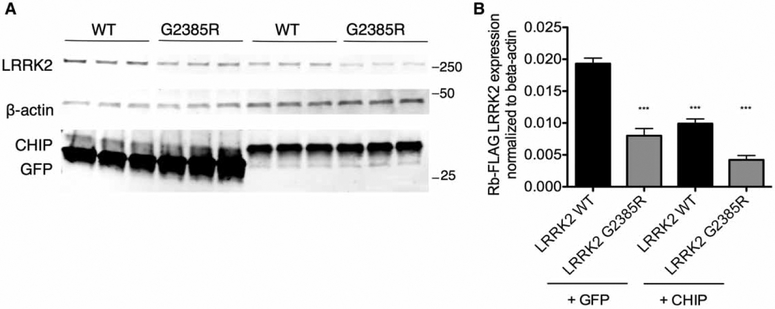
Figure 7.
CHIP overexpression decreases protein expression levels of WT and G2385R LRRK2.
HEK293FT cells were transiently co-transfected with 1 μg each of either 3xFlag-HD-human LRRK2 WT or G2385R along with either 3xFlag-GFP or 2xmyc-CHIP plasmid. Cells were collected and lysed 40 h after transfection, and immunoblotting was subsequently performed with the indicated antibodies. Note that the lowest blots were probed with CHIP and GFP antibodies simultaneously. (B) Quantifications of total LRRK2 protein are normalized to expression of β-actin. Statistical significance is indicated relative to WT LRRK2 (n = 3 independent experiments).
Discussion
Understanding how mutations and risk variants in the LRRK2 gene affect protein function has important implications for the development of potential treatments for LRRK2-associated PD. In the present study, we demonstrated that G2385R substitution in addition to reduction in kinase activity of LRRK2 [13] also destabilizes the protein and promotes its proteasomal degradation. Therefore, despite being a relatively modest substitution of one amino acid in a >250 kDa protein, this variant has profound effects on protein function.
We were able to show that the difference in WT and G2385R LRRK2 expression levels depends on the intra-cellular degradation rate of the mutant protein and nominate CHIP as a key controller of protein levels, especially for the G2385R mutant.
The outcome of whether a protein is properly folded with chaperones or destroyed by protease is known as the ‘molecular triage’ concept [23] where hydrophobic regions and other binding sites for chaperones are exposed on poorly folded or nonnative protein structures. Chaperones can successfully convert a nonnative protein into its native conformation, in which case the hydrophobic recognition sites become buried inside the protein and thus no longer can be a client for the triage machinery. However, if the protein is irreversibly damaged or if the accumulation rate of misfolded protein is high and exceeds the ability of chaperones to fold all proteins properly, then nonnative proteins will undergo degradation by proteasomes. The precise function and regulation of this complex triage machinery is not completely understood; however, age, oxidative stress, and disease could favor unfolded conformation of the protein and thus tilt the equilibrium of the system toward the degradation pathway [24,25].
CHIP participates in protein folding and degradation decisions and thus serves as a quality control regulator of the Hsp70/Hsp90 folding pathway. CHIP interacts with Hsc70/Hsp70 and Hsp90 with approximately equivalent affinity. In addition to inhibiting traditional chaperone activity, CHIP converts the chaperone complex into a chaperone-dependent ubiquitin ligase [24]. In case of ErbB2, ubiquitylation depends on a transfer of the client protein from Hsp90 to Hsp70 [26], indicating that the final ubiquitylation complex consists of CHIP and Hsp70 family members (but not Hsp90). CHIP efficiently targets client proteins, particularly when they are partially unfolded, as with many Hsp90 clients, or fully misfolded as is the case for most proteins binding to Hsp70 through exposed hydrophobic residues [24].
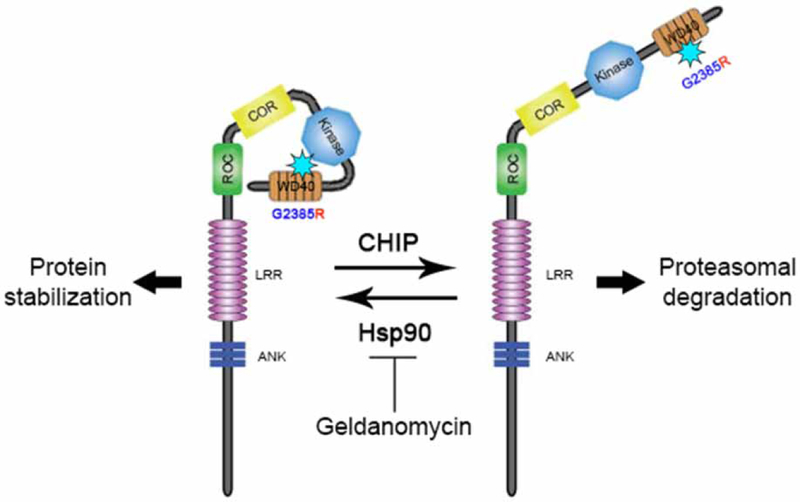
Considering that G2385R LRRK2 has stronger binding to Hsp90 compared with wild-type protein, we infer that this mutation causes partial misfolding of the protein. Hsp90 probably promotes the folding of LRRK2 to an active, stable conformation, while misfolded LRRK2 is targeted for intracellular degradation. This refolding and degradation equilibrium is probably tilted toward degradation for the G2385R mutant compared with WT LRRK2 (Figure 8). Our data demonstrate that CHIP, working with Hsp70/Hsc70, is a key player in the triage decision for G2385R LRRK2 and that the LRRK2 interactors CHIP and Hsp90 functionally antagonize each other. Intriguingly, we demonstrated an interaction of CHIP with multiple regions of LRRK2, suggesting that the large kinase might be especially dependent on the molecular triage machinery.
Figure 8.
Schematic model of regulation of G2385R LRRK2 stability.
We propose that the G2385R substitution causes misfolding of LRRK2, thereby promoting CHIP-mediated proteasomal degradation. Such misfolding can be antagonized by Hsp90 as inhibition of Hsp90 binding by Geldanamycin shifts the equilibrium toward CHIP-dependent degradation of the protein.
Although the structural basis by which CHIP recognizes mutant LRRK2 compared with wild-type protein is not fully resolved, it is important to note that G2385 is predicted to be on the exposed surface of the β-sheet-rich WD40 domain [12], and thus could indirectly interfere with inter- and intramolecular interactions of LRRK2 as well as directly affect protein binding around the 2385 residue. For example, an indirect effect of the G2358R mutation could be to misfold LRRK2 and thus increase access of CHIP to hydrophobic internal regions of the protein such as the ROC domain. The apparently higher affinity of the ΔC LRRK2 that contains the native G2385 residue supports this hypothesis. Alternatively, the G2385R mutation might additionally directly affect CHIP recruitment to LRRK2 via changes in the WD40 domain. Distinguishing these possibilities will probably require a full-length structure of LRRK2 in complex with chaperones and co-chaperones.
We attempted to confirm that endogenous CHIP controlled endogenous LRRK2 expression in neurons. Unfortunately, knockdown of CHIP with siRNA was poorly tolerated by neurons and induced cell death (Supplementary Figure S2). These results indicate that CHIP is critical for neuronal survival at least in culture and suggest that future experiments using, for example, conditional knockout of CHIP will be required to address whether LRRK2 is an authentic CHIP client in vivo.
Finally, we note that G2835R acts as a risk factor for PD rather than a penetrant mutation that runs in families. We suggest that both diminished kinase activity and lower steady-state levels of G2385R can contribute to the loss of intracellular function of mutated protein and thus represent different molecular mechanisms compared with stably expressed gain-of-function mutations (such as G2019S) in the enzymatic domains of LRRK2. This hypothesis would be important to test in patient-derived material, particularly comparing affected and at-risk but unaffected individuals.
Supplementary Material
Acknowledgements
We are thankful to Dr Jean-Marc Taymans (Jean-Pierre Aubert Research Center, Lille, France) for sharing human 3xFLAG-LRRK2 constructs in pCHMWS plasmid. We also thank Dr Leonard Petrucelli (Mayo Clinic, Jacksonville, FL, U.S.A.) for the generous gift of Myc-CHIP constructs.
Funding
This work was supported by the Intramural Research Program of the National Institute on Aging, NIH, and by The Michael J. Fox Foundation for Parkinson’s Research.
Abbreviations
- CHIP
carboxyl-terminus of Hsc70-interacting protein
- ΔC LRRK2
C-terminus-deleted LRRK2 (1–2498)
- COR
C-terminus of ROC
- DIV
days in vitro
- GA
Geldanamycin
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- HRP
horseradish peroxidase
- Hsc70
heat shock cognate 71 kDa protein
- Hsp70
heat shock protein
- 70 kDa, Hsp90
heat shock protein 90 kDa
- LRR
leucine-rich repeat
- LRRK2
leucine-rich repeat kinase 2
- MAP2
microtubule-associated protein 2
- NTC
non-targeting control
- PBS
phosphate-buffered saline
- PD
Parkinson’s disease
- ROC
Ras of complex proteins
- SiRNA
short interfering RNA
- TBS
Tris-buffered saline
Footnotes
Competing Interests
The Authors declare that there are no competing interests associated with the manuscript.
References
- 1.Singleton AB, Farrer MJ and Bonifati V (2013) The genetics of Parkinson’s disease: progress and therapeutic implications. Mov. Disord 28, 14–23 doi: 10.1002/mds.25249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cookson MR (2010) The role of leucine-rich repeat kinase 2 (LRRK2) in Parkinson’s disease. Nat. Rev. Neurosci 11, 791–797 doi: 10.1038/nrn2935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greggio E, Jain S, Kingsbury A, Bandopadhyay R, Lewis P, Kaganovich A et al. (2006) Kinase activity is required for the toxic effects of mutant LRRK2/dardarin. Neurobiol. Dis 23, 329–341 doi: 10.1016/j.nbd.2006.04.001 [DOI] [PubMed] [Google Scholar]
- 4.Jaleel M, Nichols RJ, Deak M, Campbell DG, Gillardon F, Knebel A et al. (2007) LRRK2 phosphorylates moesin at threonine-558: characterization of how Parkinson’s disease mutants affect kinase activity. Biochem. J 405, 307–317 doi: 10.1042/BJ20070209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.West AB, Moore DJ, Biskup S, Bugayenko A, Smith WW, Ross CA et al. (2005) Parkinson’s disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proc. Natl Acad. Sci. U.S.A 102, 16842–16847 doi: 10.1073/pnas.0507360102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daniëls V, Vancraenenbroeck R, Law BMH, Greggio E, Lobbestael E, Gao F et al. (2011) Insight into the mode of action of the LRRK2 Y1699C pathogenic mutant. J. Neurochem 116, 304–315 doi: 10.1111/j.1471-4159.2010.07105.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lewis PA, Greggio E, Beilina A, Jain S, Baker A and Cookson MR (2007) The R1441C mutation of LRRK2 disrupts GTP hydrolysis. Biochem. Biophys. Res. Commun 357, 668–671 doi: 10.1016/j.bbrc.2007.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li X, Tan Y-C, Poulose S, Olanow CW, Huang X-Y and Yue Z (2007) Leucine-rich repeat kinase 2 (LRRK2)/PARK8 possesses GTPase activity that is altered in familial Parkinson’s disease R1441C/G mutants. J. Neurochem 103, 238–247 doi: 10.1111/j.1471-4159.2007.04743.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taymans J-M and Greggio E (2016) LRRK2 kinase inhibition as a therapeutic strategy for Parkinson’s disease, where do we stand? Curr. Neuropharmacol 14, 214–225 doi: 10.2174/1570159X13666151030102847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Funayama M, Li Y, Tomiyama H, Yoshino H, Imamichi Y, Yamamoto M et al. (2007) Leucine-rich repeat kinase 2 G2385R variant is a risk factor for Parkinson disease in Asian population. NeuroReport 18, 273–275 doi: 10.1097/WNR.0b013e32801254b6 [DOI] [PubMed] [Google Scholar]
- 11.Fung H-C, Chen C-M, Hardy J, Singleton AB and Wu Y-R (2006) A common genetic factor for Parkinson disease in ethnic Chinese population in Taiwan. BMC Neurol. 6, 47 doi: 10.1186/1471-2377-6-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mata IF, Wedemeyer WJ, Farrer MJ, Taylor JP and Gallo KA (2006) LRRK2 in Parkinson’s disease: protein domains and functional insights. Trends Neurosci. 29, 286–293 doi: 10.1016/j.tins.2006.03.006 [DOI] [PubMed] [Google Scholar]
- 13.Rudenko IN, Kaganovich A, Hauser DN, Beylina A, Chia R, Ding J et al. (2012) The G2385R variant of leucine-rich repeat kinase 2 associated with Parkinson’s disease is a partial loss-of-function mutation. Biochem. J 446, 99–111 doi: 10.1042/BJ20120637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nichols RJ, Dzamko N, Morrice NA, Campbell DG, Deak M, Ordureau A et al. (2010) 14–3-3 binding to LRRK2 is disrupted by multiple Parkinson’s disease-associated mutations and regulates cytoplasmic localization. Biochem. J 430, 393–404 doi: 10.1042/BJ20100483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang L, Xie C, Greggio E, Parisiadou L, Shim H, Sun L et al. (2008) The chaperone activity of heat shock protein 90 is critical for maintaining the stability of leucine-rich repeat kinase 2. J. Neurosci 28, 3384–3391 doi: 10.1523/JNEUROSCI.0185-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beilina A, Rudenko IN, Kaganovich A, Civiero L, Chau H, Kalia SK et al. (2014) Unbiased screen for interactors of leucine-rich repeat kinase 2 supports a common pathway for sporadic and familial Parkinson disease. Proc. Natl Acad. Sci. U.S.A 111, 2626–2631 doi: 10.1073/pnas.1318306111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45 doi: 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohta E, Katayama Y, Kawakami F, Yamamoto M, Tajima K, Maekawa T et al. (2009) I2020T leucine-rich repeat kinase 2, the causative mutant molecule of familial Parkinson’s disease, has a higher intracellular degradation rate than the wild-type molecule. Biochem. Biophys. Res. Commun 390, 710–715 doi: 10.1016/j.bbrc.2009.10.034 [DOI] [PubMed] [Google Scholar]
- 19.Ko HS, Bailey R, Smith WW, Liu Z, Shin J-H, Lee Y-I et al. (2009) CHIP regulates leucine-rich repeat kinase-2 ubiquitination, degradation, and toxicity. Proc. Natl Acad. Sci. U.S.A 106, 2897–2902 doi: 10.1073/pnas.0810123106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding X and Goldberg MS (2009) Regulation of LRRK2 stability by the E3 ubiquitin ligase CHIP. PLoS ONE 4, e5949 doi: 10.1371/journal.pone.0005949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ballinger CA, Connell P, Wu Y, Hu Z, Thompson LJ, Yin L-Y et al. (1999) Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol. Cell. Biol 19, 4535–4545 doi: 10.1128/MCB.19.6.4535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang J, Ballinger CA, Wu Y, Dai Q, Cyr DM, Höhfeld J et al. (2001) CHIP is a U-box-dependent E3 ubiquitin ligase: identification of Hsc70 as a target for ubiquitylation. J. Biol. Chem 276, 42938–42944 doi: 10.1074/jbc.M101968200 [DOI] [PubMed] [Google Scholar]
- 23.Wickner S, Maurizi MR and Gottesman S (1999) Posttranslational quality control: folding, refolding, and degrading proteins. Science 286, 1888–1893 doi: 10.1126/science.286.5446.1888 [DOI] [PubMed] [Google Scholar]
- 24.McDonough H and Patterson C (2003) CHIP: a link between the chaperone and proteasome systems. Cell Stress Chaperones 8, 303–308 doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pratt WB, Morishima Y, Peng H-M and Osawa Y (2010) Proposal for a role of the Hsp90/Hsp70-based chaperone machinery in making triage decisions when proteins undergo oxidative and toxic damage. Exp. Biol. Med 235, 278–289 doi: 10.1258/ebm.2009.009250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu W, Marcu M, Yuan X, Mimnaugh E, Patterson C and Neckers L (2002) Chaperone-dependent E3 ubiquitin ligase CHIP mediates a degradative pathway for c-ErbB2/Neu. Proc. Natl Acad. Sci. U.S.A 99, 12847–12852 doi: 10.1073/pnas.202365899 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.