Dear Editor,
As we known, pigs play a vital role as genetic mixing vessels for human and avian influenza viruses as their tracheal epitheliums possess both sialic acid α-2,6-Gal and α-2,3-Gal receptors (Ma et al. 2008), and swine influenza viruses occasionally infect humans (Shinde et al. 2009). The Eurasian avian-like swine influenza A (H1N1) virus (EAS-H1N1) spread into China around 2007, when the first isolate (A/swine/Fujian/204/2007) from pigs in Fujian was reported via swine influenza surveillance program (Liu et al. 2009). Much later, three cases of EAS-H1N1 infection in human were reported in China and they were all children under the age of three (Yang et al. 2012; Wang et al. 2013; Zhu et al. 2016), whereas the forth case was an adult, who was confirmed in October, 2016, in Fujian province.
The patient was a 46-year-old man with fever of 39 °C and started coughing on October 19, 2016. In the day after onset, he was treated in a prefectural hospital on account of hemoptysis and dyspnea, and then he was transferred to the intensive care unit (ICU) of Fuzhou Pulmonary Hospital because of severe pneumonia on October 21. A throat-swab specimen of patient was collected and transferred to Fujian CDC rapidly with cryogenic storage container for confirming influenza virus. Clinical examination revealed white blood cell (WBC) count of 5.4 × 109/L, blood platelet count of 27 × 109/L, and creatinine 300 μmol/L. At last, the patient died on October 28 due to multi-organ failure.
A retrospective investigation was conducted to identify the potential infection source and any other possible cases. The patient lived with his wife, son, brother and sister in an argillous house, with pigs, chickens, and ducks being bred around it. The patient had not the history of feeding poultry or swine and visiting live poultry market before he got sick. A total of 9 close contacts (6 family members and 3 healthcare workers) of the patient did not develop any influenza-like symptoms during the longest incubation period. On October 23, we collected 12 specimens (including 3 poultry drinking water samples, 3 poultry cage surface wipes, 1 chicken feces sample, 3 duck feces samples and 2 pig feces samples) from the surrounding environment where the patient lived, but no influenza viruses were identified at that time.
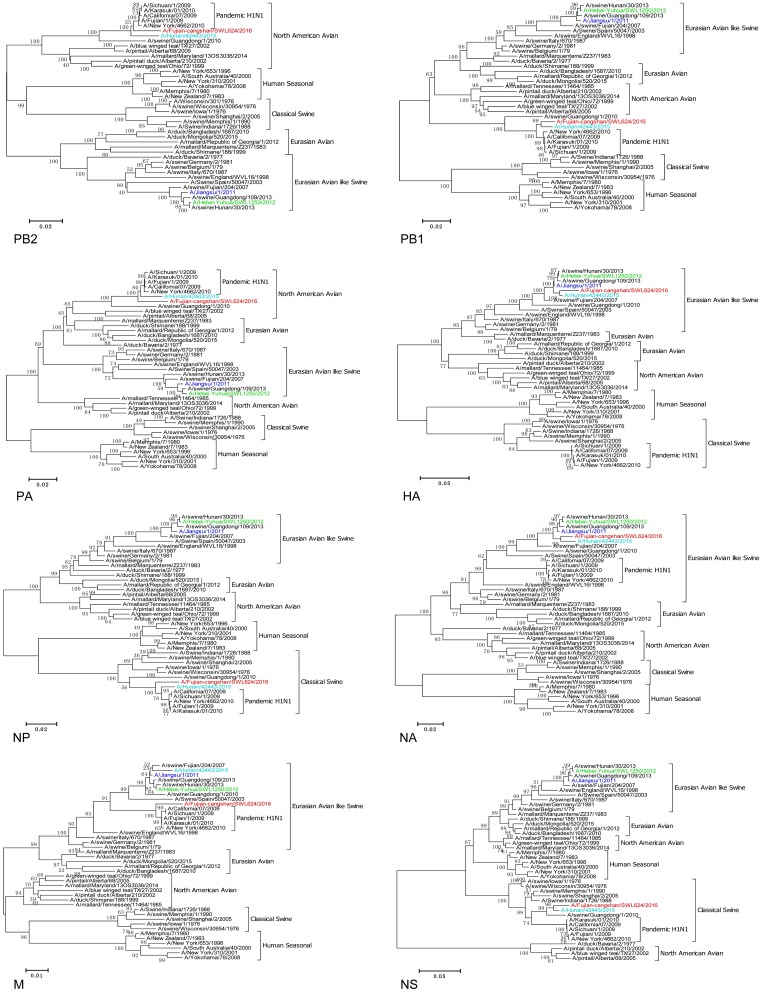
A throat-swab specimen of the patient was collected and the virus was isolated using 9-to-10-days-old specific pathogen-free (SPF) embryonated chicken eggs in the Biosafety Level 3 Laboratory, terming influenza A/Fujian-cangshan/SWL624/2016 (FJ/624/16) virus. To study the genomic characterization of FJ/624/16 virus, we sequenced the complete genomes of the viral isolate in November, 2016. Sequences were assembled using the SeqMan of the Lasergene Package (DNAStar Corporation, USA). Nucleotide alignments and phylogenetic tree were constructed using the MEGA version 6.0 software (the Biodesign Institute, USA) by the Neighbor-Joining method with 1000 bootstrap replicates as implemented. Phylogenetic analysis of genome showed that the isolate was a novel triple-reassortant H1N1 virus with genes containing from Eurasian avian-like swine (HA, NA), A (H1N1) pdm09 (PB2, PB1, PA, NP, M) and classical swine (NS) lineages (Tables 1, 2 and Fig. 1). Complete sequences of FJ/624/16 virus were 94.3%–98.2% and 91.2%–95.4% identical in all 8 gene segments with A/Hunan/42443/2015 virus and A/swine/Guangdong/1/2010 virus respectively. Nevertheless, the M gene segment of FJ/624/16 virus showed the highest homology (98.4%) with A/California/07/2009 virus than other viruses and shared the closely related with A (H1N1) pdm09 viruses via blast (the sequences of viruses from the Global Initiative on Sharing Avian Influenza Data, GISAID; Accession No. EPI_ISL_239998, EPI_ISL_206573, EPI_ISL_75321, EPI_ISL_227813, EPI_ISL_85649, EPI_ISL_138290 and EPI_ISL_33887).
Table 1.
Homology analysis of sequences of all genes identified in the A/Fujian-cangshan/SWL624/2016 virus.
| Isolates | A/Fujian-cangshan/SWL624/2016 (identities %) | |||||||
|---|---|---|---|---|---|---|---|---|
| PB2 | PB1 | PA | HA | NP | NA | M | NS | |
| A/California/07/2009 | 97.8 | 97.9 | 97.3 | 55.7 | 97.8 | 88.4 | 98.4 | 91.0 |
| A/Hunan/42443/2015 | 96.3 | 98.2 | 96.0 | 97.8 | 96.9 | 97.9 | 94.3 | 97.4 |
| A/swine/Guangdong/1/2010 | 93.4 | 93.2 | 92.5 | 91.2 | 93.1 | 92.1 | 94.1 | 95.4 |
| A/Jiangsu/1/2011 | 81.1 | 81.6 | 81.2 | 97.3 | 78.9 | 98.0 | 94.8 | 79.0 |
| A/Hebei-Yuhua/SWL1250/2012 | 80.8 | 82.0 | 80.9 | 96.8 | 79.3 | 97.7 | 94.3 | 78.2 |
| A/swine/Fujian/204/2007 | 81.3 | 81.8 | 81.9 | 95.5 | 79.4 | 96.0 | 94.4 | 78.6 |
PB, polymerase basic protein; PA, polymerase acidic protein; HA, hemagglutinin; NP, nucleocapsid protein; NA, neuraminindase; M, matrix; NS, nonstructural.
Table 2.
Genetic origin of A/Fujian-cangshan/SWL624/2016 virus isolated from human in Fujian, China.
| Isolates | Lineage assigned to gene segment | |||||||
|---|---|---|---|---|---|---|---|---|
| PB2 | PB1 | PA | HA | NP | NA | M | NS | |
| A/Fujian-cangshan/SWL624/2016a | PDM | PDM | PDM | EAS | PDM | EAS | PDM | CS |
| A/Hunan/42443/2015a | PDM | PDM | PDM | EAS | PDM | EAS | EAS | CS |
| A/swine/Guangdong/1/2010 | PDM | PDM | PDM | EAS | PDM | EAS | EAS | CS |
| A/Jiangsu/1/2011a | EAS | EAS | EAS | EAS | EAS | EAS | EAS | EAS |
| A/Hebei-Yuhua/SWL1250/2012a | EAS | EAS | EAS | EAS | EAS | EAS | EAS | EAS |
| A/swine/Fujian/204/2007 | EAS | EAS | EAS | EAS | EAS | EAS | EAS | EAS |
PDM, genes closest homology to A(H1N1)pdm09 viruses; EAS, genes with closest homology to Eurasian avian-like swine influenza viruses; CS, genes with closest homology to classical swine influenza viruses.
aHuman infection with Eurasian avian-like swine influenza virus.
Fig. 1.
Phylogenetic analysis of all eight genes on A/Fujian-cangshan/SWL624/2016 virus. Red, light blue, green, and blue are marked at the A/Fujian-cangshan/SWL624/2016, A/Hunan/42443/2015, A/Heibei-Yuhua/SWL1250/2012 and A/Jiangsu/1/2011, respectively. The reliability of the trees was assessed via bootstrap analysis with 1000 replications. The horizontal distances are proportional to the genetic distance. Scale bars indicate nucleotide substitutions per site.
We compared the newly sequenced genomes with 3 available genomes of human influenza virus strains in public databases to determine if they had attained key molecular features associated with increased virulence in mammals, mammalian transmissibility, and antiviral resistance (Table 3). In this study, FJ/624/16 possessed E190D and D225E (H3 numbering) in HA protein, suggesting the preference to α-2,6-linked sialosides and increase in the binding force of H1N1 virus in human upper respiratory tract (Matrosovich et al. 2000; Zhu et al. 2016). FJ/624/16 was found to contain an amino acid motif PSIQSR * GL at its HA cleavage sites, a characteristic of low pathogenic influenza virus (Korteweg and Gu 2008). The amino acid substitutions (H274Y, N294S, et al.) associated with neuraminindase inhibitory drugs were not observed in the NA protein of FJ/624/16, indicating sensitivity to neuramidinase inhibitors, but an S31N substitution in M2 protein suggested resistant to adamantanes. In addition, our data showed that several amino acid residues of FJ/624/16 might increase viral virulence or adaptation in mammals, such as 89V and 591R in the PB2, 336M, 356R and 409N in the PA, 215A in the M1, and 42S in the NS1 (Nguyen et al. 2012; Gao et al. 2013; Yamada et al. 2010; Wang et al. 2016; Fan et al. 2009; Jiao et al. 2008).
Table 3.
Molecular analysis of A/Fujian-cangshan/SWL624/2016 virus.
| Gene | Function | Mutation | FJ/624/16 | HN/42443/15 | HB/1250/12 | JS/1/11 | FJ/204/07 | GD/1/10 | CA/07/09 |
|---|---|---|---|---|---|---|---|---|---|
| HA | Altered receptor specificity | E190D | D | D | D | D | D | V | D |
| D225E | E | E | E | E | G | E | D | ||
| NA | Antiviral resistance (Oseltamivir) | H274Y | H | H | H | H | H | H | H |
| N294S | N | N | N | N | N | N | N | ||
| PB2 | Enhanced polymerase activity | L89 V | V | V | V | V | V | V | V |
| Virus replication in mammals | Q591R | Q | R | Q | Q | Q | R | R | |
| E627 K | E | E | E | E | E | E | E | ||
| D701 N | D | D | N | N | N | D | D | ||
| PA | Increased polymerase activity in mice | L336 M | M | M | L | L | L | L | M |
| Species-associated signature positions | K356R | R | R | K | K | K | K | R | |
| S409 N | N | N | N | N | N | N | N | ||
| M1 | Increased virulence in mice | T215A | A | A | A | A | A | A | A |
| M2 | Antiviral resistance (Amantadine) | S31 N | N | N | N | N | N | N | N |
| NS1 | Increased virulence in mice | P42S | S | S | S | S | S | S | S |
FJ/624/16, A/Fujian-cangshan/SWL624/2016; HN/42443/15, A/Hunan/42443/2015; HB/1250/12, A/Hebei-Yuhua/SWL1250/2012; JS/1/11, A/Jiangsu/1/2011; FJ/204/07, A/swine/Fujian/204/2007; GD/1/10, A/swine/Guangdong/1/2010; CA/07/09, A/California/07/2009.
So far, a total of 4 human cases of Eurasian avian-like swine influenza A (H1N1) virus infection had been reported in China. All three cases reported previously were children under 3 years old, but the first adult case-patient was confirmed in China in this study, signifying the EAS-H1N1 virus infection was not confined to children. The 8 gene segments of human infection with EAS-H1N1 virus reported in China during 2011–2012 originated from Eurasian avian influenza virus, however, the two EAS-H1N1 viruses reported in China lately differed from those: HA and NA gene clustered with Eurasian avian influenza virus, and other 6 genes shared the closely related with A (H1N1) pdm09 virus or classical swine influenza virus by phylogenetic analysis, implying that the genes of A (H1N1) pdm09 virus might have been recombined in swine since 2009. According to previous reports and this study, human infections with EAS-H1N1 virus might have been underestimated in China.
To analyze the virulence, transmissibility and drug-resistance of the EAS-H1N1 virus from the adult patient, we performed an in-depth analysis of genetic characteristics of the FJ/624/16 virus. The virus might preferentially bind to influenza virus-binding receptor of human upper respiratory tract due to the amino acid mutations E190D and D225E within HA protein, hinting the potential risk of human-to-human transmission. In addition, several amino acid residues of FJ/624/16 virus might increase viral virulence or adaptation in mammals, so the severity of disease caused by EAS-H1N1 should be valued in future studies. In our study, we found that the virus was resistant to adamantanes and sensitive to neuramidinase inhibitors, suggesting that early administration of oseltamivir or peramivir may help to reduce the severity of the disease.
In summary, human infection with the EAS-H1N1 virus could lead to severe clinical syndrome, even death. However, increasing evidences show that the EAS-H1N1 virus becomes more adaptative to mammal infection, and was equipped with viral molecular basis of cross-species transmission (Yang et al. 2012; Wang et al. 2013; Zhu et al. 2016; Qi et al. 2012). Thus, it is a great significance to strengthen the surveillance for EAS-H1N1 virus among swine and humans, and virologic analyses to assess genetic changes of EAS-H1N1 virus also highlighted the great value to determine the viral transmissibility among humans and pandemic potential.
Acknowledgements
We thank Fuzhou Center for Disease Control and Prevention and Fuzhou Pulmonary Hospital for providing the sample. We acknowledge the contributions of authors and laboratories for sharing the influenza virus sequences in GISAID’s EpiFlu Database. This study was supported by the fund of Natural Science Foundation of Fujian Province (2015J01294), the Youth Backbone Talents Cultivation Program of Health System in Fujian Province (2015-ZQN-ZD-10) and the National Science and Technology Major Project (2017ZX10103008).
Conflict of interest
The authors declare that they have no conflict of interest.
Animal and Human Rights Statement
The study was approved by the ethics committees of Fujian Provincial Center for Disease Control and Prevention. All participants provided written informed consent. Written consents were obtained from all participants involved in the study.
References
- Fan S, Deng G, Song J, Tian G, Suo Y, Jiang Y, Guan Y, Bu Z, Kawaoka Y, Chen H. Two amino acid residues in the matrix protein M1 contribute to the virulence difference of H5N1 avian influenza viruses in mice. Virology. 2009;384:28–32. doi: 10.1016/j.virol.2008.11.044. [DOI] [PubMed] [Google Scholar]
- Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, Xu X, Lu H, Zhu W, Gao Z, Xiang N, Shen Y, He Z, Gu Y, Zhang Z, Yang Y, Zhao X, Zhou L, Li X, Zou S, Zhang Y, Li X, Yang L, Guo J, Dong J, Li Q, Dong L, Zhu Y, Bai T, Wang S, Hao P, Yang W, Zhang Y, Han J, Yu H, Li D, Gao GF, Wu G, Wang Y, Yuan Z, Shu Y. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med. 2013;368:1888–1897. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- Jiao P, Tian G, Li Y, Deng G, Jiang Y, Liu C, Liu W, Bu Z, Kawaoka Y, Chen H. A single amino acid substitution in the NS1 protein changes the pathogenicity of H5N1 avian influenza viruses in mice. J Virol. 2008;82:1146–1154. doi: 10.1128/JVI.01698-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korteweg C, Gu J. Pathology, molecular biology, and pathogenesis of avian influenza A (H5N1) infection in humans. Am J Pathol. 2008;172:1155–1170. doi: 10.2353/ajpath.2008.070791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Bi Y, Qin K, Fu G, Yang J, Peng J, Ma G, Liu Q, Pu J, Tian F. Emergence of European avian influenza virus-like H1N1 swine influenza A viruses in China. J Clin Microbiol. 2009;47:2643–2646. doi: 10.1128/JCM.00262-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Kahn RE, Richt JA. The pig as a mixing vessel for influenza viruses: human and veterinary implications. J Mol Genet Med. 2008;3:158–166. [PMC free article] [PubMed] [Google Scholar]
- Matrosovich M, Tuzikov A, Bovin N, Gambaryan A, Klimov A, Castrucci MR, Donatelli I, Kawaoka Y. Early alterations of the receptor-binding properties of H1, H2, and H3 avian influenza virus hemagglutinins after their introduction into mammals. J Virol. 2000;74:8502–8512. doi: 10.1128/JVI.74.18.8502-8512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HT, Fry AM, Gubareva LV. Neuraminidase inhibitor resistance in influenza viruses and laboratory testing methods. Antivir Ther. 2012;17:159–173. doi: 10.3851/IMP2067. [DOI] [PubMed] [Google Scholar]
- Qi X, Pan Y, Qin Y, Zu R, Tang F, Zhou M, Wang H, Song Y. Molecular characterization of avian-like H1N1 swine influenza a viruses isolated in Eastern China, 2011. Virol Sin. 2012;27:292–298. doi: 10.1007/s12250-012-3262-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinde V, Bridges CB, Uyeki TM, Shu B, Balish A, Xu X, Lindstrom S, Gubareva LV, Deyde V, Garten RJ, Harris M, Gerber S, Vagasky S, Smith F, Pascoe N, Martin K, Dufficy D, Ritger K, Conover C, Quinlisk P, Klimov A, Bresee JS, Finelli L. Triple-reassortant swine influenza A (H1) in humans in the United States, 2005–2009. N Engl J Med. 2009;360:2616–2625. doi: 10.1056/NEJMoa0903812. [DOI] [PubMed] [Google Scholar]
- Wang DY, Qi SX, Li XY, Guo JF, Tan MJ, Han GY, Liu YF, Lan Y, Yang L, Huang WJ, Cheng YH, Zhao X, Bai T, Wang Z, Wei HJ, Xiao N, Shu YL. Human infection with Eurasian avian-like influenza A (H1N1) virus, China. Emerg Infect Dis. 2013;19:1709–1711. doi: 10.3201/eid1910.130420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Yang L, Zhu W, Zhang Y, Zou S, Bo H, Gao R, Dong J, Huang W, Guo J, Li Z, Zhao X, Li X, Xin L, Zhou J, Chen T, Dong L, Wei H, Li X, Liu L, Tang J, Lan Y, Yang J, Shu Y. Two outbreak sources of influenza a (H7N9) viruses have been established in China. J Virol. 2016;90:5561–5573. doi: 10.1128/JVI.03173-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada S, Hatta M, Staker BL, Watanabe S, Imai M, Shinya K, Sakai-Tagawa Y, Ito M, Ozawa M, Watanabe T, Sakabe S, Li C, Kim JH, Myler PJ, Phan I, Raymond A, Smith E, Stacy R, Nidom CA, Lank SM, Wiseman RW, Bimber BN, O'Connor DH, Neumann G, Stewart LJ, Kawaoka Y. Biological and structural characterization of a host-adapting amino acid in influenza virus. PLoS Pathog. 2010;6:e1001034. doi: 10.1371/journal.ppat.1001034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HL, Qiao CL, Tang X, Chen Y, Xin XG, Chen HL. Human infection from avian-like influenza A (H1N1) viruses in pigs, China. Emerg Infect Dis. 2012;18:1144–1146. doi: 10.3201/eid1807.120009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Zhang H, Xiang X, Zhong L, Yang L, Guo J, Xie Y, Li F, Deng Z, Feng H, Huang Y, Hu S, Xu X, Zou X, Li X, Bai T, Chen Y, Li Z, Li J, Shu Y. Reassortant Eurasian avian-like influenza A(H1N1) virus from a severely Ill child, Hunan province, China. Emerg Infect Dis. 2016;22:1930–1936. doi: 10.3201/eid2211.160181. [DOI] [PMC free article] [PubMed] [Google Scholar]